Abstract
Poplar is an important fast-growing tree, and its photosynthetic capacity directly affects its vegetative growth. Stomatal density is closely related to photosynthetic capacity and growth characteristics in plants. Here, we isolated PagSTOMAGEN from the hybrid poplar (Populus alba × Populus glandulosa) clone 84K and investigated its biological function in vegetative growth. PagSTOMAGEN was expressed predominantly in young tissues and localized in the plasma membrane. Compared with wild-type 84K poplars, PagSTOMAGEN-overexpressing plants displayed an increased plant height, leaf area, internode number, basal diameter, biomass, IAA content, IPR content, and stomatal density. Higher stomatal density improved the net photosynthetic rate, stomatal conductance, intercellular CO2 concentration, and transpiration rate in transgenic poplar. The differential expression of genes related to stomatal development showed a diverged influence of PagSTOMAGEN at different stages of stomatal development. Finally, transcriptomic analysis showed that PagSTOMAGEN affected vegetative growth by affecting the expression of photosynthesis and plant hormone-related genes (such as SAUR75, PQL2, PSBX, ERF1, GNC, GRF5, and ARF11). Taken together, our data indicate that PagSTOMAGEN could positively regulate stomatal density and increase the photosynthetic rate and plant hormone content, thereby promoting vegetative growth in poplar. Our study is of great significance for understanding the relationship between stoma, photosynthesis, and yield breeding in poplar.
Keywords: PagSTOMAGEN, stomatal density, photosynthesis, plant hormone, vegetative growth, poplar
1. Introduction
Stomata, composed of a pair of guard cells, are channels on the surface of plant leaves [1,2] which regulate gas exchange between plants and the atmosphere and play an important role in maintaining plant photosynthesis and water-use efficiency [3,4,5,6]. The formation of mature stomata undergoes several intermediate steps, termed the stomatal lineage, from meristematic cells (MMC) to stomatal lineage ground cells (SLGC), guard cell mother cells (GMC), and finally, the formation of guard cells (GC) [7,8,9]. During stomatal development, stomata are specified and positioned nonrandomly through asymmetric cell division and the integration of intercellular signaling [10].
Stomatal density is regulated by positive and negative genetic factors [11,12]. Negative regulators include the cell surface receptor TOO MANY MOUTHS (TMM) [13], ERECTA family receptor-like kinases (ER, ERL1, and ERL2) [14] and their ligands EPF1 and EPF2 [15,16,17], as well as STOMATAL DENSITY AND DISTRIBUTION 1 (SDD1) functioning in a TMM-dependent manner [18,19]. Positive regulators include the three core bHLH transcriptional regulators SPEECHLESS, MUTE, and FAMA [20,21,22], and STOMAGEN/EPFL9 (STOMAGEN) belonging to the EPF/EPFL family [23].
Some genes have pleiotropic functions. Stomatal density regulators have been reported to affect plant growth and development or stress resistance. For example, EPF1 of the EPF/EPFL family not only regulates stomatal development, but also affects plant biomass in Arabidopsis [24]. EPFL6 of the EPF/EPFL family affects stomatal density and drought resistance in poplar [25]. However, STOMAGEN, which is also a member of the EPF/EPFL family, has only been reported to affect the stomatal development of plants, but it has not been found to affect the biomass of plants [26]. STOMAGEN encodes a cysteine-rich precursor that is a signaling factor for stomatal differentiation [26,27,28]. STOMAGEN overexpression can increase stomatal density in Arabidopsis, which improves the photosynthetic rate and transpiration rate. However, vegetative growth is not affected because the reduced water-use efficiency causes water stress in overexpressing plants [23,26,27].
Vegetative growth is very important for poplar [29]. The vegetative growth of plants is affected by many factors, such as photosynthesis [30], plant hormones [31], environmental factors [31], etc. Photosynthesis is a prerequisite for biomass production [30], and is affected by a large number of key genes, such as the photosystem key factors PSAB [32], PSAN [33], and PSBX [34], as well as some important growth regulators, such as GRF5 [35], GNC [36], and PIFs [37]. Auxin-responsive factors, such as ARFs and SAUR-like auxin-responsive protein [38,39,40], are all differentially expressed when stimulated by auxin. Cytokinins are mainly affected by CKX family genes [41]. The differential expression of these genes is the reason for the difference in the vegetative growth of different plants.
As an important woody plant, it is unclear whether stomatal density and vegetative growth are regulated by STOMAGEN in poplar. Thus, we identified and cloned PagSTOMAGEN from ‘84K’ poplar. Phylogeny and sequence analysis showed that PagSTOMAGEN was homologous with STOMAGENs from Arabidopsis thaliana, Populus trichocarpa, and rice [42]. The overexpression of PagSTOMAGEN in 84K poplar increased stomatal density, which further affected photosynthetic capacity, enzymatic activity, plant hormone content, and biomass. Moreover, differentially expressed genes (DEGs) related to stomatal development in overexpressing poplar showed that PagSTOMAGEN was involved in various stages of stomatal development [43]. Based on these experimental data, we preliminarily explored the biological function of PagSTOMAGEN, which provided theoretical support for high yield by increasing the stomatal density in poplar.
2. Results
2.1. Isolation and Identification of PagSTOMAGEN
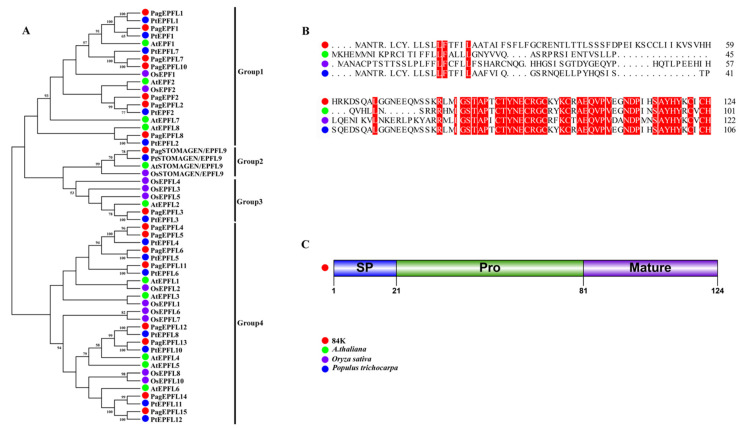
To illustrate the phylogeny and sequence characteristics of STOMAGEN in poplar, the EPF/EPFL gene family was retrieved from the 84K protein database using Arabidopsis AtEPF/EPFL proteins as queries. An unrooted neighbor-joining tree was constructed using 54 full-length protein sequences from four plant species (Figure 1A, Table S2). The results show that PagSTOMAGEN/EPFL9 was highly homologous with STOMAGEN/EPFL9s from other plants, which was further confirmed by amino acid sequence alignment and three-dimensional structural models (Figure 1B and Figure S1).
Figure 1.
Isolation and identification of PagSTOMAGEN. (A) Phylogenetic tree of EPF/EPFL gene family. The bootstrap values are shown. (B) Amino acid sequence alignment of STOMAGEN/EPFL9 proteins in 84K, Arabidopsis thaliana, Oryza sativa, and Populus trichocarpa. Identical amino acids are highlighted in red. (C) Schematic representation of the domain structure of PagSTOMAGEN/EPFL9. PagSTOMAGEN/EPFL9 is composed of the signal peptide (SP), the propeptide (Pro), and the mature domain. Amino acid positions are indicated by numbers.
The PagSTOMAGEN/EPFL9 gene encodes a small polypeptide of 124 amino acids with a putative signal peptide (SP) at the N-terminus (Figure 1C). The mature form of the PagSTOMAGEN/EPFL9 gene product, stomagen, is a peptide consisting of the C-terminal 43 amino acids of the precursor protein. It is probable that the PagSTOMAGEN/EPFL9 peptide is also subject to post-translational processing, which needs to be further determined in 84K.
2.2. Expression Patterns and Subcellular Localizations of PagSTOMAGEN
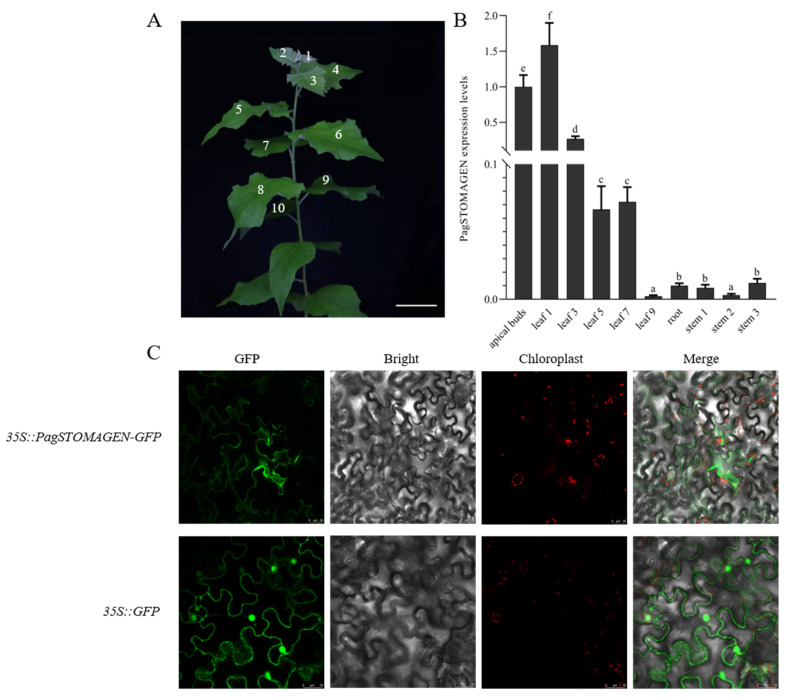
To preliminarily explore the biological roles of PagSTOMAGEN, the expression of PagSTOMAGEN was detected in the apical buds, in the first, third, fifth, seventh, and ninth leaf blades, in stems from the second to fifth internodes, and in the roots using RT-qPCR. Significant differential expression patterns were observed (Figure 2A,B). PagSTOMAGEN showed high expression in apical the buds and young leaves; its expression was low among the other tissues analyzed (Figure 2B). Therefore, PagSTOMAGEN may play a significant role in apical buds and young leaves.
