Significance
Pannexin-1 (Panx1) channels contribute to neurological disorders, including stroke and epilepsy, where their function has been linked to N-methyl D-aspartate (NMDA) receptors (NMDARs). We discovered that Ca2+ entry via NMDARs recruits endoplasmic reticulum–resident STIM proteins to activate Panx1 by binding to a hydrophobic region localized to the Panx1 N terminus. Using loss-of-function approaches, combined with molecular replacement and use of a STIM/Panx1 function–blocking antibody, we demonstrate that disrupting the STIM/Panx1 interaction prevents Panx1 activation by NMDARs, but not by hypotonic stimuli. Thus, our findings serve as a basis for the design of modality-specific inhibitors against STIM-dependent Panx1 activation that will aid in understanding the multimodal functions of Panx1 and their contribution to physiology and pathology.
Keywords: pannexin 1, NMDA receptor, STIM protein, calcium signaling, ER signaling
Abstract
Pannexin-1 (Panx1) is a large-pore ion and solute permeable channel highly expressed in the nervous system, where it subserves diverse processes, including neurite outgrowth, dendritic spine formation, and N-methyl D-aspartate (NMDA) receptor (NMDAR)-dependent plasticity. Moreover, Panx1 dysregulation contributes to neurological disorders, including neuropathic pain, epilepsy, and excitotoxicity. Despite progress in understanding physiological and pathological functions of Panx1, the mechanisms that regulate its activity, including its ion and solute permeability, remain poorly understood. In this study, we identify endoplasmic reticulum (ER)-resident stromal interaction molecules (STIM1/2), which are Ca2+ sensors that communicate events within the ER to plasma membrane channels, as binding and signaling partners of Panx1. We demonstrate that Panx1 is activated to its large-pore configuration in response to stimuli that recruit STIM1/2 and map the interaction interface to a hydrophobic region within the N terminus of Panx1. We further characterize a Panx1 N terminus–recognizing antibody as a function-blocking tool able to prevent large-pore Panx1 activation by STIM1/2. Using either the function-blocking antibody or re-expression of Panx1 deletion mutants in Panx1 knockout (KO) neurons, we show that STIM recruitment couples Ca2+ entry via NMDARs to Panx1 activation, thereby identifying a model of NMDAR-STIM-Panx1 signaling in neurons. Our study highlights a previously unrecognized and important role of the Panx1 N terminus in regulating channel activation and membrane localization. Considering past work demonstrating an intimate functional relation between NMDARs and Panx1, our study opens avenues for understanding activation modality and context-specific functions of Panx1, including functions linked to diverse STIM-regulated cellular responses.
Glutamatergic signaling plays a critical role in diverse processes linked to learning and memory formation. Ca2+ signals generated by the N-methyl D-aspartate (NMDA) subtype of glutamate receptors (NMDARs) are indispensable for several forms of synaptic plasticity, including long-term potentiation (LTP), a prototypic form of plasticity linked to memory formation (1–3). NMDAR-initiated Ca2+ signals (e.g., time course, amplitude, and spatial spread) are shaped by secondary events, including those engendered via the endoplasmic reticulum (ER) (4, 5). Ca2+ entry via NMDARs can promote Ca2+-induced Ca2+ release from ER stores by stimulating ryanodine (RyRs) (6–8) and/or IP3 receptors (IP3Rs) (9). In turn, NMDAR-initiated Ca2+ store depletion recruits ER-resident and Ca2+-sensing STIM proteins (10) to negatively regulate L-type voltage-gated Ca2+ channels (VGCCs) (13). This establishes the notion that Ca2+ entry via NMDARs can stimulate ER- and STIM-dependent cascades that regulate secondary routes of Ca2+ entry, thereby sculpting intracellular Ca2+ dynamics and in turn the cellular functions influenced by them. As part of a broader search to identify candidate Ca2+ channels able to respond to ER signaling dynamics, we found that Pannexin-1 (Panx1) can be activated through ER-based signaling following sarcoendoplasmic reticulum calcium adenosine triphosphatase (ATPase) (SERCA) pump inhibition by thapsigargin. This led us to consider the role of STIM1/2 as a candidate Panx1 activation mechanism.
Panx1 is a large-pore nonselective ion and solute permeable channel with prominent central nervous system (CNS) expression (14, 15). Panx1 activation has been linked to pathophysiological disorders, such as excitotoxicity, stroke, migraine, chronic pain, and epilepsy (16–18). However, Panx1 also mediates physiological processes in the CNS, including contributions to neural development (19, 20), spine formation (21, 22), and NMDAR-dependent synaptic plasticity (23, 24). In this context, there remains an important gap in understanding the mechanisms by which Panx1 can mediate such disparate physiological and pathological functions. Intriguingly, evidence suggests that Panx1 ion versus solute permeability may be mediated by distinct channel pore configurations (i.e., small anion vs. large solute permeable) recruited via distinct activation modalities (25). Thus, identifying novel activation mechanisms is fundamental to understanding context- and modality-specific channel function.
Here, we uncover a mechanism by which Panx1 is activated in response to ER-initiated signaling, which we demonstrate is dependent on Panx1 interaction with ER-resident STIM1/2. STIM1/2 recruitment and activation stimulates large-pore Panx1 opening, evident on the basis of increased permeability to Ca2+ and the large inorganic ion N-methyl-D-glucamine (NMDG). We map the STIM1/2 binding interface to a hydrophobic region in the N terminus of Panx1, a region not previously linked to channel gating. Our detailed structure-function analysis reveals that the Panx1 N-terminal region is necessary for its STIM1/2 responsiveness, but not for its responsiveness to hypotonic stress, demonstrating that this region mediates modality-specific regulation of Panx1 function. Using reverse genetics, ectopic rescue with Panx1 N-terminal deletion mutants, as well as a function inhibiting antibody targeting the critical N-terminal region of Panx1 identified by us, we demonstrate that NMDARs activate Panx1 in hippocampal neurons in a manner contingent upon ER-initiated signaling and reliant upon STIM proteins. Collectively, our data reveal the molecular mechanism by which STIM1/2 activates Panx1 and establishes a previously unrecognized essential role of its N-terminal region in regulating the transition of Panx1 to its large-pore solute permeable state. Our work will benefit studies aimed at understanding diverse functions of Panx1, including those linked to NMDAR-dependent signaling, stimulated in a modality- and context-specific manner by STIM proteins.
Results
ER Store Depletion Activates Panx1 in Hippocampal Neurons.
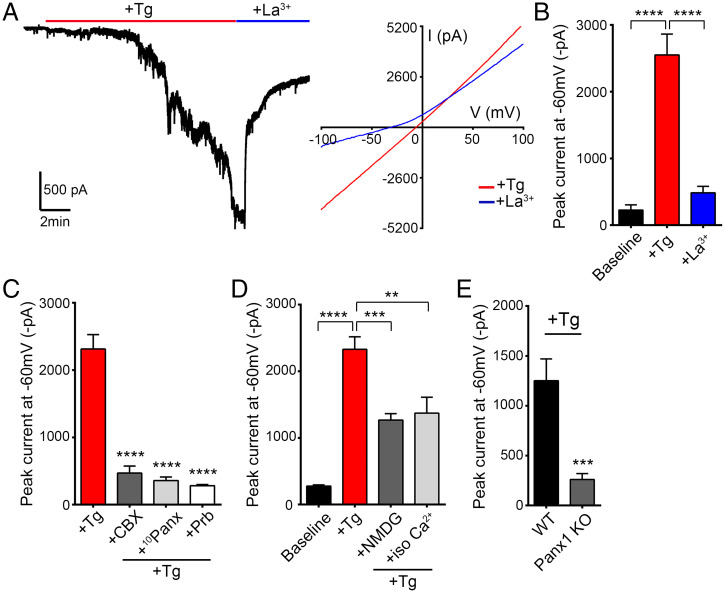
To identify whether Panx1 can respond to ER-dependent signaling, we stimulated hippocampal neurons with thapsigargin (3 μM), a SERCA pump inhibitor. In voltage-clamped neurons, thapsigargin reliably evoked slowly developing, large inward currents with linear current-voltage (IV) and reversal near 0 mV (Fig. 1A), suggesting activation of a nonselective ion conductance. Thapsigargin-stimulated inward currents were sensitive to Panx1 blockers, including lanthanum (La3+) (100 μM), carbenoxolone (CBX) (100 μM), probenecid (2 mM), and the Panx1 inhibitory peptide (100 μM) (Fig. 1 A–C). To examine the ion permeability of the thapsigargin-evoked conductance, we performed ion-substitution experiments. Despite being attenuated, substantial thapsigargin-evoked Panx1 currents (> 1,000 pA) were maintained in solutions without added Na+, where only NMDG+ or Ca2+ was available to conduct inward current (Fig. 1D). Thus, thapsigargin-induced currents are permeable to both NMDG+ and Ca2+, and these findings are consistent with opening of Panx1 to its large-pore, solute-permeable configuration (26, 27). More definitively, comparable currents were abrogated when thapsigargin was applied to hippocampal neurons derived from Panx1 knockout (KO) mice (Fig. 1E and SI Appendix, Fig. S1). Together, these results demonstrate that Panx1 channels can be activated in response to ER-dependent signaling in hippocampal neurons.
Fig. 1.
