Abstract
The Salmonella PmrA-PmrB system controls the expression of genes necessary for polymyxin B resistance. Four loci were previously identified as part of the regulon, and interaction of PmrA with the promoter region of three of them was observed. Here we characterized the interaction of PmrA with the promoter region of ugd, previously suggested to be regulated indirectly by PmrA. Our results indicate that PmrA controls the expression of ugd by interacting with a specific sequence in the promoter region of this gene.
The two-component regulatory system PmrA-PmrB of Salmonella enterica serovar Typhimurium controls the expression of genes that mediate resistance to polymyxin B and other cationic antimicrobial polypeptides (6, 8, 18, 22). This is accomplished by modifications of the lipopolysaccharide that result in both an increased substitution of the ester-linked phosphate at position 4′ of the lipid A with 4-amino-4-deoxy-l-arabinose (4-ARA) and also larger amounts of 2-aminoethanol esterifying the diphosphates of the core oligosaccharide (11), decreasing the binding of the antimicrobial compounds (11, 19, 20). Four different loci have been identified to be under control of the PmrA-PmrB system: the ugd (22) and pmrG genes (7) and the pmrCAB (22) and pmrF operons (7). ugd, also known as pmrE (7), was previously identified as pagA (17). It encodes a putative UDP-glucose dehydrogenase homologous to the products of Streptococcus pneumoniae cap3A (3) and Streptococcus pyogenes hasB (4) genes, which is responsible for the conversion of UDP-glucose into UDP-glucuronate. The pmrG gene is homologous to the ais gene of Escherichia coli (7), whose expression is aluminum induced. The pmrCAB operon encodes both the two-component system PmrA-PmrB (18) and a hydrophobic polypeptide not required for polymyxin resistance (6, 21, 22). The pmrF operon encodes seven polypeptides, some of which have homology to oxidoreductases, decarboxylases, and aminotransferases, that are predicted to be part of the UDP-glucuronate→4-ARA pathway (1, 7). Previous work using lacZ fusions strongly indicated that the regulation of the four PmrA-PmrB-dependent loci was direct (7, 22). Recently, it was demonstrated that the expression of pmrCAB, pmrF, and pmrG is directly controlled by PmrA, while indirect regulation of ugd by the response regulator was suggested (24).
Here, we characterized the interaction of PmrA with the promoter region of ugd. We determined that PmrA recognizes a specific sequence in the promoter region of ugd and that phosphorylation of the response regulator stimulates the interaction.
Interaction of PmrA with the promoter region of ugd.
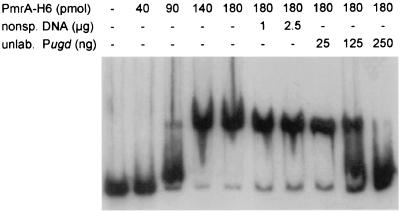
To determine if PmrA interacts with the promoter region of ugd (Pugd), a 354-bp DNA fragment that encompasses and extends 204 bp upstream from the transcriptional start site (24) was amplified by PCR using the PROM UGD-F (5′-CTGAATTCAGGCGCAGCGTG-3′) and the PROM UGD-R (5′-AACCCGTCCCGGATATCGTG-3′) oligonucleotides as primers. For DNA shift mobility assays, we constructed a pmrA His tag fusion gene. This fusion gene was generated by PCR using primers PmrA-NTF (5′-GAGGATCCATATGAAGATACTGATTG-3′) and PmrA-H6-CTR (5′-TCCAAGCTTAGTGGT GGTGGTGGTGGTGGCTTTCCTCAGTGGCAACC-3′) and then cloned between the NdeI and HindIII sites of plasmid pT7-7 to obtain plasmid pPB1022. The His-tagged PmrA protein was purified using a Ni2+-nitrilotriacetic acid-agarose affinity chromatography column according to the QIAexpressionist purification protocol (Qiagen) and exhaustively dialyzed against 20 mM Tris-HCl (pH 7.9)–50 mM KCl. The protein concentration was determined by the bicinchoninic acid assay (Bio-Rad), and the protein profile of the purified PmrA-H6 protein was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The gel mobility assays were performed essentially as described elsewhere (12) by incubating 40 to 180 pmol of purified PmrA-H6 with 1 ng of the 354-bp ugd fragment in 1× Sp1 binding buffer without NP-40 (12) in a 40-μl assay. We detected a shift of the DNA fragment when we used 90 pmol of unphosphorylated PmrA-H6 (Fig. 1). The interaction was specific, as demonstrated by the efficient competition by unlabeled homologous DNA. On the other hand, addition of nonspecific competitor (up to 2.5 μg of salmon sperm DNA) did not affect the shift. This result demonstrates that PmrA interacts directly with the promoter region of ugd.