Figure 2.
Expression patterns and subcellular protein localization of PagSTOMAGEN. (A) Definition of leaf numbers. Bar = 5 cm. (B) Expression patterns of PagSTOMAGEN in different tissues. PagACTIN was used as a reference. Error bars represent the standard deviations (SD) of three biological replicates. p < 0.05 was considered statistically significant and is represented by different letters. (C) Subcellular localization of the PagSTOMAGEN protein. Laser confocal microscopy was used to obtain images of tobacco leaves with transient expression of 35S::PagSTOMAGEN-GFP and 35S::GFP. Bars = 25 μm.
To evaluate the subcellular localization of PagSTOMAGEN, 35S::PagSTOMAGEN-GFP and 35S::GFP constructs were generated and transiently expressed in Nicotiana benthamiana. Fluorescence from 35S::PagSTOMAGEN-GFP was detected only on the plasma membrane (Figure 2C), while 35S::GFP fluorescence was detected on the cell membrane and in the nucleus (Figure 2C). The results show that PagSTOMAGEN might function on the plasma membrane.
2.3. PagSTOMAGEN Positively Regulates the Vegetative Growth of Poplar
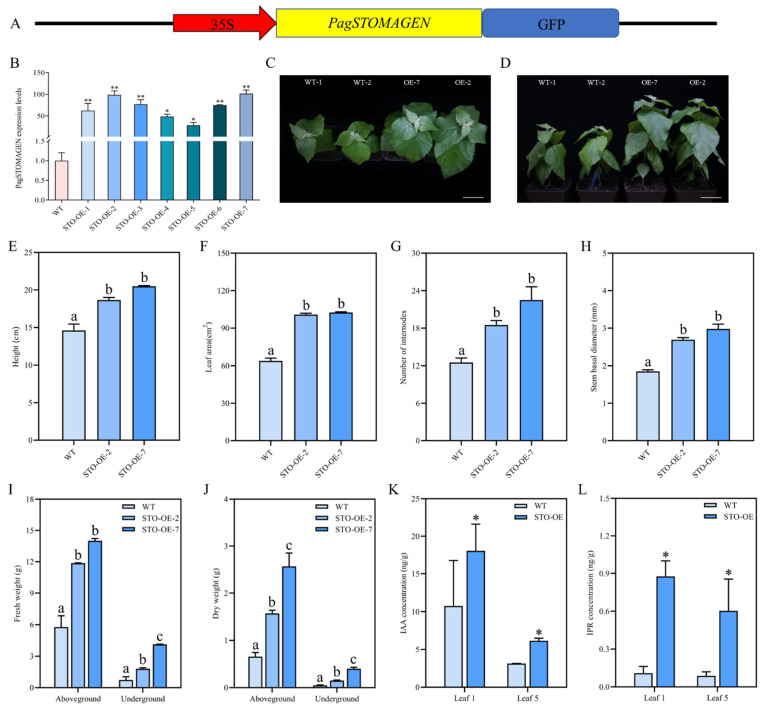
To further elucidate the biological function of PagSTOMAGEN in poplar, STO-OE lines were generated (Figure 3A–D and Figure S2). All the STO-OE lines showed similar phenotypes (Figure S3), of which STO-OE-2 and STO-OE-7 had the highest PagSTOMAGEN expression levels and were selected for further analysis (Figure 3B–D). We investigated the phenotypes of the STO-OE and wild-type (WT) lines using two-month-old plants grown in the greenhouse. Compared with the WT plants, the plant height, leaf area, number of internodes, and stem diameter were significantly increased in the STO-OE lines (Figure 3E–H). Our investigation showed increases in plant height of 27.8 and 40.9%, in leaf area of 58.2 and 80%, in number of internodes of 48 and 60.8%, and stem diameter of 45.4 and 61.1%, respectively, in STO-OE plants compared to the WT (Figure 3E–H). These growth changes indicate that STO-OE might have a higher biomass than the WT.
Figure 3.
Generation and phenotypic changes in PagSTOMAGEN-overexpressing (STO-OE) lines. (A) Schematic diagram of the PagSTOMAGEN overexpression vector. (B) Expression levels of PagSTOMAGEN in the STO-OE lines. Values represent the mean ± SD (n = 5). (C,D) Two-month-old plants. WT: non-transgenic control; OE: PagSTOMAGEN-overexpressing lines. Bars = 5 cm. (E) Measurement of plant height. (F) Measurement of leaf area. (G) Measurement of the number of internodes. (H) Measurement of stem basal diameter. (I) Aboveground and underground fresh weight of STO-OE and WT lines. (J) Aboveground and underground dry weight of STO-OE and WT lines. (K) IAA concentration in different leaf positions of STO-OE and WT plants. (L) IPR concentration in different leaf positions of STO-OE and WT plants. Error bars represent the SD for each genotype with nine plants. Asterisks indicate significant differences in the Student’s t-test. ** p < 0.01; * p < 0.05. p < 0.05 was considered statistically significant and is represented by different letters.
To determine whether biomass was increased in STO-OE, whole STO-OE and WT plants were divided into aboveground and underground parts, and their fresh and dry weights were measured (Figure 3I,J). The results show that the aboveground and underground biomass of STO-OE were both larger than those of the WT. The aboveground fresh weight of STO-OE-2 and STO-OE-7 was 2.06 and 2.43 times higher than that of the WT, respectively (Figure 3I). The underground fresh weight of STO-OE-2 and STO-OE-7 was 2.46 and 5.61 times higher than that of the WT, respectively (Figure 3I). At the same time, the dry weight of the aboveground parts of STO-OE-2 and STO-OE-7 was 2.39 and 3.90 times higher than that of the WT (Figure 3J). The underground dry weight of STO-OE-2 and STO-OE-7 was 2.97 and 7.92 times higher than that of the WT, respectively (Figure 3J).
In order to explore the reasons for the increase in plant biomass after the overexpression of PagSTOMAGEN, we measured the contents of auxin IAA and cytokinin IPR in STO-OE and WT plants. The results show that the IAA content of STO-OE plants was 1.68 and 1.96 times higher than that of the WT at the first and fifth leaf positions, respectively (Figure 3K). At the first and fifth leaf positions, the IPR content of STO-OE plants was 8.04 and 6.85 times higher than that of the WT, respectively (Figure 3L).
2.4. Involvement of PagSTOMAGEN in Stomatal Density, Size, and Stomatal Index
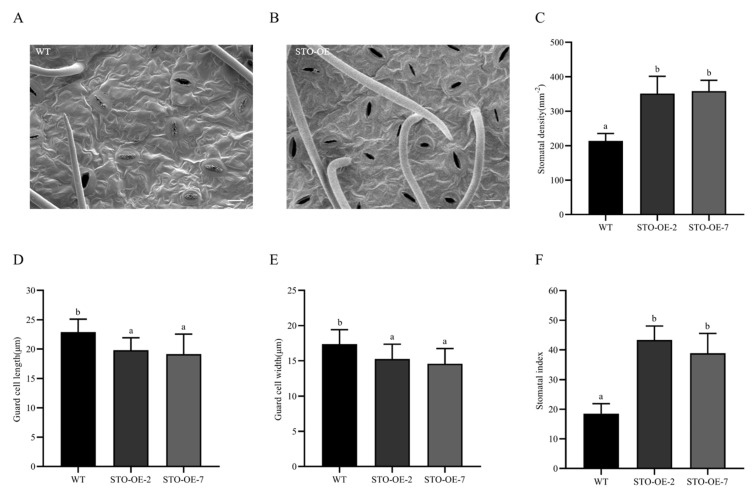
To determine whether PagSTOMAGEN was involved in the regulation of stomatal development as Arabidopsis, stomatal density and size were detected in the STO-OE lines. STO-OE showed higher stomatal density, ranging from 351 to 358 mm−2, compared to 213 mm−2 in WT plants (Figure 4A–C). However, the stomatal size of STO-OE was significantly smaller than that of the WT (Figure 4D,E). The stomatal index and the number of stomata showed similar variations to stomatal density in the STO-OE lines (Figure 4F and Figure S4). In summary, our results suggest that PagSTOMAGEN was involved in the regulation of stomatal density, size, and stomatal index.
Figure 4.
Stomatal density, size, and stomatal index in STO-OE and WT poplar. (A) Abaxial leaf epidermis of WT lines. (B) Abaxial leaf epidermis of STO-OE lines. (C) Stomatal density of STO-OE and WT lines. (D) Guard cell length of STO-OE and WT lines. (E) Guard cell width of STO-OE and WT lines. (F) Stomatal index of STO-OE and WT lines. Error bars are indicated by the SD of five biological replicates. p < 0.05 was considered significantly different and is shown by different letters. Bars = 20 μm.
2.5. PagSTOMAGEN Enhances Photosynthetic Capacity by Increasing Stomatal Density
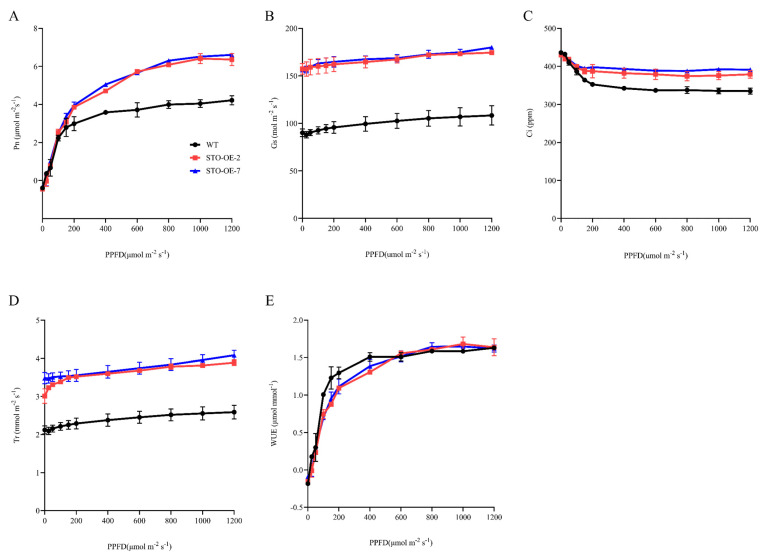
To determine whether stomatal density is involved in plant photosynthesis, we measured the light-response curves of two-month-old WT, STO-OE-2, and STO-OE-7 plants. The net photosynthetic rate (Pn), stomatal conductance (Gs), intracellular CO2 concentration (Ci), and transpiration rate (Tr) were significantly higher in STO-OE than in the WT (Figure 5A–D), which agreed with the stomatal density changes (Figure 4A–C). However, there was no significant difference in water-use efficiency (WUE) between the WT and STO-OE plants (Figure 5E).
