Abstract
We have cloned an Enterococcus faecalis gene cluster, cydABCD, which when expressed in Bacillus subtilis results in a functional cytochrome bd terminal oxidase. Our results indicate that E. faecalis V583 cells have the capacity of aerobic respiration when grown in the presence of heme.
Enterococcus faecalis, a gram-positive bacterium of low G+C content, is normally found in the human intestine. It is an opportunistic pathogen which can cause severe nosocomial infections (11, 16). Enterococci are generally considered to be facultative anaerobes that mainly use a homolactic fermentative pathway for energy production (3). However, the presence of cytochromes and a capacity for oxidative phosphorylation in E. faecalis strains have been reported in earlier studies (20, 22). The identity and function of these membrane-bound cytochromes have remained unknown. In this study, we demonstrate that E. faecalis contains a cytochrome bd-type oxidase which is expressed under some growth conditions.
Cytochrome bd terminal oxidase complexes are widely distributed in prokaryotes (13, 17). They are membrane-bound enzymes that comprise two subunits and three heme prosthetic groups. Cytochrome bd catalyzes the two-electron oxidation of quinol and the four-electron reduction of dioxygen to make water. The protons produced upon quinol oxidation are released on the outside of the cytoplasmic membrane, and the protons consumed in water production are taken up from the inside of the cell. This results in the production of an electrochemical gradient across the membrane (21). The cytochrome bd structural genes, cydA and cydB, have been cloned from different bacteria (6, 10, 14, 24, 28). In Escherichia coli and in Bacillus subtilis, two additional genes, cydC and cydD, have been shown to be required for expression of cytochrome bd (5, 18, 28). The cydC and cydD genes encode a putative heterodimeric ATP binding cassette (ABC) type of transporter (18). The cytochrome bd quinol oxidases contain two b-type cytochromes (a low-spin and a high-spin heme b) and one cytochrome d (15). The dioxygen reduction site of the enzyme is probably formed by the high-spin heme b together with heme d (8). Here we report the cloning of an E. faecalis gene cluster which encodes a cytochrome bd terminal oxidase.
E. faecalis can express a cytochrome of the bd type.
For membrane preparation, E. faecalis V583 was grown in indented flasks on a rotary shaker (200 rpm) at 37°C in a medium containing tryptone (15 g/liter), soy peptone (5 g/liter) (both from Lab M, Bury, England), NaCl (5 g/liter), and 1% (wt/vol) glucose. The medium was buffered with 30 mM sodium morpholinic propane sulfonic acid buffer (MOPS), pH 7.4, and 5 mM potassium phosphate buffer, pH 7.0. When indicated, 8 μM hemin (Sigma) was added to the medium. Twelve hours after inoculation, cells were harvested by centrifugation and washed in 20 mM sodium MOPS buffer, pH 7.4. All subsequent steps were done at 4°C or on ice. Cells were suspended in MOPS buffer containing DNase (0.1 mg/ml) (bovine pancreas, type 1; Sigma), 0.5 mM phenylmethylsulfonylfluoride and 5 mM MgSO4 and broken using a French pressure cell. After a centrifugation at 5,000 × g, for 15 min, membranes were harvested from the supernatant by centrifugation at 200,000 × g, for 90 min, washed once, and then suspended in MOPS buffer. Protein concentrations were determined using the bicinchoninic acid assay (Pierce) with bovine serum albumin as the standard. Light absorption spectra were recorded as described previously (28).
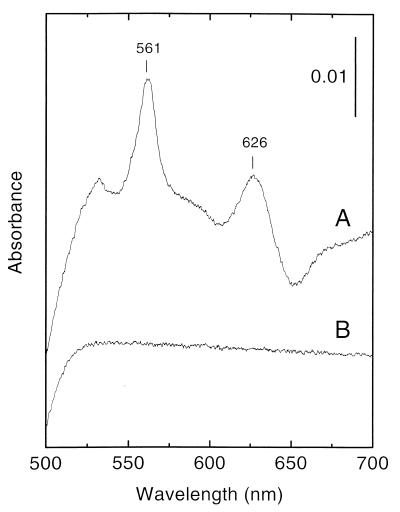
Isolated membranes from E. faecalis cells grown aerobically in the presence of hemin demonstrated light absorption difference spectra with features characteristic for cytochrome bd, with absorption peaks at 561 nm (cytochrome b) and 626 nm (cytochrome d) (Fig. 1, spectrum A). The trough at about 650 nm indicates the presence of a stable oxygenated cytochrome d species [Fe(II)-O2] (19). Membranes from cells grown without hemin lacked spectroscopically detectable cytochromes (Fig. 1, spectrum B).
