The nine decades since Smith and Townsend demonstrated that Agrobacterium tumefaciens causes plant tumors (95) have been marked by a series of surprises. Among the most important of these was the report in 1958 that these tumors could be excised and propagated in vitro without exogenous plant hormones (7). Equally important were a series of reports beginning about the same time that tumors released compounds that agrobacteria could use as nutrients (24). Perhaps the most exciting discoveries, reported in the 1970s and 1980s, were that tumorigenesis required the transfer of fragments of oncogenic DNA to infected plant cells (10), that this process evolved from a conjugal transfer system (99), and that the genes that direct this process are expressed in response to host-released chemical signals (47). This DNA transfer process has become a cornerstone of plant molecular genetics. The genus Agrobacterium also has provided excellent models for several aspects of host-pathogen interactions, including intercellular transport of macromolecules (11), bacterial detection of host organisms (47), targeting of proteins to plant cell nuclei (3), and interbacterial chemical signaling via autoinducer-type pheromones (120).
Most of the genes required for tumorigenesis are found on large extrachromosomal elements called Ti plasmids. Indeed, transfer of Ti plasmids into certain nonpathogenic bacteria converts them into tumorigenic pathogens (43). Ti plasmids are generally referred to by the types of opines whose catabolism they direct (see below). However, this nomenclature is becoming less satisfactory as we discover that all known Ti plasmids direct the catabolism of more than one opine and that opine catabolic genes are found in a variety of combinations in different plasmids. The Ti plasmids pTiA6NC, pTi15955, pTiAch5, pTiR10, and pTiB6S3, which are widely considered to be functionally identical, are generally referred to as octopine-type Ti plasmids (or, less frequently, octopine, mannityl opine-type Ti plasmids). The DNA sequencing of these plasmids was initiated almost 20 years ago (21) and was recently completed in our three laboratories. The resulting 194,140-nucleotide sequence is a composite assembly of sequences from all of the plasmids listed above. The close similarity of these plasmids is exemplified by the sequence of a 42-kb segment of the vir regions of pTiA6NC and pTi15955. These sequences differ at only one base, and this polymorphism is silent at the amino acid level. We have no evidence for polymorphisms elsewhere except for a large deletion that is unique to pTiA6NC (Fig. 1). The restriction map deduced from this sequence agrees almost perfectly with the published restriction map of pTiAch5 (25). All known and suspected genes are depicted in Fig. 1, and their demonstrated or putative functions are described in Table 1. The DNA sequence of this Ti plasmid provides a useful framework to review the roles of this plasmid in the biology of plant infection and colonization.
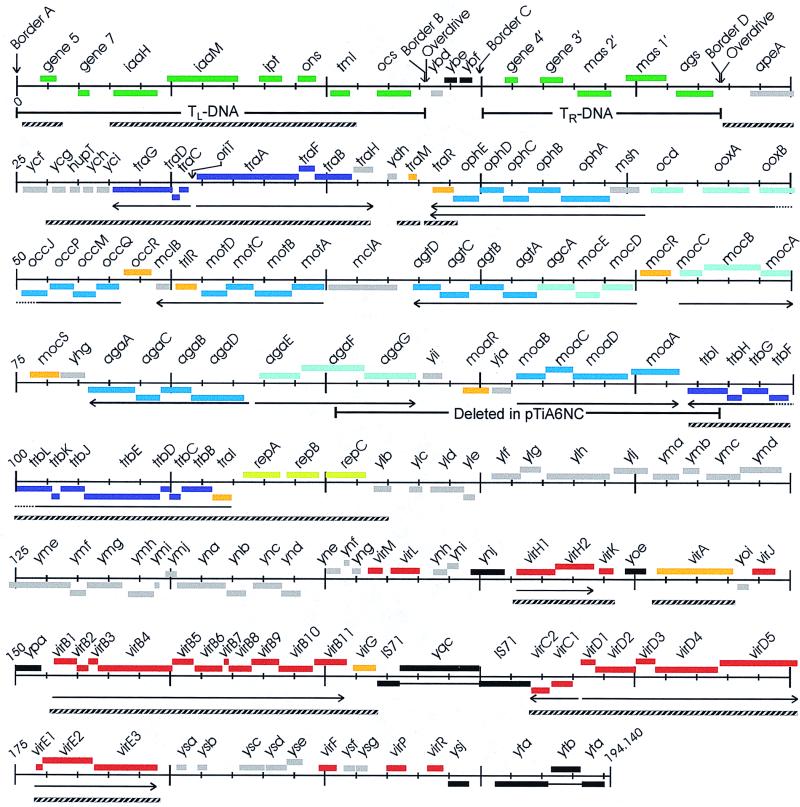
FIG. 1.
Genetic map of the octopine-type Ti plasmid. Nucleotide 1 is taken as the left end of the left border of TL. Genes transcribed from left to right are shown above the scale, while genes transcribed from right to left are shown below the scale. Known or suspected polycistronic operons are indicated with horizontal arrows. A 12.6-kb region deleted in the widely studied pTiA6NC is indicated by a horizontal bar on line 4. Regions that can form heteroduplexes with orthologous sequences of pTiC58 (27) are indicated with crosshatched bars. Green bars indicate genes that are located on the TL region or TR region and transferred to plant cells. Red bars indicate genes in the vir regulon, all of which are regulated by VirA and VirG, and many of which are required for T-strand processing and transfer. Purple bars indicate genes required for interbacterial conjugal transfer of the Ti plasmid. Light green bars indicate genes required for vegetative replication and partitioning of the Ti plasmid. Dark blue bars indicate genes encoding ATP binding cassette-type opine permeases, while light blue bars indicate opine catabolic genes. Orange bars indicate regulatory genes. Black bars indicate suspected IS elements (in this case, bars represent DNA sequence similarities rather than ORFs), while grey bars indicate ORFs of miscellaneous or unknown function. Gene names beginning with the letter “y” indicate the position of the gene, such that the second letter represents position in tens of kilobases, and the third letter indicates position in single kilobases. During the analysis of all these sequences, we found that tml, mas2′, mas1′, and ags had sequence discrepancies relative to orthologous genes from other Ti or Ri plasmids. These regions were resequenced after PCR amplification, and three errors in the original sequence (4) were detected and corrected. The resulting DNA sequence of the Ti plasmid has been deposited in the GenBank DNA sequence database (accession no. AF242881). Most Ti plasmid sequences were previously deposited in DNA sequence databases, and the original sources of these sequences are provided in the annotations of our compiled sequence.
TABLE 1.
