Abstract
Defensins play an important role in fighting bacteria, and are a good candidate for bactericidal agents. However, the function and mechanism of defensins in regulating host responses against bacteria is unclear. In this study, transcriptome analysis was used to study the comprehensive functions of pBD2 in IPEC-J2 cells against E. coli. In total, 230 differentially expressed genes (DEGs) were identified in IPEC-J2 cells between the control and E. coli groups, and were found by KEGG analysis to be involved in many signaling pathways related to immunity. Furthermore, 812 DEGs were observed between E. coli and E. coli +pBD2 groups, involved in the ribosome, oxidative phosphorylation, and certain disease pathways. Among these, 94 overlapping DEGs were in the two DEG groups, and 85 DEGs were reverse expression, which is involved in microRNA in cancer, while PTEN and CDC6 were key genes according to PPI net analysis. The results of qRT-PCR verified those of RNA-seq. The results indicated that pBD2 plays an important role against E. coli by acting on the genes related to immune response, cell cycle, ribosomes, oxidative phosphorylation, etc. The results provide new insights into the potential function and mechanism of pBD2 against E. coli. Meanwhile, this study provides a certain theoretical basis for research and the development of novel peptide drugs.
Keywords: defensins, transcriptome analysis, E. coli infection, functions, immune response
1. Introduction
In the past, the long-term use and abuse of antibiotics in animal husbandry has resulted in serious antibiotic residues and increase of drug-resistant strains, which is currently considered a great threat to public health [1]. Although the use of antibiotics in livestock has been limited as much as possible in recent years, this has brought some other challenges, such as the increase of intestinal diseases and the weakening of growth performance, especially at the weaning stage in the pig industry. It is urgent to develop new antimicrobial agents; host defensins may be one of the methods to solve this problem [2].
Defensins, members of the antimicrobial peptide family, exist widely in nature. They have shown bactericidal activity against gram-positive bacteria, gram-negative bacteria, and even multidrug-resistant bacteria; moreover, they have unique antibacterial mechanisms which make it difficult to induce bacterial resistance [3]. Defensins are part of the innate defense system and play an important role in immune response [2,4,5]. In addition, defensins could regulate the proliferation and migration of epithelial cells, which is very important for overcoming injury, infection and inflammation [4]. These advantages of defensins make them ideal antibiotic substitutes.
Porcine beta defensin 2 (pBD2) is an antibacterial peptide secreted by pigs. pBD2 has shown good antibacterial activity and immunomodulatory function in vitro [6,7,8,9]. pBD2 could inhibit E. coli and S. aureus, as well as some clinically isolated multidrug-resistant bacteria [9]. pBD2 has the strongest antimicrobial activity among the porcine beta defensins (pBDs) detected, including pBD1, pBD2, pBD114, and pBD129 [9,10,11,12]. In addition, pBD2 could alleviate the inflammatory response induced by exogenous stimulation in mice [8]. As a feed additive, pBD2 showed a certain effect on disease resistance and growth promotion, related to the reduction of numbers of harmful bacteria [13]. pBD2 may be a suitable antibiotic substitutes in the pig industry. However, the function and mechanism of how pBD2 protects the host from bacteria remains unclear.
In order to explain the functions and mechanism of pBD2 in the host cells against bacteria, Escherichia coli was selected as an example and intestinal porcine enterocyte cells (IPEC-J2) were selected as the infected model. E. coli is a gram-negative bacteria that exists widely in the intestinal tracts of animals. It is one of the most important pathogenic bacteria causing diarrhea in weaned piglets, which leads to considerable economic losses [14]. The IPEC-J2 cell line was originally isolated from the jejunum of a neonatal unsuckled piglet, and is a non-transformed, permanent intestinal cell line. IPEC-J2 cells are ideal for studying the antibacterial effect of pBD2 on cells [15]. In this experiment, IPEC-J2 cells were challenged with E. coli, and the transcriptome method was used to detect the effect of pBD2 on the gene expression of cells infected by E. coli. Our study helps to understand the functions and mechanisms of defensin in host cells against E. coli.
2. Results
2.1. Analysis of Bactericidal Activity
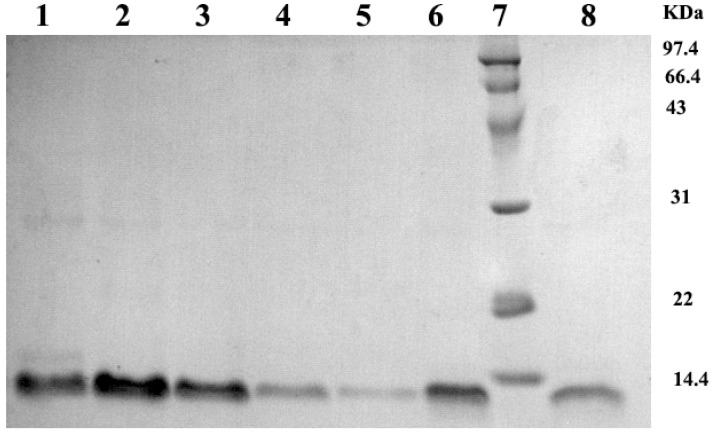
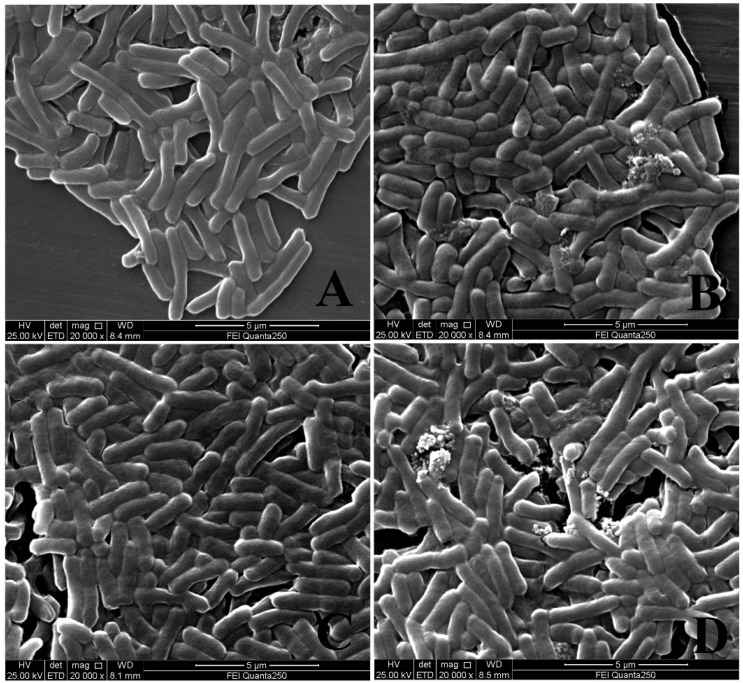
The recombinant pBD2 was induced and purified following our previous study [9]. The purified pBD2 showed high purity, and the molecular weight was approximately 12 kDα as expected (Figure 1). The recombinant pBD2 showed high bactericidal activity. E. coli were cracked and dead after incubation with 20 μg/mL pBD2 for 1 h and 4 h, the debris was observed, and the bactericidal effect was more obvious at 4 h than those at 1 h (Figure 2).
Figure 1.
Analysis of purified pBD2 by SDS-PAGE. Lanes 1, 2, 3, 4, 5, 6, 8 indicate the purified protein from different collected tubes; Lane 7 indicates the protein marker.
Figure 2.
Morphological changes of E. coli treated with pBD2, viewed by scanning electron microscopy. (A,B) indicate the E. coli treated with 0 and 20 μg/mL pBD2 for 1 h, respectively; (C,D) indicated the E. coli treated with 0 and 20 μg/mL pBD2 for 4 h, respectively.
2.2. Transcriptome Profiling
cDNA libraries were sequenced on the Illumina high-throughput platform, generating significant amounts of high-quality raw data. After removal of adaptor sequences, low-quality, and contaminated reads, only the resulting clean reads were assembled to build transcripts. In these results, 20,721,673–35,369,943 clean data were obtained for samples, and the minimum Q30 was 93.23% (Table 1). The percentage of reads mapped to the reference genome was 95.84–96.55% (Table 2).
Table 1.