FIG. 1.
Light absorption difference (dithionite-reduced minus air-oxidized) spectra of E. faecalis membranes (3 mg of protein per ml). (A) Membranes from cells grown in the presence of 8 μM hemin; (B) membranes from cells grown in the absence of added hemin. The vertical bar indicates the absorption scale. Difference spectra obtained after oxidation by potassium ferricyanide were identical to those obtained by air oxidation.
A cyd gene cluster is present in E. faecalis.
The amino acid sequence of B. subtilis CydA was used to search for related sequences in the preliminary release of the E. faecalis genomic data obtained from The Institute for Genomic Research (TIGR). The BLAST search (1) resulted in the identification of a contig containing four putative genes, similar to B. subtilis cydA, cydB, cydC, and cydD. Alignments of the B. subtilis and E. faecalis amino acid sequences showed a sequence identity of 56% for CydA, 46% for CydB, 51% for CydC, and 49% for CydD.
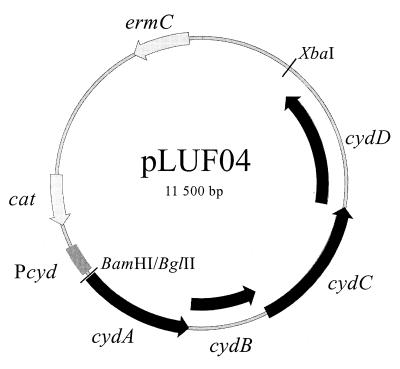
To clone the E. faecalis cydABCD genes, the DNA sequence obtained from the TIGR E. faecalis database was used to design two primers, ECYD1 (5′ GGAGATCTAATGGAAATGAACAATTCAGGTAAG-3′) (BglII restriction site underlined) and ECYD2 (5′-GGTCTAGACTATCATGGCGTTACAGAAGCAC-3′) (XbaI restriction site underlined). The BglII restriction site is located 59 nucleotides upstream of the putative translational initiation site of cydA. These primers were used in a long-range PCR (Expand High Fidelity PCR system; Roche) with 500 ng of E. faecalis chromosomal DNA (prepared essentially as described by Hoch [9]) as the template. The amplified 6.2-kb fragment was cut with restriction enzymes BglII and XbaI and ligated into plasmid pCYD26, cut with BamHI and XbaI. Plasmid pCYD26 contains the B. subtilis cyd promoter region (nucleotides −192 to +199 with respect to the transcription start site) (28) in the low-copy-number vector pHPSK. The ligate was used to transform B. subtilis 168A to chloramphenicol resistance, resulting in plasmid pLUF04 (Fig. 2). Restriction site mapping and partial DNA sequence analysis confirmed the identity of the cloned fragment.
FIG. 2.
Map of plasmid pLUF04, carrying the E. faecalis cydABCD genes. Pcyd indicates the B. subtilis cyd promoter region. BamHI/BglII shows where the PCR fragment (cut with BglII) was ligated to pCYD26 (cut with BamHI). The chloramphenicol and erythromycin resistance genes are indicated by cat and ermC, respectively.
Expression of E. faecalis cydABCD in B. subtilis.