Genes encoded by the octopine-type Ti plasmida
| Genetic locus | Description | Reference(s) |
|---|---|---|
| T-DNA genes | ||
| ags | Agropine synthase, lactonization of mannopine | 24, 40 |
| Gene 5 | Synthesis of indole-3-lactate, an auxin antagonist | 57 |
| iaaH and iaaM | Conversion of tryptophan to indole acetic acid (auxin) | 55 |
| ipt | Condensation of AMP and isopentenylpyrophosphate to form isopentenyl-AMP, a cytokinin | 66 |
| mas1′ and mas2′ | Mannopine synthase; condensation of glucose with glutamine or glutamate followed by reduction | 24 |
| ocs | Octopine synthase, reductive condensation of pyruvate with four basic amino acids | 21 |
| ons | Opine export from plant cells | 75 |
| tml (gene 6b) | Auxin sensitivity | 108 |
| Borders A, B, C, D | cis-acting sites required for T-DNA processing, functionally equivalent to conjugal origins of transfer | 125 |
| Overdrive | cis-acting site for optimal T-DNA transfer; VirC1 binding site | 110, 113 |
| vir loci | ||
| virB1-11 | Type IV transport system to transfer T-DNA and Vir proteins from bacteria to host cytoplasm | 106, 116 |
| virC and -D | T-DNA processing. VirD1 and VirD2 nick at T-DNA borders; VirC1 binds overdrive | 111, 122, 123 |
| virE | Nuclear transport of T-DNA. VirE2 binds single-stranded DNA and has nuclear localization sites; VirE1 is a chaperone for VirE2 transport | 14, 101 |
| virF | Host range factor | 73 |
| virH1-2 | P450-type oxidases; VirH2 O demethylates phenolic inducers | 51 |
| virM, -L, -K, -J, -F, -P, -R, -D3, -D5, and -E3 | Other members of the vir regulon; VirP resembles phosphatases | 50, 52 |
| Interbacterial conjugation genes | ||
| traAFB, traCDG | Ti plasmid conjugal DNA processing | 1 |
| trbB-I | Type IV transfer system required for Ti plasmid conjugation | 1 |
| oriT | cis-acting site required for conjugation | 1 |
| Vegetative replication genes | ||
| repAB | Putative partitioning system | 104 |
| repC | Essential for vegetative replication | 104 |
| Opine uptake genes | ||
| agaDBCA | Agropinic acid permease | 70 |
| agtABCD | Agropine permease | Unpublished data |
| motABCD | Mannopine permease | 80 |
| moaBCDA | Mannopinic acid permease | 70 |
| occQMPJ | Octopine permease | 112 |
| ophABCDE | Putative permease for an unknown substrate | 33 |
| Opine catabolism genes | ||
| agaE | Conversion of agropinic acid to mannopinic acid | 70 |
| agaFG | Conversion of mannopinic acid to glutamic acid and mannose | 70 |
| agcA | Catabolic mannopine cyclase, for conversion of agropine to mannopine, related to ags | 54 |
| mocAB | Oxidoreductase, and dehydratase? | 54 |
| mocCD | Conversion of mannopine to glutamine and glucose | 54 |
| mocE | Kinase? | 54 |
| ocd | Ornithine cyclodeaminase for conversion of ornithine to proline | 114 |
| ooxAB | Oxidoreductase for conversion of octopine-type opines to pyruvate and corresponding basic amino acid | 114 |
| Transcriptional regulation genes | ||
| moaR | Repressor of agaD-A, agaE-G, and moaB-A operons | 70 |
| mocR | Probable regulator of the mocD-agtD and mocC-A operons | 54 |
| mocS | Resembles MocR, function unknown | 54 |
| occR | LysR-type octopine-responsive regulator of the occQ-traR operon | 115 |
| traR and traI | LuxR-LuxI-type quorum sensing regulators; TraI synthesizes 3-oxooctanoylhomoserine lactone; TraR is a transcriptional activator | 36, 77, 128 |
| traM | TraR antagonist | 32 |
| trlR | TraR antagonist; TrlR resembles TraR but is truncated and may inhibit TraR by forming inactive heteromultimers | 80, 127 |
| virA and virG | Two-component regulators of vir regulon; VirA is a transmembrane histidine kinase; VirG is an OmpR-type response regulator | 61 |
| IS elements | ||
| IS71L and IS71R | Apparent IS element, interrupted by insertion of yqc IS element | Unpublished data |
| ybe, yoe, and yqc | Resemble IS66 of A. tumefaciens | Unpublished data |
| ybf | Resembles IS1203 of E. coli | Unpublished data |
| ynj | Resembles IS1313 of A. tumefaciens | Unpublished data |
| ypa | Resembles IS869 of A. tumefaciens | Unpublished data |
| ysj | Resembles IS492 of Pseudomonas sp. | Unpublished data |
| yta | Resembles IS21 of E. coli, disrupted by IS element ytb | Unpublished data |
| ytb | Resembles IS1111a of Coxiella burnetii | Unpublished data |
| Genes with miscellaneous and unknown functions | ||
| apeA | Exclusion of bacteriophage Ap1 | 1 |
| hupT | Resembles HNS-type proteins | 1 |
| mclA and -B | Resembles methyl-accepting chemotaxis proteins; possible role in chemotaxis to opines. MclB is severely truncated and inhibits chemotaxis in A. tumefaciens | 80, 127 |
| msh | Resembles methionine synthase | 33 |
| yhg | Resembles oxidoreductases; possible role in opine catabolism | 70 |
| ylb | Resembles DNA invertases; possible role in plasmid maintenance | Unpublished data |
| yld | Resembles DNA invertases; possible role in plasmid maintenance | Unpublished data |
| yle | Resembles plasmid stability locus; possible role in plasmid maintenance | Unpublished data |
| ylf-yng | Functions unknown | Unpublished data |
| ysa | Resembles integration host factor; weakly induced by vir inducing stimuli | 52 |
| ysb | Resembles cold shock proteins | 52 |
| ysc, ysd, and yse | ysc and ysd resemble 3′ end of traA of Ti plasmid; yse resembles Ti plasmid traF; none is detectably expressed | 29, 52 |
Types of genes correspond to bars in Fig. 1 as follows: T-DNA genes, dark green bars; vir loci, red bars; interbacterial conjugation genes, purple bars; vegetative replication genes, light green bars; opine uptake genes, dark blue bars; opine catabolism genes, light blue bars; transcriptional regulation genes, orange bars; IS elements, black bars; and genes with miscellaneous and unknown functions, grey bars.
This Ti plasmid contains 155 open reading frames (ORFs), almost all of which are likely to encode functional proteins (Fig. 1 and Table 1). The overall G+C composition of this plasmid is 55%, although a few segments are considerably richer in A's and T's, particularly in the T region (see below). Overall, the Ti plasmid exhibits a modular structure with genes of similar function or purpose grouped together. Thus, we can define five components: (i) the T region, which codes for sequences that are transferred to the plant host; (ii) the vir region, which directs the processing and transfer of the T-DNA; (iii) the rep region, which is required for replication of the Ti plasmid; (iv) the tra and trb loci, which direct the conjugal transfer of the Ti plasmid; and (v) genes that direct uptake and catabolism of opines. An exception to this clustering is the tra and trb loci, the two gene sets required for conjugal transfer, which are separated from each other by 60 kb.