Sequencing data statistics.
| Samples | Clean Reads | Clean Bases | GC Content | %≥Q30 |
|---|---|---|---|---|
| Control-1 | 35,369,943 | 10,533,069,694 | 52.27% | 94.10% |
| Control-2 | 32,617,012 | 9,703,386,646 | 52.26% | 94.14% |
| Control-3 | 31,534,327 | 9,375,721,120 | 52.35% | 94.31% |
| E. coli-1 | 24,055,348 | 7,170,857,754 | 52.64% | 93.23% |
| E. coli-2 | 25,929,754 | 7,718,143,996 | 52.37% | 93.55% |
| E. coli-3 | 22,754,029 | 6,773,668,654 | 52.28% | 93.90% |
| pBD2-1 | 20,721,673 | 6,176,881,268 | 51.76% | 93.46% |
| pBD2-2 | 23,894,463 | 7,107,703,946 | 52.28% | 94.16% |
| pBD2-3 | 21,228,755 | 6,327,976,062 | 51.94% | 93.67% |
Notes: Control, E. coli and pBD2 indicate the IPEC-J2 in the control, E. coli and E. coli +pBD2 groups, and 1–3 indicate the three replicates, respectively.
Table 2.
Gene comparison efficiency statistics.
| Samples | Mapped Reads | Unique Mapped Reads | Multiple Map Reads |
|---|---|---|---|
| Control-1 | 67,898,715 (95.98%) | 66,065,506 (93.39%) | 1,833,209 (2.59%) |
| Control-2 | 62,849,152 (96.34%) | 61,097,600 (93.66%) | 1,751,552 (2.69%) |
| Control-3 | 60,714,671 (96.27%) | 59,081,017 (93.68%) | 1,633,654 (2.59%) |
| E. coli-1 | 46,168,875 (95.96%) | 44,891,836 (93.31%) | 1,277,039 (2.65%) |
| E. coli-2 | 49,843,598 (96.11%) | 48,508,765 (93.54%) | 1,334,833 (2.57%) |
| E. coli-3 | 43,776,016 (96.19%) | 42,624,022 (93.66%) | 1,151,994 (2.53%) |
| pBD2-1 | 39,790,781 (96.01%) | 38,760,294 (93.53%) | 1,030,487 (2.49%) |
| pBD2-2 | 46,057,704 (96.38%) | 44,834,945 (93.82%) | 1,222,759 (2.56%) |
| pBD2-3 | 40,692,378 (95.84%) | 39,483,201 (92.99%) | 1,209,177 (2.85%) |
Notes: Control, E. coli and pBD2 indicate the IPEC-J2 in the control, E. coli and E. coli +pBD2 groups, and 1–3 indicate the three replicates, respectively.
2.3. Characterization of Differentially Expressed Genes by RNA Sequencing
There were 230 differential expressed genes (DEGs) between the control and E. coli treatments, including 145 significantly upregulated genes and 85 significantly downregulated genes. There were 812 DEGs were between the E. coli and E. coli +pBD2 groups, including 431 significantly upregulated genes and 381 significantly downregulated genes. There were 94 overlapping genes observed by Venn diagram analysis between the two DEG groups.
2.4. KEGG Analysis Revealed That Immune Responses of Cells Were Trigger by E. coli
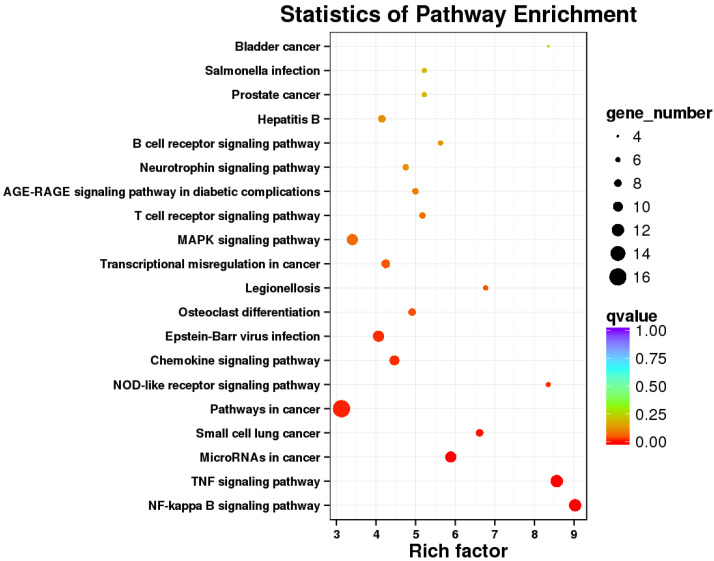
To determine the functions of the DEGs involved, Kyoto Encyclopedia of Genes and Genomes (KEGG) annotation and corresponding enrichment analysis were performed. KEGG pathway analysis of the DEGs between the E. coli and control groups revealed that the immune pathways were significantly enriched, including NF-kappa B signaling pathway, TNF signaling pathway, NOD-like receptor signaling pathway, Chemokine signaling pathway, MAPK signaling pathway, etc., suggesting that E. coli caused immune responses of IPEC-J2 cells to eliminate the invading microbes (Figure 3). These results clearly indicate that the transcriptome of IPEC-J2 cells was changed after infection. Genes involved in the immune signaling pathways are listed in Table 3. The genes related to the immune responses include CXCL2 (chemokine (C-X-C motif) ligand 2), PTGS2 (prostaglandin G/H synthase 2), TNFAIP2 (tumor necrosis factor alpha-induced protein 2), NFKB2 (nuclear factor NF-kappa-B p100 subunit), NFKBIZ (NF-kappa-B inhibitor zeta), IL11 (interleukin-11), CCL20 (C-C motif chemokine ligand 20), etc. According to the PPI (protein–protein interaction) net analysis of the 230 DEGs, NFKB1(Nuclear factor NF-kappa-B p105 subunit), TP53 (tumor protein p53), PTEN (Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase) and CDC6 (cell division control protein 6) may be the key genes in IPEC-J2 cells in response to E. coli (Figure 4).
Figure 3.
KEGG pathway analysis of DEGs between the control and E. coli groups.
Table 3.
DEGs involved in the immune signaling pathways caused by E. coli.
| KEGG Pathway | ID | Gene Name | Corrected p-Value |
|---|---|---|---|
| NF-kappa B signaling pathway | ko04064 | TNFAIP3, BIRC3, CXCL8, TICAM1, NFKBIA, ICAM1, PTGS2, BCL10, PLAU, NFKB1, RELB, NFKB2 | 0.00000127 |
| TNF signaling pathway | ko04668 | TNFAIP3, MAP3K8, BIRC3, EDN1, NFKBIA, ICAM1, PTGS2, ENSSSCG00000008954, CXCL2, NFKB1, CCL20, BCL3 | 0.00000233 |
| NOD-like receptor signaling pathway | ko04621 | TNFAIP3, BIRC3, CXCL8, NFKBIA, NFKBIB, NFKB1 | 0.014302364 |
| Chemokine signaling pathway | ko04062 | FOXO3, RAC1, CXCL8, NFKBIA, NFKBIB, ENSSSCG00000008954, CXCL2, NFKB1, GNAI1, CCL20 | 0.014397446 |
Figure 4.
PPI analysis of DEGs in the control and E. coli groups.