Panx1 is activated in response to thapsigargin treatment of hippocampal neurons. (A) Representative whole-cell recording (Left) shows activation of a large inward current upon 3 µM thapsigargin (Tg) application. This current is inhibited by application of 100 µM lanthanum (La3+). IV curves (Right) before (+Tg) and after application of La3+ (+La3+) averaged from a series of recordings (n = 3). (B) Summary of currents recorded from cultured CD1 neurons (n = 9). (C) Thapsigargin-induced currents in cultured CD1 neurons are inhibited by application of Panx1 blockers carbenoxolone (CBX) (100 µM), Panx1 inhibitory peptide (10Panx) (100 µM), or probenecid (Prb) (2 mM) (n = 5 for each) when compared with +Tg (n = 5). (D) Summary of baseline (n = 6) and peak thapsigargin-evoked currents recorded in extracellular fluid (ECF) (n = 5), NMDG-Cl (140 mM; n = 6), or CaCl2 (120 mM; n = 5) solutions. (E) Summary of thapsigargin-stimulated currents recorded in Panx1 KO (n = 13) or WT neurons (n = 13). Data are represented as mean ± SEM (B–E). **P < 0.01, ***P < 0.001, and ****P < 0.0001, one-way ANOVA with post hoc Bonferroni test (B–D). ***P < 0.001, unpaired t test (E).
Panx1 Channels Are Activated in a STIM-Dependent Manner.
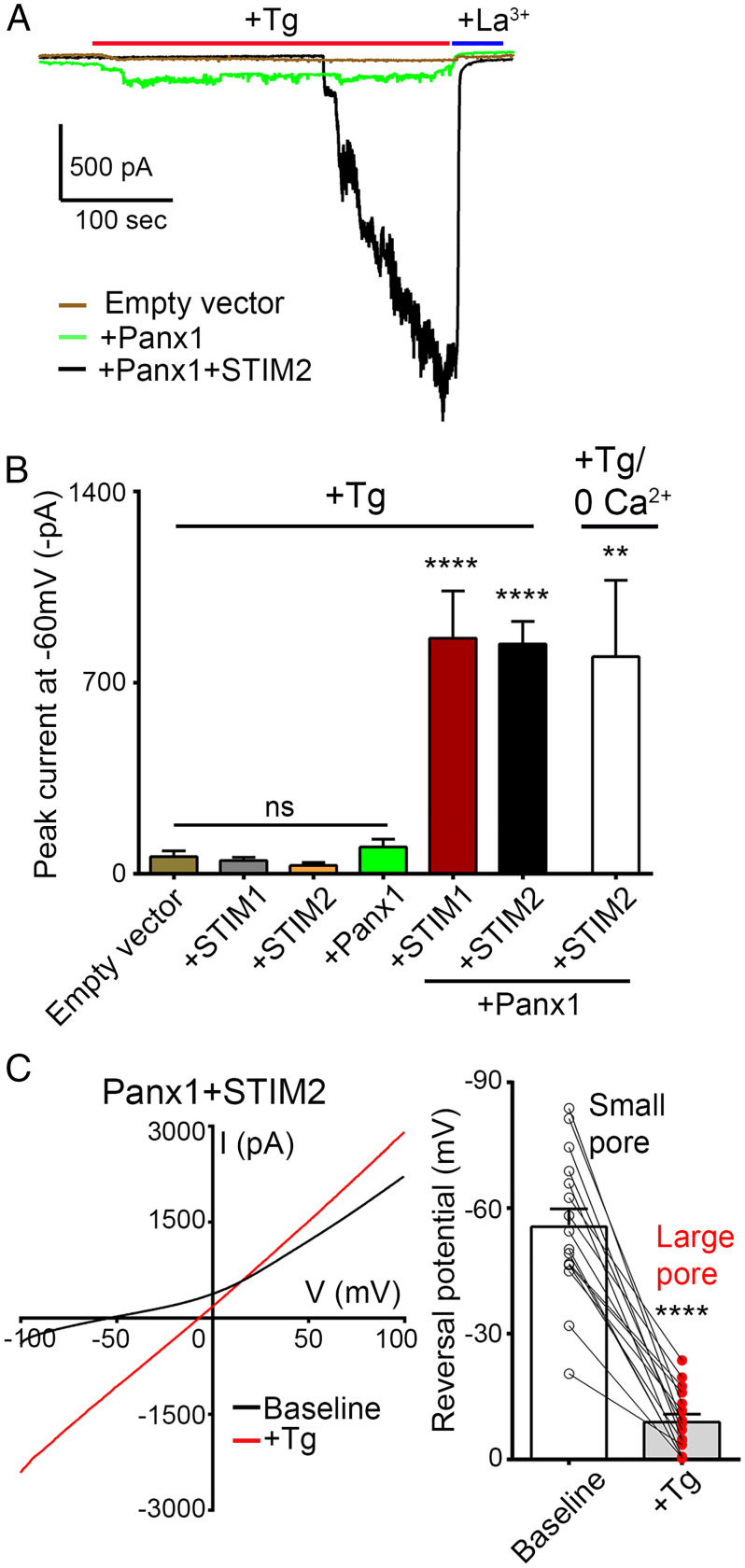
Given the role of STIM proteins in relaying ER-dependent signaling to plasma membrane channels in response to thapsigargin stimulation, we sought to determine whether Panx1 activation by thapsigargin is STIM dependent. To do so, we used a functional screen to examine whether coexpression of STIM in human embryonic kidney 293T (HEK) cells is necessary to impart thapsigargin responsiveness to Panx1. Notably, thapsigargin (3 μM) does not elicit a response when applied to mock transfected HEK cells (Fig. 2 A and B). In addition to its large-pore state, with nonselective ion permeability and linear IV characteristics, Panx1 can reside in a small-pore state with outwardly rectifying current primarily carried by Cl− and readily blocked by CBX (28, 29). Consistent with this, CBX-sensitive outwardly rectifying currents could be recorded in HEK cells that express Panx1, but not in mock transfected control cells (SI Appendix, Fig. S2 A and B). However, when Panx1, STIM1, or STIM2 were expressed alone, treatment with thapsigargin did not elicit any measurable Panx1-mediated inward current (Fig. 2 A and B). Rather, coexpression of Panx1 with either STIM1 or STIM2 was necessary to reconstitute thapsigargin-stimulated currents comparable to those recorded in neurons (Fig. 2 A and B). Additionally, applying thapsigargin (3 μM) in a solution devoid of extracellular Ca2+ and supplemented with 1 mM ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA) (known to stimulate robust STIM activation) resulted in similar Panx1 inward currents (Fig. 2B). These currents could be inhibited by application of La3+ (100 μM) and exhibited linear IV characteristics (Fig. 2 A and C). The onset of thapsigargin-stimulated currents was associated with a pronounced depolarizing shift in current reversal from ∼−60 mV to ∼−9 mV (Fig. 2C), consistent with Panx1 transitioning from a small-pore, primarily Cl− permeable state to a large-pore nonselective ion channel configuration. These findings support that thapsigargin-induced Panx1 activation is STIM dependent.
Fig. 2.
Panx1 activation in response to thapsigargin treatment is STIM dependent. (A) Representative whole-cell recordings from HEK 293T cells transfected with empty vector, Panx1 alone (+Panx1), or +Panx1+STIM2. Tg-stimulated currents (3 µM), recorded from cells coexpressing Panx1 and STIM2, were inhibited upon application of La3+ (100 µM). (B) Summary of peak currents recorded from empty vector (n = 14), +STIM1 (n = 10), +STIM2 (n = 12), +Panx1 (n = 21), Panx1+STIM1 (n = 17), and Panx1+STIM2 (n = 17), where Tg was applied in solutions containing Ca2+ (+Tg). Additionally, currents from Panx1+STIM2 (n = 9) were recorded with thapsigargin applied in solutions devoid of extracellular Ca2+ (+Tg/0 Ca2+). Data are represented as mean ± SEM ****P < 0.0001 and **P < 0.01, one-way ANOVA with post hoc Bonferroni test when compared with empty vector or +Panx1. ns, not significant. (C) IV curves (Left) and current reversal potential (Right), averaged from a series of recordings, before (baseline) and after application of thapsigargin (3 µM). ****P < 0.0001, paired t test (n = 16).
Panx1 Interacts with ER-Resident STIM1/2.
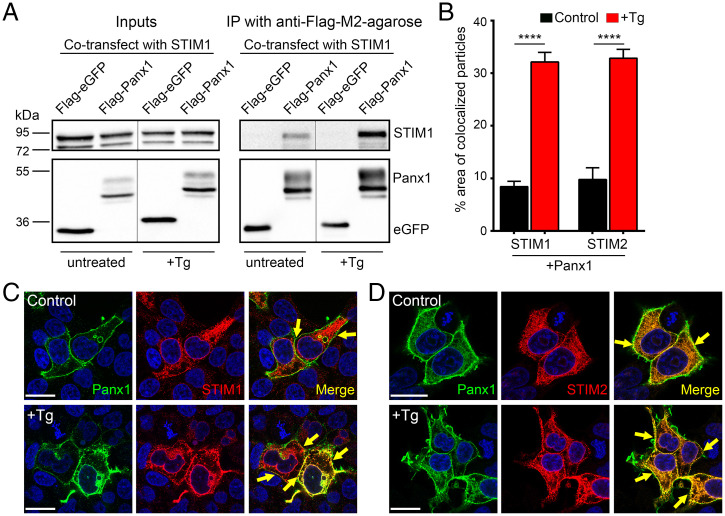
To determine the nature of STIM/Panx1 interaction, we performed immunoprecipitation from lysates of HEK cells expressing STIM1 and Flag-Panx1. STIM1 could be detected in Flag-Panx1 immunoprecipitates from untreated HEK cells (Fig. 3A). Moreover, STIM1/Panx1 biochemical association was increased in cells treated with thapsigargin (Figs. 3A and 4 C and D). Similar results were observed when we examined the subcellular distribution profiles of Panx1 and STIM1/2 by immunofluorescence microscopy (Fig. 3 B–D). STIM1 and STIM2 were primarily distributed in a reticular manner, reflecting their predominant ER localization, while Panx1 channels were highly enriched at the plasma membrane (Fig. 3 C and D). Consistent with our biochemical findings, colocalization between Panx1 and STIM1/2 could be observed near the plasma membrane in untreated control cells (Fig. 3B). In cells stimulated with thapsigargin (3 μM), both STIM1 and STIM2 adopted a more punctate appearance with increased colocalization with Panx1 evident at the plasma membrane (>30%, Fig. 3 B–D). Importantly, although preformed STIM/Panx1 complexes are present in untreated cells at rest, expression of STIM1/2 by itself does not promote large-pore Panx1 opening, nor does it influence basal current amplitude, current reversal potential, outward rectification, or sensitivity to CBX (SI Appendix, Fig. S2 A and B). Further, we saw no change in STIM1/2 expression in brain lysates from Panx1 KO mice (SI Appendix, Fig. S1 A and B). Accordingly, our results are consistent with a model in which STIMs activate Panx1 via direct binding. Moreover, thapsigargin stimulation is strictly necessary to promote STIM-dependent, large-pore Panx1 activation.