TRANSFER OF TWO DNA FRAGMENTS TO HOST PLANT CELLS
During infection, A. tumefaciens strains carrying an octopine-type Ti plasmid transfer two fragments of DNA to the nuclei of host plants by a mechanism that requires cell-cell contact and resembles plasmid conjugation. These fragments are designated the TL-DNA and TR-DNA (Fig. 1, top line), and are 13 and 7.8 kb in length, respectively (4, 105). The corresponding segments of the Ti plasmid are called T regions, and each is flanked by cis-acting, 25-bp direct repeats, called border sequences (121, 125). The left border of the TL-DNA is dispensable for T-DNA transfer, while the right border is essential and acts in a polar fashion, suggesting that transfer may initiate at the right border and proceed leftward (76). Inversion of the right border leads to attenuated tumorigenesis, and tumors made by such mutants contain extremely long T-DNA fragments consisting of virtually the entire Ti plasmid (76). Adjacent to the right border of TL is another cis-acting site called overdrive (94), which is required for wild-type transfer efficiency and provides a binding site for the VirC1 protein (see below). A second possible overdrive sequence is located adjacent to the right border of TR, though the role of this sequence in T-DNA transfer has not been studied.
In the presence of proteins encoded by the vir region (see below), the DNA within the T regions undergoes several processing steps (Fig. 2). Each border is cleaved on the bottom DNA strand at a site exactly 4 nucleotides from its left end. This reaction is catalyzed by the VirD2 protein (see below), which remains covalently bound to the 5′ end of each cleaved strand. While the top strand remains in duplex form, approximately half of the bottom strands can be recovered in a single-stranded linear form, referred to as T strands (97). These T strands are thought to represent the transferred form of the T-DNA and are probably formed by displacement during rolling-circle DNA synthesis that initiates from the 3′ ends of each right border. At an early stage of transformation, T strands can be detected in plant cells (124), showing that the T-DNA is transferred in a single-stranded form. T strands are integrated into the host genome at apparently random sites by illegitimate recombination (72) and are stably transmitted to daughter plant cells upon mitotic cell division, and during meiosis and syngamy.
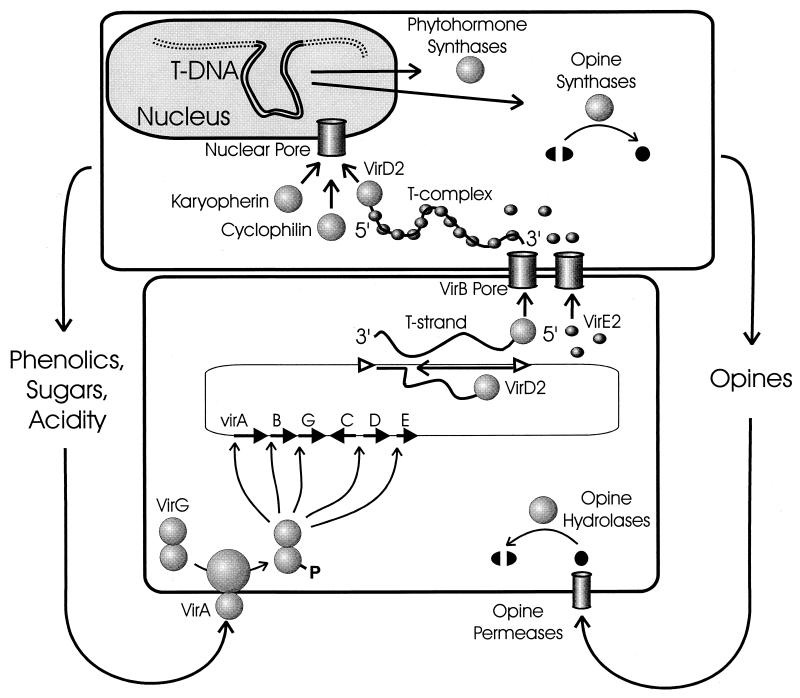
FIG. 2.
Two-way exchange of chemical signals between A. tumefaciens and host plants. Wound-released chemical stimuli are perceived by the VirA to VirG proteins, which leads to transcription of vir promoters. T-DNA is processed by the VirD2 protein, and single-stranded linear T strands are formed by strand displacement. T strands and VirE2 are translocated from the bacteria via a pore encoded by the virB operon and form a T complex within the plant cytoplasm. T complexes are transported into the nucleoplasm via the host protein karyopherin alpha, and the T-DNA is integrated into genomic DNA. Transferred genes encode phytohormone synthases that lead to plant cell proliferation and opine synthases that provide nutrients to the colonizing bacteria. Opines are released from the plant cell, enter the bacteria via dedicated opine permeases, and are catabolized via opine-specific catabolic proteins. Opine permeases and catabolic enzymes are encoded by the Ti plasmid. For the sake of clarity, the relative orientations of vir genes and T-DNA have been inverted.
EXPRESSION AND FUNCTIONS OF TRANSFERRED GENES
Collectively, TL-DNA and TR-DNA encode 13 proteins (Fig. 1, dark green bars). The nontranscribed regions of each transferred gene possess many of the features of plant genes, including typical eukaryotic TATA and CAAT boxes, transcriptional enhancers, and poly(A) addition sites (6). No introns have been reported for any of the A. tumefaciens transferred genes, although at least one T-DNA gene in Agrobacterium rhizogenes contains an intron in its 5′ nontranslated region (71). The coding regions of the T-DNAs have a G+C content of approximately 50%. However, the intergenic regions, especially the 3′ nontranslated regions, are extremely poor in G's and C's, approximately 20 to 30%.
One group of T-DNA genes directs the production of plant growth hormones that are responsible for the proliferation of the transformed plant cells (6). The iaaM and iaaH products direct the conversion of tryptophan via indoleacetamide to indoleacetic acid (auxin). The ipt product condenses isopentenyl pyrophosphate and AMP (6), and host enzymes are presumed to convert the resulting isopentenyl-AMP into the cytokinin zeatin by removal of the phosphoribosyl group and hydroxylation of one methyl group of the isopentenyl moiety. Two other T-DNA genes are thought to play ancillary roles in tumorigenesis. The gene 5 product directs the synthesis of indole-3-lactate, an antagonistic auxin analogue (57), while tml (also designated gene 6b) increases the sensitivity of plant cells to phytohormones by a mechanism that remains to be discovered (108). This gene can provoke tumors in certain host plants in the absence of the other oncogenes (42).
A second set of transferred genes directs the production of bacterial nutrients called opines. Octopine-type Ti plasmids direct their hosts to synthesize at least eight opines. The ocs gene encodes octopine synthase, which reductively condenses pyruvate with either arginine, lysine, histidine, or ornithine to produce octopine, lysopine, histopine, or octopinic acid, respectively, all of which can be detected in crown gall tumors (24). The mas2′ product is thought to condense glutamine or glutamic acid with glucose (although this has not been experimentally demonstrated), while the mas1′ product reduces these intermediates, forming mannopine and mannopinic acid, respectively. The ags product catalyzes the lactonization of mannopine to form agropine. Mannopine and agropine also can spontaneously lactamize to form agropinic acid (24). Thus, tumors induced by strains harboring octopine-type Ti plasmids can produce as many as four members of the octopine family and four members of the mannityl opine family.