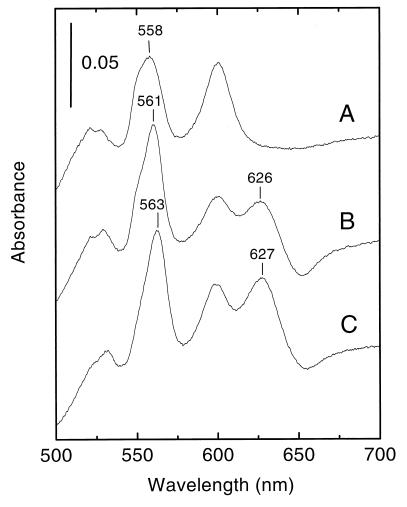
To facilitate characterization of E. faecalis cytochrome bd, we aimed to find a method for overproduction of the enzyme complex. Since cytochrome bd from B. subtilis and E. faecalis appeared to be closely related, we choose to use B. subtilis as our expression host. Strains and plasmids used in this study are listed in Table 1. B. subtilis strain LUW20 lacks cytochrome bd, and hence membranes from this strain lack the spectroscopic features of cytochrome bd (28). LUW20/pLUF04 (E. faecalis cydABCD), LUW20/pCYD23 (B. subtilis cydABCD) (28), and LUW20/pCYD26 (vector only) were grown at 37°C in nutrient sporulation medium with phosphate (4) supplemented with 0.5% glucose (NSMPG) and chloramphenicol (5 mg/liter). The cultures were harvested in the stationary phase. Membranes were prepared as described previously (7) and suspended in 20 mM sodium MOPS buffer, pH 7.4. Light absorption difference spectra of membranes from LUW20/pLUF04 showed an increased absorption at 561 nm and a peak at 626 nm, due to expression of a cytochrome bd (Fig. 3, spectrum B). Membranes from LUW20/pCYD23 showed a spectrum with an increased absorption at 563 nm and a peak at about 627 nm (Fig. 3, spectrum C), whereas membranes from the control, LUW20/pCYD26, lacked the peaks characteristic for cytochrome bd (Fig. 3, spectrum A), as expected. The absorption peak at about 600 nm in the spectra is mainly due to cytochrome a of the cytochrome aa3 oxidase (27). These results show that the cydABCD genes of E. faecalis V583 can be expressed in B. subtilis, resulting in the formation of a spectroscopically detectable cytochrome bd.
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| E. faecalis V583 | 23 | |
| B. subtilis | ||
| 168A | trpC2 | Laboratory stock |
| LUW20 | trpC2 ΔcydABCD::tet | 28 |
| LUH14 | trpC2 ΔqoxABCD::kan | Δqox→168Ab |
| Δqox | trpC2 ΔqoxABCD::kan | 26 |
| LUW173 | trpC2 ΔqoxABCD::kan ΔcydABCD::tet pCYD23 | This work |
| LUW174 | trpC2 ΔqoxABCD::kan ΔcydABCD::tet pLUF04 | This work |
| Plasmids | ||
| pHPSK | Cmr Emr | 12 |
| pCYD23 | B. subtilis cydABCD in pHP13 | 28 |
| pCYD26 | B. subtilis cyd promoter in pHPSK | This work |
| pLUF04 | E. faecalis cydABCD in pCYD26 | This work |
| pDG148 | Kmr Emr | 25 |
Cmr, Emr, and Kmr indicates resistance to chloramphenicol, erythromycin, and kanamycin, respectively.
An arrow indicates transformation and point from donor to recipient.
FIG. 3.
Light absorption difference (dithionite-reduced minus ferricyanide-oxidized) spectra of B. subtilis membranes (2.5 mg of protein per ml). (A) LUW20/pCYD26; (B) LUW20/pLUF04 (E. faecalis cydABCD); (C) LUW20/pCYD23 (B. subtilis cydABCD). The vertical bar indicates the absorption scale.
The E. faecalis cydABCD genes can complement a B. subtilis cytochrome bd-deficient mutant.