Figure 5.
Overexpression of PagSTOMAGEN enhances photosynthesis in poplar. Light-response curves were obtained for STO-OE and WT. (A) Net photosynthetic rate (Pn)–light curve. (B) Stomatal conductance (Gs)–light curve. (C) Intracellular CO2 concentration (Ci)–light curve. (D) Transpiration rate (Tr)–light curve. (E) Water-use efficiency (WUE)–light curve. Error bars are indicated by the SD of three biological replicates.
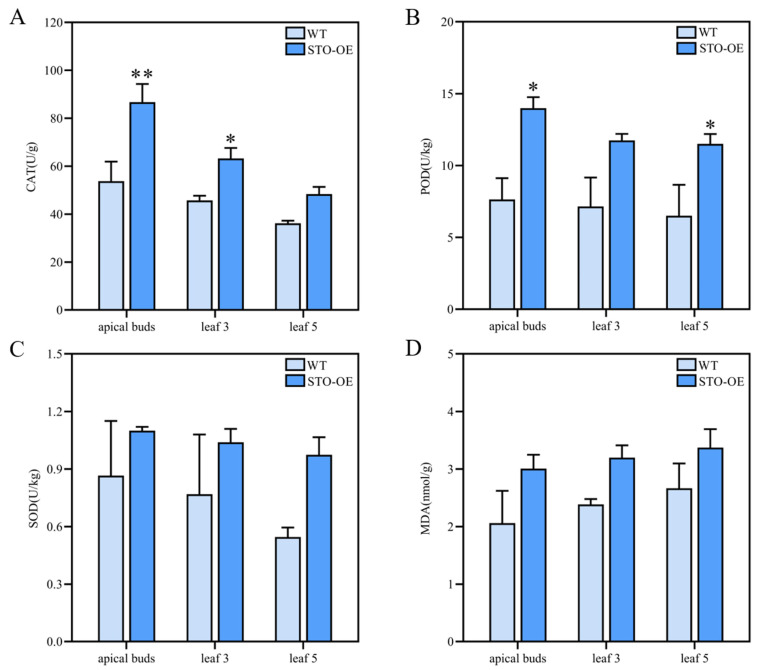
2.6. Influence of PagSTOMAGEN on Enzyme Activity and Malondialdehyde Content
Due to the involvement of CAT, POD, and SOD activities and MDA content in plant growth, these physiological indexes were determined. The CAT activity of STO-OE was significantly higher than that of the WT in apical buds and the third leaf position, while a significant difference was not observed in the fifth leaf position (Figure 6A). POD activity was significantly higher in STO-OE in the apical buds and the fifth leaf position, but there was no significant difference in the third leaf position (Figure 6B). SOD activity and MDA content exhibited no significant differences in any of the detected tissues between the STO-OE and WT lines (Figure 6C,D). Taken together, PagSTOMAGEN overexpression increased CAT and POD activities but had a little influence on SOD activity and MDA content.
Figure 6.
Enzyme activities and malondialdehyde content of STO-OE and WT poplar. (A) Activity of catalase (CAT) in STO-OE and WT lines. (B) Activity of peroxidase (POD) in STO-OE and WT lines. (C) Activity of superoxide dismutase (SOD) in STO-OE and WT lines. (D) Malondialdehyde (MDA) content of STO-OE and WT lines. Values are the mean ± SD (n > 3). Asterisks denote significant differences: ** p < 0.01; * p < 0.05.
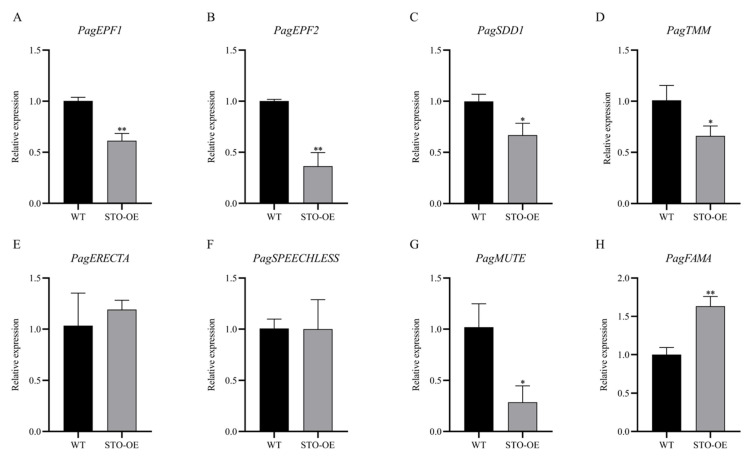
2.7. PagSTOMAGEN Modulates the Expression of Genes Related to Stomatal Development and Patterning
In Arabidopsis, AtSTOMAGEN affects the expression of genes involved in stomatal development and patterning. To determine whether PagSTOMAGEN modulates genes related to stomatal development and patterning in poplar, the expression of these genes was examined in the STO-OE and WT lines. The expression levels of PagEPF1 and PagEPF2, two negative regulators of stomatal formation, were significantly reduced in the STO-OE lines (Figure 7A,B). The expression of the stomatal density inhibitor PagSDD1 was also significantly decreased (Figure 7C). PagTMM and PagERECTA, two putative receptors of PagSTOMAGEN, showed decreased and unchanged expression, respectively, in STO-OE poplar (Figure 7D,E). Three bHLH transcription factors, PagSPEECHLESS, PagFAMA, and PagMUTE, exhibited diverse expression patterns in STO-OE. No changes were observed for the expression of PagSPEECHLESS (Figure 7F), which is involved in the first step of stomatal development by regulating the differentiation of protodermal cells. PagSTOMAGEN overexpression decreased the expression of PagMUTE (Figure 7G). The expression level of PagFAMA was significantly increased by PagSTOMAGEN overexpression (Figure 7H).
Figure 7.
PagSTOMAGEN affects the expression of genes involved in stomatal development and patterning. (A) Relative expression level of PagEPF1 in STO-OE and WT lines. (B) Relative expression level of PagEPF2 in STO-OE and WT lines. (C) Relative expression level of PagSDD1 in STO-OE and WT lines. (D) Relative expression level of PagTMM in STO-OE and WT lines. (E) Relative expression level of PagERECTA in STO-OE and WT lines. (F) Relative expression level of PagSPEECHLESS in STO-OE and WT lines. (G) Relative expression level of PagMUTE in STO-OE and WT lines. (H) Relative expression level of PagFAMA in STO-OE and WT lines. Expression was estimated using RT-qPCR normalized to PagACTIN expression. Error bars represent the SD (data are the means of three biological replicates). Asterisks indicate significant differences in the Student’s t-test. ** p < 0.01; * p < 0.05.
2.8. PagSTOMAGEN Promotes the Accumulation of Plant Biomass by Positively Regulating the Expression of Photosynthesis and Growth-Related Genes
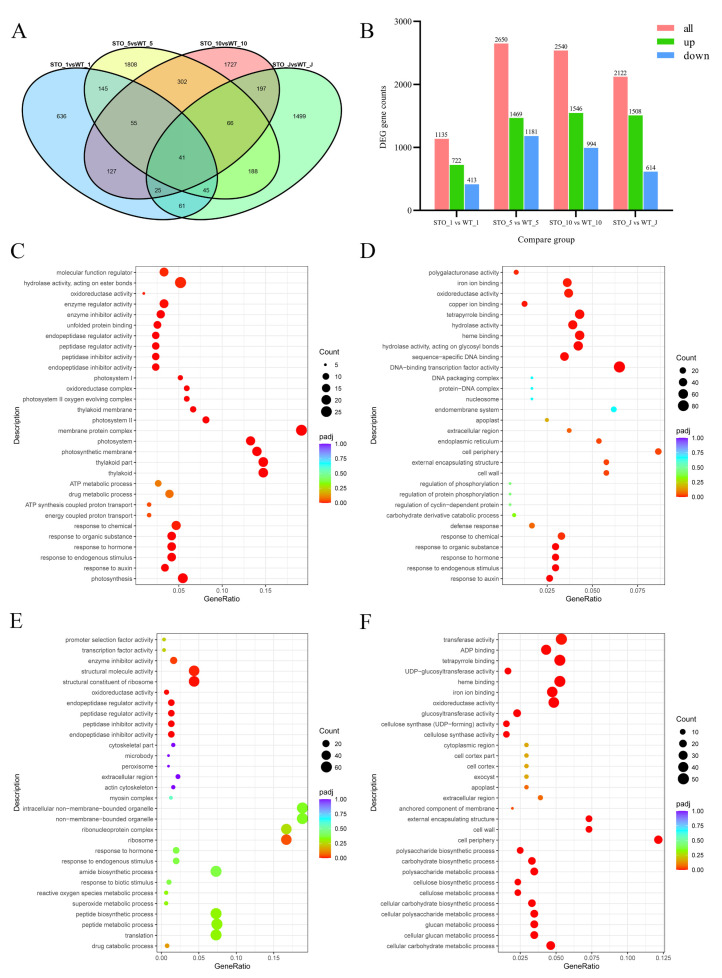
The phenotypic changes, particularly the higher biomass in STO-OE lines, prompted us to identify DEGs whose transcription was changed in the leaves and stems. To achieve this, we ran RNA-seq experiments with three STO-OE lines (35S::PagSTOMAGEN-GFP-nos) and three WT 84K poplar replicates. We compared transgenic lines to WT for DEG identification (Figure 3B, Table S3). We identified 1135 DEGs (corrected p < 0.05) by comparing the transgenic and WT 84K poplar lines in the first leaf position (Figure 8A,B, Table S4). Of these DEGs, 722 were up-regulated while 413 were down-regulated (Figure 8A,B, Table S4). In the fifth leaf position, 2650 DEGs were identified, with 1469 genes up-regulated and 1181 down-regulated, respectively (Figure 8A,B). In the 10th leaf position, there were 2540 DEGs, of which 1546 genes were up-regulated and 994 genes were down-regulated (Figure 8A,B, Table S4). In young stem segments, there were 2122 DEGs, of which 1508 genes were up-regulated and 614 genes were down-regulated (Figure 8A,B, Table S4).
