Abbreviations
- EBV
Epstein‐Barr virus
- GO
gene ontology
- GWAS
genome‐wide association analysis
- HBV
hepatitis B virus
- HCV
hepatitis C virus
- HLA
human leukocyte antigen
- KEGG
Kyoto encyclopedia of genes and genomes
- LD
linkage disequilibrium
- NPC
nasopharyngeal carcinoma
- PIP
posterior inclusion probability
- QC
quality control
- TWAS
transcriptome‐wide association analysis
- ZEBRA
BamHI Z EBV replication activator
Dear Editor,
1.
Nasopharyngeal carcinoma (NPC) is a common malignancy in East and Southeast Asia, especially in South China. The etiology of NPC has been linked to genetic susceptibility, Epstein‐Barr virus (EBV) infection, and environmental factors. Accumulated evidence including multiple genome‐wide association studies (GWASs) has revealed robust genetic predisposition of NPC. However, GWAS‐identified genetic variants collectively account for only 8.2% of NPC heritability [1]. The underlying inherited predisposition is largely undetermined. The strongest genetic signal for NPC consistently hits the human leukocyte antigen (HLA) region on 6p21 [2]. However, the highly polymorphic nature and complicated long‐range linkage disequilibrium (LD) in the HLA region particularly obscure the causal variants driving the association. In addition, most genetic variants located in introns or intergenic regions. The causal genes mediating genetic effects on NPC risk have rarely been ascertained by GWAS alone.
Recently, transcriptome‐wide association study (TWAS) has been proposed as an attractive approach to identify novel gene‐trait associations and prioritize causal genes for complex traits [3]. By integrating GWAS and gene expression data, TWAS can effectively and economically assess associations between genetically predicted gene expression levels and disease risks in large populations. Hence, using the cis‐regulated expression in addition to genetic variants to explore NPC susceptibility genes could be promising and reasonable for mechanistic and functional inference. Nevertheless, neither public data of nasopharyngeal tissue were available, nor TWAS for NPC had been conducted.
Herein, we integrated genome and transcriptome data of 89 nasopharyngeal tumor tissues and investigated the associations between predicted gene expression levels and NPC risk using multicenter GWAS data involving 4506 NPC cases and 5384 cancer‐free subjects (defined as controls) from South China. Given the close relationship between EBV infection and NPC, a cis‐regulated expression weight matrix from EBV‐transformed lymphocytes (n = 117) in the GTEx project was used for further evaluation. Study populations and detailed methodology are described in the Supplementary file of methods.
We predicted the expression levels of 2505 and 2411 genes in the GWAS population by constructing the models for the prediction of gene expression in nasopharyngeal tissues (NP models) and EBV‐transformed lymphocytes (lymphocyte models), respectively (Supplementary Table S1), and 377 genes overlapped (Supplementary Figure S1). Thirty‐three genes were associated with NPC at a Bonferroni‐corrected threshold, and all were located in the HLA region (Figure 1A). Among them, 11 of 13 previously reported genes were replicated. Our results were consistent with the studies focusing on the HLA region in South China [4, 5], where most of the reported genes available in TWAS were replicated. The predicted expression levels of ZFP57 (NP models), MICA (both models), and HLA‐C (lymphocyte models) were significantly higher in cases than in controls, while the expression levels of MOG, HCG27, HLA‐DQB1, HLA‐H, HLA‐U (NP models), HLA‐F (both models), HLA‐A, and HLA‐DRB1 (lymphocyte models) were lower in cases than in controls. The two overlapping genes showed similar associations with NPC (HLA‐F: Z score = ‐10.28 and ‐8.95; MICA: Z score = 7.82 and 6.60, for NP and lymphocyte models, respectively) (Supplementary Table S2). Interestingly, half of the previously reported genes belonged to HLA class I. Most of them showed lower levels of predicted expression in cases than in controls, possibly because EBV transcripts in NPC tumors were involved in the inhibition of HLA class I gene expression [6]. It is rational to assume that the low expression levels of these genes may affect the anti‐EBV immune response in presenting peptides to cytotoxic T cells, facilitating immune evasion of tumor cells or EBV‐mediated oncogenic action.
FIGURE 1.