FIG. 1.
PmrA interacts with the promoter region of ugd. The electrophoretic shift mobility assay was performed using the 32P-, 3′-end-labeled 354-bp PCR fragment of the promoter region of ugd, incubated with purified PmrA-H6, in the absence or presence of different amounts of either salmon sperm DNA as a nonspecific competitor (nonsp. DNA) or the unlabeled promoter region of ugd (unlab. Pugd).
The PmrA-H6 concentration necessary to induce a shift in our experiment was comparable to the maximal concentration reported previously (24). Indeed, when we used DNA fragments from the pmrCAB and pmrF promoter regions, a shift was observed at PmrA-H6 concentrations similar to those required for the shift of the ugd promoter fragment (our unpublished observations). This finding is consistent with previous in vivo observations that showed that the PmrA-dependent expression of ugd did not require a different level of activation of the PmrA-PmrB regulatory system (7, 22). The larger amounts of PmrA-H6 required to induce a shift under our experimental conditions could be explained by the 1 mM dithiothreitol (DTT) that was included in the binding buffer. Reducing agents such as DTT and β-mercaptoethanol have been demonstrated to affect the oligomeric status of PmrA, reducing its binding capabilities (24) (see below). Furthermore, while we used a 354-bp fragment that included 326 bp upstream and 25 bp downstream from the initiation codon of ugd, Wösten and Groisman used a 268-bp fragment that included 203 bp upstream and 54 bp downstream from the ugd ATG start codon (24). This suggests that the additional 123-bp sequence used in our assay may play a role in the protein-DNA interaction.
Determination of the PmrA-binding site.
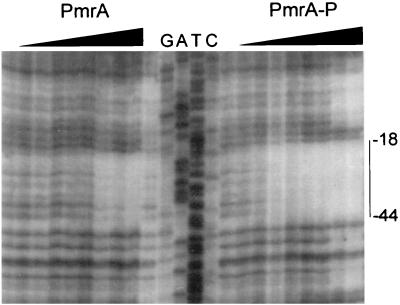
Since the ugd promoter region did not contain the imperfect inverted repeat previously described as the PmrA binding site (24), we decided to determine in this promoter the DNA sequence recognized by PmrA. DNA-footprinting analysis was performed on both the coding and noncoding strands. Protein-DNA complexes were allowed to form for 20 min at 25°C in a solution containing 40 mM Tris-HCl (pH 7.5), 0.1 mM EDTA, 10 mM MgCl2, 50 mM KCl, 0.5 mM phenylmethylsulfonyl fluoride, 30 μg of bovine serum albumin/ml, 4 μg of salmon sperm DNA/ml, 20% glycerol, and 1 mM DTT (FTB buffer) in a total volume of 40 μl, after which DNase I digestion and DNA purification were performed as described previously (2). Using the purified PmrA-H6 protein, we observed a protected sequence from −18 to −44, and from −16 to −44, relative to the transcriptional start site in the positive and negative strands, respectively (see Fig. 3, below; other data not shown). Because it has been demonstrated that addition of reducing agents like DTT or β-mercaptoethanol favor the dissociation of a dimeric form of PmrA (24), the footprinting analysis was carried out in the absence of DTT. We observed that protection of the same region was detected with 24 pmol of PmrA, 1/10 of the amount of the response regulator needed in the presence of the reducing agent, corroborating the result reported previously (24). The footprinting analysis findings are consistent with the large amounts of PmrA that were required to observe a protein-DNA interaction in the shift assay, since DTT was included in the binding buffer.