Fig. 3.
Panx1 and STIM1/2 interaction in Tg-stimulated and unstimulated HEK 293T cells. (A) Representative blot from lysates (Left) and immunoprecipitates (IP) (Right) from HEK 293T cells cotransfected with STIM1 and Flag-eGFP (negative control) or Flag-Panx1. Beads coupled to anti-Flag coimmunoprecipitated STIM1 in HEK 293T cells coexpressing Flag-Panx1, but not in cells coexpressing Flag-eGFP. Treatment with Tg (3 µM) for 15 min resulted in increased STIM1 pulldown in Flag-Panx1–expressing cells. Note, stippled lines between untreated versus Tg-treated (+Tg) indicates noncontiguous lanes from the same blot. (B–D) Summary of percent area of colocalized particles at plasma membrane (B) for Panx1+STIM1 in control (n = 56) or +Tg (n = 44) and Panx1+STIM2 in control (n = 36) or +Tg (n = 43). When treated with Tg (3 µM in 0 Ca2+ ECF) for 15–20 min, increased STIM1/2 colocalization with Panx1 was observed. ****P < 0.0001, one-way ANOVA with post hoc Bonferroni test when compared to respective controls. Representative images showing the subcellular distribution of Panx1 (green) with STIM1 (red, C) or STIM2 (red, D) in HEK 293T cells under control and thapsigargin-treated conditions. Colocalization between Panx1 and STIM1/2 can be observed at plasma membrane/ER junction (merge image, marked with arrows). Scale bars, 20 µm. Data are represented as mean ± SEM (B).
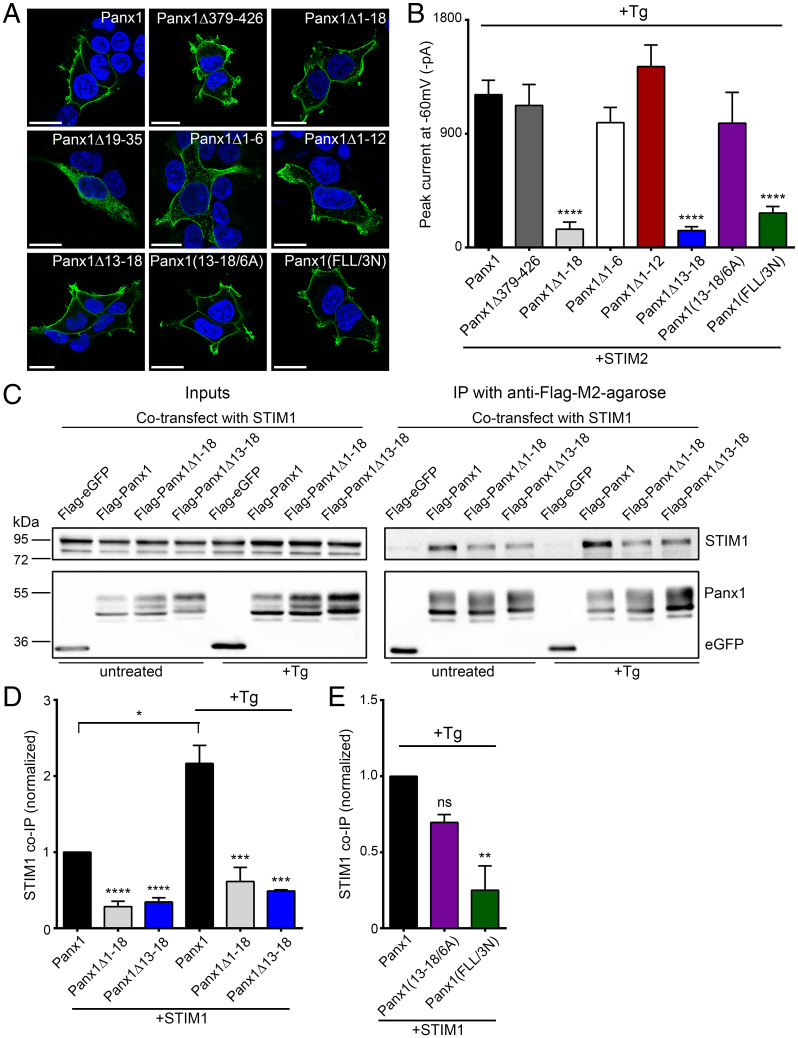
Fig. 4.
Panx1-STIM1/2 interaction domain lies within first 18 amino acids of Panx1 N terminus. (A) Representative images showing localization of Flag-tagged Panx1, carboxyl-terminal deletion and N-terminal deletion, or substitution mutants expressed in HEK 293T cells. Cell nuclei were stained using DAPI; scale bars, 20 µm. (B) Summary of peak +Tg (3 μM)-stimulated currents recorded from HEK 293T cells coexpressing STIM2 and full-length Panx1 (n = 29), Panx1Δ379–426 (n = 14), Panx1Δ1–18 (n = 14), Panx1Δ1–6 (n = 12), Panx1Δ1–12 (n = 12), Panx1Δ13–18 (n = 16), Panx1(13-18/6A) (n = 14), or Panx1(FLL/3N) (n = 11) . ****P < 0.0001, one-way ANOVA with post hoc Bonferroni test when compared with Panx1. No significant difference was seen between other deletions and mutants. (C) Representative blot from lysates (Left) and anti-Flag immunoprecipitates (IP) (Right) in Tg-treated (3 μM, 15 min) and untreated HEK cells coexpressing STIM1 with either Flag-Panx1, Flag-Panx1 deletion mutants, or Flag-eGFP (negative control). (D) Quantified data for coimmunoprecipitation (co-IP) experiments in HEK cells coexpressing STIM1 with Panx1 (n = 4), Panx1Δ1–18 (n = 4), or Panx1Δ13–18 (n = 3) under untreated or +Tg-treated conditions. ****P < 0.0001 and ***P < 0.001, one-way ANOVA with post hoc Bonferroni test when compared with Panx1 or Panx1+Tg, respectively.*P < 0.05, when Panx1 was compared with Panx1+Tg using paired t test. No significant difference was seen between Panx1Δ1–18 and Panx1Δ1–18+Tg or Panx1Δ13–18 and Panx1Δ13–18+Tg with paired t test. (E) Quantified data for coimmunoprecipitation in Tg-treated HEK cells coexpressing STIM1 with Panx1 (n = 3), Panx1(13-18/6A) (n = 3), or Panx1(FLL/3N) (n = 3). ns P > 0.05 and **P < 0.01, one-way ANOVA with post hoc Bonferroni test when compared with Panx1. Data are represented as mean ± SEM (B, D, and E).
Mapping the STIM1/2-Panx1 Interaction Interface.
To identify the STIM-interacting domain of Panx1, we generated Panx1 mutants in which most of the N or carboxyl terminus was deleted (SI Appendix, Fig. S3). Previous reports show that deletion of the entire carboxyl terminus (Panx1Δ300–426) causes intracellular retention of Panx1 (30), which we likewise observed (SI Appendix, Fig. S4A). We therefore assessed the consequence of deleting the region distal to the carboxyl-terminal caspase cleavage site (Panx1Δ379–426; with amino acids 379–426 deleted, inclusively) (31). Panx1Δ379–426 expresses well at the plasma membrane (Fig. 4A) and generates robust outwardly rectifying currents with negative reversal potential (SI Appendix, Fig. S4 B and C). When coexpressed with STIM2, Panx1Δ379–426 was activated upon thapsigargin (3 μM) application, with no difference in current amplitude when compared with cells expressing full-length Panx1 (Fig. 4B). This suggests that at least the distal portion of the carboxyl terminus is dispensable for STIM-dependent Panx1 activation.