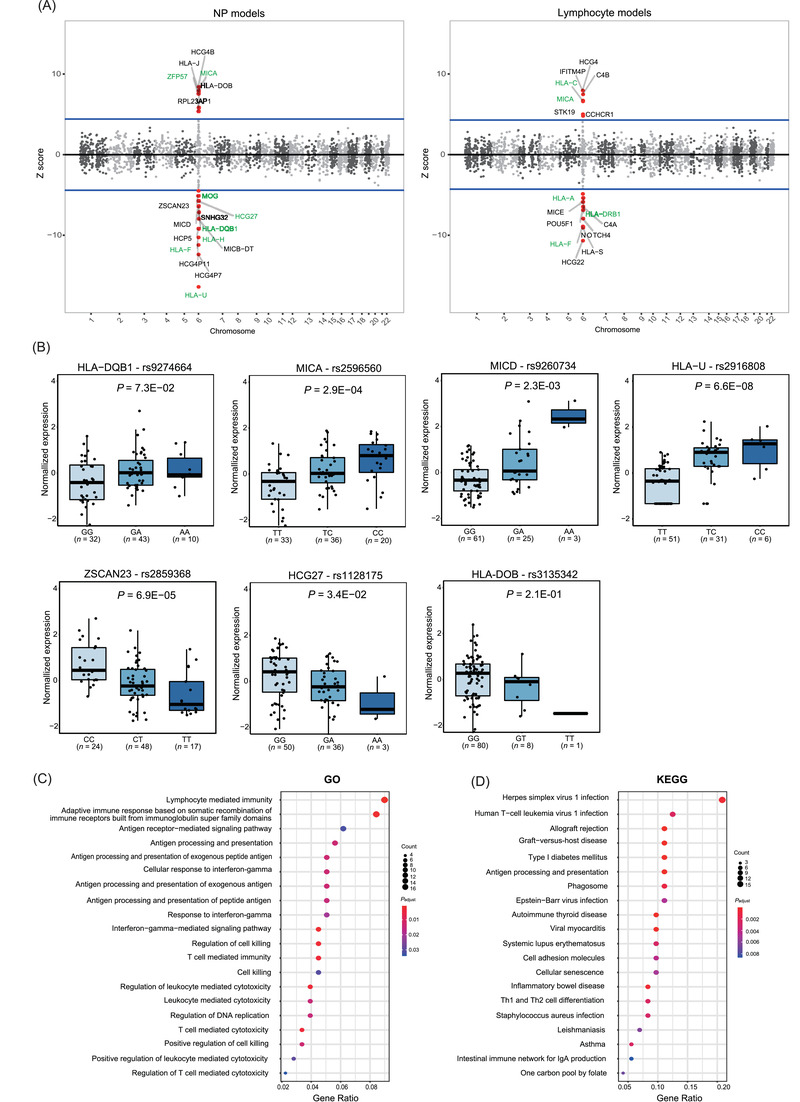
TWAS‐identified susceptibility genes and pathways for NPC. (A) Manhattan plot of TWAS in NP models and lymphocyte models. The blue lines represent the Bonferroni‐corrected significance threshold. The red dots above or below the blue line represent the genes passed the Bonferroni threshold in the association analysis. The genes with green labels have been reported to be associated with NPC by previous genome‐wide or candidate pathway association studies. The genes with black labels were newly identified as NPC susceptibility genes by our study. The genes in different chromosomes were exhibited in light and dark grey dots. (B) Expression quantitative trait locus analysis for the seven putative causal genes in the expression data of 89 nasopharyngeal tissue samples. The Kruskal‐Wallis test was used to compare medians among three genotypes for most of the variants. In a certain homozygote group, the P values were recalculated using only the wild‐type and heterozygous groups for the expression of MICD, HCG27 and HLA‐DOB by excluding the groups with a sample size less than 5. (C) GO pathway enrichment analysis of NPC. (D) KEGG pathway enrichment analysis of NPC. “Gene Ratio” refers to the percentage of total significant genes in the given pathway. All 354 significant genes (P < 0.05) in TWAS were used in the enrichment analysis.
