Abstract
A carotenoid biosynthesis gene cluster involved in canthaxanthin production was isolated from the photosynthetic Bradyrhizobium sp. strain ORS278. This cluster includes five genes identified as crtE, crtY, crtI, crtB, and crtW that are organized in at least two operons. The functional assignment of each open reading frame was confirmed by complementation studies.
Bradyrhizobium strains isolated from Aeschynomene stem nodules are photosynthetic (4; see reference 6 for a review), which is a rare trait in Rhizobium bacteria. These strains exhibit a photoheterotrophic and strictly aerobic photosynthesis (6; A. Vermeglio, personal communication). In culture, most of these stem isolates show the same pink coloration, while a few strains produce orange pigmentation (12, 16). Pigment analyses showed that bacteriochlorophyll and spirilloxanthin, two pigments of the light harvesting system, are common to all of these photosynthetic Bradyrhizobium strains, whereas orange strains produce an additional bicyclic carotenoid, canthaxanthin (4,4′-diketo-β-carotene) (12). This was the first report on the presence of this carotenoid in photosynthetic bacteria. Bradyrhizobium sp. strain ORS278 produces the highest quantity of canthaxanthin of all tested photosynthetic bacteria; canthaxanthin represents 85% of its total carotenoid content (12).
Contrary to anaerobic purple phototrophic bacteria, aerobic phototrophic bacteria synthesize an unusually diverse variety of carotenoids, including photosynthetic carotenoids such as spirilloxanthin or spheroidenone, and often a large amount of bicyclic carotenoid (β-carotene and hydroxyl derivatives) (24; see reference 26 for a review). These carotenoids were shown to not be bound to the photosynthetic apparatus of these aerobic bacteria (17, 27) and their function is still unclear—they could have a protective role against photo-oxidative damage, as already observed for several carotenoids (19, 25).
Synthetic canthaxanthin is applied for both direct and indirect food coloring (10, 23). In cosmetology and pharmacology, it is also combined with β-carotene for use as a dermal photoprotector (8). Canthaxanthin is, therefore, a pigment of high economic value, but its level in Bradyrhizobium sp. strain ORS278 (1.43 mg/g of dry cell weight) remains insufficient for this organism to be a realistic candidate for natural canthaxanthin production (12). However, it could be possible to enhance the production of canthaxanthin by cloning carotenoid biosynthesis genes of this strain.
In this paper, we describe the cloning and characterization of the canthaxanthin gene cluster of Bradyrhizobium sp. strain ORS278.
Isolation of a carotenoid gene cluster.
The genes crtB and crtI, encoding, respectively, phytoene synthase and phytoene desaturase, two enzymes involved in the initial steps of carotenoid biosynthesis (Fig. 1), have been isolated and characterized in various microorganisms (1, 9, 11, 14, 15, 21). In all of these cases, these genes were found to be adjacent and oriented in the same direction. Comparison of the deduced amino acid sequences of the CrtI and CrtB proteins from Erwinia uredovora, Erwinia herbicola, Flavobacterium sp. strain ATCC 21588, Rhodobacter sphaeroides, and Agrobacterium aurantiacum revealed well-conserved domains at the C-terminal end of CrtI (LVGAGTHPG) and in the central region of CrtB (QLTNIARD). These motifs were chosen for designing the degenerated primers CrtIf (5′-GTNGGNGCRGGCACNCAYCC-3′) and CrtBr (5′-TCGCGRGCRATRTTSGTSARRTG-3′). PCR amplification was performed with a Perkin-Elmer model 2400 thermocycler in a 50-μl (total volume) reaction mixture containing 100 ng of strain ORS278 genomic DNA, each deoxynucleotide triphosphate (200 μM), primers (0.8 μM each), MgCl2 (1.5 mM), 1.25 U of Taq DNA polymerase (Promega, Charbonières, France), and the buffer supplied with the enzyme. A touchdown PCR (3) was done as follows: initial denaturation at 94°C for 5 min followed by 20 cycles consisting of a 30-s denaturation at 94°C, 30 s at an annealing temperature of 60 to 50°C, and a 1-min primer extension at 72°C, followed by 15 cycles consisting of a 30-s denaturation at 94°C, 30 s at an annealing temperature at 50°C, and a 1-min primer extension at 72°C. After the final elongation step at 72°C for 7 min, the amplified 620-bp fragment obtained (probe A) was purified by a Wizard procedure and was ligated into a pGEM-T vector (Promega). The ABI Prism BigDye Terminator Cycle Sequence Kit (Applied Biosystems, Foster City, Calif.) was used to sequence the cloned PCR product with the universal oligonucleotides M13 forward and M13 reverse. Sequencing reactions were analyzed on an Applied Biosystems model 310 DNA sequencer. The sequence of the amplified 620-bp fragment was highly similar to known CrtB sequences at the amino acid level.