Next, we examined the consequence of deleting the Panx1 N terminus in its entirety (Panx1Δ1–35) or partially (Panx1Δ1–18 and Panx1Δ19–35; SI Appendix, Fig. S3). When expressed in HEK cells, Panx1Δ1–35 was retained intracellularly (SI Appendix, Fig. S4A), as was Panx1Δ19–35 where it codistributed with calnexin, consistent with its retention within the ER (Fig. 4A and SI Appendix, Fig. S4 D and E). In contrast, Panx1Δ1–18 was extensively localized at the plasma membrane. However, when coexpressed with STIM2, Panx1Δ1–18 did not respond to thapsigargin treatment (Fig. 4B). Consistent with our electrophysiology results, the association of STIM1 with Panx1Δ1–18 was markedly reduced in untreated cells and no longer responsive to thapsigargin treatment (Fig. 4 C and D). These results suggest that the 1–18 region of the N terminus is necessary for Panx1 activation by STIM. To define the minimal interacting region, we created sequential deletion mutants targeting the Panx1 1–18 region (SI Appendix, Fig. S3). Deletion of amino acids 1–6 (Panx1Δ1–6) and 1–12 (Panx1Δ1–12) did not affect membrane localization or thapsigargin-induced Panx1 activation (Fig. 4 A and B). However, deletion of amino acids 13–18 (Panx1Δ13–18) rendered the resulting Panx1 mutant unresponsive to thapsigargin, without altering its surface expression (Fig. 4 A and B). Consistent with this, the basal association of STIM1 with Flag-Panx1Δ13–18 was reduced (untreated in Fig. 4 C and D) and was not increased in response to thapsigargin treatment. These findings support that STIM recruitment activates Panx1 via interaction with amino acids 13–18 of Panx1 (13-SDFLLK-18).
Hydrophobic Amino Acids within Panx1 13–18 Are Required for Panx1-STIM1/2 Interaction.
To identify key residues in Panx1 required for Panx1-STIM binding, we generated a series of point mutants targeting the STIM1/2-interacting region of Panx1 (13-SDFLLK-18) that includes amino acids with side chains of differing size and properties (e.g., charge, hydropathy, etc.) (SI Appendix, Fig. S3). We first substituted all of these residues with the small, uncharged, hydrophobic amino acid alanine (Panx1(13-18/6A)), thus eliminating residues with charged hydrophilic (D and K) or bulky hydrophobic side chains (L and F). Panx1 (13-18/6A) localized at the plasma membrane and was able to bind STIM, and its STIM-dependent activation in response to thapsigargin (3 µM) was indistinguishable from Panx1 (Fig. 4 A, B, and E and SI Appendix, Fig. S5A). Thus, neither charged hydrophilic nor bulky hydrophobic residues are required for STIM to interact and activate Panx1. One consequence of the all-alanine substitution is that the overall hydrophobic character within the 13–18 region was increased. Given past evidence supporting an important role of hydrophobic interactions in allowing STIM-dependent Orai activation (32–36), we generated a mutant with L and F (bulky hydrophobic side chains) replaced by N (positively charged, hydrophilic), termed Panx1(FLL/3N) (SI Appendix, Fig. S3). Panx1(FLL/3N) localized to the plasma membrane but was not responsive to thapsigargin treatment when coexpressed with STIM2 (Fig. 4 A and B). Moreover, Panx1(FLL/3N) binding to STIM1 in thapsigargin-treated HEK cells was reduced (Fig. 4E and SI Appendix, Fig. S5A). Thus, STIM binding and Panx1 activation occurs through a short string of amino acids in Panx1 N terminus (15FLL17) with predominant hydrophobic character.
N-Terminal Regulation of Panx1 Channel Function is Modality Specific.
In addition to abrogating thapsigargin responsiveness, deletions or mutations targeting the 1–18 region of the Panx1 N terminus displayed reduced basal outwardly rectifying currents without impacting surface localization (Fig. 4A and SI Appendix, Fig. S4 B and C). This was observed whether they interact with STIM1/2 (i.e., Panx1Δ1–6, Panx1Δ1–12, and Panx1(13-18/6A)) or not (i.e., Panx1Δ1–18, Panx1Δ13–18, and Panx1(FLL/3N). This is in keeping with our finding that basal outwardly rectifying Panx1 currents are not affected by coexpression of STIM1 or STIM2 (SI Appendix, Fig. S2), which supports that reduced basal currents in Panx1 N-terminal mutants is not due to altered STIM1/2 interaction. To rule out whether this effect was due to reduced protein expression, we assessed the expression levels of Panx1 mutants. We observed no change in protein levels of Panx1Δ379–426, Panx1Δ1–18, Panx1Δ13–18, or Panx1(13-18/6A) when compared with Panx1 (SI Appendix, Fig. S5B). Interestingly, Panx1(FLL/3N), though more highly expressed than Panx1, nevertheless exhibited reduced basal outwardly rectifying currents. Similarly, Panx1Δ1–18, Panx1Δ13–18, and Panx1(FLL/3N) showed higher levels of the mature Gly-2 species (known to be expressed at the cell surface (37, 38)) compared with Panx1 (SI Appendix, Fig. S5C) with reduced basal currents. Note, Gly-2 was the only Panx1 species observed in brain lysates from wild-type (WT) mice (SI Appendix, Fig. S1C). Thus, mutations or deletions within Panx1 N terminus do not reduce surface expression of Panx1, and the effects we observed are solely due to functional importance of Panx1 N terminus. This implies that the 1–18 region plays an important role in maintaining basal, small-pore, constitutive Panx1 currents.
To examine whether N-terminal deletions disrupt channel gating by alternate modalities, we assessed their impact on the sensitivity of Panx1 to mechanical stimulation in response to hypotonic conditions (26, 39–41). Using whole-cell voltage-clamp, outward rectifying Panx1 currents were substantially increased (greater than twofold) upon application of a hypotonic solution (250 mOsmol/kg, from a resting value of 310 mOsmol/kg) (SI Appendix, Fig. S6 A–C). Mediation by Panx1 was confirmed with reduced currents in presence of CBX (100 µM). Notably, the fractional response of Panx1Δ1–18 and Panx1Δ13–18 to application of a hypotonic solution was identical to that of full-length Panx1 (SI Appendix, Fig. S6B). Thus, the distal portion of the Panx1 N terminus regulates channel properties in a modality-specific manner, as it is required for activation by STIM, but not in response to a hypotonic stimulus.
Ca2+ Entry via NMDARs Activates Panx1 in a STIM-Dependent Manner.
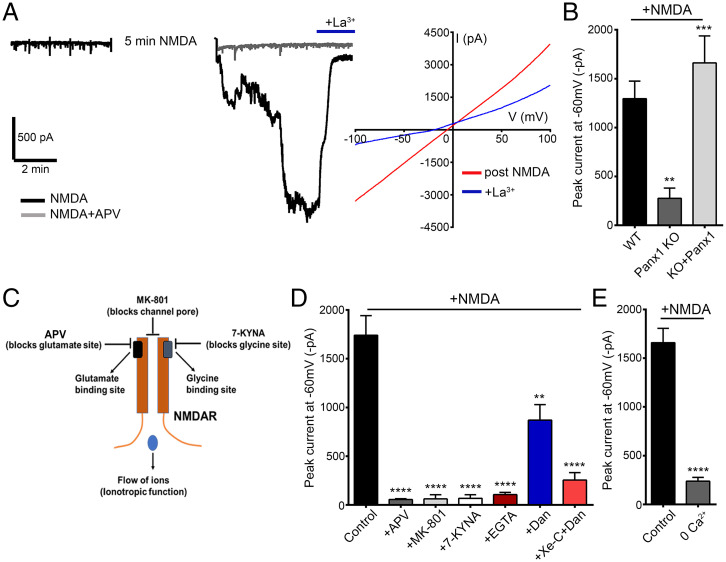
STIM can be recruited in response to NMDAR stimulation to regulate VGCCs (13). We therefore assessed whether STIM can likewise participate in activating Panx1 in response to NMDAR stimulation of hippocampal neurons. NMDAR stimulation can activate Panx1-mediated inward currents in a graded manner, contingent on the duration of NMDAR stimulation (27), and we therefore reasoned that NMDAR-initiated STIM recruitment may represent one candidate mechanism for activating Panx1. Bath application of NMDA (100 µM) reliably evoked large inward currents mediated by Panx1 on the basis of their linear IV, block by La3+, and absence of similar current in cultured neurons derived from Panx1 KO mice (Fig. 5 A and B). We confirmed that STIM1/2 expression was not altered in Panx1 KO (SI Appendix, Fig. S1 A and B). Further, NMDAR-stimulated Panx1 currents were rescued upon lentivirus-based re-expression of Panx1 in Panx1 KO neurons (Fig. 5B). Next, we tested whether Ca2+ entry via NMDARs is necessary to promote downstream Panx1 activation. NMDA-stimulated Panx1 currents were prevented in neurons with NMDARs inhibited by structurally dissimilar agents that prevent glutamate (D-2-amino-5-phosphonovaleric acid [APV], 100 µM) or glycine (7-kynurenic acid [KYNA], 10 µM) binding, as well as by MK-801 (20 µM), a NMDAR pore blocker (Fig. 5C and D). As MK-801 obstructs ion flow, but not agonist or coagonist binding, blockade of NMDAR-stimulated Panx1 by MK-801 implies that the ionotropic rather than metabotropic function of NMDARs is required in this instance, most likely due to the entry of Ca2+. Confirming this requirement, NMDA application in extracellular solution devoid of added Ca2+, or during recordings with elevated intracellular Ca2+ buffering (with 11 mM EGTA), failed to evoke Panx1-mediated currents (Fig. 5 D and E). Together, these data support that postexposure currents stimulated by NMDA are Panx1 in nature and Ca2+ influx through NMDARs is necessary for Panx1 activation. Consistent with recruitment of an ER-dependent activation mechanism, Panx1 currents induced in response to NMDA treatment were completely abolished when RyRs and IP3Rs were blocked by dantrolene (50 or 10 µM) and xestospongin-C (2.5 µM), respectively (Fig. 5D).
Fig. 5.