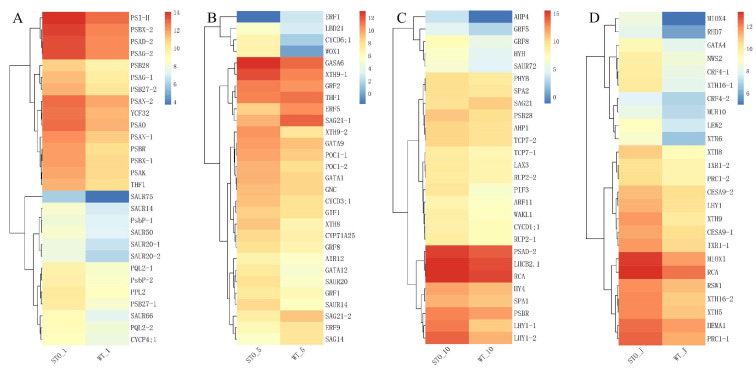
Figure 8.
Identification of DEGs in PagSTOMAGEN-OE 84K poplar transgenic lines and GO enrichment analysis. (A) Venn plots for different sample combinations. (B) DEG gene counts for different sample combinations. Includes all up-regulated and down-regulated gene counts. GO enrichment analysis of 1st leaf position (C), 5th leaf position (D), 10th leaf position (E), and young stem (F) in STO-OE and WT lines. The Y-axis indicates the GO pathway; the X-axis indicates the gene ratio. The dot size indicates the number of DEGs in the pathway, and the dot color indicates the p-value.
The Gene Ontology (GO) enrichment of differential genes in different tissues was not the same. In the first leaf position, differential genes were mainly enriched in pathways such as photosynthesis, the response to auxin, and the photosystem (Figure 8C, Table S5). In the fifth leaf position, differential genes were mainly enriched in response to auxin, the response to hormones, and cell wall pathways (Figure 8D, Table S6). In the 10th leaf position, differential genes were mainly enriched in pathways such as the response to hormones, transcription factor activity, and structural molecule activity (Figure 8E, Table S7). In young stem segments, differential genes were mainly enriched in the cellular carbohydrate metabolic process, the cellular carbohydrate biosynthetic process, and cell wall pathways (Figure 8F, Table S8).
Differential genes between the WT and STO-OE lines were enriched in photosynthesis and hormone-related pathways in different tissues. In the first leaf position, the auxin-responsive genes SAUR75, SAUR20, SAUR66, SAUR14, SAUR50, etc. were significantly up-regulated in STO-OE plants (Figure 9A, Table S9). At the same time, the expression levels of photosystem-related genes such as PQL2, PSBX, PSAG, and PSAN in overexpressed plants were significantly higher than those in WT (Figure 9A, Table S9). These results may be the reason the overexpressed plants have stronger photosynthesis than the WT, thereby promoting biomass accumulation (Figure 9A, Figure 3). In the fifth leaf position, the expression levels of ethylene-responsive factors ERF1, ERF5, and ERF9 and the senescence-related genes SAG14, SAG21, etc. in STO-OE were significantly lower than those in the WT, while the expression levels of the growth regulators GNC, GRF8, GRF2, GRF1, etc. were significantly higher than those in the WT, suggesting that these genes may be responsible for the growth advantage of overexpressed plants (Figure 9B, Table S10). The expression levels of the auxin-responsive genes SAUR20 and SAUR14 in the overexpressed plants at this leaf position were significantly higher than those in the WT, which further confirmed that the auxin content of the overexpressed plants was higher than that of the WT (Figure 9B, Figure 3K). At the same time, the expression levels of the cyclins CYCD3;1 and CYCD6;1 in the overexpressed PagSTOMAGEN plants were also significantly higher than those in the WT. This shows that the high expression of these genes in the fifth leaf position promoted cell proliferation and differentiation, making the overexpressed plants appear bigger leaf size (Figure 9B, Figure 3F). In the 10th leaf position, the growth regulators (GRF8 and GRF5) and the auxin response factor (ARF11) were significantly up-regulated in the overexpressed plants (Figure 9C, Table S11). The senescence-associated gene SAG21 was significantly down-regulated (Figure 9C, Table S11). In the young stem segment, the internode near the terminal bud, the cell wall expansion-related gene XTH9 was significantly up-regulated in overexpressing plants, which promoted cell expansion (Figure 9D, Table S12).
Figure 9.
The analysis of changes in DEGs involved in photosynthesis and growth. (A) Heatmap of DEGs of STO-OE and WT lines in 1st leaf position. (B) Heatmap of DEGs of STO-OE and WT lines in 5th leaf position. (C) Heatmap of DEGs of STO-OE and WT lines in 10th leaf position. (D) Heatmap of DEGs of STO-OE and WT lines in young stem. The color scale represents the FPKM value.
We performed a correlation analysis between STOMAGEN and some differentially expressed genes related to growth, and then, constructed a co-expression network. The pearson correlation coefficient was used to evaluate the gene pair’s expression correlation. The screening threshold for high correlation was r > 0.85 and p < 0.05. The larger nodes had stronger connectivity degrees, indicating that the genes may be more important. We identified 314 pairs correlated with STOMAGEN and 27 genes in this co-expression network (Figure S5). The results show that the reason STOMAGEN overexpression in poplar affected plant photosynthesis and biomass might be that it affected the expression of a large number of photosynthesis- and growth-related genes. Through correlation analysis, we found that STOMAGEN had the highest correlation with photosynthesis-related genes, such as TMP14, PQL2, PDE332, THF1, PSAD, PSAN, and PSAK (Figure S5). Among them, PQL2 functions in the chloroplast NAD (P) H dehydrogenase (NDH) complex to provide additional ATP for CO2 assimilation, followed by HEMA1, WOX1, PIF4, and other genes with strong correlations (Figure S5). These genes are closely related to plant growth and can directly or indirectly promote the biomass accumulation of plants, which is also the reason for the increased biomass in STO-OE poplar, which promotes the vegetative growth of poplar. By analyzing the transcriptome data of PagSTOMAGEN-overexpressing plants, we showed that PagSTOMAGEN altered the expression of genes related to photosynthesis and plant hormone response, which, in turn, promoted poplar growth and development.
3. Discussion
3.1. Overexpression of PagSTOMAGEN Increases Stomatal Density in Poplar
This study was the first to demonstrate, in woody plants, that the overexpression of PagSTOMAGEN could increase the stomatal density of poplar by 64.8–68.1% (Figure 4A–C). Although this was consistent with the results in Arabidopsis, we found that the mechanism by which PagSTOMAGEN affected stomatal development in poplar was different to that in Arabidopsis.
The mature form of STOMAGEN contains 45 amino acids and positively regulates stomatal density in Arabidopsis [23,27,44], while EPF1 and EPF2 negatively regulate stomatal density [42,45,46,47]. In this study, PagSTOMAGEN from 84K poplar was homologous with STOMAGEN in Arabidopsis, rice, and Populus trichocarpa (Figure 1A,B), and its mature form contained 43 amino acids (Figure 1C). Since stomata are mainly formed in young tissues, the tissue-specific expression patterns of PagSTOMAGEN mainly being expressed in apical buds and young leaves showed that PagSTOMAGEN may directly regulate stomatal formation (Figure 2A,B).
The three bHLH family transcription factors—SPEECHLESS, MUTE, and FAMA—are involved in the sequential regulation of the stomatal lineage [7,20,48]. Previous studies have demonstrated that STOMAGEN acts in the early stage of stomatal formation, which is the MMC to the SLGC stage [49]. STOMAGEN acts in the early stage of stomatal development by inhibiting the expression of its receptors to trigger the MAPK cascade, which, in turn, inhibits the expression of SPEECHLESS [49]. We detected the expression levels of stomatal development-related genes using RT-qPCR and found that three bHLH transcription factors showed inconsistent expression changes in STO-OE plants. The expression level of PagSPEECHLESS was basically unchanged; the expression level of PagFAMA, which functions in the final step of stomatal formation [11], was significantly increased; and the expression level of PagMUTE, which limits the frequency of asymmetric division and promotes the differentiation of meristemoids to GMCs [50], was significantly reduced (Figure 7F–H). Therefore, we speculated that PagSTOMAGEN played an important role in the middle and later stages of stomatal development in poplar, which is from the SLGC to the GC stage. This was a new finding based on previous research results.
At the same time, we also found that PagSTOMAGEN can inhibit the expression of the stomatal density suppressors PagEPF1 and PagEPF2 (Figure 7A,B), which indicates that PagSTOMAGEN could improve stomatal density by reducing the expression of its antagonists in poplar. A similar trend was observed for the PagSDD1 gene (Figure 7C), which encodes a stomatal density suppressor [18,51,52,53]. The expression of its receptor PagTMM was also inhibited (Figure 7D). It has been shown that PagSTOMAGEN increases stomatal density because it competes with PagEPF1 and PagEPF2 to bind to the receptor-like protein PagTMM and inhibit the expression of PagTMM [44,54]. However, the expression of PagERECTA, which is also a receptor for PagSTOMAGEN, did not change significantly between WT and PagSTOMAGEN-overexpressing plants (Figure 7E). It was speculated that there may be a certain competition between PagERECTA and PagTMM, which resulted in the combination of less PagSTOMAGEN with PagERECTA. The transcriptional changes in these genes suggest that PagSTOMAGEN was directly or indirectly involved in the diverse stages of the stomatal lineage.