TI PLASMID-ENCODED PROTEINS REQUIRED FOR T-DNA PROCESSING AND TRANSFER
Proteins responsible for T-DNA processing and transfer are encoded by the vir region of the Ti plasmid. Twenty genes in this region are essential for wild-type levels of pathogenesis on most host plants and are expressed in six operons, virA, -B, -C, -D, -E, and -G. The proteins required for border cleavage are encoded by virD1 and virD2, with the VirD2 protein remaining covalently bound to the 5′ end of the T-strands (98, 123). Purified VirD2 cleaves single-stranded oligonucleotides containing border sequences at the same site, creating a covalent bond between the 5′ phosphate and tyrosine 29 (86). This reaction is fully reversible, indicating that the DNA-protein phosphodiester linkage is a high-energy bond and suggesting that a reverse reaction might be important for the integration of T-DNA into the plant genome (109). VirD2 alone was not able to cleave the same sequence in double-stranded form but was able to do so in the presence of VirD1 (90). VirC1 binds to the overdrive site, which lies adjacent to the left border (111). VirC1 and VirC2 are not required for T-region processing but are required for efficient T-strand transfer into most host plants, suggesting that they play a role in T-strand export.
The T-DNA transfer apparatus is encoded by the virB operon, which contains 11 genes (11). Each except VirB1 is essential for tumorigenesis (5). All 10 essential proteins have been localized to the inner or outer membrane, and most appear either to be integral membrane proteins or to be exported from the cytoplasm (107). Two VirB proteins, VirB4 and VirB11, are peripherally bound to the others and located primarily in the cytoplasm, although a small part of VirB4 may span the inner membrane (19). VirB4 and VirB11 have ATPase activity and are thought to provide the energy required for export of other protein subunits, for T-strand transport, or both (12, 93). VirB proteins direct the production of pili that resemble conjugative pili (31), and VirB2 is the major subunit of these pili (58). VirB2 is processed to a 7.2-kDa product that is cyclized such that the amino terminus is linked to the carboxyl terminus via an amide bond (26). Cyclization does not require any Ti plasmid-encoded proteins but does not occur in Escherichia coli, suggesting that this reaction requires a protein encoded elsewhere in the A. tumefaciens genome. VirB7 may help to anchor this pilus to the bacterial cell, as it is an outer membrane lipoprotein that forms disulfide bonds with the periplasmically localized VirB9 (2, 96). The VirB mating bridge is thought to be coupled to the T-strand complex by the VirD4 protein, which is located in the inner membrane and is absolutely required for transfer (67, 82). VirB1 possesses sequence motifs found in bacterial transglycosylases and eukaryotic lysozymes, suggesting a role in the localized digestion of the peptidoglycan (78).
The VirB apparatus delivers T strands to the plant cell cytoplasm, where additional steps are required to transport this DNA to the nucleoplasm and to integrate it into host DNA. The carboxyl terminus of VirD2 contains a nuclear localization signal that is thought to guide nuclear targeting by interacting with the karyopherin alpha and cyclophilin proteins (3, 23, 44). The VirE2 protein appears also to play a role in nuclear import. This protein binds tightly and cooperatively to single-stranded nucleic acids, forming coiled, cylindrical filaments (14). Like VirD2, VirE2 contains nuclear localization sites that mediate transport of the T-DNA from the cytoplasm to the nucleoplasm (15).
Transgenic plants expressing VirE2 can be transformed by virE2 mutants of A. tumefaciens, indicating that VirE2 is required only in plant cells (15). Similar data have been obtained for another protein, VirF, which is required for tumorigenesis on such plants as tomato and Nicotiana glauca (87). Mutations in either virE2 or virF can be complemented extracellularly, that is, by coinfection with a helper strain possessing the vir region but lacking an oncogenic T-DNA (73, 84). Initially, it was thought that such complexes were formed within the bacterium, but more recent genetic evidence suggests that VirE2 and T-DNA are transferred separately and form complexes in the plant cell cytoplasm (101). Transfer of VirE2 requires VirE1, while transfer of T-DNA does not, suggesting that VirE1 acts as an export chaperone for VirE2 (22, 101). Conversely, transfer of T-DNA requires VirC1 and VirC2 while transfer of VirE2 does not require either protein (13). These studies provide the best evidence that T strands and VirE2 are transferred independently, although biochemical evidence addressing this hypothesis will await future studies. These data indicate that the virB-encoded transfer system, in addition to transferring T-DNA, can carry out contact-dependent translocation of at least three proteins, VirD2, VirE2, and VirF. This property of protein transport is highly reminiscent of the family of type III protein translocation systems of plant and animal pathogens, although these systems have independent ancestries (18, 37).
Many aspects of T-DNA transfer resemble interbacterial conjugal transfer of plasmid DNA (63). In both processes, transfer is initiated by single-stranded scissions at specific cis-acting sites. Moreover, the protein that catalyzes the scission remains bound to the 5′ end of the cleaved strand, and in both cases, DNA is transferred in a single-stranded form. The most direct evidence that the T-DNA transfer apparatus evolved from a conjugal transfer system is the extensive sequence similarities between Vir proteins and certain Tra proteins. For example, all 11 VirB proteins resemble the mating pair formation (Mpf) subset of Tra proteins encoded by the IncN plasmid pKM101 and show a lower degree of similarity to the Tra proteins of IncW, IncP, and IncF plasmids (48, 62). Similarly, the VirD1, VirD2, and VirD4 proteins resemble the donor transfer and replication (Dtr) subset of Tra proteins. In fact, the virB and virD operons together would constitute a complete set of conjugation proteins. The T-DNA border resembles the oriT sites of IncP plasmids, and nicking occurs at identical positions in the two transfer systems (117). The gene family of Vir and Tra proteins also includes the Ptl proteins of Bordetella pertussis, which direct the export of pertussis toxin; the VirB proteins of Brucella spp., which are required for intracellular survival; and other protein translocators of bacterial pathogens, collectively referred to as type IV export systems (18). The VirC and VirE proteins do not significantly resemble known transfer proteins, and VirC1 resembles a plasmid partitioning protein (38).
Some members of the vir regulon are not essential for tumorigenesis on all hosts and may be required only in specific hosts or may play other roles in pathogenesis. These include virD5, -E3, -F, -H, -J, -K, -L, -M, P, and -R (50, 52). However, the lack of an apparent role in tumorigenicity could be a consequence of functional redundancy. For example, virJ is essential for tumorigenicity, but only in the absence of the homologous chromosomal gene acvB (49). The virH operon consists of two genes whose products resemble the family of P450 monooxygenases (51). VirH2 chemically modifies certain phenolic vir gene inducers by O demethylation, converting them to noninducers (51). For example, the inducer ferrulic acid is O demethylated to create the noninducer caffeic acid. This finding suggests that VirH2 acts as a regulatory governor.