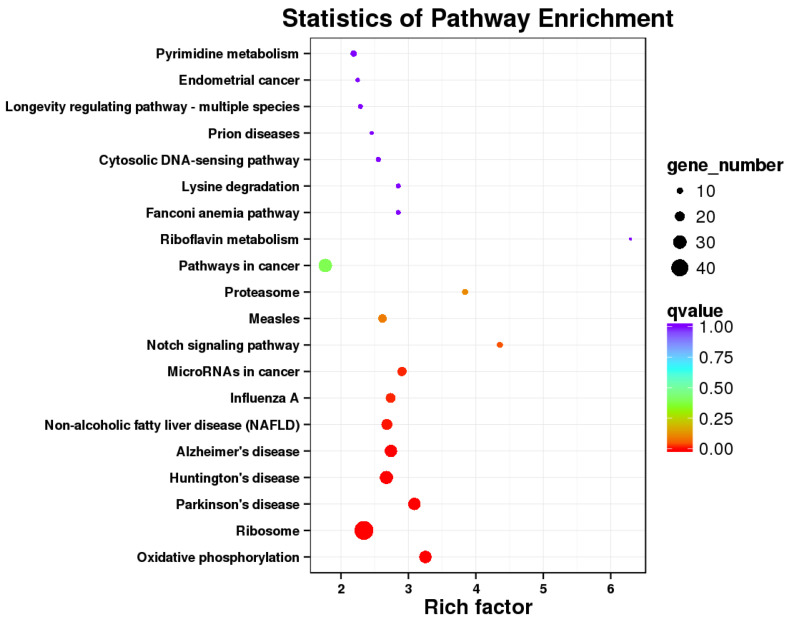
2.5. KEGG Pathway Analysis Revealed That pBD2 Has Multiple Functions in IEPC-J2 Cells against E. coli
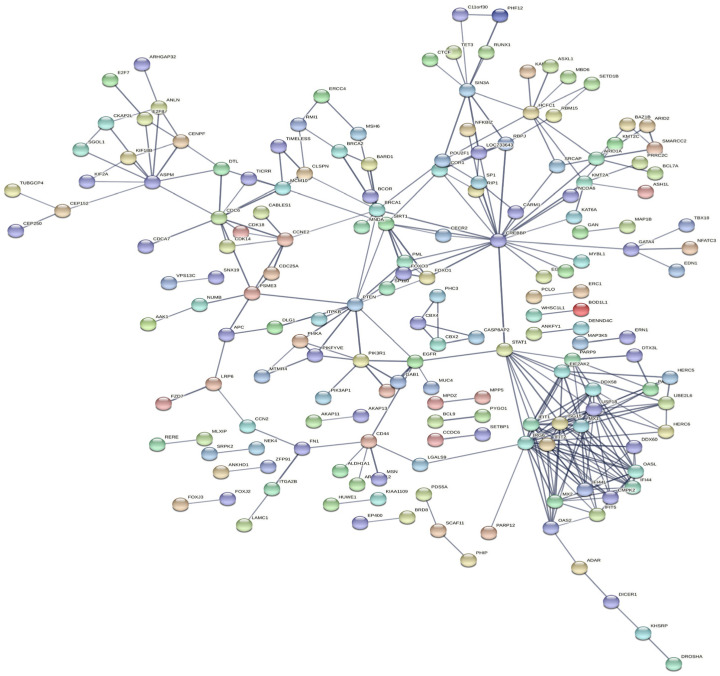
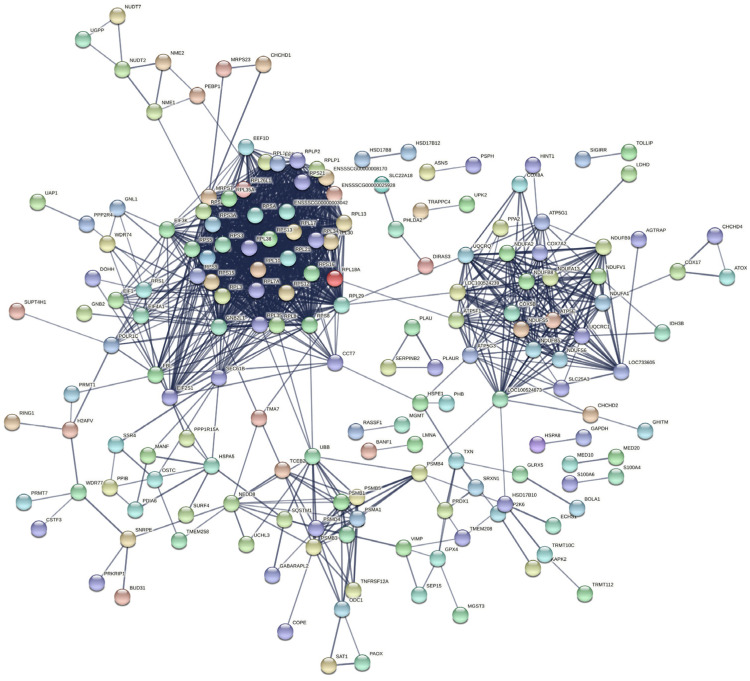
There were 812 DEGs between the E. coli and E. coli +pBD2 groups. KEGG pathway analysis revealed DEGs involved in ribosomes, oxidative phosphorylation, and some diseases including Parkinson’s disease, Huntington’s disease, Alzheimer’s disease, Non-alcoholic fatty liver disease (NAFLD), and others (Figure 5). These results clearly indicate that the transcriptome of IPEC-J2 cells was regulated by pBD2. The 431 upregulated DEGs and 381 downregulated DEGs were analyzed by PPI net, respectively (Figure 6 and Figure 7). The results showed that the DEGs related to immune responses were obvious in the upregulated DEGs by PPI analysis, including MX1 (interferon-induced GTP-binding protein Mx1), EIF2AK2 (eukaryotic translation initiation factor 2 alpha kinase 2), OAS2 (2′-5′-oligoadenylate synthetase 2), IFIT1 (interferon-induced protein with tetratricopeptide repeats 3), IFIT2 (interferon-induced protein with tetratricopeptide repeats 2), IFIT5 (interferon-induced protein with tetratricopeptide repeats 5), IF144 (interferon-induced protein 44), etc. STAT1 (signal transducer and activator of transcription 1) and CREBBP (CREB binding protein) were the key genes (Figure 6). The downregulated DEGs were clustered into three groups (ribosome, oxidative phosphorylation, and proteasome) (Figure 7). These results indicate that pBD2 has multiple functions in cells against E. coli.
Figure 5.
KEGG pathway analysis of DEGs in the E. coli and E. coli +pBD2 groups.
Figure 6.
PPI analysis of the upregulated DEGs in the E. coli and E. coli +pBD2 groups.
Figure 7.
PPI analysis of the downregulated DEGs in the E. coli and E. coli +pBD2 groups.
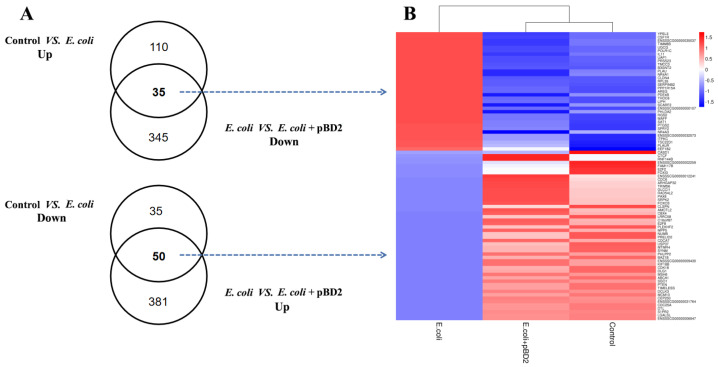
2.6. Comparison of pBD2 Effect with E. coli-Induced Transcriptome Changes
There were 94 overlapping DEGs by Venn diagram analysis, which enabled us to identify transcripts specifically regulated by pBD2. Among these 94 DEGs, 85 DEGs were reverse expressed including 35 DEGs upregulated by E. coli and downregulated by pBD2 under E. coli stimulation (E. coli + pBD2 group). The other 50 DEGs were reverse expressed conversely to the above genes in both DEG groups (Figure 8).
Figure 8.
Overview of the DEGs after different treatments. (A) Venn diagrams of known DEGs based on comparisons of different groups. (B) Hierarchical clustering of the overlapping DEGs among different groups. The red indicates upregulated genes and the blue indicates downregulated genes.