3.2. Overexpression of PagSTOMAGEN Promotes the Vegetative Growth of Poplar
Poplar is one of the most widely planted tree species in the world due to its important greening function and economic value [55]. It is also an important raw material for paper, construction and the bioenergy industry [56]. STO-OE poplar displayed an increased plant height, leaf area, number of internodes, stem basal diameter, and biomass compared to the WT (Figure 3E–J and Figure S3). This indicates that PagSTOMAGEN overexpression can promote vegetative growth in poplar. However, AtSTOMAGEN overexpression cannot enhance vegetative growth in Arabidopsis under constant light conditions [26]. It was unclear why poplar overexpressing PagSTOMAGEN behaved differently to Arabidopsis in vegetative growth.
Firstly, we analyzed the reasons for the growth advantage of STO-OE poplar on the phenotypic and physiological levels. PagSTOMAGEN overexpression enhanced the photosynthetic rate in poplar. The enhanced photosynthetic rate was caused by increased stomatal conductance and intercellular carbon dioxide concentration in STO-OE plants (Figure 5A–C). This indicates that increased stomatal density enhanced the photosynthetic rate by regulating the diffusion process of CO2 gas [26,57,58], which is consistent with Arabidopsis.
Although the photosynthetic rate and transpiration rate were enhanced in the STO-OE lines for poplar and Arabidopsis (Figure 5A,D), WUE displayed diverse changes. In Arabidopsis, WUE decreased in AtSTOMAGEN-overexpressing lines, which subjected the plants to water stress, thereby resulting in no significant change in vegetative growth [26]. In poplar, there was no significant difference in WUE and MDA content between the STO-OE and WT lines (Figure 3E and Figure 6D), which indicates that PagSTOMAGEN-overexpressing poplar was not subjected to water stress, further enhancing vegetative growth (Figure 3I,J). These results clearly show that increased stomatal density enhanced photosynthetic capacity but had no effects on WUE in STO-OE poplar. At the same time, studies have shown that an increase in CAT and POD enzyme activities can promote biomass accumulation [4]. CAT and POD enzyme activities were significantly increased in PagSTOMAGEN-overexpressing poplar compared with the WT (Figure 6A,B), which may also explain the vegetative growth advantage in STO-OE poplar.
Then, we explored the reasons for the increase in the biomass of STO-OE poplar at the level of molecular mechanisms. Changes in biomass are affected by many factors. Plant hormones such as auxin and cytokinin play vital roles in regulating the growth and development of poplar, and they control plant height, leaf area, and cell size [35,59]. We assessed the auxin and cytokinin contents of the WT and STO-OE and found that the auxin and cytokinin contents were substantially increased in STO-OE poplar (Figure 3K,L). These results verified that PagSTOMAGEN promoted vegetative growth by promoting the accumulation of auxin and cytokinin in poplar.
Through the transcriptome analysis of different tissues of the WT and STO-OE, we found that auxin pathway-related genes were significantly highly expressed in STO-OE at the 1st, 5th, and 10th leaf positions and young stem segments, such as SAUR75, SAUR14, SAUR50, SAUR20, SAUR66, SAUR72, and ARF11 (Figure 9). SAUR50 is involved in the de-etiolation of Arabidopsis cotyledons [60]. Auxin response factor (ARF) family genes play an important role in controlling sensitivity to the plant hormone auxin [61]. This result indicates that different tissues in the STO-OE lines were affected by high auxin content, compared to WT, and auxin response-related genes could regulate auxin sensitivity, thereby promoting poplar growth.
We also compared the expression level of the genes involved in photosynthesis signaling pathways in the leaf and stem of the WT and STO-OE lines. As shown in Figure 9, the expression levels of PSBX, PSAD, PSAG, PSAN, THF1, PQL2, POC1, GIF1, GNC, and GRF5 in STO-OE were increased compared to those in the WT. THF1 is involved in the metabolic pathway that controls the assembly of the PS II complex [62]. GNC-overexpressing poplars exhibited a higher biomass accumulation, chlorophyll content, photosynthetic rate, and plant height, compared with the WT [36]. GRF5 significantly enlarged the leaf size in GRF5-overexpressing transgenic poplars by enhancing both cell division and cell expansion [35]. These examples demonstrate that the high expression of these transcription factors in the STO-OE lines was responsible for the increase in plant photosynthetic rate and biomass.
At the same time, the expression levels of ERF1, ERF5 and ERF9 involved in the ethylene pathway were reduced in the STO-OE lines, compared with the WT (Figure 9). The constitutive expression of ERF1 phenocopies exhibited ethylene over-production [63]. Thus, their low expression inhibited the accumulation of ethylene in STO-OE poplar, thereby delaying plant senescence and prolonging photosynthetically active periods. In addition, the senescence-related transcription factor SAG14, which promotes plant senescence [64], was also expressed at a lower level in STO-OE plants than in the WT. These results are consistent with the plant hormone and photosynthetic capacity analyses (Figure 3K and Figure 5A), suggesting that PagSTOMAGEN gene expression might affect hormone content and photosynthetic rate and that these changes affected leaf and stem development in poplar.
The above findings clarify that the overexpression of PagSTOMAGEN could promote the photosynthetic rate and transpiration rate by increasing stomatal density in poplar. This resulted in increased biomass in poplar. Our study indicates that PagSTOMAGEN overexpression had positive effects on the stomatal development and vegetative growth of poplar. Therefore, selecting a gene to improve multiple traits in poplar simultaneously could be an effective strategy. Whether the PagSTOMAGEN gene is also involved in the regulation of other important growth and development traits and the pathways that encode the signal effect remains to be explored in the future.
4. Materials and Methods
4.1. Sequence Retrieval and Gene Identification
The deduced amino acid sequences from Arabidopsis AtEPF/EPFL proteins were downloaded from the TAIR database (http://www.arabidopsis.org, accessed on 15 December 2021). Arabidopsis AtEPF/EPFL proteins were blast analyzed against the 84K protein sequences using the BLASTp algorithm [65,66]. The protein sequences of EPF/EPFLs from Oryza sativa and Populus trichocarpa were downloaded from Phytozome (http://phytozome.jgi.doe.gov/pz/portal.html, accessed on 15 December 2021) (Table S2). The neighbor-joining (NJ) tree was constructed via the maximum likelihood method using MEGA 7 with 1000 bootstrap replicates [67]. Multiple sequence alignment was performed for STOMAGEN/EPFL9 (STOMAGEN) from four species using ClustalW (http://www.ebi.ac.uk/Tools/clustalw/, accessed on 15 December 2021) [68]. The protein structure was predicted using I-TASSER (https://zhanggroup.org/I-TASSER/, accessed on 19 December 2021).
4.2. Plant Material and Growth Conditions
The poplar line ‘84K’ (Populus alba × P. glandulosa cv.) was used for gene cloning, expression pattern analysis, and genetic transformation. The tissue cultures of 84K poplar were grown on 1/2 Murashige and Skoog (MS) agar medium at 25 °C under a photoperiod of 16 h light and 8 h dark [69]. The seeds of Nicotiana benthamiana were sown in pots filled with a 3:1 mixture of soil and perlite in a growth chamber at 22 °C with a 16 h/d light photoperiod. They were grown for 30 days, and then, used for subcellular localization assays.
4.3. RNA Extraction and Gene Cloning
Total RNA was isolated from the collected plant tissues using a plant total RNA extraction kit (Tiangen, China, Cat DP432) in accordance with the manufacturer’s instructions, and treated with DNase I (Tiangen, China, Cat DP432). The quality and quantity of RNA were measured using a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). First-strand cDNA was synthesized from the total RNA using a cDNA Synthesis Kit (Tiangen, China, Cat KR106) as per the manual. The PagSTOMAGEN coding sequence was cloned using the PrimeStar® High-fidelity Thermostable DNA Polymerase Reagent Kit (Takara Biotechnology Co., Ltd., Dalian, Liaoning, China), and the primers are shown in Supplementary Table S1.
4.4. Quantitative Real-Time Polymerase Chain Reaction Analysis
Total RNA extracted from various poplar tissues was reverse-transcribed into cDNA using a cDNA Synthesis Kit (Tiangen, China, Cat KR106). Quantitative real-time polymerase chain reaction (RT-qPCR) assays were performed using TransStart Top Green qPCR SuperMix (TRANSGEN, China, cat AQ132-22) on the Applied Biosystems 7500 real-time PCR system according to the manufacturer’s manual. Three technical replicates and three biological replicates were performed for each tissue sample. PagACTIN was used as an internal control, as described previously [35]. Gene-specific primers were obtained by directly querying the primer data from qPrimerDB (https://biodb.swu.edu.cn/qprimerdb, accessed on 13 December 2021) [70]. The 2−ΔΔCt method was used for calculating gene relative expression levels. The primers are listed in Supplementary Table S1.
4.5. Subcellular Localization
The ORF of PagSTOMAGEN without the termination codon was PCR-amplified and inserted into pBI121-GFP to yield 35S::PagSTOMAGEN-GFP (Figure 3A), which had a cauliflower mosaic virus 35S promotor upstream of the cloning site. The 35S::PagSTOMAGEN-GFP and 35S::GFP were transformed into Agrobacterium tumefaciens strain GV3101, and then, injected into the leaf lamina of Nicotiana benthamiana plants as described previously [35]. Two days after injection, transformed tobacco leaves were observed using laser confocal fluorescence microscopy (Leica TCS SP8; Leica, Wetzlar, Germany). An argon ion wavelength of 488 nm was employed for GFP and chlorophyll. Fluorescence was detected at 495–515 nm for GFP and at 650 nm for chlorophyll [68].