NMDAR stimulation activates downstream Panx1 currents in hippocampal neurons. (A) Representative trace (Left) from whole-cell recordings in hippocampal neurons treated for 5 min with NMDA (100 µM) in the presence or absence of APV (100 µM). The postexposure current, recorded after NMDA treatment, was inhibited by application of La3+ (100 µM). IV curves (Right), averaged from a series of recordings from NMDA-treated neurons (n = 16), before (post-NMDA) and after application of La3+. (B) Summary data for recordings in hippocampal neurons cultured from Panx1 KO (n = 12) or WT (n = 20), as well as in neurons from Panx1 KO, in which Panx1 was re-expressed (eGFP tagged; KO+Panx1 [n = 19]). **P < 0.01 and ***P < 0.001, one-way ANOVA with post hoc Bonferroni test when compared with WT or Panx1 KO, respectively. No significant difference was seen between KO+Panx1 and WT. (C) A cartoon figure illustrating binding sites for NMDAR antagonists APV (blocks glutamate binding), MK-801 (blocks channel pore and Ca2+ influx through NMDARs), or 7-KYNA (blocks glycine binding). (D) Summary of NMDA treatment–induced Panx1 currents in cultured CD1 neurons. Treatment-induced Panx1 currents were blocked when NMDA was applied in the presence of APV (100 µM, n = 9), MK-801 (20 µM, n = 8), 7-KYNA (10 µM, n = 9), or EGTA (11 mM in intracellular solution, n = 6) and reduced in the presence of dantrolene (Dan) alone (50 µM, n = 8) or Dan (10 µM) with xestospongin-C (Xe-C) (2.5 µM, n = 11). ****P < 0.0001 and **P < 0.01, one-way ANOVA with post hoc Bonferroni test when compared with control (n = 21). (E) Summary data from a series of comparable recordings show that Panx1 activation is reduced when NMDA is applied in solution containing 0 Ca2+ (n = 8) when compared with control (n = 8). For this set of recordings, NMDA was applied for a duration of 10 min. ****P < 0.0001, unpaired t test. Data are represented as mean ± SEM (B, D, and E).
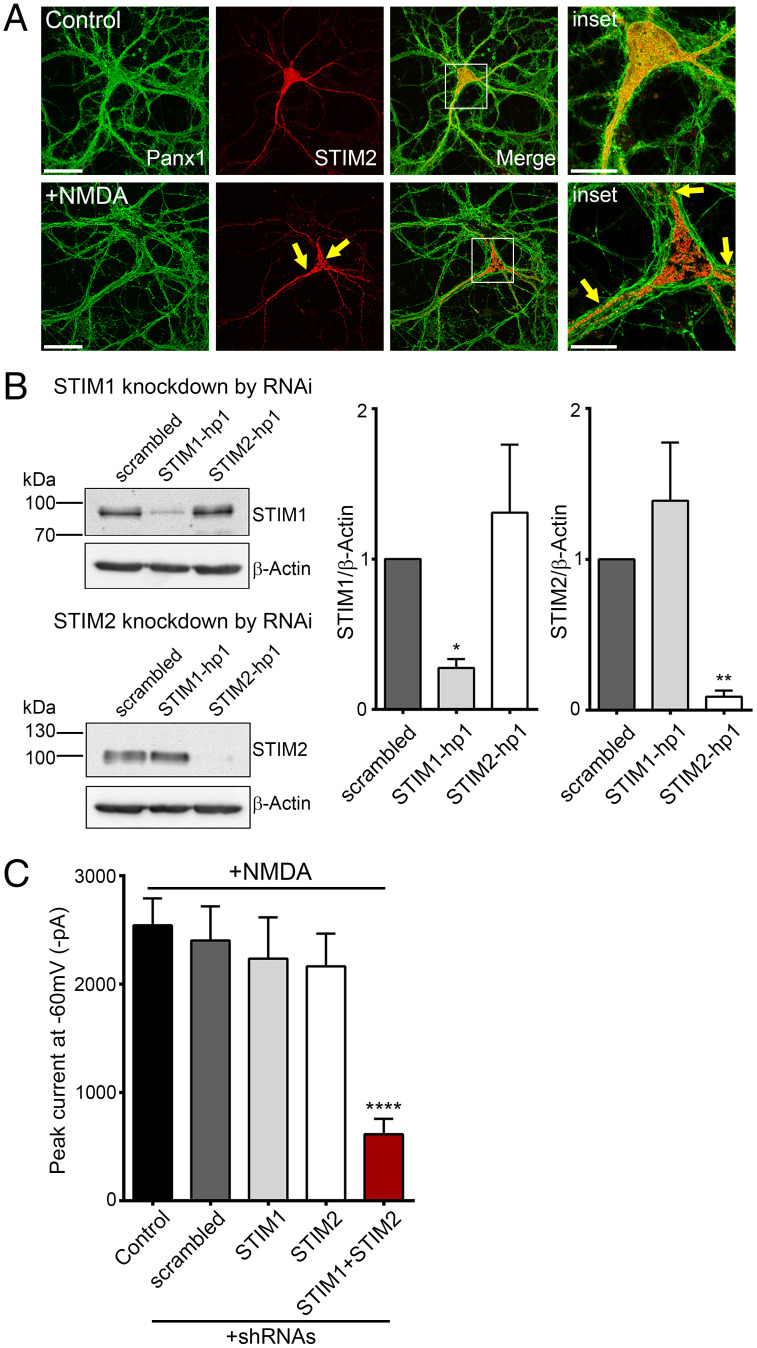
Since Panx1 activation in response to NMDAR stimulation relies on ER-dependent signaling, we asked whether this proceeds via increased association between STIM and Panx1. Thus, Panx1-KO neurons were transduced with lentivirus containing mCherry-STIM2 and Panx1-eGFP viral expression vectors, and their subcellular localization was examined. In untreated neurons, STIM2 was present in a reticular manner, but in NMDA-treated neurons, STIM2 appeared in a more punctate form (Fig. 6A). This is consistent with a model whereby NMDAR stimulation recruits STIM proteins to in turn activate Panx1 channels. To substantiate such a mechanism, we knocked down STIM1/2 in cultured neurons using lentiviral-mediated expression of short hairpin RNA (shRNA) sequences targeting STIM1 and STIM2. When STIM proteins were knocked down, Panx1 activation in response to NMDA treatment was abrogated (Fig. 6B and C). In contrast, NMDAR-stimulated Panx1 activation was not abolished when STIM1 or STIM2 were individually knocked down, hinting at some functional redundancy by the two STIM isoforms. Together, this supports that Panx1-STIM interaction, stimulated downstream of NMDAR stimulation, can promote Panx1 activation.
Fig. 6.
NMDAR-stimulated Panx1 currents require STIM1/2 in hippocampal neurons. (A) Representative image confirming expression of Panx1-eGFP and mCherry-STIM2 in lentivirus-infected Panx1 KO neurons. Compared with control, NMDA-treated neurons (100 µM, 5 min) show change in STIM2 distribution and puncta formation (marked with arrows). Scale bars, 50 µm; inset scale bars, 20 µm. (B) Representative blots (Left) showing expression of STIM1, STIM2, or β-actin in neurons expressing shRNAscrambled, shRNASTIM1-hp1, or shRNASTIM2-hp1. Results from three experiments were quantified densitometrically, and band intensities were normalized to β-actin (Right). *P < 0.05 and **P < 0.01, RM one-way ANOVA with post hoc Bonferroni test compared to scrambled. (C) Summary data from a series of recordings demonstrating that NMDA treatment–induced Panx1 currents are inhibited when both STIM1 and STIM2 are knocked down in neurons coexpressing shRNASTIM1-hp1 and shRNASTIM2-hp1. ****P < 0.0001, one-way ANOVA with post hoc Bonferroni test when STIM1+STIM2 (n = 28) were compared with control (n = 16) and scrambled (n = 17). No significant difference was seen when STIM1 (n = 8) and STIM2 (n = 8) were compared with control and scrambled. Data are represented as mean ± SEM (B and C).
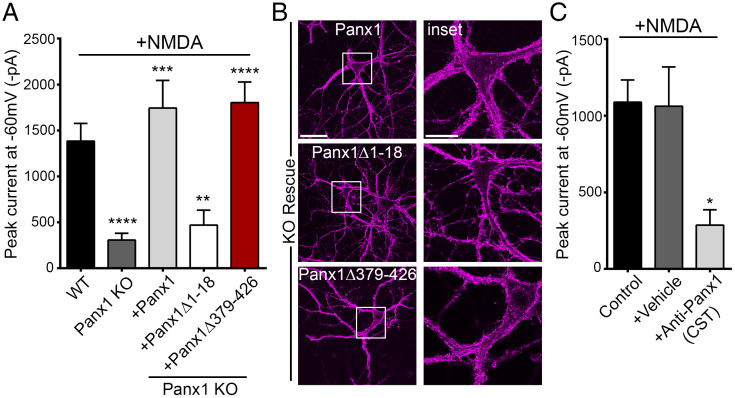
To establish that NMDAR-stimulated, STIM-dependent Panx1 activation likewise requires the 1–18 region of Panx1, we utilized two approaches. Firstly, we transduced Panx1 KO neurons to express Panx1Δ1–18 and show that NMDAR-initiated Panx1 activation was abolished in neurons expressing Panx1Δ1–18, but not when neurons expressed Panx1 or Panx1Δ379–426 (Fig. 7A and B). Secondly, we tested whether intracellular application of an antibody recognizing the N-terminal region of Panx1 could prevent NMDAR-initiated Panx1 activation. We identified this antibody (Cell Signaling Technology [CST], product no. 91137S; with epitope not disclosed) in the course of our studies mapping the subcellular localization of Panx1 and its deletion mutants. Anti-Panx1 (CST) recognizes Panx1, but not Panx1Δ1–18 (SI Appendix, Fig. S7A). We therefore reasoned that anti-Panx1 (CST) could hinder STIM-dependent Panx1 activation and thus act as a function-inhibiting antibody. Indeed, inclusion of anti-Panx1 (CST) in the patch pipette (1:400 dilution) inhibited thapsigargin (3 µM)-induced Panx1 currents in HEK cells (SI Appendix, Fig. S7B). Interestingly, anti-Panx1 (CST) does not interfere with outward-rectifying basal Panx1 currents in HEK cells, indicating that the antibody prevents large-pore activation in response to thapsigargin stimulation but does not interfere with small-pore function (SI Appendix, Fig. S7 C and D). Confirming the importance of the 1–18 region to Panx1 activation in response to neuronal NMDAR stimulation, Panx1 currents were inhibited when antibody, but not vehicle control, was included in the patch pipette (Fig. 7C). Thus, Panx1 activation downstream of NMDARs is regulated by the Panx1-STIM interaction domain and is dependent on Ca2+ entry via the NMDARs.