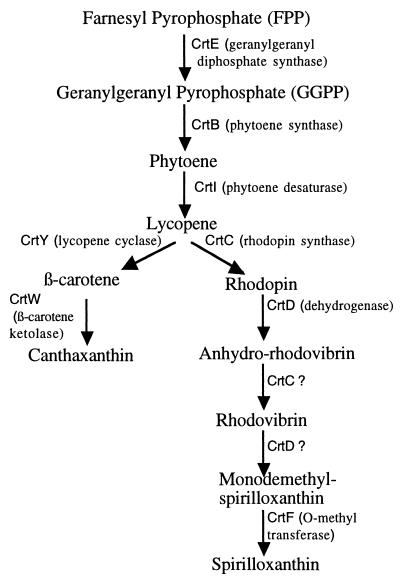
FIG. 1.
Scheme of the canthaxanthin (15) and spirilloxanthin (20) biosynthesis pathways.
Two specific primers, CrtIBfow.ORS278 (5′-ATTCGCAGCGGCTCGAAGAG-3′) and CrtIBrev.ORS278 (5′-GATCGCCGACATCATCACGC-3′), based on the sequence of the amplified DNA fragment, were designed for PCR screening of a library of the ORS278 strain constructed with the SuperCos I cosmid vector kit (Stratagene, La Jolla, Calif.), as instructed by the manufacturer. Four positive clones were isolated and confirmed by Southern blot analysis by using the 620-bp fragment as a probe. Clone pSTM73, containing an insert of approximately 35 kb, was used to characterize this crt gene cluster.
Structure of the canthaxanthin crt gene cluster.
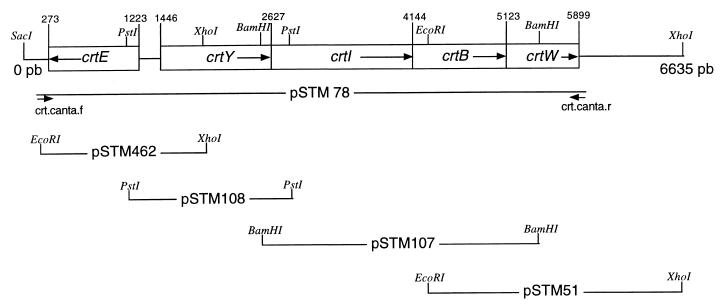
A 6.5-kb region in the inserted DNA fragment of the pSTM73 cosmid, showing a positive hybridization signal to probe A, was sequenced and analyzed as shown in Fig. 2. This nucleotide sequence had five open reading frames (ORFs) encoding proteins with similarity to known Crt enzymes (Fig. 3). Based on sequence similarity (45% amino acid identity with CrtY of E. herbicola), one of these ORFs was assigned to a crtY gene which encodes lycopene cyclase, a key enzyme that converts lycopene into the cyclic carotenoid β-carotene. Another ORF was similar in sequence to a crtW gene encoding a β-carotene ketolase that synthesizes canthaxanthin from β-carotene via echinenone (15). This indicated that we had isolated a crt gene cluster involved in canthaxanthin biosynthesis. Four of the five ORFs, identified as crtY, crtI, crtB, and crtW, were found to be clustered in this order in the same orientation, whereas the ORF crtE preceded these four but was oriented in the opposite direction (Fig. 2). The crtY, crtI, and crtB genes are closely linked physically; i.e., the stop codons of crtY and crtI overlap the start codon of the following ORF, suggesting that these genes are translationally coupled (18). Note that the crtY, crtI, and crtB genes always occurred in this order and were oriented in the same direction in the other cyclic carotenoid biosynthesis clusters described previously (see Fig. 3).
FIG. 2.