Abbreviations: TWAS, Transcriptome‐wide association analysis; NPC, Nasopharyngeal carcinoma; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes
Although the significant signals consistently hit the HLA region, 22 additional genes not previously reported were identified in TWAS. Among them, the predicted expression levels of 9 genes were significantly higher in NPC cases than in controls, including HLA‐DOB, HCG4B, RPL23AP1, HLA‐J in NP models and HCG4, CCHCR1, STK19, C4B, IFITM4P in lymphocyte models, while 13 other genes showed significantly lower expression levels in cases than in controls, including HCP5, ZSCAN23, HCG4P11, HCG4P7, MICD, MICB‐DT, SNHG32 in NP models and NOTCH4, C4A, HCG22, POU5F1, MICE, HLA‐S in lymphocyte models (Figure 1A). We performed conditional analyses to determine whether the associations between predicted gene expression levels and NPC were influenced by the GWAS signals. After conditioning on the respective GWAS index SNP, the associations for HLA‐DOB, NOTCH4, ZSCAN23, STK19, C4B, HLA‐J, HLA‐S, and MICB‐DT remained significant. After conditioning on all previously reported SNPs, NOTCH4, HCG4, HCG22, POU5F1, HCG4B, HCG4P11, MICB‐DT, STK19 and IFITM4P remained significant. It indicated that their associations were partially independent of the GWAS signals (Supplementary Table S3).
Due to the complicated structure with high LD and co‐expression networks in the HLA region, we conducted fine‐mapping analyses to prioritize the causal genes. Using posterior inclusion probability (PIP) analysis, we prioritized 7 causal genes: MICA, HLA‐DQB1, HLA‐DOB, ZSCAN23, HCG27, MICD, and HLA‐U. HLA‐DOB, ZSCAN23, and MICD were newly identified as NPC susceptibility genes (Supplementary Table S4). Furthermore, we conducted expression quantitative trait locus (eQTL) analyses to identify whether the genetic variants could influence the expression levels of these genes. We found that individuals with relevant risk SNPs (the GWAS index SNPs) exhibited higher expression of HLA‐DQB1, MICA, MICD and HLA‐U, or lower expression levels of ZSCAN23, HCG27, and HLA‐DOB. These results indicated that the risk alleles affected the expression levels of the causal genes (Figure 1B). Two HLA class II genes (HLA‐DQB1 and HLA‐DOB) were prioritized as causal genes. Both genes were associated with other virus‐associated cancers, such as cervical cancer [7]. A comprehensive TWAS exploring genetic susceptibility for antiviral immune response using 7924 subjects from the UK Biobank cohort revealed that the genetic determinants for EBV infection were predominantly located on HLA class II genes. The most significant signals associated with the antibody level of BamHI Z EBV replication activator (ZEBRA) hit HLA‐DQB1 [8]. HLA‐DOB may impact viral clearance capacity and persistent infection of hepatitis B virus (HBV) and hepatitis C virus (HCV) [9]. Since EBV reactivation with elevated EBV DNA load or antibodies was observed at the preclinical phase of NPC, we hypothesized that HLA class II genes, especially HLA‐DQB1and HLA‐DOB, participate in the early stage of NPC tumorigenesis by influencing EBV infection. Besides, some identified pseudogenes, such as IFITM4P [10], may function by regulating their parental genes. However, their biological mechanisms remain unclear, and further researches are needed.
Gene Ontology (GO) enrichment analysis confirmed that TWAS‐identified genes (354 genes with P < 0.05) were enriched in the pathways of cell‐mediated immune response, antigen processing and presentation (Figure 1C). Similarly, the top pathways annotated with the Kyoto Encyclopedia of Genes and Genomes (KEGG) database focused on infection of herpes simplex virus type 1, human T‐cell leukemia virus type 1, EBV, and autoimmune disorders such as graft‐versus‐host disease (Figure 1D).
In summary, using a TWAS approach, we corroborated the central role of HLA genes in NPC susceptibility. Apart from HLA class I genes, we propose critical roles of HLA class II genes and other nonclassical HLA genes. Seven genes, including HLA‐DQB1 and HLA‐DOB, were prioritized as causal genes. Recent evidence indicated that these genes are pivotal in the metastable equilibrium between host and virus. Our findings provide additional evidence for a better understanding of the genetic etiology of NPC and clues to further advance this field.
DECLARATIONS
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to declare.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The Institutional Review Board of Sun Yat‐sen University Cancer Center approved this study. Informed consent was obtained from all study participants.
AUTHOR CONTRIBUTIONS
WHJ and YQH devised the project and the main conceptual ideas; YQH, WQX, DHL, and TMW wrote the original draft; DHL and TMW performed the computational analyses; TMW, DWY, CMD, and WLZ contributed to implementation of data processing and analyses; DWY, CMD, YL, WLZ, RWX, LL, HD, XT, YW, TZ, XZL, PFZ, XHZ, SDZ, YZH, MT, YZ, YC and JBZ contributed to the sample preparation; TMW and WLZ contributed to the RNA‐seq quantification and quality control pipeline; ETC, ZZ, GH, SMC, QL, LF, YS, MLL, HOA, WY, and THL contributed to the interpretation of the results; YQH, WQX, and TMW revised and wrote the final version of the manuscript; verified the analytical methods; WHJ supervised the project. All authors read and approved the final manuscript.