Fig. 7.
Functional importance of Panx1 N terminus. (A) Summary data from whole-cell recordings in neurons from Panx1 KO (n = 19) or WT (n = 28) mice and, additionally, in Panx1 KO neurons expressing Flag-Panx1 (n = 9), Flag-Panx1Δ1–18 (n = 15), or Flag-Panx1Δ379–426 (n = 13). For this set of recordings, NMDA (100 μM) was applied for a duration of 3–5 min. **P < 0.01, ***P < 0.001, and **** P < 0.0001, one-way ANOVA with post hoc Bonferroni test when compared with WT or Panx1 KO. No significant difference was seen between WT, KO+Panx1, and KO+Panx1Δ379–426. (B) Representative images confirming expression and cell surface localization (magenta) of Flag-Panx1, Flag-Panx1Δ1–18, and Flag-Panx1Δ379–426 expressed ectopically in neurons derived from Panx1 KO. Scale bars, 50 µm; inset scale bars, 20 µm. (C) Whole-cell recordings from neurons with control intracellular fluid (ICF) (control) (n = 13) or ICF supplemented with either vehicle (+vehicle) (n = 12) or anti-Panx1 (CST) (+Anti-Panx1 [CST]; 1:400 dilution) (n = 8) applied through patch pipette. For this set of recordings, NMDA was applied for a duration of 3 min. *P < 0.05, one-way ANOVA with post hoc Bonferroni test when anti-Panx1 (CST) was compared with control or vehicle. No significant difference was seen between vehicle and control. Data are represented as mean ± SEM (A and C).
Discussion
Here, we show that STIM1/2 is a new Panx1 binding and signaling partner. STIM1/2 enables Panx1 activation in response to ER-dependent signaling, including downstream of NMDAR-induced Ca2+ entry. While past studies established the important role of the carboxyl terminus in regulating Panx1 activity, notably in response to caspase-mediated cleavage in cells undergoing apoptosis (31), a prospective role of the N terminus was not known. We now demonstrate that STIM-dependent activation is entirely contingent on binding of STIM to a region of the Panx1 N terminus we now define. We also find that the N terminus regulates constitutive channel function but does not influence the mechanical response of Panx1 to a hypotonic stimulus. Activation by stimuli that recruit STIM provokes opening of Panx1 to its large-pore Ca2+ and solute-permeable state from a resting, primarily anion-permeable constitutively active state. Thus, STIM is a modality-specific activator of the Panx1 large pore, which we show binds to a stretch of hydrophobic amino acids in the Panx1 N terminus (15FLL17). The involvement of hydrophobic interactions is consistent with the requirements for Orai coupling of STIM (32–36). Notably, combining use of reverse genetics (i.e., STIM1/2 RNAi and Panx1 KO), molecular replacement of Panx1 with Panx1 N-terminal deletion mutants in Panx1 KO neurons, and a newly identified STIM-Panx1 function blocking antibody, we show that STIM1/2 couples Ca2+ entry via NMDARs to Panx1 opening. Our identification of this ER-based and STIM-dependent signaling cascade is a distinct mechanism that drives large-pore Panx1 activation. Our studies have therefore identified molecular determinants that are potential therapeutic targets in neurological disorders associated with dysregulated Panx1 function.
Past work has established an important role for the distal Panx1 carboxyl terminus in regulating Panx1 (31, 42, 43). This region interacts with the channel pore to regulate permeability through a ball-and-chain mechanism (42, 44). Considering this, we first focused on the Panx1 carboxyl terminus as a candidate STIM interaction domain. We found that thapsigargin-stimulated, STIM-dependent Panx1 activation was not altered when the distal carboxyl terminus was deleted (Panx1Δ379–426). Rather, Panx1 activation by STIM required the N terminus, more precisely the 13–18 region. Although only partially resolved, recent Panx1 cryoelectron microscopy (cryo-EM) structures (45–47) suggest that its N terminus likely orients toward the channel pore, where it may form a constriction of the permeation pathway. Considered in this light, STIM-dependent Panx1 activation can be conceived to occur through a rearrangement of its N terminus, which is permissive toward nonselective ion flux. Our identification of an antibody that recognizes a region of the Panx1 N terminus that includes the 13–18 region as a function-blocking antibody provides additional validation that the Panx1 N terminus is a required interaction site for STIM-dependent activation. Using this STIM/Panx1 function-blocking antibody, as well as N-terminal Panx1 deletion mutants re-expressed in neurons derived from Panx1 KO mice, we deciphered an exciting role for the Panx1 N terminus in regulating large-pore Panx1 opening, including in response to NMDAR stimulation.
Long considered to function primarily as a large-pore nonselective ion channel (26, 27), Panx1 is now recognized to exhibit multiple conductance states with varying permeability to ions and solutes. In the absence of activating stimuli, Panx1 exhibits a smaller outwardly rectifying conductance (∼50–80 pS), dubbed the small-pore state, with predominant Cl− permeability and corresponding negative reversal potential (28, 29, 48). In heterologous unstimulated cells expressing Panx1, coexpression of STIM proteins did not alter small-pore properties of Panx1; outward rectification, reversal potential, and peak current amplitudes were comparable with or without STIM expression. Rather, STIM-dependent Panx1 activation required thapsigargin stimulation, which then promoted Panx1 opening to its large-pore state evident on the basis of its linear IV characteristic, positive shift in current reversal potential from ∼−60 mV to ∼−9 mV, and permeability to Ca2+ and NMDG. Given our evidence of STIM1/2-Panx1 interaction at rest, these findings suggest that Panx1 regulation by STIM proteins requires a permissive conformational change in STIMs from a quiescent state at rest. Moreover, this permissive STIM1/2 conformation interacts more readily with Panx1, evident on the basis of increased STIM puncta formation in proximity to Panx1 and coimmunoprecipitation with Panx1. Altogether, this activation scheme is consistent with the established model for STIM-induced Orai1 activation underlying store-operated Ca2+ entry (SOCE) (32, 49). Given the broad ion and solute permeability of Panx1 as well as the requirement for robust NMDAR stimulation to trigger its STIM-dependent activation, it seems unlikely that Panx1 would be a major contributor to SOCE, where highly Ca2+-selective Orai channels are specialized for this task. Importantly, although STIM proteins were first identified as sensors of ER Ca2+ store depletion, subsequent studies have established that they also can be activated by temperature (50) or by oxidative stress-induced S-glutathionylation (51). These alternate STIM-activating modalities may provide additional means by which STIMs could be recruited to stimulate Panx1 large-pore activation.
Panx1 activity is subject to multimodal regulation by both cell-intrinsic and extrinsic mechanisms. Intrinsic mechanisms include activation by positive membrane potentials (≥ +20 mV), mechanical stimuli (e.g., hypotonicity-evoked changes in cell volume), and caspase-mediated carboxyl-terminal cleavage. Cell-extrinsic activation mechanisms include elevated extracellular levels of K+ (≥ 20 mM) or ATP as well as receptor-based mechanisms, for example, in response to α1-adrenergic, P2X7, or NMDAR stimulation. Activation by extreme membrane depolarization, pronounced accumulation of extracellular K+, and recruitment of executioner caspases, though potentially relevant in the context of seizure spread or ischemic insult, are difficult to reconcile with reported contributions of Panx1 to neurodevelopment, synaptic plasticity, and learning and memory. In this context, coupling of Panx1 downstream of NMDAR stimulation is more highly relevant, and multiple lines of evidence lend credence to the importance of this activation mechanism. Panx1 activation by NMDARs was first demonstrated in the context of anoxic depolarization (52), whereby activation proceeds via recruitment of Src tyrosine kinase and phosphorylation of Y308 localized to the Panx1 carboxyl terminus (52, 53). This scheme is notably distinct from STIM-dependent Panx1 activation, which proceeds via molecular determinants localized to the N terminus. Further, while Src-mediated Panx1 activation requires the metabotropic function of NMDARs, STIM-dependent Panx1 activation requires the ionotropic function of NMDARs. The occurrence of distinct molecular mechanisms that converge to regulate Panx1 activity downstream of NMDARs, paired with evidence that Panx1 regulates bidirectional NMDAR-dependent plasticity (23, 24, 54), highlights the need to understand the dynamics responsible for coordinating the contribution of distinct NMDAR-dependent mechanisms in driving Panx1 activation. One possibility is that each mechanism may be recruited separately by distinct populations of NMDARs localized within distinct subcellular compartments (e.g., synaptic vs. extrasynaptic), where NMDARs may vary in terms of their association within protein complexes composed of STIM1/2 and/or Src. Alternatively, each mechanism may be favored by activation of NMDARs having distinct GluN2 subunit composition (e.g., GluN2A vs. GluN2B). More recently, Panx1 activation in response to NMDAR stimulation has been implicated in crosstalk between glutamatergic and electrical transmission via gap junctions (55). Collectively, this suggests an important role of Panx1 as a connecting point and downstream effector of NMDAR-mediated physiological and pathological functions.