Organization of the canthaxanthin biosynthesis gene cluster of Bradyrhizobium sp. strain ORS278 and locations of various subcloned fragments. The restriction fragments are inserted into pUC18 (pSTM108, pSTM107, and pSTM51) or pUC19 (pSTM462), the crt genes are transcribed from the lac promoter of the vector. In the plasmid pSTM78, the insert was obtained by Long PCR using the primers Crt.canta.f (5′-GCAACCGGTACCCGAGTTAATTCGCTCGGAATG-3′) and Crt.canta.r (5′-ATGGTGAAGCTTATGCGGCAGCGGGTTTAGTC-3′) and was cloned into pGEM-T (Promega). In pSTM78, the crtY, crtI, crtB, and crtW genes are under lac promoter control.
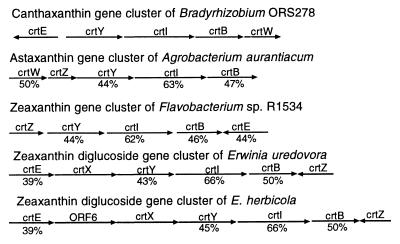
FIG. 3.
Comparison of the organization of the cyclic carotenoid gene clusters of Bradyrhizobium sp. strain ORS278, A. aurantiacum (15), Flavobacterium sp. strain R1534 (21), E. uredovora (14), and E. herbicola (9). Arrows represent the orientations of ORFs. The percentage values below the genes indicate the percentages of amino acid identity compared to Bradyrhizobium sp. strain ORS278.
Phylogenetic trees were constructed with available CrtI and CrtB sequences. The CrtB proteins (Fig. 4) and the CrtI proteins (data not shown) formed two distinct clusters which do not correlate with the taxonomical position of the strains, but rather with the nature of the carotenoid (cyclic or noncyclic) synthesized under the control of crtI and crtB. The fact that α- and γ-Proteobacteria and Flavobacteria group in the same cluster (Fig. 4) suggests that lateral gene transfer has occurred between these phylogenetically unrelated bacteria. Moreover, the fact that strain ORS278 genes are clustered with crtI and crtB genes from nonphotosynthetic strains producing cyclic carotenoids rather than with photosynthetic strains producing photosynthetic carotenoids raises the question of whether an additional copy of the crtI and crtB genes involved in the biosynthesis of spirilloxanthin does exist. The crtC and crtD genes, which were reported to be involved in the biosynthesis of spirilloxanthin from lycopene (20), have just been isolated in another cosmid which did not overlap the pSTM73 cosmid (E. Giraud and B. Dreyfus, unpublished data). We are currently investigating if another copy of the crtI and crtB genes is physically linked to these crtC and crtD genes, as has been found in photosynthetic bacteria (1, 11).
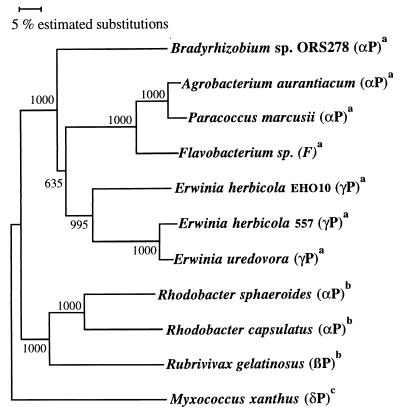
FIG. 4.
Phylogenetic tree based on the CrtB sequences and constructed by using the neighbor-joining method (22). Bootstrap values (5), expressed as percentages of 1,000 replications, are given at the branching points. P, Proteobacteria; F, Flavobacteria; a, bicyclic carotenoid; b, acyclic carotenoid; c, monocyclic carotenoid. GenBank accession numbers are as follows: AF218415, Bradyrhizobium sp. strain ORS278; D58420, A. aurantiacum; Y15112, Paracoccus marcusii; U62808, Flavobacterium sp.; M87280, E. herbicola EHO10; M90698, E. herbicola 557; D90087, E. uredovora; AF195122, R. sphaeroides; X52291, Rhodobacter capsulatus; U87626, Rubrivivax gelatinosus; Z211955, Myxococcus xanthus.
Carotenoid production in Escherichia coli transformants.