UPTAKE AND CATABOLISM OF OPINES
As described above, several T-DNA-encoded genes direct the synthesis of opines, which serve the bacteria as nutrient sources. Over 40 genes are devoted to opine uptake and catabolism. These include no fewer than six ATP binding cassette-type permeases (Fig. 1, dark blue bars) and 12 opine catabolic enzymes (light blue bars), whose functions are summarized in Table 1. These opine permeases are only distantly related to each other, suggesting that they were adapted from diverse sources. An additional gene (mclA) could encode a protein that resembles methyl-accepting chemoreceptors. A. tumefaciens strains are chemotactic toward opines, and chemotaxis requires the cognate periplasmic opine binding proteins (each a component of an opine uptake system) but does not require Ti plasmid-encoded methyl-accepting chemotaxis proteins (53). It seems likely that this is another example of redundancy in which these periplasmic binding proteins can interact either with chromosomally encoded or with Ti plasmid-encoded methyl-accepting chemotaxis proteins.
Characteristically, Ti plasmids code only for the opine catabolism systems that correspond to the set of opine biosynthesis genes located in the T regions. This presents the interesting problem of how these paired gene systems arise and how they remain grouped together despite the fact that they are located in different segments of the plasmid. Sequence analysis of the mannopine-agropine catabolic loci indicated that certain of these genes resemble the cognate opine biosynthetic genes. The catabolic protein AgcA, which interconverts mannopine and agropine, resembles the agropine synthase protein Ags, which carries out the same reaction. In fact, ags can complement an agcA mutant for catabolism of agropine (40). Similarly, MocC and MocD, which together degrade mannopine, resemble Mas1′ and Mas2′, which synthesize mannopine (54). Based on these comparisons, we have suggested that the T-region genes coding for mannityl opine synthesis by the transformed plant cells arose by gene duplication from bacterial genes required for catabolism of these or closely related substrates (54). However, not all opine synthases resemble their corresponding catabolic enzymes. For example, the octopine and nopaline synthases do not resemble their cognate catabolic enzymes.
REPLICATION FUNCTIONS
A DNA fragment containing just repA, repB, and repC provides all functions required for stable replication in A. tumefaciens (104). Only repC is critical for vegetative replication, while repA or repB is required for stable plasmid inheritance. RepA and RepB resemble a family of plasmid partitioning systems that are thought to ensure that during cell division each daughter cell inherits at least one copy of the plasmid. All three genes resemble replication genes of other large, low-copy-number plasmids present in members of the family Rhizobiaceae (64). Incompatibility functions also are determined by the DNA fragment containing repABC (56). The octopine-type Ti plasmid is incompatible with nopaline-type Ti plasmids (41), but in spite of the relatedness of their replicators, the octopine Ti plasmid is compatible with Ri plasmids (17).
INTERBACTERIAL CONJUGATION OF TI PLASMIDS
The octopine Ti plasmid is capable of interbacterial conjugation (28) and contains a complete transfer system (Fig. 1, purple bars). On the basis of similarity to other conjugation systems, the cluster of tra genes probably is required for DNA transfer and replication, while the trb gene cluster is probably required for mating pair formation and could direct the synthesis of conjugal pili. In the closely related conjugation system of pTiC58, traB is not essential for transfer, although it is required for maximal efficiency, while traH is not required for efficient transfer (29) and is here designated a tra gene simply because it lies in the tra regulon (see below). All other tra and trb genes of pTiC58 are known or thought to be required for efficient conjugation (29, 65), with the exception of trbK, which is probably not required for conjugation but may mediate entry exclusion (28). The three operons of the conjugal transfer system are strongly conserved among all of the Ti plasmids analyzed to date (although one report [103] claimed otherwise, that study was based upon an incorrect DNA sequence). These genes also resemble the tra genes of at least one symbiosis megaplasmid, pNGR234a of Rhizobium sp. strain NGR234 (30). The Tra system functions independently of the T-DNA transfer system described above (16).
The tra genes appear to have diverse origins. TraG, TraF, and all 11 Trb proteins closely resemble IncP-type Tra proteins. In contrast, TraA, the putative nickase-helicase of this system, does not closely resemble any IncP-type Tra protein. Instead, the amino-terminal domain of TraA, which should contain oriT nicking activity, resembles the strand transferase of the IncQ plasmid RSF1010, while its carboxyl-terminal domain, which contains a possible helicase, resembles Tra proteins of IncN, IncW, and IncF plasmids. The oriT also resembles the corresponding site in RSF1010. Interestingly, the Vir system seems also to have chimeric origins, as all 11 VirB proteins resemble IncN Tra proteins, while two VirD proteins, VirD2 and VirD4, resemble IncP-type Tra proteins. The T-DNA borders resemble the oriT site of IncP plasmids (117). In all cases, sequence similarities between Ti plasmid Tra proteins and corresponding Vir proteins are relatively weak.
REGULATED EXPRESSION OF TI PLASMID-ENCODED GENES
Virtually all of the genes described above are tightly regulated by proteins that also are encoded on the Ti plasmid (Fig. 1, orange bars). For example, the vir regulon is coordinately induced in response to host-released phenolic compounds in combination with monosaccharides and extracellular acidity in the range of pH 5.0 to 5.5 (47). This acidity may be necessary to protonate phenolic compounds, which would increase their membrane permeability. These chemical stimuli are detected by the transmembrane two-component sensor kinase VirA, which phosphorylates the response regulator VirG. Phospho-VirG positively regulates all vir promoters, including those of virA and virG, which results in positive autoregulation of this regulon (100, 119).
VirA contains four functional domains, designated the periplasmic, linker, kinase, and receiver domains (9), and exists as a dimer both in the presence and in the absence of inducing stimuli (85). The periplasmic domain is required for detection of a sugar binding protein called ChvE (8, 92), while the linker domain is required for detection of phenolic compounds, and the receiver plays an inhibitory role in vir gene expression (9). VirA can undergo autophosphorylation in vitro and transfers its phosphoryl group to Asp52 of VirG (46). The carboxyl-terminal domain of VirG binds to sequences called vir boxes that are found near all VirG-regulated promoters (88, 118). While there is still some controversy about whether phenolic inducers bind directly to VirA (59), genetic evidence suggests that this is so, since virA genes from different strains of A. tumefaciens, when introduced into an isogenic background, encode proteins that are stimulated by different types of inducers (60).
All opine uptake and catabolic systems are induced by their cognate substrates. For example, octopine induces transcription of a 14-kb occQ-traR operon via the OccR protein, a LysR-type regulator. OccR binds to its binding site, which lies directly upstream of the occQ promoter, in the presence or absence of octopine but undergoes a conformational change in response to octopine (115). The mannopine and agropine permeases and catabolic enzymes also are induced by the cognate opines, probably via the MocR protein, which resembles the LacI repressor of E. coli. Similarly, regulated expression of the aga and moa genes by the cognate opines requires the MoaR repressor, which resembles yet another family of regulators, including the galacticol repressor of E. coli (70). Expression of the opine catabolism gene sets also is influenced by global control systems. Transcription of the mannityl opine catabolism genes, while inducible by their cognate substrates, also is controlled by catabolite repression (39, 127), since these genes are not induced by mannopine or agropine when favored carbon sources such as glutamate or succinate also are provided. Furthermore, these catabolic genes are part of the nitrogen assimilation regulon, since catabolite repression by succinate is not observed when mannopine is provided as the sole source of nitrogen (39).