Our characterization of N-terminal deletion mutants has revealed an important role of this region in maintaining constitutive outwardly rectifying Panx1 currents, which are evident upon application of voltage ramps (±100 mV). Deletion of the distal Panx1 N terminus reduced the amplitude of CBX-sensitive, outwardly rectifying currents in HEK cells expressing Panx1 with deletions or mutations within the 1–18 region. These mutants express well and do not differ from full-length Panx1 with respect to their enrichment at the plasma membrane. The reduced amplitude of constitutive currents in Panx1 N-terminal mutants was observed irrespective of their ability to interact with STIM1/2. Moreover, the amplitude of outwardly rectifying, small-pore Panx1 currents in HEK cells was not influenced by coexpression of STIM1 or STIM2. Collectively, these findings suggest that STIM1/2 does not influence the constitutive small-pore state. Of note, independent experiments, published during review of our manuscript, likewise show a loss of small-pore function when deleting the first 20 residues of the Panx1 N terminus (Panx1Δ2–20) (56). Using cryo-EM, the authors compared the structure of Panx1Δ2–20 to full-length Panx1 (with and without probenecid present). Their functional and structural analysis suggests that partial N-terminal deletion (or presence of probenecid) induces a conformational change in which the N terminus assumes a “closed” position associated with reduced channel activity due to phospholipid migration and obstruction of the pore. Note, however, that cryo-EM approaches have yet to resolve distinct conformations induced by distinct activation modalities involving protein complexes or posttranslational modifications that likely vary with cellular and/or subcellular context. Also, much of the carboxyl terminus remains unresolved in cryo-EM structures reported to date, and thus, its distinct contributions to channel gating must be born in mind. Thus, our findings take on added importance in complementing those from Kuzuya et al. by revealing that Panx1 deletion mutants with the N terminus in a closed position may nevertheless transition to an opened state, given that mutants with deletions within the 1–18 region (Panx1Δ1–18 and Panx1Δ13–18) responded to application of a hypotonic solution with robust (greater than twofold increase at +100 mV) outwardly rectifying and CBX-sensitive currents. Note, these Panx1 deletions are unresponsive to thapsigargin and have reduced constitutive function. Collectively, this indicates that Panx1 possesses distinct permeation modalities and functions that are separable, given that they are regulated via distinct molecular determinants, and can thus be stimulated in a context-dependent manner (i.e., constitutive [small pore] vs. osmotic stress induced [small pore] vs. STIM dependent [large pore]).
In summary, our work identifies STIM proteins as binding and gating partners of Panx1. In this respect, STIM1 and STIM2 are each able to activate Panx1 in response to stimuli that induce a permissive conformational change. This was evident from ectopic expression experiments in HEK cells and suggested from experiments in hippocampal neurons where knockdown of both STIM1 and STIM2 was necessary to abolish Panx1 activation in response to NMDAR stimulation. This supports functional redundancy but does not necessarily imply promiscuity or interchangeability with regards to Panx1 activation. Indeed, STIM1 and STIM2 display overlapping but also distinct subcellular expression patterns in neurons. While both localize to the cell body, evidence suggests that STIM2 is more highly expressed at distal processes, including dendritic spines (57–59). Each isoform also differs with regards to their Ca2+ binding affinity and thus their activation threshold in response to changes in ER luminal Ca2+. Evidence supports that Panx1 is broadly distributed within neuronal compartments, including dendritic spines, suggesting that STIM1/2 may regulate Panx1 within discrete compartments in response to distinct signaling events that alter luminal Ca2+ to varying degrees. When considering the consequence of Panx1 activation in response to NMDAR stimulation, several studies have shown that inhibition or deletion of Panx1 is associated with altered learning and memory, perhaps as a consequence of imbalanced NMDAR-dependent bidirectional plasticity. This follows from evidence that suppressing Panx1 function is associated with enhanced LTP at the expense of long-term depression (23, 24, 54). The extent to which Panx1 regulates synaptic plasticity as a consequence of activity- and NMDAR-dependent Panx1 activation, for example, during the plasticity-inducing repetitive stimulation, remains to be established. But of note, a recent study has linked augmented Panx1 activity, associated with amyloid-beta accumulation, to LTP deficits in a mouse model of Alzheimer’s disease (60). Intriguingly, double KO of STIM1/2 phenocopies Panx1 deletion, at least in terms of the observed markedly enhanced LTP (61). Lastly, given the broad cellular distribution of STIM and Panx1, it is likely that the mechanism identified by us is of relevance not only in neurons but in other cell types as well. In nonneuronal cells, STIM recruitment would likely be initiated through processes independent of NMDARs, of which many have been identified. This could provide a means for modality-specific and cellular-context-dependent Panx1 functions.
Materials and Methods
Primary cultures of mouse hippocampal neurons were prepared from embryonic day 17–18 CD1, Panx1 KO, or WT mice, according to previously described procedures (62). Panx1 KO first mice (Panx1tm1a(KOMP)Wtsi) were purchased from Knockout Mouse Project at University of California, Davis. For knockdown or ectopic expression experiments, CD1 or Panx1 KO neurons were transduced with lentivirus, as previously described (27), to allow expression of shRNAs (targeting STIM1 or STIM2) or Flag-tagged Panx1 (full-length or deletion mutants). HEK 293T cells were transiently transfected to express mouse Panx1 (full length, deletions, or mutations) with or without mouse STIM1 or STIM2. Whole-cell, voltage-clamp recordings were performed in cultured hippocampal neurons or transfected HEK 293T cells to study Panx1 channel activity, as described before (27, 63, 64). Further, western blotting, immunoprecipitation, and immunocytochemistry assays were performed to study Panx1 and STIM1/2 protein expression, interaction, and distribution. Detailed materials and methods for cell culture, transfections, whole-cell recordings, western blotting, immunoprecipitation, immunocytochemistry, and deglycosylation assays are provided in SI Appendix, SI Materials and Methods.
Data Analysis and Statistics.
All data consist of a minimum of three independent experiments. For electrophysiology, n denotes no. of cells recorded per condition, whereas for biochemistry, n denotes no. of replicates from separate culture preparations. All data are represented as mean ± SEM unless stated in figure legends. Averaged IV curves are represented as mean only. All graphs were prepared using Prism 6 (GraphPad Software). Statistical analysis was performed using Prism 6, and differences were considered significant when P < 0.05 with following definitions: *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Statistical tests used were specified in each figure legend.
Supplementary Material
Acknowledgments
We wish to thank Drs. Dale Laird and Silvia Penuela from Western University for their kind gift of anti-Panx1 antibody (Panx1CT395). We also acknowledge that all fluorescence images were generated using a Zeiss LSM880 with AiryScan located in the Kleysen Institute for Advanced Medicine Live Cell Imaging Facility. This work was supported by grants from the Canadian Institutes of Health Research to M.F.J. (MOP‐125901) and the Natural Sciences and Engineering Research Council of Canada to M.F.J. (RGPIN-05477-2017) and T.J.S. (RGPIN-2015-05994). C.S.P. was supported by studentships awarded by Research Manitoba, the Alzheimer Societies of Canada and Manitoba, and the McCrorie-West Family Fellowship for Alzheimer Research. R.S. was supported by a postdoctoral fellowship from Research Manitoba.
Footnotes
Competing interest statement: C.S.P., N.E.L., and M.F.J. are inventors on a provisional patent application (filed by the University of Manitoba) for a peptide targeting Panx1 N terminus–interacting partners.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2112870119/-/DCSupplemental.
Data, Materials, and Software Availability
All data are included in the manuscript and/or SI Appendix.