E. coli transformants carrying the entire crt gene cluster of canthaxanthin from strain ORS278, cloned in pGEMT (pSTM78) or SuperCosI (pSTM73), did not produce any carotenoids (Table 1), suggesting that these genes are not expressed or that their products are not functional in E. coli. Misawa et al. (15) constructed E. coli transformants which accumulate each precursor of the zeaxanthin biosynthesis pathway by introducing various combinations of E. uredovora crt genes. To check the functionality of the different ORFs identified in strain ORS278, we complemented several carotenoid-accumulating E. coli transformants with plasmids carrying various crt genes of strain ORS278 and analyzed carotenoids synthesized by high-pressure liquid chromatography (Table 1). The conditions were as follows: 5-μm Hypersil C18 column (250 by 4.6 mm; Alltech, Templemars, France), eluent of acetonitrile-methanol-isopropanol (85/10/5, vol/vol/vol), flow rate of 1 ml/min, and detection at 470 nm (450 nm for β-carotene). Peaks were compared and coeluted with standard compounds then identified by their visible spectra and partition coefficients (12).
TABLE 1.
Analysis of carotenoids accumulated in E. coli transformants carrying various combinations of crt genes from E. uredovora and Bradyrhizobium sp. strain ORS278a
|
E. coli host strain characteristics
|
E. coli transformant characteristics after complementation
|
||
|---|---|---|---|
| Plasmid (crt genes of E. uredovora carried) | Carotenoid accumulatedc | Plasmid introducedd (crt genes of ORS278 carried) | Carotenoid accumulatedef |
| None | −g | pSTM73 (crtE crtY crtI crtB crtW) | − |
| None | − | pSTM78 (crtE crtY crtI crtB crtW) | − |
| pACCRT-Eb (crtE) | GGPP | pSTM78 (crtE crtY crtI crtB crtW) | Canthaxanthin (100%) [95.4] |
| pSTM420 (crtI crtB crtY) | − | pSTM462 (crtE) | β-Carotene (98%), nich (2%) |
| pACCRT-Eb (crtE) | GGPP | pSTM107 (crtI crtB) | Lycopene (100%) |
| pACCRT-EBb (crtE crtB) | Phytoene | pSTM107 (crtI crtB) | Lycopene (100%) |
| pACCRT-EIBb (crtE crtI crtB) | Lycopene | pSTM108 (crtY) | β-Carotene (100%) |
| pACCRT-EIBYb (crtE crtI crtB crtY) | β-Carotene | pSTM51 (crtW) | Canthaxanthin (90%) [800], echinenone (2%), nic (8%) |
Transformants were grown in Luria-Bertani medium for 36 h in the presence of ampicillin (50 μg/ml), chloramphenicol (30 μg/ml), and 0.125 mM isopropyl-1-thio-β-d-galactopyranoside. The carotenoids were extracted according to the method of Lorquin et al. (12).
The plasmids used were described by Misawa et al. (15). Plasmid pSTM420 was obtained after deletion by SalI digestion of the crtE gene from the plasmid pACCRT-EIBY.
The carotenoids found in E. coli host strains are in accordance with the findings of Misawa et al. (15).
Details on insertion of the various constructed plasmids are presented in Fig. 2.
The percentage of the accumulated carotenoid of the total carotenoid content is indicated in parentheses.
In square brackets, total (cis plus trans) canthaxanthin level is indicated in micrograms per gram of dry cell weight.
−, carotenoids not detected.
nic, nonidentified compound.
When plasmid pSTM78 carrying the complete crt cluster of Bradyrhizobium sp. strain ORS278 was introduced into the E. coli transformant that had accumulated geranylgeranyl pyrophosphate (GGPP) as a result of the presence of the crtE gene of E. uredovora, the new transformant obtained was shown to accumulate canthaxanthin. This result indicates that the crtY, crtI, crtB, and crtW genes are functional and allow the production of canthaxanthin in E. coli. Nevertheless, the amount of canthaxanthin produced remains lower than in the wild-type strain ORS278. When plasmid pSTM462 carrying the crtE gene of Bradyrhizobium sp. strain ORS278 under the lac promoter was introduced into an E. coli transformant containing the crtI, crtB, and crtY genes of E. uredovora, the new transformant accumulated β-carotene, showing the functionality of the crtE gene.