The TraR-TraI system positively controls expression of the tra and trb operons (Fig. 1, purple bars). Transcription of this regulon is controlled by a regulatory cascade that is initiated by octopine acting through OccR, which leads to expression of traR (33). TraR in turn is a direct positive regulator of the tra and trb genes (36). TraR is a member of the LuxR family of quorum-sensing transcriptional regulators (35), and its activity requires N-3-oxooctanoyl-l-homoserine lactone (126). Synthesis of this compound, called an autoinducer, is directed by the TraI protein, which utilizes 3-oxooctanoyl-acyl carrier protein and S-adenosylmethionine as substrates (36, 77). This compound is synthesized in the bacterial cytoplasm but diffuses across the cell envelope and acts as a bacterial pheromone, providing a mechanism for the bacteria to estimate their population densities (35). Since the Ti plasmid encodes both TraI and TraR, each conjugal donor takes a census of other donors rather than of recipients (34). Purified TraR binds one molecule of this compound per protein monomer and binds directly to dyad symmetrical DNA sequences called tra boxes, which are found directly upstream of the traA, traC, and traI promoters (34, 128). TraR stimulates transcription of tra promoters in vitro on supercoiled templates but is largely inactive on linear templates (128). DNA binding by TraR requires the autoinducer (68).
TraR activity is antagonized by two proteins encoded by the traM and trlR genes. Interestingly, the traM gene is positively regulated by TraR, thereby creating a negative autoregulatory loop (32). TraM is an antiactivator and directly interacts with the carboxyl terminus of TraR (45, 69). This interaction rapidly inhibits TraR activity and disrupts TraR-DNA complexes (69). TrlR is very closely related to TraR in its autoinducer binding domain but lacks a DNA binding domain (80, 127) and is thought to form inactive heterodimers with TraR. The trlR gene is positively regulated by mannopine. Consistent with this, mannopine inhibits tra gene expression, while inhibition is abolished by a trlR mutation (80, 127).
INSERTION SEQUENCES (ISS) AND UNCHARACTERIZED ORFS
A number of possible ISs are present on the Ti plasmid (Fig. 1, black bars), although transposition of these elements has not been detected experimentally. Three of these resemble IS66, which was originally found inserted in the iaaH gene of a strain of an A. tumefaciens mutant that causes shooty teratomas rather than tumors. However, several of these IS66-like elements are considerably shorter than IS66, suggesting that they may be defective remnants of the original element. The other possible IS-like elements resemble a wide variety of IS elements in many eubacteria.
While most of the genes in the Ti plasmid have been ascribed functions, a contiguous 24-kb region (coordinates 112 to 136) contains 25 ORFs that have no known function (Fig. 1, grey bars). Most of the ORFs in this region are at least 100 codons in length and have moderately strong translation initiation motifs, and many of these ORFs appear to be translationally coupled to adjacent ORFs. All of these considerations suggest that these ORFs are expressed genes, but the functions of their products are at present unknown. Approximately half of these genes resemble genes identified in genome sequencing projects, though none of these homologous genes has been characterized genetically or biochemically.
RELATION OF THE OCTOPINE-TYPE TI PLASMIDS TO OTHER PLASMIDS OF THE RHIZOBIACEAE
The modular structure of this Ti plasmid is entirely in keeping with the model presented by Otten and colleagues in which these elements evolve by IS-mediated intramolecular rearrangements and by recombination with other plasmids (83). Signs of such recombination events are scattered over the length of this plasmid. For example, trlR may well have arisen by a recombination event that fused the mannityl opine catabolism region with its attendant mannopine-regulated traR allele to the region just upstream of the octopine catabolism locus (80, 127). Similarly, the structural and regulatory association of the functional traR allele with the occ operon arose from a fortuitous recombination event that fused traR to occ. Interestingly, such associations of traR with various opine catabolism operons are a consistent feature of Ti plasmids (28, 33, 80, 127).
Despite this plasticity, certain gene associations seem to be strongly conserved among these elements. Most notably, the repABC complex is tightly linked with the trb operon in all Ti and opine catabolism plasmids that have been examined to date (pTiC58, pTi-SAKURA, and pAtK84b) (64). This conservation extends to at least four plasmids present in members of the genus Rhizobium (64), suggesting that this linkage is strongly selected. Consistent with this interpretation, the intergenic region separating the two divergently oriented gene systems on many of these plasmids, including all Ti plasmids, contains two copies of the tra box sequence. Coupled with the recent observation that copy number of the nopaline-type Ti plasmid is positively enhanced by TraR in a quorum-dependent fashion (64), it is reasonable to conclude that conjugal transfer and plasmid replication are functionally linked.
CONCLUSIONS
As described above, most of the genes of the Ti plasmid play direct or indirect roles in some aspect of tumorigenesis or tumor colonization. We understand the roles of most of the T-DNA-encoded genes, although the functions of some remain mysterious. We have some insights about the processing and transfer of the T-DNA, although our understanding of the VirB-encoded pore is rudimentary, as are the steps involved in nuclear transport and integration. At least three Vir proteins are thought to be transferred from the bacterium into plant cells during infection, though the physical detection of these proteins in plant cells remains a goal for future studies. It will be interesting to identify any additional translocated proteins and to elucidate their functions. VirA and VirG remain important paradigms for host detection, and the multidomain structure of VirA remains fertile ground for future work. Future studies will decide once and for all whether VirA binds phenolic inducers directly or through an accessory phenolic binding protein. Of the 34 known VirG-regulated genes, one-third do not seem essential for tumorigenesis (at least on certain host plants), suggesting that plant-released vir-inducing signals elicit multiple bacterial responses that remain to be described.
Another challenge lies in comparative analysis of the many different Ti plasmids that have been isolated, as well as other plasmids found in members of the Rhizobiaceae. We know that approximately 65 kb of octopine-type plasmids are conserved in the nopaline-type Ti plasmid pTiC58 (Fig. 1, crosshatched boxes), including part of the T-DNA and the tra, trb, rep, and vir regions (27), while the remaining 130 kb are not conserved. As more Ti plasmids and related plasmids are characterized (102), it will be possible to refine our insights about the evolution of these genetic elements.
The use of A. tumefaciens to create transgenic plants has become routine for many dicots as well as for some monocots, and yet new insights about fundamental aspects of Agrobacterium-plant interactions will lead to improved technologies in plant transformation. Future work will lead, for example, to further expansion of the organism's host range, to new approaches to transferring extremely long fragments of DNA, and to new approaches to using T-DNA to disrupt plant genes.
It is striking that such a large portion of the Ti plasmid is devoted to opine uptake and catabolism, and few of these systems have been studied in any depth. Studies of opine chemotaxis, uptake, and catabolism will continue. In addition, one challenge for the next 10 years will be to apply these insights about opines to agriculture. Several reports have already appeared showing that bacteria that utilize a particular opine enjoy a competitive advantage in colonizing transgenic plants that produce the same opine (81, 89). We suspect that this technology may revolutionize efforts to foster beneficial plant-microbe associations.
ACKNOWLEDGMENTS
We thank our many colleagues for their encouragement and their willingness to share unpublished data, insights, and ideas, which has helped to make this field so rewarding.