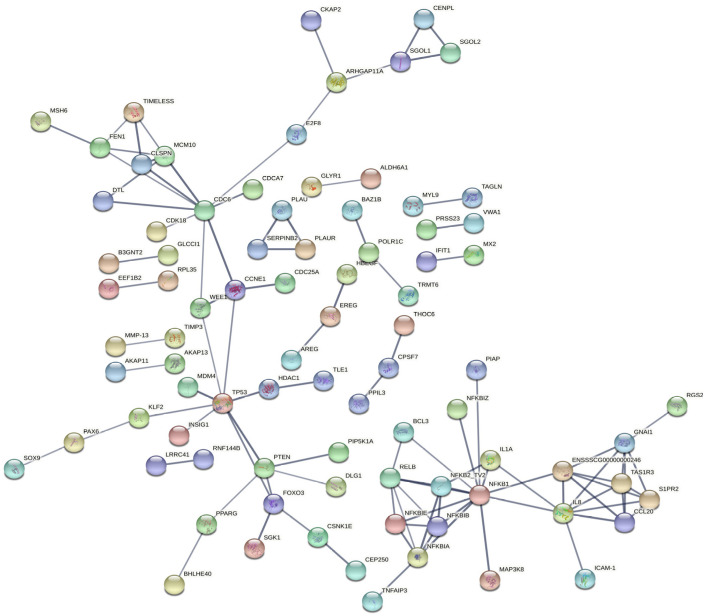
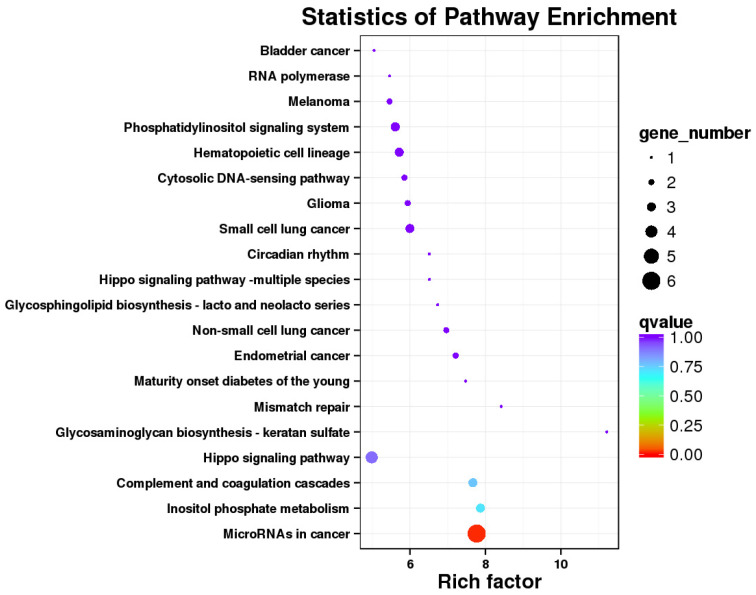
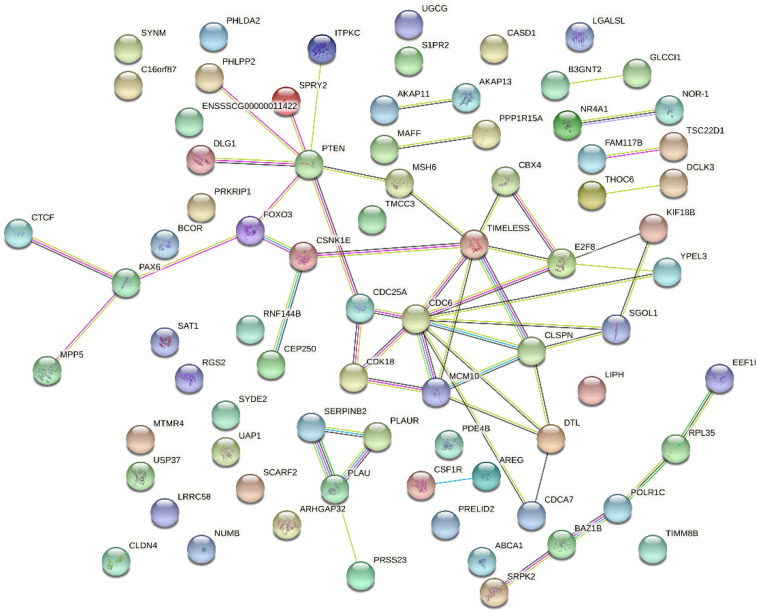
The KEGG pathway analysis of 85 DEGs with reverse expression showed that microRNA in cancer was the only significantly different signaling pathway, including PTEN, CDC25A (cell division cycle 25A), SPRY2 (protein sprouty homolog 2), E2F2 (E2F2 transcription factor), PTGS2, and PLAU (urokinase-type plasminogen activator) (Figure 9). In addition, PTEN and CDC6 were the hub genes in the PPI network for pBD2 to regulate IPEC-J2 cell responses to E. coli (Figure 10). In addition, immune-response-related genes among the 85 reverse expressed genes were considered. The immune-response-related genes IL11, PTGS2 and PLAU were all upregulated in the E. coli group compared with the control group, and pBD2 significantly downregulated the expression of these genes compared with the E. coli group.
Figure 9.
KEGG enrichment analysis of the DEGs overlapping between the two DEGs groups.
Figure 10.
PPI analysis of the DEGs overlapping between the two DEG groups.
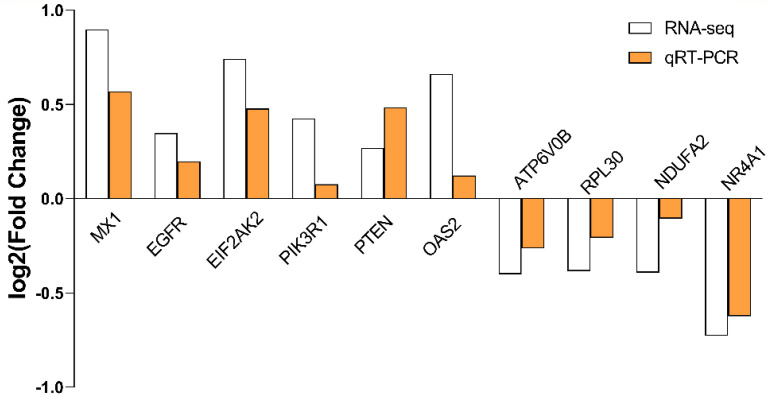
2.7. Validation of RNA-seq Data by qRT-PCR
Ten genes were selected for validation of RNA-seq, and the results of qRT-PCR were consistent with those of transcriptomics (Figure 11).
Figure 11.
The verification of RNA-Seq results by qRT-PCR, between E. coli and E. coli +pBD2 groups. The samples were analyzed in triplicate by qRT-PCR, and fold-changes in gene expression were calculated by 2−ΔΔCT methods with TUBA1B (tubulin alpha 1b) as a reference gene. MX1: Interferon-induced GTP-binding protein Mx1; EGFR: Epidermal growth factor receptor; EIF2AK2: Eukaryotic translation initiation factor 2 alpha kinase 2; PIK3R1: Phosphoinositide-3-kinase regulatory subunit alpha; PTEN: Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase; OAS2: 2′-5′-oligoadenylate synthetase 2; ATP6V0B: ATPase H+ transporting V0 subunit b; RPL30: Ribosomal protein L30; NDUFA2: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 2; NR4A1: Nuclear receptor subfamily 4, group A, member 1.
3. Discussion
Defensins, a kind of antimicrobial peptide, play an active role in resisting the invasion of pathogenic microorganisms [16]. pBD2 is one of the porcine beta defensins and shows strong antimicrobial activity against E. coli, S. aureus and isolated multidrug-resistant bacteria, with the advantages of high salt-tolerance, thermal stability, and low hemolytic activity, as demonstrated in our previous studies [9]. In addition, pBD2 has an immunomodulatory function [8]. However, the function and mechanisms of defensins in protecting the host from bacteria are still unclear.
In this study, we explored by transcriptomic analyses the effects of defensins on IPEC-J2 cells against E. coli. A total of 230 DEGs were obtained between the control and E. coli groups. The significant signaling pathways were mainly involved in immune responses, including NF-κB signaling pathway, TNF signaling pathway, NOD-like receptor signaling pathway, chemokine signaling pathway, etc., as demonstrated by KEGG enrichment analysis. The DEGs in these signaling pathways were upregulated, including NFKB1, NFKB2, NFKBIA (NF-kappa-B inhibitor alpha), NFKBIB (NF-kappa-B inhibitor beta), cytokines CXCL2, CCL20, IL1α (Interleukin 1 alpha), CXCL8 (C-X-C motif chemokine ligand 8), etc. Furthermore, NFKB1, TP53, PTEN, and CDC6 were key genes against E. coli, identified in the cells by PPI net analysis. NFKB1 is a key gene in the NF-κB signaling pathway, and PTEN is an important gene in the PI3K phosphatidyl inositol 3-kinase (PI3K)–protein kinase B (PKB/Akt) signaling pathway, which is involved in cell proliferation, differentiation, apoptosis, and immunity [17]. TP53 and PTEN are essential for initiating apoptosis and inflammatory response [18,19]. NFKB1, TP53, and PTEN were all upregulated, and were closely related to inflammatory response. The transcriptomic analyses indicated that immune responses of IPEC-J2 cells were triggered by E. coli, by which cells could eliminate invading pathogens for survival. It has been reported that multiple signaling pathways related to inflammation were activated by E. coli or LPS, resulting in the release of inflammatory cytokines [20,21,22,23,24,25]; those results were consistent with ours. CDC6 was a key gene for cell proliferation by PPI net analysis (Figure 4). In addition to CDC6, CDK18 (cyclin-dependent kinase 18), E2F8 (E2F8 transcription factor), CDCA7 (cell division cycle-associated protein 7), MCM10 (minichromosome maintenance 10 replication initiation factor) and TIMELESS (protein timeless homolog) are all related to cell proliferation according to their functions (Figure 4). It has been reported that CDC6 and CDK18 promoted proliferation of epithelial cells and inhibited apoptosis [26]. These genes were all decreased in the E. coli group, which perhaps suggest that cell proliferation was inhibited by E. coli.