In this study, we cloned and characterized all of the crt genes of Bradyrhizobium sp. strain ORS278 necessary for canthaxanthin biosynthesis. This is the first report of a cyclic carotenoid biosynthesis gene cluster in a photosynthetic bacterium. It would be interesting to determine the genetic links of this canthaxanthin crt gene cluster to the photosynthetic gene cluster. In Bradyrhizobium sp. strain ORS278, canthaxanthin production is stimulated by light (13), suggesting that the expression of canthaxanthin biosynthesis genes is regulated by photoinduction, as already reported for other pigments in different organisms (2, 7). Production of this pigment could be optimized by identifying the signal transduction system controlling canthaxanthin biosynthesis. However, characterization of the entire crt gene cluster necessary for canthaxanthin biosynthesis already provides a basis for the construction of a recombinant strain that could overproduce this carotenoid.
Nucleotide sequence accession number.
The DNA sequence obtained in this study has been deposited in the GenBank database under accession no. AF218415.
Acknowledgments
We thank N. Misawa for kindly providing the plasmids carrying the crt genes of E. uredovora used in this study.
REFERENCES
- 1.Armstrong A G, Alberti M, Leach F, Hearst J E. Nucleotide sequence, organization, and nature of the protein products of the carotenoid biosynthesis gene cluster of Rhodobacter capsulatus. Mol Gen Genet. 1989;216:254–268. doi: 10.1007/BF00334364. [DOI] [PubMed] [Google Scholar]
- 2.Bramley P M, Mackenzie A. Regulation of carotenoid biosynthesis. Curr Top Cell Regul. 1988;29:291–343. doi: 10.1016/b978-0-12-152829-4.50009-4. [DOI] [PubMed] [Google Scholar]
- 3.Don R H, Cox P T, Wainwright B, Baker K, Mattick J S. “Touchdown” PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eaglesham A R J, Ellis J M, Evans W R, Fleischman D E, Hungria M, Hardy R W F. The first photosynthetic N2-fixing Rhizobium: characteristics. In: Gresshoff P M, Roth L E, Stacey G, Newton W L, editors. Nitrogen fixation: achievements and objectives. New York, N.Y: Chapman and Hall; 1990. pp. 805–811. [Google Scholar]
- 5.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 6.Fleischman D, Kramer D. Photosynthetic rhizobia. Biochim Biophys Acta. 1998;1364:17–36. doi: 10.1016/s0005-2728(98)00011-5. [DOI] [PubMed] [Google Scholar]
- 7.Fontes M, Ruiz-Vázquez R, Murillo F J. Growth phase dependence of the activation of a bacterial gene for carotenoid synthesis by blue light. EMBO J. 1993;12:1265–1275. doi: 10.1002/j.1460-2075.1993.tb05771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goralczyk R, Buser S, Bausch J, Bee W, Zühlke U, Barker F M. Occurrence of birefringent retinal inclusions in cynomolgus monkeys after high doses of canthaxanthin. Investig Ophthalmol Vis Sci. 1997;38:741–752. [PubMed] [Google Scholar]
- 9.Hundle B, Alberti M, Nievelstein V, Beyer P, Kleinig H, Armstrong G A, Burke D H, Hearst J E. Functional assignment of Erwinia herbicola Eho10 carotenoid genes expressed in Escherichia coli. Mol Gen Genet. 1994;245:406–416. doi: 10.1007/BF00302252. [DOI] [PubMed] [Google Scholar]
- 10.Kläui H. Industrial and commercial uses of carotenoids. In: Britton G, Goodwin T W, editors. Carotenoid chemistry and biochemistry. Oxford, England: Pergamon Press, Inc.; 1982. pp. 309–328. [Google Scholar]
- 11.Lang H P, Cogdell R J, Gardiner A T, Hunter C N. Early steps in carotenoid biosynthesis: sequence and transcriptional analysis of crtI and crtB genes of Rhodobacter sphaeroides and overexpression and reactivation of crtI in Escherichia coli and R. sphaeroides. J Bacteriol. 1994;176:3859–3869. doi: 10.1128/jb.176.13.3859-3869.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lorquin J, Molouba F, Dreyfus B L. Identification of the carotenoid canthaxanthin from photosynthetic Bradyrhizobium strains. Appl Environ Microbiol. 1997;63:1151–1154. doi: 10.1128/aem.63.3.1151-1154.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lorquin J, Molouba F, Dupuy N, Ndiaye S, Alazard D, Gillis M, Dreyfus B L. Diversity of photosynthetic Bradyrhizobium strains from stem nodules of Aeschynomene species. In: Palacios R, Mora J, Newton W E, editors. New horizons in nitrogen fixation. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1993. pp. 683–689. [Google Scholar]