4.6. Plant Transformation and Molecular Identification of Transgenic Poplar
The constructed 35S::PagSTOMAGEN-GFP was introduced into the Agrobacterium strain GV3101 using the freeze–thaw method [71]. The Agrobacterium tumefaciens-mediated transformation of 84K poplar was performed as described previously [35]. The infected leaves were transplanted to MS agar medium containing 30 mg∙L−1 kanamycin and 200 mg∙L−1 timentin for screening. Regenerating buds were transferred to 1/2 MS agar medium with 30 mg∙L−1 kanamycin and 200 mg∙L−1 timentin until rooting.
Genomic DNA was extracted from each presumptive transgenic line and the WT plant using a Plant Genome Extraction Kit (Tiangen, China, Cat DP320). Transformation was verified using primers the 35S-F and PagSTOMAGEN-R (Table S1). The expression level of PagSTOMAGEN was detected using RT-qPCR. The 84K transgenic lines identified via PCR and RT-qPCR were used for propagation. One-month-old transgenic plants were transplanted to the greenhouse (16 h light/8 h dark, 22–25 °C, relative humidity: 40–45%).
4.7. Evaluation of Plant Growth
The plant height, leaf area, number of internodes, and stem diameter were measured in two-month-old transgenic and WT plants with nine individual plants per line. Aboveground and underground plant materials were collected and weighed to obtain the fresh weight for the WT and PagSTOMAGEN-overexpressing (STO-OE) lines. The above materials were dried to a constant weight at 80 °C to obtain their dry weight using the gravimetric method.
4.8. Detection of Plant Hormones
The plant samples (leaves) were harvested from the WT and STO-OE plants, frozen in liquid nitrogen, and stored at −80 °C. Following the manufacturer’ s protocols of Convinced (Nanjing Convinced-test Technology Co., Ltd., Nanjing, China), the samples were dissolved in methanol/water/formic acid (15:4:1, v/v/v). A total of 10 μL of 100 ng/mL internal standard mixed solution (IS) was added into the extract to allow for quantification. The extract was then evaporated to dryness, dissolved in methanol, and filtered. Auxin and cytokinin were detected using a liquid chromatography-tandem mass spectrometry (LC-MS/MS) system (AB Sciex QTRAP 6500) from Nanjing Convinced-test Technology Co., Ltd. Three biological replicates were analyzed.
4.9. Determination of Stomatal Density, Guard Cell Length and Width, and Stomatal Index
Scanning electron microscopy analysis of the leaves was carried out for two-month-old WT and STO-OE lines according to the previous procedure, with minor modifications [72]. The leaves were fixed in 2.5% glutaraldehyde at 4 °C. The tissue blocks were washed with 0.1 M Phosphate Buffer (PB, pH 7.4) three times and transferred to 1% osmium tetroxide (OsO4) made in 0.1 M PB (pH 7.4) for 1–2 h at room temperature. Subsequently, the tissue blocks were washed three times using 0.1 M PB (pH 7.4). The washed tissue blocks were dehydrated in a graded ethanol series and immersed in tert-butanol for 30 min. The samples were dried in the K850 Critical Point Dryer (Quorum Technologies Ltd., Lewes, UK), and then, attached to metallic stubs using carbon stickers and sputter-coated with gold for 30 s. The images were observed and taken with a scanning electron microscope. Stomatal density and guard cell length and width were determined from the images. A stomatal index (SI) was calculated using the following formula: SI = (number of stomata) /(number of stomata + number of other epidermal cells) × 100.
4.10. Gas Exchange Analysis
A LI-COR 6400 portable photosynthesis meter was used to measure the net photosynthetic rate (Pn), stomatal conductance (Gs), intercellular CO2 concentration (Ci), and transpiration rate (Tr) in two-month-old WT and STO-OE plants for 4–5 h after the beginning of the photoperiod. The seventh and eighth leaves were used because they represented mature leaves with complete stomatal development, and their photosynthetic parameters were relatively stable. Photosynthetic light-response curves were determined at photosynthetic photon flux densities (PPFD) of 1200, 1000, 800, 600, 400, 200, 150, 100, 50, 25, and 0 μmol m−2⋅s−1 with 400 μmol mol−1 external CO2. WUE was calculated as the ratio Pn/Tr.
4.11. Measurement of Catalase, Peroxidase, and Superoxide Dismutase Activities
Catalase (CAT), peroxidase (POD), and superoxide dismutase (SOD) activities were measured according to the manufacturer’s instructions (Nanjing Mofan Biotechnology Co., Ltd., Nanjing, China). Briefly, 0.1 g of leaves from two-month-old plants was ground into fine powder. The powder was added to a 1 mL extract, and then, extracted for 3–5 min in an ice bath using a tissue crusher. The extracted solution was centrifuged at 8000× g for 10 min at 4 °C, and the supernatant was collected. The absorbance was measured at 405, 470, and 560 nm for CAT, POD, and SOD, respectively, using a SpectraMax 190 full-wavelength microplate reader (Molecular Devices, Sunnyvale, CA, USA) [73].
4.12. Measurement of Malondialdehyde Content
Malondialdehyde (MDA) concentration was measured using the thiobarbituric acid method, with minor modifications [74]. Leaves (0.2 g) from two-month-old plants were homogenized with 3 mL of 10% thiobarbituric acid at 4 °C. The supernatant was collected after centrifugation at 8000× g for 10 min at 4 °C. Equal volumes of the supernatant and 0.6% thiobarbituric acid were mixed. The obtained mixture was reacted in a boiling water bath for 20 min, and then, cooled down immediately. The cooled mixture was centrifuged at 4000 rpm for 10 min, and the supernatant was collected. The light absorbance (A) of the supernatant was measured at 450, 532, and 600 nm using an MD SpectraMax 190 full-wavelength microplate reader [75]. MDA content (nmol/g) = MDA concentration × volume of extracted liquid (mL)/fresh weight (g).
4.13. Transcriptome Analysis
The 1st, 5th, and 10th leaf positions and the young stems samples used for genome-wide transcriptome sequencing in the WT and STO-OE plants were harvested, and then, quickly frozen in liquid nitrogen. A TRIzol reagent kit (Invitrogen, Carlsbad, CA, USA) was used to extract the total RNA from the samples, and an RNase-Free DNaseSet (Qiagen China, Shanghai, China) was used to purify the RNA. To detect the integrity of the RNA, we performed agarose gel electrophoresis, and a NanoDrop 2000 biological analyzer (Thermo Fisher Scientific Inc., Wilmington, DE, USA) was used to detect the concentration of RNA. High-quality RNA was used for subsequent sequencing. Transcriptome sequencing of the samples was performed by Nuohezhiyuan Technology Co., Ltd. (Beijing, China). Then, the FPKM of each gene was calculated based on the length of the gene and the read counts mapped to this gene. The DESeq R package was used for differential gene expression analysis [76]. Genes for which an adjusted p-value ≤ 0.05 was found by DESeq were assigned as differentially expressed. Information on gene annotation was acquired from the NCBI (http://www.ncbi.nlm.nih.gov/), JGI (http://jgi.doe.gov/), PopGenie (http://popgenie.org/), Tair (http://www.arabidopsis.org/), and KEGG (http://www.kegg.jp/) databases.
4.14. Statistical Analyses
SPSS software (IBM Corp., Armonk, NY, USA) was used to perform the statistical analyses, and the data are represented as the mean ± SD. The Student’s t-test and one-way ANOVA were used to determine variance among the mean values [77].
Supplementary Materials
The supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms231710165/s1.