B. subtilis 168A cannot grow aerobically if both its quinol oxidases, cytochrome bd and cytochrome aa3, are absent (L. Winstedt and C. von Wachenfeldt, unpublished data). The cytochrome aa3 is encoded by the qoxABCD operon (27). To determine if E. faecalis cytochrome bd functions as a terminal oxidase, we examined whether a B. subtilis strain devoid of both cytochrome bd and cytochrome aa3, but carrying pLUF04, could grow under aerobic conditions. Chromosomal DNA from LUH14 (ΔqoxABCD::kan), prepared as described by Hoch (9), was used to transform LUW20/pLUF04, LUW20/pCYD23, and LUW20/pCYD26 to kanamycin resistance. The same limiting amount of LUH14 DNA (0.2 mg/liter of competent cells) was used for all three strains. Transformants were selected on tryptose blood agar base plates supplemented with 1% (wt/vol) glucose and containing chloramphenicol (5 mg/liter) and kanamycin (5 mg/liter). To verify that the transformants obtained still lacked the chromosomal copy of the B. subtilis cydABCD operon, they were streaked on plates containing tetracycline (15 mg/liter). As shown in Table 2, kanamycin- and tetracycline-resistant transformants, i.e., transformants deleted for both the qoxABCD and the cydABCD operons in the B. subtilis chromosome, were obtained only with LUW20/pLUF04 and LUW20/pCYD23. One transformant from each strain was kept and designated LUW174 and LUW173, respectively. The few transformants obtained with LUW20/pCYD26 were all sensitive to tetracycline; i.e., the tetracycline resistance marker in LUW20 had been substituted with the B. subtilis cydABCD operon from the LUH14 chromosomal DNA.
TABLE 2.
Functional complementation of B. subtilis cytochrome bd-deficient mutants
| Recipient straina | No. of transformants (Kmr)bc | % of transformants which were also Tetrc |
|---|---|---|
| LUW20/pCYD23 | 108 | 99 |
| LUW20/pLUF04 | 130 | 98 |
| LUW20/pCYD26 | 3 | 0 |
B. subtilis LUW20, carrying a ΔcydABCD::tet mutation in the chromosome and one of three plasmids, was transformed with chromosomal DNA from LUH14, carrying a ΔqoxABCD::kan mutation. All strains showed a similar degree of competence as tested by transformation with plasmid pDG148.
Transformants were selected on plates containing kanamycin and chloramphenicol and then restreaked on plates containing tetracycline.
Kmr and Tetr indicate resistance to kanamycin and tetracycline, respectively.
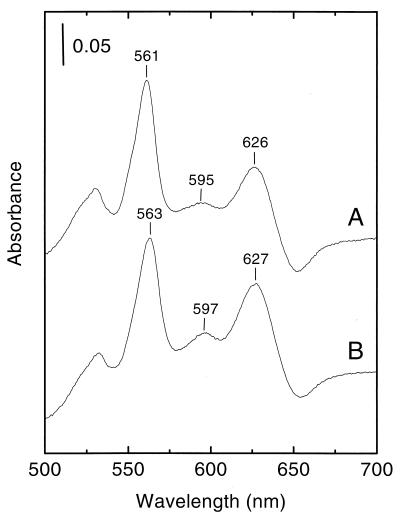
To further characterize LUW174 and LUW173 and to compare the spectroscopic features of E. faecalis V583 cytochrome bd and B. subtilis cytochrome bd in more detail, the two strains were grown in NSMPG and membranes were prepared as described above. The growth properties of LUW174 did not differ from those of LUW173. Light absorption difference spectra of membranes from LUW174 (containing E. faecalis cytochrome bd) showed peaks at about 561, 595, and 626 nm (Fig. 4, spectrum A), indicating the presence of three prosthetic groups (low-spin heme b, high-spin heme b, and heme d). Membranes from LUW173 (containing B. subtilis cytochrome bd) showed peaks at about 563, 597, and 627 nm (Fig. 4, spectrum B). The absence of a peak at about 600 nm confirmed that the strains lack cytochrome aa3.
FIG. 4.
Light absorption difference (dithionite-reduced minus ferricyanide-oxidized) spectra of B. subtilis membranes (2.5 mg of protein per ml). (A) LUW174 (ΔqoxABCD::kan ΔcydABCD::tet pLUF04), (B) LUW173 (ΔqoxABCD::kan ΔcydABCD::tet pCYD23). The vertical bar indicates the absorption scale.
Conclusion.
In this work, we show that E. faecalis V583 contains a cydABCD gene cluster and that membranes from this strain grown in the presence of heme contain a cytochrome bd. Under these growth conditions, cytochrome bd is the major (and possibly the only) membrane-bound cytochrome in E. faecalis. The cloned E. faecalis cydABCD gene cluster expressed in B. subtilis resulted in a cytochrome bd which showed a spectrum indistinguishable from that of the cytochrome bd found in E. faecalis. The E. faecalis cytochrome bd can functionally complement a B. subtilis cytochrome bd-deficient mutant, indicating that the E. faecalis cytochrome bd is a menaquinol oxidase. E. faecalis and B. subtilis both contain naphthoquinones in the cytoplasmic membrane: demethylmenaquinone and menaquinone, respectively (2). Thus, a specific electron donor for cytochrome bd is present in E. faecalis. These results indicate that E. faecalis is capable of aerobic respiration. The physiological importance of the identified respiratory system and its role in pathogenesis remain to be determined.