AVAILABILITY OF DATA AND MATERIALS
Methods and materials are available in the supplementary file. The datasets generated and used during the current study are available at Research Data Deposit (RDD) public platform (www.researchdata.org.cn) with the approval RDD number of RDDB2021406340.
Supporting information
Supporting information
ACKNOWLEDGEMENTS
We thank the staffs from Sun Yat‐sen University Cancer Center biorepository. We thank all the study participants and research staff who recruited participants and collected samples in this study. This study was funded by the National Key Research and Development Program of China (2021YFC2500400), the Basic and Applied Basic Research Foundation of Guangdong Province, China (2021B1515420007), Sino‐Sweden Joint Research Programme (81861138006), the Science and Technology Planning Project of Guangzhou, China (201804020094), the Special Support Program for High‐level Professionals on Scientific and Technological Innovation of Guangdong Province, China (2014TX01R201), National Natural Science Foundation of China (81973131, 81903395, 81803319, 82003520), National Science Fund for Distinguished Young Scholars of China (81325018).
Yong‐Qiao He, Wen‐Qiong Xue, Dan‐Hua Li, and Tong‐Min Wang have contributed equally to this work.
REFERENCES
- 1. Dai J, Shen W, Wen W, Chang J, Wang T, Chen H, et al. Estimation of heritability for nine common cancers using data from genome‐wide association studies in Chinese population. Int J Cancer. 2017;140(2):329–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bei JX, Li Y, Jia WH, Feng BJ, Zhou G, Chen LZ, et al. A genome‐wide association study of nasopharyngeal carcinoma identifies three new susceptibility loci. Nat Genet. 2010;42(7):599–603. [DOI] [PubMed] [Google Scholar]
- 3. Wainberg M, Sinnott‐Armstrong N, Mancuso N, Barbeira AN, Knowles DA, Golan D, et al. Opportunities and challenges for transcriptome‐wide association studies. Nat Genet. 2019;51(4):592–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ning L, Ko JM, Yu VZ, Ng HY, Chan CK, Tao L, et al. Nasopharyngeal carcinoma MHC region deep sequencing identifies HLA and novel non‐HLA TRIM31 and TRIM39 loci. Commun Biol. 2020;3(1):759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tian W, Zhu F, Cai J, Li L, Jin H, Wang W. Multiple low‐frequency and rare HLA‐B allelic variants are associated with reduced risk in 1,105 nasopharyngeal carcinoma patients in Hunan province, southern China. Int J Cancer. 2020;147(5):1397–1404. [DOI] [PubMed] [Google Scholar]
- 6. Sengupta S, den Boon JA, Chen IH, Newton MA, Dahl DB, Chen M, et al. Genome‐wide expression profiling reveals EBV‐associated inhibition of MHC class I expression in nasopharyngeal carcinoma. Cancer Res. 2006;66(16):7999–8006. [DOI] [PubMed] [Google Scholar]
- 7. Peng S, Trimble C, Wu L, Pardoll D, Roden R, Hung CF, et al. HLA‐DQB1*02‐restricted HPV‐16 E7 peptide‐specific CD4+ T‐cell immune responses correlate with regression of HPV‐16‐associated high‐grade squamous intraepithelial lesions. Clin Cancer Res. 2007;13(8):2479–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kachuri L, Francis SS, Morrison ML, Wendt GA, Bosse Y, Cavazos TB, et al. The landscape of host genetic factors involved in immune response to common viral infections. Genome Med. 2020;12(1):93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Denzin LK, Khan AA, Virdis F, Wilks J, Kane M, Beilinson HA, et al. Neutralizing Antibody Responses to Viral Infections Are Linked to the Non‐classical MHC Class II Gene H2‐Ob. Immunity. 2017. 47(2):310–22 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xiao M, Chen Y, Wang S, Liu S, Rai KR, Chen B, et al. LncRNA IFITM4P regulates host antiviral responses by acting as a ceRNA. J Virol. 2021;JVI0027721. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information
Data Availability Statement
Methods and materials are available in the supplementary file. The datasets generated and used during the current study are available at Research Data Deposit (RDD) public platform (www.researchdata.org.cn) with the approval RDD number of RDDB2021406340.