Research in our laboratories is supported by NIH grant GM42893 (S.C.W.), NSF grant MCB-9904917 (S.C.W.), NIH grant GM52465 (S.K.F.), USDA grant AG92-3312-8231 (S.K.F.), and C-FAR grant 99K-059-4 (S.K.F.) and by the Netherlands Foundations of Biological Research and Chemical Research and by the Technology Foundation (P.J.J.H.).
REFERENCES
- 1.Alt-Mörbe J, Stryker J L, Fuqua C, Li P-L, Farrand S K, Winans S C. The conjugal transfer system of Agrobacterium tumefaciens octopine-type Ti plasmids is closely related to the transfer system of an IncP plasmid and distantly related to Ti plasmid vir genes. J Bacteriol. 1996;178:4248–4257. doi: 10.1128/jb.178.14.4248-4257.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson L B, Hertzel A V, Das A. Agrobacterium tumefaciens VirB7 and VirB9 form a disulfide-linked protein complex. Proc Natl Acad Sci USA. 1996;93:8889–8894. doi: 10.1073/pnas.93.17.8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballas N, Citovsky V. Nuclear localization signal binding protein from Arabidopsis mediates nuclear import of Agrobacterium VirD2 protein. Proc Natl Acad Sci USA. 1997;30:10723–10728. doi: 10.1073/pnas.94.20.10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barker R F, Idler K B, Thompson D V, Kemp J D. Nucleotide sequence of the T-DNA region from the Agrobacterium tumefaciens octopine Ti plasmid pTi15955. Plant Mol Biol. 1983;2:335–350. doi: 10.1007/BF01578595. [DOI] [PubMed] [Google Scholar]
- 5.Berger B R, Christie P J. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J Bacteriol. 1994;176:3646–3660. doi: 10.1128/jb.176.12.3646-3660.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Binns A N, Castantino P. The Agrobacterium oncogenes. In: Spaink H P, Kondorosi A, Hooykaas P J J, editors. The Rhizobiaceae: molecular biology of model plant-associated bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 251–266. [Google Scholar]
- 7.Braun A C. A physiological basis for autonomous growth of the crown-gall tumor cell. Proc Natl Acad Sci USA. 1958;44:344–349. doi: 10.1073/pnas.44.4.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cangelosi G A, Ankenbauer R G, Nester E W. Sugars induce the Agrobacterium virulence genes through a periplasmic binding protein and a transmembrane signal protein. Proc Natl Acad Sci USA. 1990;87:6708–6712. doi: 10.1073/pnas.87.17.6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang C H, Winans S C. Functional roles assigned to the periplasmic, linker, and receiver domains of the Agrobacterium tumefaciens VirA protein. J Bacteriol. 1992;174:7033–7039. doi: 10.1128/jb.174.21.7033-7039.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chilton M-D, Drummond M H, Merlo D J, Sciaky D, Montoya A L, Gordon M P, Nester E W. Stable incorporation of plasmid DNA into higher plant cells: the molecular basis of crown gall tumorigenesis. Cell. 1977;11:263–271. doi: 10.1016/0092-8674(77)90043-5. [DOI] [PubMed] [Google Scholar]
- 11.Christie P J. Agrobacterium tumefaciens T-complex transport apparatus: a paradigm for a new family of multifunctional transporters in eubacteria. J Bacteriol. 1997;179:3085–3094. doi: 10.1128/jb.179.10.3085-3094.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christie P J, Ward J E, Jr, Gordon M P, Nester E W. A gene required for transfer of T-DNA to plants encodes an ATPase with autophosphorylating activity. Proc Natl Acad Sci USA. 1989;86:9677–9681. doi: 10.1073/pnas.86.24.9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christie P J, Ward J E, Winans S C, Nester E W. The Agrobacterium tumefaciens virE2 gene product is a single-stranded-DNA-binding protein that associates with T-DNA. J Bacteriol. 1988;170:2659–2667. doi: 10.1128/jb.170.6.2659-2667.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Citovsky V, Guralnick B, Simon M N, Wall J S. The molecular structure of Agrobacterium VirE2-single stranded DNA complexes involved in nuclear import. J Mol Biol. 1997;5:718–727. doi: 10.1006/jmbi.1997.1230. [DOI] [PubMed] [Google Scholar]
- 15.Citovsky V, Zupan J, Warnick D, Zambryski P. Nuclear localization of Agrobacterium VirE2 protein in plant cells. Science. 1992;256:1802–1805. doi: 10.1126/science.1615325. [DOI] [PubMed] [Google Scholar]
- 16.Cook D M, Li P-L, Ruchaud F, Padden S, Farrand S K. Ti plasmid conjugation is independent of vir: reconstitution of the tra functions from pTiC58 as a binary system. J Bacteriol. 1997;179:1291–1297. doi: 10.1128/jb.179.4.1291-1297.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Costantino P, Hooykaas P J, den Dulk-Ras H, Schilperoort R A. Tumor formation and rhizogenicity of Agrobacterium rhizogenes carrying Ti plasmids. Gene. 1980;11:79–87. doi: 10.1016/0378-1119(80)90088-8. [DOI] [PubMed] [Google Scholar]
- 18.Covacci A, Telford J L, Del Giudice G, Parsonnet J, Rappuoli R. Helicobacter pylori virulence and genetic geography. Science. 1999;284:1328–1333. doi: 10.1126/science.284.5418.1328. [DOI] [PubMed] [Google Scholar]
- 19.Dang T A, Christie P J. The VirB4 ATPase of Agrobacterium tumefaciens is a cytoplasmic membrane protein exposed at the periplasmic surface. J Bacteriol. 1997;179:453–462. doi: 10.1128/jb.179.2.453-462.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Greve H, Decraemer H, Seurinck J, Van Montagu M, Schell J. The functional organization of the octopine Agrobacterium tumefaciens plasmid pTiB6S3. Plasmid. 1981;6:235–248. doi: 10.1016/0147-619x(81)90069-x. [DOI] [PubMed] [Google Scholar]
- 21.De Greve H, Dhaese P, Seurinck J, Lemmers M, Van Montagu M, Schell J. Nucleotide sequence and transcript map of the Agrobacterium tumefaciens Ti plasmid-encoded octopine synthase gene. J Mol Appl Genet. 1982;1:499–511. [PubMed] [Google Scholar]
- 22.Deng W, Chen L, Peng W T, Liang X, Sekiguchi S, Gordon M P, Comai L, Nester E W. VirE1 is a specific molecular chaperone for the exported single-stranded-DNA-binding protein VirE2 in Agrobacterium. Mol Microbiol. 1999;31:1795–1807. doi: 10.1046/j.1365-2958.1999.01316.x. [DOI] [PubMed] [Google Scholar]
- 23.Deng W, Chen L, Wood D W, Metcalfe T, Liang X, Gordon M P, Comai L, Nester E W. Agrobacterium VirD2 protein interacts with plant host cyclophilins. Proc Natl Acad Sci USA. 1998;95:7040–7045. doi: 10.1073/pnas.95.12.7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dessaux Y, Petit A, Farrand S K, Murphy P J. Opines and opine-like molecules involved in plant-Rhizobiaceae interactions. In: Spaink H P, Kondorosi A, Hooykaas P J J, editors. The Rhizobiaceae: molecular biology of model plant-associated bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 173–197. [Google Scholar]