Author Contributions
Conceptualization, Y.X., W.W., J.L. and X.K.; methodology, Y.X., K.D. and J.L.; formal analysis, Y.X., J.L. and A.L.; writing—original draft preparation, Y.X.; writing—review and editing, Y.X., W.W., J.L. and X.K. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was supported by the National Key R&D Program of China during the 14th Five-year Plan Period (2021YFD2200105).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hetherington A.M., Woodward F.I.J.N. The role of stomata in sensing and driving environmental change. Nature. 2003;424:901–908. doi: 10.1038/nature01843. [DOI] [PubMed] [Google Scholar]
- 2.Guo H., Xiao C., Liu Q., Li R., Yan Z., Yao X., Hu H. Two galacturonosyltransferases function in plant growth, stomatal development, and dynamics. Plant Physiol. 2021;187:2820–2836. doi: 10.1093/plphys/kiab432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Durand M., Brendel O., Bure C., Le Thiec D. Altered stomatal dynamics induced by changes in irradiance and vapour-pressure deficit under drought: Impacts on the whole-plant transpiration efficiency of poplar genotypes. New Phytol. 2019;222:1789–1802. doi: 10.1111/nph.15710. [DOI] [PubMed] [Google Scholar]
- 4.Li N.N., Shi F., Gao H.Y., Khan A., Wang F.Y., Kong X.H., Luo H.H. Improving photosynthetic characteristics and antioxidant enzyme activity of capsule wall and subtending leaves increases cotton biomass under limited irrigation system. Photosynthetica. 2021;59:215–227. doi: 10.32615/ps.2021.012. [DOI] [Google Scholar]
- 5.Liu Q., Wang Z., Yu S., Li W., Zhang M., Yang J., Li D., Yang J., Li C. Pu-miR172d regulates stomatal density and water-use efficiency via targeting PuGTL1 in poplar. J. Exp. Bot. 2021;72:1370–1383. doi: 10.1093/jxb/eraa493. [DOI] [PubMed] [Google Scholar]
- 6.Shen C., Zhang Y., Li Q., Liu S., He F., An Y., Zhou Y., Liu C., Yin W., Xia X. PdGNC confers drought tolerance by mediating stomatal closure resulting from NO and H2 O2 production via the direct regulation of PdHXK1 expression in Populus. New Phytol. 2021;230:1868–1882. doi: 10.1111/nph.17301. [DOI] [PubMed] [Google Scholar]
- 7.Simmons A.R., Bergmann D.C. Transcriptional control of cell fate in the stomatal lineage. Curr. Opin. Plant. Biol. 2016;29:1–8. doi: 10.1016/j.pbi.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ha Y., Shang Y., Nam K.H. Brassinosteroids modulate ABA-induced stomatal closure in Arabidopsis. J. Exp. Bot. 2016;67:6297–6308. doi: 10.1093/jxb/erw385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fanourakis D., Nikoloudakis N., Pappi P., Markakis E., Doupis G., Charova S.N., Delis C., Tsaniklidis G. The Role of Proteases in Determining Stomatal Development and Tuning Pore Aperture: A Review. Plants. 2020;9:340. doi: 10.3390/plants9030340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bergmann D.C., Lukowitz W., Somerville C.R. Stomatal development and pattern controlled by a MAPKK kinase. Science. 2004;304:1494–1497. doi: 10.1126/science.1096014. [DOI] [PubMed] [Google Scholar]
- 11.Shimada T., Sugano S.S., Hara-Nishimura I. Positive and negative peptide signals control stomatal density. Cell Mol. Life Sci. 2011;68:2081–2088. doi: 10.1007/s00018-011-0685-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J.S., Hnilova M., Maes M., Lin Y.C., Putarjunan A., Han S.K., Avila J., Torii K.U. Competitive binding of antagonistic peptides fine-tunes stomatal patterning. Nature. 2015;522:439–443. doi: 10.1038/nature14561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geisler M., Nadeau J., Sack F.D. Oriented Asymmetric Divisions That Generate the Stomatal Spacing Pattern in Arabidopsis Are Disrupted by the too many mouths Mutation. Plant Cell. 2000;12:2075. doi: 10.1105/tpc.12.11.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H., Yang Y., Wang H., Liu S., Jia F., Su Y., Li S., He F., Feng C., Niu M., et al. The Receptor-Like Kinase ERECTA Confers Improved Water Use Efficiency and Drought Tolerance to Poplar via Modulating Stomatal Density. Int. J. Mol. Sci. 2021;22:7245. doi: 10.3390/ijms22147245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nadeau J.A., Sack F.D. Control of stomatal distribution on the Arabidopsis leaf surface. Science. 2002;296:1697–1700. doi: 10.1126/science.1069596. [DOI] [PubMed] [Google Scholar]
- 16.Shpak E.D., McAbee J.M., Pillitteri L.J., Torii K.U. Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science. 2005;309:290–293. doi: 10.1126/science.1109710. [DOI] [PubMed] [Google Scholar]
- 17.Han S.K., Kwak J.M., Qi X. Stomatal Lineage Control by Developmental Program and Environmental Cues. Front. Plant Sci. 2021;12:751852. doi: 10.3389/fpls.2021.751852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schluter U., Muschak M., Berger D., Altmann T. Photosynthetic performance of an Arabidopsis mutant with elevated stomatal density (sdd1-1) under different light regimes. J. Exp. Bot. 2003;54:867–874. doi: 10.1093/jxb/erg087. [DOI] [PubMed] [Google Scholar]
- 19.Lampard G.R. Plasticity in stomatal development: What role does MAPK signaling play? Plant Signal. Behav. 2010;5:576–579. doi: 10.4161/psb.11494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacAlister C.A., Ohashi-Ito K., Bergmann D.C. Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature. 2007;445:537–540. doi: 10.1038/nature05491. [DOI] [PubMed] [Google Scholar]
- 21.Chen L., Wu Z., Hou S. SPEECHLESS Speaks Loudly in Stomatal Development. Front. Plant Sci. 2020;11:114. doi: 10.3389/fpls.2020.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han C., Liu Y., Shi W., Qiao Y., Wang L., Tian Y., Fan M., Deng Z., Lau O.S., De Jaeger G., et al. KIN10 promotes stomatal development through stabilization of the SPEECHLESS transcription factor. Nat. Commun. 2020;11:4214. doi: 10.1038/s41467-020-18048-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sugano S.S., Shimada T., Imai Y., Okawa K., Tamai A., Mori M., Hara-Nishimura I. Stomagen positively regulates stomatal density in Arabidopsis. Nature. 2010;463:241–244. doi: 10.1038/nature08682. [DOI] [PubMed] [Google Scholar]
- 24.Sakoda K., Yamori W., Shimada T., Sugano S.S., Hara-Nishimura I., Tanaka Y. Higher Stomatal Density Improves Photosynthetic Induction and Biomass Production in Arabidopsis Under Fluctuating Light. Front. Plant Sci. 2020;11:589603. doi: 10.3389/fpls.2020.589603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiao Z., Han S., Li Z., Huang M., Niu M.-X., Yu X., Liu C., Wang H.-L., Yin W., Xia X. PdEPFL6 reduces stomatal density to improve drought tolerance in poplar. Ind. Crops Prod. 2022;182:114873. doi: 10.1016/j.indcrop.2022.114873. [DOI] [Google Scholar]
- 26.Tanaka Y., Sugano S.S., Shimada T., Hara-Nishimura I. Enhancement of leaf photosynthetic capacity through increased stomatal density in Arabidopsis. New Phytol. 2013;198:757–764. doi: 10.1111/nph.12186. [DOI] [PubMed] [Google Scholar]
- 27.Hunt L., Bailey K.J., Gray J.E. The signalling peptide EPFL9 is a positive regulator of stomatal development. New Phytol. 2010;186:609–614. doi: 10.1111/j.1469-8137.2010.03200.x. [DOI] [PubMed] [Google Scholar]
- 28.Wang S., Zhou Z., Rahiman R., Lee G.S.Y., Yeo Y.K., Yang X., Lau O.S. Light regulates stomatal development by modulating paracrine signaling from inner tissues. Nat. Commun. 2021;12:3403. doi: 10.1038/s41467-021-23728-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang X., Wei H. Breeding polyploid Populus: Progress and perspective. For. Res. 2022;2:4. doi: 10.48130/FR-2022-0004. [DOI] [Google Scholar]
- 30.Heyneke E., Fernie A.R. Metabolic regulation of photosynthesis. Biochem. Soc. Trans. 2018;46:321–328. doi: 10.1042/BST20170296. [DOI] [PubMed] [Google Scholar]
- 31.Jaillais Y., Chory J. Unraveling the paradoxes of plant hormone signaling integration. Nat. Struct. Mol. Biol. 2010;17:642–645. doi: 10.1038/nsmb0610-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ji D., Li Q., Guo Y., An W., Manavski N., Meurer J., Chi W. NADP+ Supply Adjusts the Synthesis of Photosystem I in Arabidopsis Chloroplasts. Plant Physiol. 2022;4:4. doi: 10.1093/plphys/kiac161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang Y., Kang K., Gan L., Ning S., Xiong J., Song S., Xi L., Lai S., Yin Y., Gu J., et al. Drought-responsive genes, late embryogenesis abundant group3 (LEA3) and vicinal oxygen chelate, function in lipid accumulation in Brassica napus and Arabidopsis mainly via enhancing photosynthetic efficiency and reducing ROS. Plant Biotechnol. J. 2019;17:2123–2142. doi: 10.1111/pbi.13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcia-Cerdan J.G., Sveshnikov D., Dewez D., Jansson S., Funk C., Schroder W.P. Antisense inhibition of the PsbX protein affects PSII integrity in the higher plant Arabidopsis thaliana. Plant Cell Physiol. 2009;50:191–202. doi: 10.1093/pcp/pcn188. [DOI] [PubMed] [Google Scholar]
- 35.Wu W., Li J., Wang Q., Lv K., Du K., Zhang W., Li Q., Kang X., Wei H. Growth-regulating factor 5 (GRF5)-mediated gene regulatory network promotes leaf growth and expansion in poplar. New Phytol. 2021;230:612–628. doi: 10.1111/nph.17179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.An Y., Zhou Y., Han X., Shen C., Wang S., Liu C., Yin W., Xia X. The GATA transcription factor GNC plays an important role in photosynthesis and growth in poplar. J. Exp. Bot. 2020;71:1969–1984. doi: 10.1093/jxb/erz564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y., Pfeiffer A., Tepperman J.M., Dalton-Roesler J., Leivar P., Gonzalez Grandio E., Quail P.H. Central clock components modulate plant shade avoidance by directly repressing transcriptional activation activity of PIF proteins. Proc. Natl. Acad. Sci. USA. 2020;117:3261–3269. doi: 10.1073/pnas.1918317117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Galli M., Khakhar A., Lu Z., Chen Z., Sen S., Joshi T., Nemhauser J.L., Schmitz R.J., Gallavotti A. The DNA binding landscape of the maize AUXIN RESPONSE FACTOR family. Nat. Commun. 2018;9:4526. doi: 10.1038/s41467-018-06977-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J., Sun N., Zhang F., Yu R., Chen H., Deng X.W., Wei N. SAUR17 and SAUR50 Differentially Regulate PP2C-D1 during Apical Hook Development and Cotyledon Opening in Arabidopsis. Plant Cell. 2020;32:3792–3811. doi: 10.1105/tpc.20.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weeraratne G., Wang H., Weeraratne T.P., Sabharwal T., Jiang H.W., Cantero A., Clark G., Roux S.J. APYRASE1/2 mediate red light-induced de-etiolation growth in Arabidopsis seedlings. Plant Physiol. 2022;3:3. doi: 10.1093/plphys/kiac150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu W., Du K., Kang X., Wei H. The diverse roles of cytokinins in regulating leaf development. Hortic. Res. 2021;8:118. doi: 10.1038/s41438-021-00558-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takata N., Yokota K., Ohki S., Mori M., Taniguchi T., Kurita M. Evolutionary relationship and structural characterization of the EPF/EPFL gene family. PLoS ONE. 2013;8:e65183. doi: 10.1371/journal.pone.0065183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hronkova M., Wiesnerova D., Simkova M., Skupa P., Dewitte W., Vrablova M., Zazimalova E., Santrucek J. Light-induced STOMAGEN-mediated stomatal development in Arabidopsis leaves. J. Exp. Bot. 2015;66:4621–4630. doi: 10.1093/jxb/erv233. [DOI] [PubMed] [Google Scholar]