There were 812 DEGs between the E. coli and E. coli +pBD2 groups, including 431 upregulated and 381 downregulated genes. PPI net analysis confirmed that the genes in the upregulated DEGs related to immune responses and cell proliferation, and STAT1 and CREBBP were the key genes (Figure 6). These genes are involved in many signaling pathways, including JAK-STAT and PI3K-AKT, which are related to immune responses. Our results were consistent with previous findings that defensin regulates immune responses in complex ways [27]. The genes related to cell proliferation in the upregulated DEGs included CDC6, CDK18, CDK14, E2F8, CDCA7, MCM10, TIMELESS, CDC25A, CCNE2 (G1/S-specific cyclin-E2), etc., according to their functions and the PPI net analysis. These genes were all increased by pBD2, which indicates that pBD2 protected cells from E. coli infection by regulating cell proliferation and inhibiting apoptosis, which might be critical for the resolution of injury, infection, and inflammation [2]. pBD2 upregulated DEGs related to immune responses and cell proliferation, which perhaps indicates that pBD2 could promote cell proliferation and inhibit the immune responses caused by E. coli.
The downregulated DEGs were obviously clustered into three groups (ribosome, oxidative phosphorylation, and proteasome) (Figure 7), which indicates that pBD2 had multiple functions in the cells. The ribosome is a complex molecular machine composed of numerous distinct proteins and nucleic acids, and is responsible for the translation of information contained in mRNAs into functional proteins, which play an important role in the execution of gene-expression programs, regulating basic biological processes such as cell growth, cell division, and differentiation. The hyperactivation of ribosome biogenesis has a critical role in cancer initiation and progression [28], and inhibition of ribosome biogenesis represents a potential therapeutic avenue for cancer treatment [29]. pBD2 downregulated the DEGs in the ribosome, perhaps implying that pBD2 could play a role in inhibiting the occurrence of cancer. The DEGs in oxidative phosphorylation (OXPHOS) were related to many diseases, and were also downregulated. OXPHOS was another source of ATP in cells in addition to glycolysis. Although it was reported that OXPHOS is related to cancer, many highly proliferative cell types including many cancer cells can preferentially utilize glycolysis [30,31,32,33]. In addition, OXPHOS inhibitors have heralded novel uses either for treating cancers in which OXPHOS is upregulated or alleviating tumor hypoxia to improve treatment outcomes [34]. In this study, pBD2 could downregulate the DEGs related to OXPHOS, which perhaps further implies that pBD2 could treat cancer. It was reported that peptide antibiotic leucinostatins showed inhibitory action on OXPHOS [35], which is consistent with our results. It was reported that the DEGs in OXPHOS were significantly enriched and upregulated in the large yellow croaker (Larimichthys crocea) when infected by Pseudomonas plecoglossicida, and were also upregulated in the murine model when infected by R. conorii [36,37]. These findings suggest that OXPHOS was related to immune responses and contributed to anti-infection strategies in the hosts. The mechanism underlying this strengthened energy metabolism is unknown, and the DEGs in OXPHOS were not enriched between the control and E. coli infection groups in this study. pBD2 could downregulate the DEGs, perhaps implying that pBD2 could protect the host from excessive inflammatory responses.
The proteasome is a large protein complex, responsible for the degradation of intracellular proteins, which regulates cellular proteostasis through selective degradation of ubiquitylated proteins [38,39]. Thereby, it performs a crucial role in cellular regulation and homeostasis, and has an important function in a variety of basic cellular processes including regulation of cell cycle progression, signal transduction, modulation of immune and inflammatory responses, etc. Malfunction of the ubiquitin-proteasome system (UPS) contributes to various diseases including cancer, inflammation, and neurodegeneration [40]. In this study, pBD2 inhibited the DEGs in the proteasome, and even lowered some ubiquitin expression including UBB (polyubiquitin-B), NEDD8, UBE2J2 (ubiquitin-conjugating enzyme E2 J2), APC2 and APC11 (anaphase-promoting complex subunit 2 and 11), proteasomal ubiquitin receptor UCHL3 (ubiquitin carboxyl-terminal hydrolase isozyme L3), USP19 (ubiquitin carboxyl-terminal hydrolase 19) genes, etc., all perhaps further indicating that pBD2 inhibited inflammations and the occurrence of cancer, as discussed above. It was reported that PR39, one of the porcine antimicrobial peptides, was a non-competitive and reversible inhibitor of 20S proteasome, and inhibited inflammation [41,42]. Our results are consistent with that report. pBD2 downregulated DEGs related to ribosome, oxidative phosphorylation, and proteasome, indicating that pBD2 could inhibit the occurrence of cancer, which needs further research. Some reported defensins have been regarded as a potential therapeutic target for cancer treatment, and attractive novel therapeutic candidates for antimicrobial and anticancer purposes [43,44], which is also consistent with our results.
pBD2 changed the transcriptional response caused by E. coli. There were 85 genes with reverse expression in the two DEGs group; KEGG analysis showed that these overlapping genes included enrichment of the microRNA in cancer (Figure 9). In addition, PTEN and CDC6 were key genes for pBD2 according to PPI net analysis (Figure 10). It has been reported that defensin regulated the expression of microRNA, and that the expression of miR-34a-5p increased in ethanol-induced liver injury in transgenic (TG) mice with overexpressing human neutrophil peptide 1, compared with WT (wild type) mice [45]. Those results perhaps imply that pBD2 had the ability to regulate gene expression by regulating microRNA, which needs further study. The genes associated with microRNA in cancer included PTEN, CDC25A, SPY2, PLAU, E2F2, and PTGS2. It was reported that PTEN could inhibit immune response and tumors [46]. SPRY2 was upregulated in glioblastoma, and overexpression of SPRY2 is associated with human oral squamous-cell carcinogenesis [47,48]. Dysregulation of PLAU is often accompanied by various cancers, and inhibition of PLAU expression could suppress tumor growth [49]. CDC25A, E2F2, and PTGS2 are all related to the cell cycle, and their dysregulation is related to tumors [50,51,52]. In this study, pBD2 showed anti-tumor potential by upregulating the expression of PTEN and downregulating the expression of SPRY2 and PLAU, and pBD2 was also shown to promote cell proliferation by upregulating the expression of CDC25A, E2F2, and PTGS2, which perhaps might lead to tumorigenesis. Some scholars have reported that defensin had a strong inhibitory effect on cancer [53,54,55], but others had opposite view suggesting that defensin might be related to the occurrence of cancer because the expression of defensins increased abnormally in cancer [56,57]. In addition to CDC25A, E2F2, and PTGS2 relating to the cell cycle, these genes are all related to cell proliferation including CDC6, CDK18, E2F8, CLSPN, CDCA7, MCM10, TIMELESS, KIF18B, etc., as mentioned above (Figure 10). These genes were all decreased in the E. coli group, and all increased by pBD2, indicating that pBD2 regulated cell proliferation and inhibited apoptosis, playing its role in the resolution of injury, infection, and inflammation [2]. pBD3 enhanced ovarian granulosa cell proliferation and migration [58]. Human β-defensins also stimulated various cellular activities, including keratinocyte proliferation, migration, and wound healing [59]. Those results are consistent with ours, which perhaps indicate that pBD2 could inhibit the occurrence of cancer, and promote cell proliferation to protect cells from infection. The analysis of the overlapping genes with reverse expression was consistent with the above analysis of DEGs in the E. coli and E. coli +pBD2 groups. Our results provide new insights into the potential function and mechanism of pBD2 against E. coli, and the results should be confirmed by further study on mice or human cell lines.