FIG. 3.
Enhanced affinity of phosphorylated PmrA for the promoter region of ugd. The footprinting assay was performed on the coding strand of the promoter region of ugd, incubated with 0, 0.5, 1, 2.5, 5, 10, 15, 30, and 60 pmol of PmrA-H6 and 1 pmol of H6-PmrBc, in the absence or presence of 1 mM ATP and without addition of DTT. The solid line represents the PmrA-protected region.
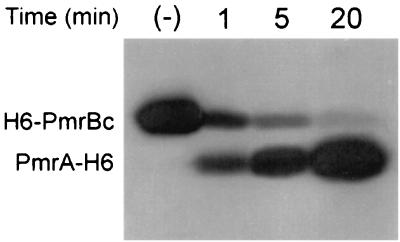
The two-component paradigm supports the concept that phosphorylation of a transcriptional regulator affects its DNA binding properties (23). In order to obtain phosphorylated PmrA, we isolated a truncated form of PmrB fused to a His tag (H6-PmrBc). This form encompasses the cytoplasmic region of PmrB and retains the autokinase and phosphotransfer activities (Fig. 2). pmrBc was amplified by PCR using the PmrB-NTF (5′-GAGGATCCATATGCGTTTTCAGCGAAG-3′) and PmrB-NTR (5′-AAGGCCTTACCGCCTGGTAACA-3′) oligonucleotides and cloned between the BamHI and HindIII sites of plasmid pQE32 (Qiagen) to obtain plasmid pPB1023. H6-PmrBc was purified by following the protocol described above for PmrA-H6. The protein profile of the purified H6-PmrBc protein was analyzed by SDS-PAGE. The autokinase and phosphotransfer activities of the H6-PmrBc protein were determined in a 40-μl total volume. One picomole of H6-PmrBc was incubated in FTB buffer (without the addition of DTT), and 1 mM [γ-32P]ATP (1,850 cpm/pmol; New England Nuclear), in the absence or presence of 30 pmol of PmrA-H6, for 1, 5, and 20 min at 37°C (Fig. 2). Maximal phosphorylation of PmrA-H6 was observed at 20 min, and it remained stable for at least an additional 20 min (data not shown).
FIG. 2.
Autokinase and phosphotransfer activities of H6-PmrBc. One picomole of the H6-PmrBc protein was incubated with 1 mM [γ-32P]ATP in the presence of 30 pmol of PmrA-H6 in FTB buffer (without DTT) for 1, 5, and 20 min at 37°C. For the negative control (−), H6-PmrB was incubated under the same conditions for 20 min without the addition of PmrA-H6. The reactions were stopped by the addition of 0.5% SDS. The samples were loaded in an SDS–12% PAGE gel, transferred to nitrocellulose, and analyzed by autoradiography.
To evaluate the effect of the phosphorylation of PmrA-H6 on its interaction with the promoter region of ugd, the footprinting assay was performed with H6-PmrBc and ATP. PmrA-H6 was incubated in FTB buffer at 37°C with 1 pmol of H6-PmrBc, in the presence or absence of 1 mM ATP. The reaction was allowed to proceed for 20 min, after which the footprinting assay was performed with the addition of the 32P-labeled ugd fragment. Phosphorylation of the PmrA-H6 protein resulted in a 10-fold increase in protection (Fig. 3). This finding indicates that phosphorylation of PmrA enhances its affinity for the sequence, as has been demonstrated for other related response regulators (10, 13, 14). Analysis of the protected region showed the presence of a direct repeat (5′-CTTAAT-N5-CTTAAT-3′).
To determine the extent of phosphorylation of PmrA under the conditions used for the footprinting assays, aliquots from a parallel assay, in which 2.5 μl of [γ-32P]ATP (3,000 Ci/mmol) was added, were analyzed by SDS-PAGE. The PmrA band was cut from the gel and the incorporation of 32P was determined using a Wallac 1209 Rackbetta liquid scintillation counter. From three independent experiments, we determined a concentration of 0.09 ± 0.02 mol of phosphate per mol of PmrA, indicating that only 7 to 11% of PmrA was phosphorylated under the conditions used for the footprinting assays. This result suggests that phospho-PmrA has an increase in affinity much higher than that calculated from the footprinting assay described above. (Additional protected areas appeared when large amounts of phospho-PmrA-H6 were used [Fig. 3]. We are currently analyzing the role of these protected areas in the PmrA-dependent regulation of the expression of ugd, using deletions and different DNA point mutations.)