- 44.Kondo T., Kajita R., Miyazaki A., Hokoyama M., Nakamura-Miura T., Mizuno S., Masuda Y., Irie K., Tanaka Y., Takada S., et al. Stomatal density is controlled by a mesophyll-derived signaling molecule. Plant Cell Physiol. 2010;51:1–8. doi: 10.1093/pcp/pcp180. [DOI] [PubMed] [Google Scholar]
- 45.Hunt L., Gray J.E. The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Curr. Biol. 2009;19:864–869. doi: 10.1016/j.cub.2009.03.069. [DOI] [PubMed] [Google Scholar]
- 46.Guo Z.F. A Review: Molecular Regulation of Stomatal Development Related to Environmental Factors and Hormones in Plants. Appl. Ecol. Environ. Res. 2019;17:12091–12109. doi: 10.15666/aeer/1705_1209112109. [DOI] [Google Scholar]
- 47.Zeng S.M., Lo E.K.W., Hazelton B.J., Morales M.F., Torii K.U. Effective range of non-cell autonomous activator and inhibitor peptides specifying plant stomatal patterning. Development. 2020;147:dev192237. doi: 10.1242/dev.192237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lopez-Anido C.B., Vaten A., Smoot N.K., Sharma N., Guo V., Gong Y., Anleu Gil M.X., Weimer A.K., Bergmann D.C. Single-cell resolution of lineage trajectories in the Arabidopsis stomatal lineage and developing leaf. Dev. Cell. 2021;56:1043–1055.e1044. doi: 10.1016/j.devcel.2021.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Torii K.U. Mix-and-match: Ligand-receptor pairs in stomatal development and beyond. Trends Plant Sci. 2012;17:711–719. doi: 10.1016/j.tplants.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 50.Kanaoka M.M., Pillitteri L.J., Fujii H., Yoshida Y., Bogenschutz N.L., Takabayashi J., Zhu J.K., Torii K.U. SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to arabidopsis stomatal differentiation. Plant Cell. 2008;20:1775–1785. doi: 10.1105/tpc.108.060848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Von Groll U., Berger D., Altmann T. The subtilisin-like serine protease SDD1 mediates cell-to-cell signaling during Arabidopsis stomatal development. Plant Cell. 2002;14:1527–1539. doi: 10.1105/tpc.001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vrablova M., Vrabl D., Hronkova M., Kubasek J., Santrucek J. Stomatal function, density and pattern, and CO2 assimilation in Arabidopsis thaliana tmm1 and sdd1-1 mutants. Plant Biol. 2017;19:689–701. doi: 10.1111/plb.12577. [DOI] [PubMed] [Google Scholar]
- 53.Morales-Navarro S., Perez-Diaz R., Ortega A., de Marcos A., Mena M., Fenoll C., Gonzalez-Villanueva E., Ruiz-Lara S. Overexpression of a SDD1-Like Gene From Wild Tomato Decreases Stomatal Density and Enhances Dehydration Avoidance in Arabidopsis and Cultivated Tomato. Front. Plant Sci. 2018;9:940. doi: 10.3389/fpls.2018.00940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee J.S., Kuroha T., Hnilova M., Khatayevich D., Kanaoka M.M., McAbee J.M., Sarikaya M., Tamerler C., Torii K.U. Direct interaction of ligand-receptor pairs specifying stomatal patterning. Genes Dev. 2012;26:126–136. doi: 10.1101/gad.179895.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu X., Lu B., Dong Y., Li Y., Yang M. Cloning and functional identification of PeWRKY41 from Populus × euramericana. Ind. Crops Prod. 2022;175:114279. doi: 10.1016/j.indcrop.2021.114279. [DOI] [Google Scholar]
- 56.Zhang Y., Lin S., Zhou Y., Wen J., Kang X., Han X., Liu C., Yin W., Xia X. PdNF-YB21 positively regulated root lignin structure in poplar. Ind. Crops Prod. 2021;168:113609. doi: 10.1016/j.indcrop.2021.113609. [DOI] [Google Scholar]
- 57.Harrison E.L., Arce Cubas L., Gray J.E., Hepworth C. The influence of stomatal morphology and distribution on photosynthetic gas exchange. Plant J. 2020;101:768–779. doi: 10.1111/tpj.14560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chieppa J., Brown T., Giresi P., Juenger T.E., Resco de Dios V., Tissue D.T., Aspinwall M.J. Climate and stomatal traits drive covariation in nighttime stomatal conductance and daytime gas exchange rates in a widespread C4 grass. New Phytol. 2021;229:2020–2034. doi: 10.1111/nph.16987. [DOI] [PubMed] [Google Scholar]
- 59.Du K., Liao T., Ren Y., Geng X., Kang X. Molecular Mechanism of Vegetative Growth Advantage in Allotriploid Populus. Int. J. Mol. Sci. 2020;21:441. doi: 10.3390/ijms21020441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dong J., Sun N., Yang J., Deng Z., Lan J., Qin G., He H., Deng X.W., Irish V.F., Chen H., et al. The Transcription Factors TCP4 and PIF3 Antagonistically Regulate Organ-Specific Light Induction of SAUR Genes to Modulate Cotyledon Opening during De-Etiolation in Arabidopsis. Plant Cell. 2019;31:1155–1170. doi: 10.1105/tpc.18.00803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tian C.E., Muto H., Higuchi K., Matamura T., Tatematsu K., Koshiba T., Yamamoto K.T. Disruption and overexpression of auxin response factor 8 gene of Arabidopsis affect hypocotyl elongation and root growth habit, indicating its possible involvement in auxin homeostasis in light condition. Plant J. 2004;40:333–343. doi: 10.1111/j.1365-313X.2004.02220.x. [DOI] [PubMed] [Google Scholar]
- 62.Yabuta S., Ifuku K., Takabayashi A., Ishihara S., Ido K., Ishikawa N., Endo T., Sato F. Three PsbQ-like proteins are required for the function of the chloroplast NAD(P)H dehydrogenase complex in Arabidopsis. Plant Cell Physiol. 2010;51:866–876. doi: 10.1093/pcp/pcq060. [DOI] [PubMed] [Google Scholar]
- 63.Zhou J., Mu Q., Wang X., Zhang J., Yu H., Huang T., He Y., Dai S., Meng X. Multilayered synergistic regulation of phytoalexin biosynthesis by ethylene, jasmonate, and MAPK signaling pathways in Arabidopsis. Plant Cell. 2022;8:8. doi: 10.1093/plcell/koac139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hao C., Yang Y., Du J., Deng X.W., Li L. The PCY-SAG14 phytocyanin module regulated by PIFs and miR408 promotes dark-induced leaf senescence in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2022;119:e2116623119. doi: 10.1073/pnas.2116623119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu H., Song J., Luo H., Zhang Y., Li Q., Zhu Y., Xu J., Li Y., Song C., Wang B., et al. Analysis of the Genome Sequence of the Medicinal Plant Salvia miltiorrhiza. Mol. Plant. 2016;9:949–952. doi: 10.1016/j.molp.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Altschul S.F., Madden T.L., Schffer A.A., Jinghui Z., Zheng Z., Webb M., Lipman D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kumar S., Stecher G., Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang Y., Li H.G., Wang J., Wang H.L., He F., Su Y., Zhang Y., Feng C.H., Niu M., Li Z., et al. ABF3 enhances drought tolerance via promoting ABA-induced stomatal closure by directly regulating ADF5 in Populus euphratica. J. Exp. Bot. 2020;71:7270–7285. doi: 10.1093/jxb/eraa383. [DOI] [PubMed] [Google Scholar]
- 69.He F., Wang H.L., Li H.G., Su Y., Li S., Yang Y., Feng C.H., Yin W., Xia X. PeCHYR1, a ubiquitin E3 ligase from Populus euphratica, enhances drought tolerance via ABA-induced stomatal closure by ROS production in Populus. Plant Biotechnol. J. 2018;16:1514–1528. doi: 10.1111/pbi.12893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lu K., Li T., He J., Chang W., Zhang R., Liu M., Yu M., Fan Y., Ma J., Sun W., et al. qPrimerDB: A thermodynamics-based gene-specific qPCR primer database for 147 organisms. Nucleic Acids Res. 2018;46:D1229–D1236. doi: 10.1093/nar/gkx725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Höfgen R., Willmitzer L. Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res. 1988;16:9877. doi: 10.1093/nar/16.20.9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Irish V., Sussex I. Function of the apetala-1 Gene during Arabidopsis Floral Development. Plant Cell. 1990;2:741–753. doi: 10.1105/tpc.2.8.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Inui T., Okumura K., Matsui H., Hosoya T., Kumazawa S. Effect of harvest time on some in vitro functional properties of hop polyphenols. Food Chem. 2017;225:69–76. doi: 10.1016/j.foodchem.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 74.Dhindsa R.S., Matowe W. Drought Tolerance in Two Mosses: Correlated with Enzymatic Defence Against Lipid Peroxidation. J. Exp. Bot. 1981;32:79–91. doi: 10.1093/jxb/32.1.79. [DOI] [Google Scholar]
- 75.Liu M., Ma Y., Du Q., Hou X., Wang M., Lu S. Functional Analysis of Polyprenyl Diphosphate Synthase Genes Involved in Plastoquinone and Ubiquinone Biosynthesis in Salvia miltiorrhiza. Front. Plant Sci. 2019;10:893. doi: 10.3389/fpls.2019.00893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang L., Feng Z., Wang X., Wang X., Zhang X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 77.Kopertekh L., Reichardt S. At-CycD2 Enhances Accumulation of Above-Ground Biomass and Recombinant Proteins in Transgenic Nicotiana benthamiana Plants. Front. Plant Sci. 2021;12:712438. doi: 10.3389/fpls.2021.712438. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available in Supplementary Materials.