4. Materials and Methods
4.1. Strains and Cells
E. coli ATCC 25922 was purchased from the Beijing Ordinary Microbiology Strain Store Center (Beijing, China). The IPEC-J2 cells were a gift from Zhanyong Wei at the College of Veterinary Medicine, Henan Agricultural University.
4.2. Preparation for pBD2
pBD2 was expressed and purified in our laboratory by affinity chromatography based on the constructed strains BL21(DE3) pLysS-pET30a-pBD2 [9]. The expression products were analyzed by SDS-PAGE, and the protein concentration was determined by bicinchoninic acid (BCA) assay (CW Biotech, Beijing, China).
4.3. Bactericidal Activity
The bactericidal activity was analyzed by scanning electron microscopy, as described previously [6]. E. coli ATCC 25922 was cultured in Luria Bertani (LB) medium until logarithmic growth stage at 220 rpm, then E. coli was incubated with 20 μg/mL pBD2 for 1 h or 4 h, and harvested by centrifugation. After being washed for 3 × 10 min in 10 mM PBS buffer, E. coli was fixed with 2.5% glutaraldehyde for 4 h, then dehydrated with gradient concentrations of ethanol, and the samples were observed by scanning electron microscopy (FEI Quanta 250, FEI, Hillsboro, OR, USA).
4.4. Cell Culture and Treatment
The IPEC-J2 cells were grown in 1640 medium supplemented with 10% fetal bovine serum (TianHang Biotechnology, Zhejiang, China) and 1% penicillin/streptomycin at 37 °C in an atmosphere of 5% CO2. The cells were seeded into a six-well plate and cultured until reaching 80% confluence, then at the logarithmic period (MOI = 50:1) the IPEC-J2 cells were challenged with E. coli or E. coli and 20 μg/mL pBD2 for 2 h in 1640 medium without fetal bovine serum and 1% penicillin/streptomycin. After washing with PBS three times, the cells were cultured for an additional 2 h in the medium without fetal bovine serum and 1% penicillin/streptomycin. After washing three times with PBS, 1 mL of TRIzon was added to the six-well plate, and the cells were lysed by pipetting, then the solute was transferred to RNase-free Eppendorf tubes for RNA extraction.
4.5. Library Preparation and Quality Control
RNAs were extracted using TRIzol reagents (CW Biotech, Beijing, China). The extracted RNAs were quantified using a spectrophotometer (Nanodrop 2000, Thermo Fisher Scientific, Waltham, MA, USA). A total of 3 μg RNA was sent to Biomarker Co., Ltd. (Beijing, China) to construct the cDNA libraries. Sequencing was performed on an Illumina HiSeqTM 2500 platform. The original raw data were saved in FASTQ file format. Each sequenced sample included two FASTQ files, containing reads from either end of the cDNA fragments. Quality control was performed to remove adaptor sequences, low-quality, and contaminated reads. Then the clean reads were mapped to the reference genome by alignment software HISAT2.
4.6. Analysis of DEGs
The transcript and gene expression levels were measured by FPKM (fragments per kilobase of transcript per million fragments mapped). The DESeq2 R package was used to analyze differential gene expression. The false discovery rate (FDR) was corrected using the Benjamini–Hochberg procedure. The threshold of FDR < 0.05 was used to filter out differential expressed genes (DEGs). Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of the DEGs were performed using BMKCloud (www.biocloud.net, accessed on 5 September 2019), based on the KEGG database (http://www.genome.jp/kegg/, accessed on 5 September 2019), and protein–protein interaction (PPI) networks of the DEGs were constructed on https://www.string-db.org/, accessed on 17 August 2021.
4.7. qRT-PCR
The extracted RNAs were converted to complimentary (c) DNA by a reverse transcriptase synthesis kit (DRR047A; TaKaRa Biotechnology, Shiga, Japan). The primers were designed using Primer Premier™ 5.0 (Sigma–Aldrich, Saint Louis, MO, USA) and are detailed in Table 4. Assays were performed with SYBR green dye (QIAGEN, Dusseldorf, Germany) using a real-time PCR cycler (LightCycler 96, Roche, Basel, Switzerland). The program was 95 °C for 30 s, 95 °C for 15 s, 60 °C for 30 s, 72 °C for 15 s, for 40 cycles. The results were analyzed using the 2−ΔΔCT method with TUBA1B (tubulin alpha 1b) as a reference gene [60].
Table 4.
Primers used for qRT-PCR.
| Genes | Sequence (5′ → 3′) | Size (bp) | GenBank Number | |
|---|---|---|---|---|
| MX1 | Forward | GTTACCGGGACAGCGAGATT | 105 | NM_214061.2 |
| Reverse | CATGACTGATTCCCACGCCT | |||
| EGFR | Forward | AGGACGAAGCAACATGGTCA | 132 | NM_214007.1 |
| Reverse | TGCATAGCACAGGTTTCGGT | |||
| EIF2AK2 | Forward | CCCTGCACTTCTAGCCATCT | 121 | NM_214319.1 |
| Reverse | CGACCACTGGCCATTTCTTTC | |||
| PIK3R1 | Forward | CTTGAGTCGGGTGCTGGAAC | 164 | XM_021076847.1 |
| Reverse | AACGCGTCCCTAACCGATTC | |||
| PTEN | Forward | TGCAATCCTCAGTTTGTGGT | 224 | NM_001143696.1 |
| Reverse | TCCTCTGGTCCTGGTATGAAG | |||
| OAS2 | Forward | AGCCAGAGCAATGGGAAACT | 228 | NM_001031796.1 |
| Reverse | GAGTTGCCCCTCAAGACTGT | |||
| ATP6VOB | Forward | AACCCCAGCCTCTTCGTAAA | 100 | XM_021096834.1 |
| Reverse | TCACTCTGGAGGTCTGAAGG | |||
| RPL30 | Forward | GACAAGGTCCAATGTTCCCA | 110 | NM_001190178.1 |
| Reverse | CCAACCTCTTTTGTAGCCGT | |||
| NDUFA2 | Forward | TGCTAAGTGGCAAAGCCTG | 167 | XM_003124046.4 |
| Reverse | GGTAGAGGGTGGAACAAGGAA | |||
| NR4A1 | Forward | TGAGAAGGTTCCCGGCTTTG | 196 | ENSSSCG00000031321 |
| Reverse | GATGCTGTCGATCCAGTCCC | |||
| TUBA1B | Forward | TACTCACCTCGACTCTTAGC | 103 | NM_001044544.1 |
| Reverse | GATGCACTCACGCATGG | |||
4.8. Statistical Analysis
All experiments were conducted with three biological replicates. Data were assessed by analysis of variance using SPSS Statistics 24 (IBM, Armonk, NY, USA).
5. Conclusions
In this study, IPEC-J2 cells were challenged with E. coli, and the effect of pBD2 on the gene expression of cells infected by E. coli was detected by transcriptome analyses. KEGG enrichment analysis revealed that DEGs between the control and E. coli groups were involved in signaling pathways related to immune responses. The DEGs between the E. coli and E. coli +pBD2 groups were involved in the ribosome, oxidative phosphorylation, and some disease pathways. These results indicate that pBD2 can play an active role in many biological processes including cancer, cell proliferation, and immunity. Furthermore, analysis of the overlapping genes with reverse expression in two DEG groups further revealed that pBD2 was involved in multiple biological processes to protect cells from E. coli. Our results provide new insights into the potential function and mechanism of pBD2 against E. coli. Meanwhile, this study provides a certain theoretical basis for the research and development of novel peptide drugs.
Author Contributions
C.L. designed the experiments and supervised the whole study. S.L. and X.L. performed the cell experiment; S.L. and X.S. performed the RNA extract and qRT-PCR. X.S., Y.L. and F.Z. analyzed the data of RNA-seq. C.L. and X.L. analyzed the data and wrote the paper. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated during this study are included in this article. RNA-Seq data have been deposited in the National Center for Biotechnology Information database under accession number (PRJNA862330).
Conflicts of Interest
The authors declare no competing interest.