References
- 1.Whitlock J. R., Heynen A. J., Shuler M. G., Bear M. F., Learning induces long-term potentiation in the hippocampus. Science 313, 1093–1097 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Nabavi S., et al. , Engineering a memory with LTD and LTP. Nature 511, 348–352 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gruart A., Muñoz M. D., Delgado-García J. M., Involvement of the CA3-CA1 synapse in the acquisition of associative learning in behaving mice. J. Neurosci. 26, 1077–1087 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verkhratsky A., Physiology and pathophysiology of the calcium store in the endoplasmic reticulum of neurons. Physiol. Rev. 85, 201–279 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Padamsey Z., Foster W. J., Emptage N. J., Intracellular Ca2+ release and synaptic plasticity: A tale of many stores. Neuroscientist 25, 208–226 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Alford S., Frenguelli B. G., Schofield J. G., Collingridge G. L., Characterization of Ca2+ signals induced in hippocampal CA1 neurones by the synaptic activation of NMDA receptors. J. Physiol. 469, 693–716 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Emptage N., Bliss T. V., Fine A., Single synaptic events evoke NMDA receptor-mediated release of calcium from internal stores in hippocampal dendritic spines. Neuron 22, 115–124 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Goussakov I., Miller M. B., Stutzmann G. E., NMDA-mediated Ca(2+) influx drives aberrant ryanodine receptor activation in dendrites of young Alzheimer’s disease mice. J. Neurosci. 30, 12128–12137 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holbro N., Grunditz A., Oertner T. G., Differential distribution of endoplasmic reticulum controls metabotropic signaling and plasticity at hippocampal synapses. Proc. Natl. Acad. Sci. U.S.A. 106, 15055–15060 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soboloff J., Rothberg B. S., Madesh M., Gill D. L., STIM proteins: Dynamic calcium signal transducers. Nat. Rev. Mol. Cell Biol. 13, 549–565 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park C. Y., Shcheglovitov A., Dolmetsch R., The CRAC channel activator STIM1 binds and inhibits L-type voltage-gated calcium channels. Science 330, 101–105 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Wang Y., et al. , The calcium store sensor, STIM1, reciprocally controls Orai and CaV1.2 channels. Science 330, 105–109 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dittmer P. J., Wild A. R., Dell’Acqua M. L., Sather W. A., STIM1 Ca2+ sensor control of L-type Ca2+-channel-dependent dendritic spine structural plasticity and nuclear signaling. Cell Rep. 19, 321–334 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.MacVicar B. A., Thompson R. J., Non-junction functions of pannexin-1 channels. Trends Neurosci. 33, 93–102 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Bond S. R., Naus C. C., The pannexins: Past and present. Front. Physiol. 5, 58 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Penuela S., Harland L., Simek J., Laird D. W., Pannexin channels and their links to human disease. Biochem. J. 461, 371–381 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Sanchez-Arias J. C., et al. , Purinergic signaling in nervous system health and disease: Focus on pannexin 1. Pharmacol. Ther. 225, 107840 (2021). [DOI] [PubMed] [Google Scholar]
- 18.Yeung A. K., Patil C. S., Jackson M. F., Pannexin-1 in the CNS: Emerging concepts in health and disease. J. Neurochem. 154, 468–485 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Wicki-Stordeur L. E., Swayne L. A., Panx1 regulates neural stem and progenitor cell behaviours associated with cytoskeletal dynamics and interacts with multiple cytoskeletal elements. Cell Commun. Signal. 11, 62 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wicki-Stordeur L. E., et al. , Pannexin 1 differentially affects neural precursor cell maintenance in the ventricular zone and peri-infarct cortex. J. Neurosci. 36, 1203–1210 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanchez-Arias J. C., et al. , Pannexin 1 regulates network ensembles and dendritic spine development in cortical neurons. eNeuro 6, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanchez-Arias J. C., Candlish R. C., van der Slagt E., Swayne L. A., Pannexin 1 regulates dendritic protrusion dynamics in immature cortical neurons. eNeuro 7, ENEURO.0079-20.2020 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ardiles A. O., et al. , Pannexin 1 regulates bidirectional hippocampal synaptic plasticity in adult mice. Front. Cell. Neurosci. 8, 326 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prochnow N., et al. , Pannexin1 stabilizes synaptic plasticity and is needed for learning. PLoS One 7, e51767 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J., et al. , The membrane protein Pannexin1 forms two open-channel conformations depending on the mode of activation. Sci. Signal. 7, ra69 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bao L., Locovei S., Dahl G., Pannexin membrane channels are mechanosensitive conduits for ATP. FEBS Lett. 572, 65–68 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Thompson R. J., et al. , Activation of pannexin-1 hemichannels augments aberrant bursting in the hippocampus. Science 322, 1555–1559 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Ma W., et al. , Pannexin 1 forms an anion-selective channel. Pflugers Arch. 463, 585–592 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Romanov R. A., et al. , The ATP permeability of pannexin 1 channels in a heterologous system and in mammalian taste cells is dispensable. J. Cell Sci. 125, 5514–5523 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Epp A. L., et al. , A novel motif in the proximal C-terminus of Pannexin 1 regulates cell surface localization. Sci. Rep. 9, 9721 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chekeni F. B., et al. , Pannexin 1 channels mediate ‘find-me’ signal release and membrane permeability during apoptosis. Nature 467, 863–867 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muik M., et al. , Dynamic coupling of the putative coiled-coil domain of ORAI1 with STIM1 mediates ORAI1 channel activation. J. Biol. Chem. 283, 8014–8022 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Stathopulos P. B., et al. , STIM1/Orai1 coiled-coil interplay in the regulation of store-operated calcium entry. Nat. Commun. 4, 2963 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X., et al. , Distinct Orai-coupling domains in STIM1 and STIM2 define the Orai-activating site. Nat. Commun. 5, 3183 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frischauf I., et al. , Molecular determinants of the coupling between STIM1 and Orai channels: Differential activation of Orai1-3 channels by a STIM1 coiled-coil mutant. J. Biol. Chem. 284, 21696–21706 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Derler I., et al. , The extended transmembrane Orai1 N-terminal (ETON) region combines binding interface and gate for Orai1 activation by STIM1. J. Biol. Chem. 288, 29025–29034 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boassa D., et al. , Pannexin1 channels contain a glycosylation site that targets the hexamer to the plasma membrane. J. Biol. Chem. 282, 31733–31743 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Penuela S., et al. , Pannexin 1 and pannexin 3 are glycoproteins that exhibit many distinct characteristics from the connexin family of gap junction proteins. J. Cell Sci. 120, 3772–3783 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Okada S. F., Nicholas R. A., Kreda S. M., Lazarowski E. R., Boucher R. C., Physiological regulation of ATP release at the apical surface of human airway epithelia. J. Biol. Chem. 281, 22992–23002 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ransford G. A., et al. , Pannexin 1 contributes to ATP release in airway epithelia. Am. J. Respir. Cell Mol. Biol. 41, 525–534 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seminario-Vidal L., et al. , Rho signaling regulates pannexin 1-mediated ATP release from airway epithelia. J. Biol. Chem. 286, 26277–26286 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sandilos J. K., et al. , Pannexin 1, an ATP release channel, is activated by caspase cleavage of its pore-associated C-terminal autoinhibitory region. J. Biol. Chem. 287, 11303–11311 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang J., Dahl G., SCAM analysis of Panx1 suggests a peculiar pore structure. J. Gen. Physiol. 136, 515–527 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chiu Y.-H., et al. , A quantized mechanism for activation of pannexin channels. Nat. Commun. 8, 14324 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deng Z., et al. , Cryo-EM structures of the ATP release channel pannexin 1. Nat. Struct. Mol. Biol. 27, 373–381 (2020). [DOI] [PubMed] [Google Scholar]
- 46.Michalski K., et al. , The Cryo-EM structure of pannexin 1 reveals unique motifs for ion selection and inhibition. eLife 9, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruan Z., Orozco I. J., Du J., Lü W., Structures of human pannexin 1 reveal ion pathways and mechanism of gating. Nature 584, 646–651 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Locovei S., Wang J., Dahl G., Activation of pannexin 1 channels by ATP through P2Y receptors and by cytoplasmic calcium. FEBS Lett. 580, 239–244 (2006). [DOI] [PubMed] [Google Scholar]
- 49.Calloway N., Vig M., Kinet J.-P., Holowka D., Baird B., Molecular clustering of STIM1 with Orai1/CRACM1 at the plasma membrane depends dynamically on depletion of Ca2+ stores and on electrostatic interactions. Mol. Biol. Cell 20, 389–399 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao B., Coste B., Mathur J., Patapoutian A., Temperature-dependent STIM1 activation induces Ca2+ influx and modulates gene expression. Nat. Chem. Biol. 7, 351–358 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hawkins B. J., et al. , S-glutathionylation activates STIM1 and alters mitochondrial homeostasis. J. Cell Biol. 190, 391–405 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weilinger N. L., Tang P. L., Thompson R. J., Anoxia-induced NMDA receptor activation opens pannexin channels via Src family kinases. J. Neurosci. 32, 12579–12588 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weilinger N. L., et al. , Metabotropic NMDA receptor signaling couples Src family kinases to pannexin-1 during excitotoxicity. Nat. Neurosci. 19, 432–442 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Gajardo I., et al. , Lack of pannexin 1 alters synaptic glun2 subunit composition and spatial reversal learning in mice. Front. Mol. Neurosci. 11, 114 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Siu R. C. F., Kotova A., Timonina K., Zoidl C., Zoidl G. R., Convergent NMDA receptor-Pannexin1 signaling pathways regulate the interaction of CaMKII with Connexin-36. Commun. Biol. 4, 702 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kuzuya M., et al. , Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids. Sci. Signal. 15, eabg6941 (2022). [DOI] [PubMed] [Google Scholar]
- 57.Sun S., et al. , Reduced synaptic STIM2 expression and impaired store-operated calcium entry cause destabilization of mature spines in mutant presenilin mice. Neuron 82, 79–93 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang H., et al. , Store-operated calcium channel complex in postsynaptic spines: A new therapeutic target for Alzheimer’s disease treatment. J. Neurosci. 36, 11837–11850 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pchitskaya E., et al. , Stim2-Eb3 association and morphology of dendritic spines in hippocampal neurons. Sci. Rep. 7, 17625 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Flores-Muñoz C., et al. , Acute Pannexin 1 blockade mitigates early synaptic plasticity defects in a mouse model of Alzheimer’s disease. Front. Cell. Neurosci. 14, 46 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garcia-Alvarez G., et al. , Impaired spatial memory and enhanced long-term potentiation in mice with forebrain-specific ablation of the Stim genes. Front. Behav. Neurosci. 9, 180 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Belrose J. C., Xie Y. F., Gierszewski L. J., MacDonald J. F., Jackson M. F., Loss of glutathione homeostasis associated with neuronal senescence facilitates TRPM2 channel activation in cultured hippocampal pyramidal neurons. Mol. Brain 5, 11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shao Q., et al. , A germline variant in the PANX1 gene has reduced channel function and is associated with multisystem dysfunction. J. Biol. Chem. 291, 12432–12443 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nouri-Nejad D., et al. , Pannexin 1 mutation found in melanoma tumor reduces phosphorylation, glycosylation, and trafficking of the channel-forming protein. Mol. Biol. Cell 32, 376–390 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in the manuscript and/or SI Appendix.