Acknowledgments
We thank Lars Rutberg for reading the manuscript. E. faecalis genome sequence data was obtained from The Institute for Genomic Research website at http://www.tigr.org.
This work was in part supported by grants from the Crafoordska stiftelsen, the Emil och Wera Cornells stiftelse (to C.V.W.), and by the Swedish Natural Science Research Council (to L.H.).
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Collins M D, Jones D. Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implication. Microbiol Rev. 1981;45:316–354. doi: 10.1128/mr.45.2.316-354.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Devriese L A, Collins M D, Wirth R. The genus Enterococcus. In: Balows A, Truper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. (2nd ed.) New York, N.Y: Springer-Verlag; 1992. pp. 1465–1481. [Google Scholar]
- 4.Fortnagel P, Freese E. Analysis of sporulation mutants. II. Mutants blocked in the citric acid cycle. J Bacteriol. 1968;95:1431–1438. doi: 10.1128/jb.95.4.1431-1438.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Georgiou C D, Fang H, Gennis R B. Identification of the cydC locus required for expression of the functional form of the cytochrome d terminal oxidase complex in Escherichia coli. J Bacteriol. 1987;169:2107–2112. doi: 10.1128/jb.169.5.2107-2112.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Green G N, Kranz J E, Gennis R B. Cloning the cyd gene locus coding for the cytochrome d complex of Escherichia coli. Gene. 1984;32:99–106. doi: 10.1016/0378-1119(84)90037-4. [DOI] [PubMed] [Google Scholar]
- 7.Hederstedt L. Molecular properties, genetics, and biosynthesis of Bacillus subtilis succinate dehydrogenase complex. Methods Enzymol. 1986;126:399–414. doi: 10.1016/s0076-6879(86)26040-1. [DOI] [PubMed] [Google Scholar]
- 8.Hill J J, Alben J O, Gennis R B. Spectroscopic evidence for a heme-heme binuclear center in the cytochrome bd ubiquinol oxidase from Escherichia coli. Proc Natl Acad Sci USA. 1993;90:5863–5867. doi: 10.1073/pnas.90.12.5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoch J A. Genetic analysis in Bacillus subtilis. Methods Enzymol. 1991;204:305–320. doi: 10.1016/0076-6879(91)04015-g. [DOI] [PubMed] [Google Scholar]
- 10.Howitt C A, Vermaas W F. Quinol and cytochrome oxidases in the cyanobacterium Synechocystis sp. PCC 6803. Biochemistry. 1998;37:17944–17951. doi: 10.1021/bi981486n. [DOI] [PubMed] [Google Scholar]
- 11.Huycke M M, Sahm D F, Gilmore M S. Multiple-drug resistant enterococci: the nature of the problem and an agenda for the future. Emerg Infect Dis. 1998;4:239–249. doi: 10.3201/eid0402.980211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johansson P, Hederstedt L. Organization of genes for tetrapyrrole biosynthesis in gram-positive bacteria. Microbiology. 1999;145:529–538. doi: 10.1099/13500872-145-3-529. [DOI] [PubMed] [Google Scholar]
- 13.Jünemann S. Cytochrome bd terminal oxidase. Biochim Biophys Acta. 1997;1321:107–127. doi: 10.1016/s0005-2728(97)00046-7. [DOI] [PubMed] [Google Scholar]
- 14.Kelly M J, Poole R K, Yates M G, Kennedy C. Cloning and mutagenesis of genes encoding the cytochrome bd terminal oxidase complex in Azotobacter vinelandii: mutants deficient in the cytochrome d complex are unable to fix nitrogen in air. J Bacteriol. 1990;172:6010–6019. doi: 10.1128/jb.172.10.6010-6019.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller M J, Gennis R B. The purification and characterization of the cytochrome d terminal oxidase complex of the Escherichia coli aerobic respiratory chain. J Biol Chem. 1983;258:9159–9165. [PubMed] [Google Scholar]