PmrA is a member of the OmpR family of response regulators in which the DNA binding motif is a winged helix-turn-helix (15, 16), and members of this family are known for binding direct repeats (5, 9, 14). Since a similar sequence is also present in the protected promoter regions of the pmrCAB and pmrF operons (Fig. 4), we propose that the direct repeat (YTTAAK) is the target site for the PmrA-controlled expression of this regulon.
FIG. 4.
Alignment of the promoter regions of ugd, pmrCAB, and the pmrF operon. The sequences from the ugd, pmrCAB, and pmrF promoter regions were piled up from the transcriptional start site. The YTTAAK direct repeats are boxed, and the proposed −10 regions are marked in bold. The arrows indicate the transcription start sites identified previously (24).
Acknowledgments
We thank Esteban Serra for comments on the manuscript.
This work was supported by grants from CONICET (Proyecto PIP 0849) and from ANPCyT (Proyecto 01-0000-00409) to F.C.S. and a grant from the Third World Academy of Sciences (Trieste, Italy) to E.G.V. E.G.V. is a career investigator of the National Research Council (CONICET, Argentina), and S.L. is a fellow of the same institution. F.C.S. is a member of the Rosario National University Research Council (CIUNR) and CONICET and is also an International Research Scholar of the Howard Hughes Medical Institute.
REFERENCES
- 1.Baker S J, Gunn J S, Morona R. The Salmonella typhi melittin resistance gene pqaB affects intracellular growth in PMA-differentiated U937 cells, polymyxin B resistance and lipopolysaccharide. Microbiology. 1999;145:367–378. doi: 10.1099/13500872-145-2-367. [DOI] [PubMed] [Google Scholar]
- 2.Cullen P J, Bowman W C, Kranz R G. In vitro reconstitution and characterization of the Rhodobacter capsulatus NtrB and NtrC two-component system. J Biol Chem. 1996;271:6530–6536. doi: 10.1074/jbc.271.11.6530. [DOI] [PubMed] [Google Scholar]
- 3.Dillard J P, Vandersea M W, Yother J. Characterization of the cassette containing genes for type 3 capsular polysaccharide biosynthesis in Streptococcus pneumoniae. J Exp Med. 1995;181:973–983. doi: 10.1084/jem.181.3.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dougherty B A, van de Rijn I. Molecular characterization of hasB from an operon required for hyaluronic acid synthesis in group A streptococci. J Biol Chem. 1993;268:7118–7124. [PubMed] [Google Scholar]
- 5.Eder S, Liu W, Hulett F M. Mutational analysis of the phoD promoter in Bacillus subtilis: implications for PhoP binding and promoter activation of Pho regulon promoters. J Bacteriol. 1999;181:2017–2025. doi: 10.1128/jb.181.7.2017-2025.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Groisman E A, Kayser J, Soncini F C. Regulation of polymyxin resistance and adaptation to low-Mg2+ environments. J Bacteriol. 1997;179:7040–7045. doi: 10.1128/jb.179.22.7040-7045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gunn J S, Lim K B, Krueger J, Kim K, Guo L, Hackett M, Miller S I. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol Microbiol. 1998;27:1171–1182. doi: 10.1046/j.1365-2958.1998.00757.x. [DOI] [PubMed] [Google Scholar]
- 8.Gunn J S, Miller S I. PhoP-PhoQ activates transcription of pmrAB, encoding a two-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J Bacteriol. 1996;178:6857–6864. doi: 10.1128/jb.178.23.6857-6864.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harlocker S L, Bergstrom L, Inouye M. Tandem binding of six OmpR proteins to the ompF upstream regulatory sequence of Escherichia coli. J Biol Chem. 1995;270:26849–26856. doi: 10.1074/jbc.270.45.26849. [DOI] [PubMed] [Google Scholar]