- 14.Misawa N, Nagakawa M, Kobayashi K, Yamano S, Izawa Y, Nakamura K, Harashima K. Elucidation of the Erwinia uredovora carotenoid biosynthetic pathway by functional analysis of gene products expressed in Escherichia coli. J Bacteriol. 1990;172:6704–6712. doi: 10.1128/jb.172.12.6704-6712.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Misawa N, Satomi Y, Kondo K, Yokoyama A, Kajiwara S, Saito T, Ohtani T, Miki W. Structure and functional analysis of a marine bacterial carotenoid biosynthesis gene cluster and astaxanthin biosynthetic pathway proposed at the gene level. J Bacteriol. 1995;177:6575–6584. doi: 10.1128/jb.177.22.6575-6584.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Molouba F, Lorquin J, Willems A, Hoste B, Giraud E, Dreyfus B, Gillis M, de Lajudie P, Masson-Boivin C. Photosynthetic bradyrhizobia from Aeschynomene spp. are specific to stem nodulated species and form a separate 16S ribosomal DNA restriction fragment length polymorphism group. Appl Environ Microbiol. 1999;6:3084–3094. doi: 10.1128/aem.65.7.3084-3094.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noguchi T, Hayashi H, Shimada K, Takaichi S, Tasumi M. In vivo states and function of carotenoids in an aerobic photosynthetic bacterium, Erythrobacter longus. Photosynth Res. 1992;31:21–30. doi: 10.1007/BF00049533. [DOI] [PubMed] [Google Scholar]
- 18.Oppenheim D S, Yanofsky C. Translational coupling during expression of the tryptophan operon of Escherichia coli. Genetics. 1980;95:785–795. doi: 10.1093/genetics/95.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ouchane S, Picaud M, Vernotte C, Astier C. Photooxidative stress stimulates illegitimate recombination and mutability in carotenoid-less mutants of Rubrivivax gelatinosus. EMBO J. 1997;16:4777–4787. doi: 10.1093/emboj/16.15.4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ouchane S, Picaud M, Vernotte C, Reiss-Husson F, Astier C. Pleiotropic effects of puf interposon mutagenesis on carotenoid biosynthesis in Rubrivivax gelatinosus. J Biol Chem. 1997;17:1670–1676. doi: 10.1074/jbc.272.3.1670. [DOI] [PubMed] [Google Scholar]
- 21.Pasamontes L, Hug D, Tessier M, Hohmann H P, Schierle J, van Loon A P G M. Isolation and characterization of the carotenoid biosynthesis genes of Flavobacterium sp. strain R1534. Gene. 1997;185:35–41. doi: 10.1016/s0378-1119(96)00624-5. [DOI] [PubMed] [Google Scholar]
- 22.Saitou R R, Nei M. A neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;44:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 23.Simpson K L, Katayama T, Chichester C O. Carotenoids in fish feeds. In: Bauernfeind J C, editor. Carotenoids as colorants and vitamin A precursors. New York, N.Y: Academic Press, Inc.; 1981. pp. 463–538. [Google Scholar]
- 24.Takaichi S, Shimada K, Ishidsu J I. Carotenoids from the aerobic photosynthetic bacterium, Erythrobacter longus: β-carotene and its hydroxyl derivatives. Arch Microbiol. 1990;153:118–122. [Google Scholar]
- 25.Tuveson R W, Larson R A, Kagan J. Role of cloned carotenoid genes expressed in Escherichia coli in protecting against inactivation by near-UV light and specific phototoxic molecules. J Bacteriol. 1988;170:4675–4680. doi: 10.1128/jb.170.10.4675-4680.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yurkov V V, Beatty J T. Aerobic anoxygenic phototrophic bacteria. Microbiol Mol Biol Rev. 1998;62:695–724. doi: 10.1128/mmbr.62.3.695-724.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yurkov V V, Gad'on N, Drews G. The major part of polar carotenoids of the aerobic bacteria Roseococcus thiosulfatophilus, RB3 and Erythromicrobium ramosum, E5 is not bound to the bacteriochlorophyll a complexes of the photosynthetic apparatus. Arch Microbiol. 1993;160:372–376. [Google Scholar]