- 16.Murray B E, Weinstock G M. Enterococci: new aspects of an old organism. Proc Assoc Am Physicians. 1999;111:328–334. doi: 10.1046/j.1525-1381.1999.99241.x. [DOI] [PubMed] [Google Scholar]
- 17.Osborne J P, Gennis R B. Sequence analysis of cytochrome bd oxidase suggests a revised topology for subunit I. Biochim Biophys Acta. 1999;1410:32–50. doi: 10.1016/s0005-2728(98)00171-6. [DOI] [PubMed] [Google Scholar]
- 18.Poole R K, Hatch L, Cleeter M W J, Gibson F, Cox G B, Wu G. Cytochrome bd biosynthesis in Escherichia coli: the sequences of the cydC and cydD genes suggest that they encode components of an ABC membrane transporter. Mol Microbiol. 1993;10:421–430. [PubMed] [Google Scholar]
- 19.Poole R K, Kumar C, Salmon I, Chance B. The 650 chromophore in Escherichia coli is an ‘oxy-’ or oxygenated compound, not the oxidized form of cytochrome oxidase d: an hypothesis. J Gen Microbiol. 1983;129:1335–1344. doi: 10.1099/00221287-129-5-1335. [DOI] [PubMed] [Google Scholar]
- 20.Pritchard G G, Wimpenny J W. Cytochrome formation, oxygen-induced proton extrusion and respiratory activity in Streptococcus faecalis var. zymogenes grown in the presence of haematin. J Gen Microbiol. 1978;104:15–22. doi: 10.1099/00221287-104-1-15. [DOI] [PubMed] [Google Scholar]
- 21.Puustinen A, Finel M, Haltia T, Gennis R B, Wikstrom M. Properties of the two terminal oxidases of Escherichia coli. Biochemistry. 1991;30:3936–3942. doi: 10.1021/bi00230a019. [DOI] [PubMed] [Google Scholar]
- 22.Ritchey T W, Seely H W., Jr Distribution of cytochrome-like respiration in streptococci. J Gen Microbiol. 1976;93:195–203. doi: 10.1099/00221287-93-2-195. [DOI] [PubMed] [Google Scholar]
- 23.Sahm D F, Kissinger J, Gilmore M S, Murray P R, Mulder R, Solliday J, Clarke B. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob Agents Chemother. 1989;33:1588–1591. doi: 10.1128/aac.33.9.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakamoto J, Koga E, Mizuta T, Sato C, Noguchi S, Sone N. Gene structure and quinol oxidase activity of a cytochrome bd-type oxidase from Bacillus stearothermophilus. Biochim Biophys Acta. 1999;1411:147–158. doi: 10.1016/s0005-2728(99)00012-2. [DOI] [PubMed] [Google Scholar]
- 25.Stragier P, Bonamy C, Karmazyn-Campelli C. Processing of a sporulation sigma factor in Bacillus subtilis: how morphological structure could control gene expression. Cell. 1988;52:697–704. doi: 10.1016/0092-8674(88)90407-2. [DOI] [PubMed] [Google Scholar]
- 26.Villani G, Tattoli M, Capitanio N, Glaser P, Papa S, Danchin A. Functional analysis of subunits III and IV of Bacillus subtilis aa3-600 quinol oxidase by in vitro mutagenesis and gene replacement. Biochim Biophys Acta. 1995;1232:67–74. doi: 10.1016/0005-2728(95)00112-5. [DOI] [PubMed] [Google Scholar]
- 27.von Wachenfeldt C, Hederstedt L. Molecular biology of Bacillus subtilis cytochromes. FEMS Microbiol Lett. 1992;100:91–100. doi: 10.1111/j.1574-6968.1992.tb14025.x. [DOI] [PubMed] [Google Scholar]
- 28.Winstedt L, Yoshida K, Fujita Y, von Wachenfeldt C. Cytochrome bd biosynthesis in Bacillus subtilis: characterization of the cydABCD operon. J Bacteriol. 1998;180:6571–6580. doi: 10.1128/jb.180.24.6571-6580.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]