- 10.Head C G, Tardy A, Kenney L J. Relative binding affinities of OmpR and OmpR-phosphate at the ompF and ompC regulatory sites. J Mol Biol. 1998;281:857–870. doi: 10.1006/jmbi.1998.1985. [DOI] [PubMed] [Google Scholar]
- 11.Helander I M, Kilpeläinen I, Vaara M. Increased substitution of phosphate groups in lipopolysaccharides and lipid A of the polymyxin-resistant pmrA mutants of Salmonella typhimurium: a 31P-NMR study. Mol Microbiol. 1994;11:481–487. doi: 10.1111/j.1365-2958.1994.tb00329.x. [DOI] [PubMed] [Google Scholar]
- 12.Kerr L D. Electrophoretic mobility shift assay. Methods Enzymol. 1995;254:619–632. doi: 10.1016/0076-6879(95)54044-x. [DOI] [PubMed] [Google Scholar]
- 13.Liu W, Hulett F M. Bacillus subtilis PhoP binds to the phoB tandem promoter exclusively within the phosphate starvation-inducible promoter. J Bacteriol. 1997;179:6302–6310. doi: 10.1128/jb.179.20.6302-6310.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Makino K, Shinagawa H, Amemura M, Kimura S, Nakata A, Ishihama A. Regulation of the phosphate regulon of Escherichia coli, activation of pstS transcription by PhoB protein in vivo. J Mol Biol. 1988;203:85–95. doi: 10.1016/0022-2836(88)90093-9. [DOI] [PubMed] [Google Scholar]
- 15.Martínez-Hackert E, Stock A M. The DNA-binding domain of OmpR: crystal structure of a winged helix transcription factor. Structure. 1996;5:109–124. doi: 10.1016/s0969-2126(97)00170-6. [DOI] [PubMed] [Google Scholar]
- 16.Martínez-Hackert E, Stock A M. Structural relationships in the OmpR family of winged-helix transcription factors. J Mol Biol. 1997;269:109–124. doi: 10.1006/jmbi.1997.1065. [DOI] [PubMed] [Google Scholar]
- 17.Miller S I, Kukral A M, Mekalanos J J. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc Natl Acad Sci USA. 1989;86:5054–5058. doi: 10.1073/pnas.86.13.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roland K L, Martin L E, Esther C R, Spitznagel J K. Spontaneous pmrA mutants of Salmonella typhimurium LT2 define a new two-component regulatory system with a possible role in virulence. J Bacteriol. 1993;175:4154–4164. doi: 10.1128/jb.175.13.4154-4164.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roland K L, Spitznagel J K. Molecular genetics of polymyxin resistance in Salmonella typhimurium. In: Levin J, Alving C R, Munford R S, Redl H, editors. Bacterial endotoxins: lipopolysaccharides from genes to therapy. New York, N.Y: Wiley-Liss, Inc.; 1995. pp. 3–14. [PubMed] [Google Scholar]
- 20.Shafer W M, Casey S G, Spitznagel J K. Lipid A and resistance of Salmonella typhimurium to antimicrobial granule proteins of human neutrophil granulocytes. Infect Immun. 1984;43:834–838. doi: 10.1128/iai.43.3.834-838.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soncini F C, García Véscovi E, Solomon F, Groisman E A. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J Bacteriol. 1996;178:5092–5099. doi: 10.1128/jb.178.17.5092-5099.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soncini F C, Groisman E A. Two-component regulatory systems can interact to process multiple environmental signals. J Bacteriol. 1996;178:6796–6801. doi: 10.1128/jb.178.23.6796-6801.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stock J B, Surette M G, Levit M, Park P. Two-component signal transduction systems: structure-function relationships and mechanisms of catalysis. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C.: ASM Press; 1995. pp. 25–51. [Google Scholar]
- 24.Wösten M M, Groisman E A. Molecular characterization of the PmrA regulon. J Biol Chem. 1999;274:27185–27190. doi: 10.1074/jbc.274.38.27185. [DOI] [PubMed] [Google Scholar]