Funding Statement
This research was funded by Science and Technology Research Project in Henan Province, grant number 202102110089.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Shao Y., Wang Y., Yuan Y., Xie Y. A systematic review on antibiotics misuse in livestock and aquaculture and regulation implications in China. Sci. Total Environ. 2021;798:149205. doi: 10.1016/j.scitotenv.2021.149205. [DOI] [PubMed] [Google Scholar]
- 2.Blyth G.A.D., Connors L., Fodor C., Cobo E.R. The network of colonic host defense peptides as an innate immune defense against enteropathogenic bacteria. Front. Immunol. 2020;11:965. doi: 10.3389/fimmu.2020.00965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avila E.E. Functions of antimicrobial peptides in vertebrates. Curr. Protein Pept. Sci. 2017;18:1098–1119. doi: 10.2174/1389203717666160813162629. [DOI] [PubMed] [Google Scholar]
- 4.Robinson K., Deng Z., Hou Y., Zhang G. Regulation of the Intestinal Barrier Function by Host Defense Peptides. Front. Vet. Sci. 2015;2:57. doi: 10.3389/fvets.2015.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peschel A., Sahl H.G. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat. Rev. Microbiol. 2006;4:529–536. doi: 10.1038/nrmicro1441. [DOI] [PubMed] [Google Scholar]
- 6.Chen R.B., Zhang K., Zhang H., Gao C.Y., Li C.L. Analysis of the antimicrobial mechanism of porcine beta defensin 2 against E. coli by electron microscopy and differentially expressed genes. Sci. Rep. 2018;8:14711. doi: 10.1038/s41598-018-32822-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang K., Zhang H., Gao C., Chen R., Li C. Antimicrobial Mechanism of pBD2 against Staphylococcus aureus. Molecules. 2020;25:3513. doi: 10.3390/molecules25153513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han F., Zhang H., Xia X., Xiong H., Song D., Zong X., Wang Y. Porcine β-defensin 2 attenuates inflammation and mucosal lesions in dextran sodium sulfate-induced colitis. J. Immunol. 2015;194:1882–1893. doi: 10.4049/jimmunol.1402300. [DOI] [PubMed] [Google Scholar]
- 9.Li C.L., Zhao Y.C., Song X.Y., Huang X.X., Zhao W.D. Molecular cloning, expression and characterization of the porcine β defensin 2 in E. coli. Protein Pept. Lett. 2013;20:715–723. doi: 10.2174/0929866511320060010. [DOI] [PubMed] [Google Scholar]
- 10.Li C.L., Xu T.T., Chen R.B., Huang X.X., Zhao Y.C., Bao Y.Y., Zhao W.D., Zheng Z.Y. Cloning, expression and characterization of antimicrobial porcine β defensin 1 in Escherichia coli. Protein Expr. Purif. 2013;88:47–53. doi: 10.1016/j.pep.2012.11.015. [DOI] [PubMed] [Google Scholar]
- 11.Su G., Xie K., Chen D., Yu B., Huang Z., Luo Y., Mao X., Zheng P., Yu J., Luo J., et al. Differential expression, molecular cloning, and characterization of porcine beta defensin 114. J. Anim. Sci. Biotechnol. 2019;10:60. doi: 10.1186/s40104-019-0367-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie K., Su G., Chen D., Yu B., Huang Z., Yu J., Zheng P., Luo Y., Yan H., Li H., et al. The immunomodulatory function of the porcine β-defensin 129: Alleviate inflammatory response induced by LPS in IPEC-J2 cells. Int. J. Biol. Macromol. 2021;188:473–481. doi: 10.1016/j.ijbiomac.2021.07.194. [DOI] [PubMed] [Google Scholar]
- 13.Peng Z., Wang A., Feng Q., Wang Z., Ivanova I.V., He X., Zhang B., Song W. High-level expression, purification and characterisation of porcine β-defensin 2 in Pichia pastoris and its potential as a cost-efficient growth promoter in porcine feed. Appl. Microbiol. Biotechnol. 2014;98:5487–5497. doi: 10.1007/s00253-014-5560-7. [DOI] [PubMed] [Google Scholar]
- 14.Vermeire B., Gonzalez L.M., Jansens R.J.J., Cox E., Devriendt B. Porcine small intestinal organoids as a model to explore ETEC-host interactions in the gut. Vet. Res. 2021;52:94. doi: 10.1186/s13567-021-00961-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vergauwen H. The IPEC-J2 cell line. In: Verhoeckx K., Cotter P., López-Expósito I., Kleiveland C., Lea T., Mackie A., Requena T., Swiatecka D., Wichers H., editors. The Impact of Food Bioactives on Health: In Vitro and Ex Vivo Models. 1st ed. Volume 12. Springer; Cham, Switzerland: 2015. pp. 125–134. [PubMed] [Google Scholar]
- 16.Holly M.K., Diaz K., Smith J.G. Defensins in viral infection and pathogenesis. Annu. Rev. Virol. 2017;4:369–391. doi: 10.1146/annurev-virology-101416-041734. [DOI] [PubMed] [Google Scholar]
- 17.Hemmings B.A., Restuccia D.F. PI3K-PKB/Akt pathway. Cold Spring Harb. Perspect. Biol. 2012;4:a011189. doi: 10.1101/cshperspect.a011189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blagih J., Buck M.D., Vousden K.H. p53, cancer and the immune response. J. Cell Sci. 2020;133:jcs237453. doi: 10.1242/jcs.237453. [DOI] [PubMed] [Google Scholar]
- 19.Nunes-Santos C.J., Uzel G., Rosenzweig S.D. PI3K pathway defects leading to immunodeficiency and immune dysregulation. J. Allergy Clin. Immunol. 2019;143:1676–1687. doi: 10.1016/j.jaci.2019.03.017. [DOI] [PubMed] [Google Scholar]
- 20.Xiao H.B., Wang C.R., Liu Z.K., Wang J.Y. LPS induces pro-inflammatory response in mastitis mice and mammary epithelial cells: Possible involvement of NF-κB signaling and OPN. Pathol. Biol. 2015;63:11–16. doi: 10.1016/j.patbio.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 21.Chen G., Goeddel D.V. TNF-R1 signaling: A beautiful pathway. Science. 2002;296:1634–1635. doi: 10.1126/science.1071924. [DOI] [PubMed] [Google Scholar]
- 22.Freche B., Reig N., van der Goot F.G. The role of the inflammasome in cellular responses to toxins and bacterial effectors. Semin. Immunopathol. 2007;29:249–260. doi: 10.1007/s00281-007-0085-0. [DOI] [PubMed] [Google Scholar]
- 23.Souvannavong V., Saidji N., Chaby R. Lipopolysaccharide from Salmonella enterica activates NF-kappaB through both classical and alternative pathways in primary B Lymphocytes. Infect. Immun. 2007;75:4998–5003. doi: 10.1128/IAI.00545-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huxford T., Hoffmann A., Ghosh G. Understanding the logic of IκB:NF-κB regulation in structural terms. Curr. Top. Microbiol. Immunol. 2011;349:1–24. doi: 10.1007/82_2010_99. [DOI] [PubMed] [Google Scholar]
- 25.Igata M., Islam M.A., Tada A., Takagi M., Kober A., Albarracin L., Aso H., Ikeda-Ohtsubo W., Miyazawa K., Yoda K., et al. Transcriptome Modifications in Porcine Adipocytes via Toll-Like Receptors Activation. Front. Immunol. 2019;10:1180. doi: 10.3389/fimmu.2019.01180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schafer K.A. The cell cycle: A review. Vet. Pathol. 1998;35:461–478. doi: 10.1177/030098589803500601. [DOI] [PubMed] [Google Scholar]
- 27.Fruitwala S., El-Naccache D.W., Chang T.L. Multifaceted immune functions of human defensins and underlying mechanisms. Semin. Cell Dev. Biol. 2019;88:163–172. doi: 10.1016/j.semcdb.2018.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orsolic I., Jurada D., Pullen N., Oren M., Eliopoulos A.G., Volarevic S. The relationship between the nucleolus and cancer: Current evidence and emerging paradigms. Semin. Cancer Biol. 2016;37–38:36–50. doi: 10.1016/j.semcancer.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 29.Pelletier J., Thomas G., Volarević S. Ribosome biogenesis in cancer: New players and therapeutic avenues. Nat. Rev. Cancer. 2018;18:51–63. doi: 10.1038/nrc.2017.104. [DOI] [PubMed] [Google Scholar]
- 30.Ganapathy-Kanniappan S. Molecular intricacies of aerobic glycolysis in cancer: Current insights into the classic metabolic phenotype. Crit. Rev. Biochem. Mol. Biol. 2018;53:667–682. doi: 10.1080/10409238.2018.1556578. [DOI] [PubMed] [Google Scholar]
- 31.Liberti M.V., Locasale J.W. The warburg effect: How does it benefit cancer cells? Trends Biochem. Sci. 2016;41:211–218. doi: 10.1016/j.tibs.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lunt S.Y., Vander Heiden M.G. Aerobic glycolysis: Meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- 33.Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ashton T.M., McKenna W.G., Kunz-Schughart L.A., Higgins G.S. Oxidative phosphorylation as an emerging target in cancer therapy. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2018;24:2482–2490. doi: 10.1158/1078-0432.CCR-17-3070. [DOI] [PubMed] [Google Scholar]
- 35.Shima A., Fukushima K., Arai T., Terada H. Dual inhibitory effects of the peptide antibiotics leucinostatins on oxidative phosphorylation in mitochondria. Cell Struct. Funct. 1990;15:53–58. doi: 10.1247/csf.15.53. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y., Lu L., Li C., Shao G., Chen X. Transcriptome analysis revealed multiple immune processes and energy metabolism pathways involved in the defense response of the large yellow croaker Larimichthys crocea against Pseudomonas plecoglossicida. Comp. Biochem. Physiol. Part D Genom. Proteom. 2021;40:100886. doi: 10.1016/j.cbd.2021.100886. [DOI] [PubMed] [Google Scholar]
- 37.Narra H.P., Sahni A., Khanipov K., Fofanov Y., Sahni S.K. Global transcriptomic profiling of pulmonary gene expression in an experimental murine model of Rickettsia conorii infection. Genes. 2019;10:204. doi: 10.3390/genes10030204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Varshavsky A. Regulated protein degradation. Trends Biochem. Sci. 2005;30:283–286. doi: 10.1016/j.tibs.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 39.Tanaka K. The proteasome: Overview of structure and functions. Proc. Jpn. Academy. Ser. B Phys. Biol. Sci. 2009;85:12–36. doi: 10.2183/pjab.85.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J., Maldonado M.A. The ubiquitin-proteasome system and its role in inflammatory and autoimmune diseases. Cell. Mol. Immunol. 2006;3:255–261. [PubMed] [Google Scholar]
- 41.Rechsteiner M., Hill C.P. Mobilizing the proteolytic machine: Cell biological roles of proteasome activators and inhibitors. Trends Cell Biol. 2005;15:27–33. doi: 10.1016/j.tcb.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 42.Anbanandam A., Albarado D.C., Tirziu D.C., Simons M., Veeraraghavan S. Molecular basis for proline- and arginine-rich peptide inhibition of proteasome. J. Mol. Biol. 2008;384:219–227. doi: 10.1016/j.jmb.2008.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li S., Li H., Xu Y., Ning W., Hu S., Wei S., Song H., Sun J., Ziebolz D., Schmalz G., et al. Implications of human antimicrobial peptide defensin beta-1 in clinical oral squamous cell carcinoma patients via an integrated bioinformatics approach. Comput. Math. Methods Med. 2022;2022:2203615. doi: 10.1155/2022/2203615. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Hein M.J.A., Kvansakul M., Lay F.T., Phan T.K., Hulett M.D. Defensin-lipid interactions in membrane targeting: Mechanisms of action and opportunities for the development of antimicrobial and anticancer therapeutics. Biochem. Soc. Trans. 2022;50:423–437. doi: 10.1042/BST20200884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ibusuki R., Uto H., Oda K., Ohshige A., Tabu K., Mawatari S., Kumagai K., Kanmura S., Tamai T., Moriuchi A., et al. Human neutrophil peptide-1 promotes alcohol-induced hepatic fibrosis and hepatocyte apoptosis. PLoS ONE. 2017;12:e0174913. doi: 10.1371/journal.pone.0174913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carnero A., Blanco-Aparicio C., Renner O., Link W., Leal J.F. The PTEN/PI3K/AKT signalling pathway in cancer, therapeutic implications. Curr. Cancer Drug Targets. 2008;8:187–198. doi: 10.2174/156800908784293659. [DOI] [PubMed] [Google Scholar]
- 47.Park J.W., Wollmann G., Urbiola C., Fogli B., Florio T., Geley S., Klimaschewski L. Sprouty2 enhances the tumorigenic potential of glioblastoma cells. Neuro-Oncol. 2018;20:1044–1054. doi: 10.1093/neuonc/noy028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liao P.H., Wang Y.Y., Wang W.C., Chen C.H., Kao Y.H., Hsu J.W., Chen C.Y., Chen P.H., Yuan S.S., Chen Y.K. Overexpression of sprouty2 in human oral squamous cell carcinogenesis. Arch. Oral Biol. 2018;87:131–142. doi: 10.1016/j.archoralbio.2017.12.021. [DOI] [PubMed] [Google Scholar]
- 49.Chen G., Sun J., Xie M., Yu S., Tang Q., Chen L. PLAU promotes cell proliferation and epithelial-mesenchymal transition in head and neck squamous cell carcinoma. Front. Genet. 2021;12:651882. doi: 10.3389/fgene.2021.651882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen T., Huang S. The role of Cdc25A in the regulation of cell proliferation and apoptosis. Anti-Cancer Agents Med. Chem. 2012;12:631–639. doi: 10.2174/187152012800617678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou Q., Zhang F., He Z., Zuo M.Z. E2F2/5/8 serve as potential prognostic biomarkers and targets for human ovarian cancer. Front. Oncol. 2019;9:161. doi: 10.3389/fonc.2019.00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hashemi Goradel N., Najafi M., Salehi E., Farhood B., Mortezaee K. Cyclooxygenase-2 in cancer: A review. J. Cell. Physiol. 2019;234:5683–5699. doi: 10.1002/jcp.27411. [DOI] [PubMed] [Google Scholar]
- 53.Uraki S., Sugimoto K., Shiraki K., Tameda M., Inagaki Y., Ogura S., Kasai C., Nojiri K., Yoneda M., Yamamoto N., et al. Human β-defensin-3 inhibits migration of colon cancer cells via downregulation of metastasis-associated 1 family, member 2 expression. Int. J. Oncol. 2014;45:1059–1064. doi: 10.3892/ijo.2014.2507. [DOI] [PubMed] [Google Scholar]
- 54.Droin N., Hendra J.B., Ducoroy P., Solary E. Human defensins as cancer biomarkers and antitumour molecules. J. Proteom. 2009;72:918–927. doi: 10.1016/j.jprot.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 55.Ghavami S., Asoodeh A., Klonisch T., Halayko A.J., Kadkhoda K., Kroczak T.J., Gibson S.B., Booy E.P., Naderi-Manesh H., Los M. Brevinin-2R(1) semi-selectively kills cancer cells by a distinct mechanism, which involves the lysosomal-mitochondrial death pathway. J. Cell. Mol. Med. 2008;12:1005–1022. doi: 10.1111/j.1582-4934.2008.00129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arimura Y., Ashitani J., Yanagi S., Tokojima M., Abe K., Mukae H., Nakazato M. Elevated serum beta-defensins concentrations in patients with lung cancer. Anticancer Res. 2004;24:4051–4057. [PubMed] [Google Scholar]
- 57.Wong C.C., Zhang L., Ren S.X., Shen J., Chan R.L., Cho C.H. Antibacterial peptides and gastrointestinal diseases. Curr. Pharm. Des. 2011;17:1583–1586. doi: 10.2174/138161211796197025. [DOI] [PubMed] [Google Scholar]
- 58.Liu C., Pan B., Yang L., Wang B., Li J. Beta defensin 3 enhances ovarian granulosa cell proliferation and migration via ERK1/2 pathway in vitro†. Biol. Reprod. 2019;100:1057–1065. doi: 10.1093/biolre/ioy246. [DOI] [PubMed] [Google Scholar]
- 59.Kiatsurayanon C., Niyonsaba F., Smithrithee R., Akiyama T., Ushio H., Hara M., Okumura K., Ikeda S., Ogawa H. Host defense (Antimicrobial) peptide, human β-defensin-3, improves the function of the epithelial tight-junction barrier in human keratinocytes. J. Investig. Dermatol. 2014;134:2163–2173. doi: 10.1038/jid.2014.143. [DOI] [PubMed] [Google Scholar]
- 60.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated during this study are included in this article. RNA-Seq data have been deposited in the National Center for Biotechnology Information database under accession number (PRJNA862330).