- 25.De Vos G, De Beuckeleer M, Van Montagu M, Schell J. Restriction endonuclease mapping of the octopine tumor-inducing plasmid pTiAch5 of Agrobacterium tumefaciens. Plasmid. 1981;6:249–253. doi: 10.1016/0147-619x(81)90070-6. [DOI] [PubMed] [Google Scholar]
- 26.Eisenbrandt R, Kalkum M, Lai E M, Lurz R, Kado C I, Lanka E. Conjugative pili of IncP plasmids, and the Ti plasmid T pilus are composed of cyclic subunits. J Biol Chem. 1999;274:22548–22555. doi: 10.1074/jbc.274.32.22548. [DOI] [PubMed] [Google Scholar]
- 27.Engler G, Depicker A, Maenhaut R, Villarroel R, Van Montagu M, Schell J. Physical mapping of DNA base sequence homologies between an octopine and a nopaline Ti plasmid of Agrobacterium tumefaciens. J Mol Biol. 1981;152:183–208. doi: 10.1016/0022-2836(81)90239-4. [DOI] [PubMed] [Google Scholar]
- 28.Farrand S K. Conjugal plasmids and their transfer. In: Spaink H P, Kondorosi A, Hooykaas P J J, editors. The Rhizobiaceae: molecular biology of model plant-associated bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 199–233. [Google Scholar]
- 29.Farrand S K, Hwang I, Cook D M. The tra region of the nopaline-type Ti plasmid is a chimera with elements related to the transfer systems of RSF1010, RP4, and F. J Bacteriol. 1996;178:4233–4247. doi: 10.1128/jb.178.14.4233-4247.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freiberg C, Fellay R, Bairoch A, Broughton W J, Rosenthal A, Perret X. Molecular basis of symbiosis between Rhizobium and legumes. Nature. 1997;387:394–401. doi: 10.1038/387394a0. [DOI] [PubMed] [Google Scholar]
- 31.Fullner K J, Lara J C, Nester E W. Pilus assembly by Agrobacterium T-DNA transfer genes. Science. 1996;273:1107–1109. doi: 10.1126/science.273.5278.1107. [DOI] [PubMed] [Google Scholar]
- 32.Fuqua C, Burbea M, Winans S C. Activity of the Agrobacterium Ti plasmid conjugal transfer regulator TraR is inhibited by the product of the traM gene. J Bacteriol. 1995;177:1367–1373. doi: 10.1128/jb.177.5.1367-1373.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fuqua C, Winans S C. Localization of OccR-activated and TraR-activated promoters that express two ABC-type permeases and the traR gene of Ti plasmid pTiR10. Mol Microbiol. 1996;20:1199–1210. doi: 10.1111/j.1365-2958.1996.tb02640.x. [DOI] [PubMed] [Google Scholar]
- 34.Fuqua C, Winans S C. Conserved cis-acting promoter elements are required for density-dependent transcription of Agrobacterium tumefaciens conjugal transfer genes. J Bacteriol. 1996;178:435–440. doi: 10.1128/jb.178.2.435-440.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fuqua C, Winans S C, Greenberg E P. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 36.Fuqua W C, Winans S C. A LuxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J Bacteriol. 1994;176:2796–2806. doi: 10.1128/jb.176.10.2796-2806.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Galan J E, Collmer A. Type III secretion machines: bacterial devices for protein delivery into host cells. Science. 1999;284:1322–1328. doi: 10.1126/science.284.5418.1322. [DOI] [PubMed] [Google Scholar]
- 38.Gallie D R, Kado C I. Agrobacterium tumefaciens pTAR parA promoter region involved in autoregulation, incompatibility and plasmid partitioning. J Mol Biol. 1987;193:465–478. doi: 10.1016/0022-2836(87)90260-9. [DOI] [PubMed] [Google Scholar]
- 39.Hong S B, Dessaux Y, Chilton W S, Farrand S K. Organization and regulation of the mannopine cyclase-associated opine catabolism genes in Agrobacterium tumefaciens 15955. J Bacteriol. 1993;175:401–410. doi: 10.1128/jb.175.2.401-410.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hong S B, Hwang I, Dessaux Y, Guyon P, Kim K S, Farrand S K. A T-DNA gene required for agropine biosynthesis by transformed plants is functionally and evolutionarily related to a Ti plasmid gene required for catabolism of agropine by Agrobacterium strains. J Bacteriol. 1997;179:4831–4840. doi: 10.1128/jb.179.15.4831-4840.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hooykaas P J, den Dulk-Ras H, Ooms G, Schilperoort R A. Interactions between octopine and nopaline plasmids in Agrobacterium tumefaciens. J Bacteriol. 1980;143:1295–1306. doi: 10.1128/jb.143.3.1295-1306.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hooykaas P J J, den Dulk-Ras H, Schilperoort R A. The Agrobacterium tumefaciens T-DNA gene 6b is an onc gene. Plant Mol Biol. 1988;11:791–794. doi: 10.1007/BF00019519. [DOI] [PubMed] [Google Scholar]
- 43.Hooykaas P J J, Klapwijk P M, Nuti M P, Schilperoort R A, Rorsch A. Transfer of the Agrobacterium tumefaciens Ti plasmid to avirulent Agrobacteria and to Rhizobium ex planta. J Gen Microbiol. 1977;98:477–484. [Google Scholar]
- 44.Howard E A, Zupan J R, Citovsky V, Zambryski P C. The VirD2 protein of A. tumefaciens contains a C-terminal bipartite nuclear localization signal: implications for nuclear uptake of DNA in plant cells. Cell. 1992;68:109–118. doi: 10.1016/0092-8674(92)90210-4. [DOI] [PubMed] [Google Scholar]
- 45.Hwang I, Smyth A J, Luo Z Q, Farrand S K. Modulating quorum sensing by antiactivation: TraM interacts with TraR to inhibit activation of Ti plasmid conjugal transfer genes. Mol Microbiol. 1999;34:282–294. doi: 10.1046/j.1365-2958.1999.01595.x. [DOI] [PubMed] [Google Scholar]
- 46.Jin S G, Prusti R K, Roitsch T, Ankenbauer R G, Nester E W. Phosphorylation of the VirG protein of Agrobacterium tumefaciens by the autophosphorylated VirA protein: essential role in biological activity of VirG. J Bacteriol. 1990;172:4945–4950. doi: 10.1128/jb.172.9.4945-4950.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson T M, Das A. Organization and regulation of expression of the Agrobacterium virulence genes. In: Spaink H P, Kondorosi A, Hooykaas P J J, editors. The Rhizobiaceae: molecular biology of model plant-associated bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 265–279. [Google Scholar]
- 48.Kado C I. Promiscuous DNA transfer system of Agrobacterium tumefaciens: role of the virB operon in sex pilus assembly and synthesis. Mol Microbiol. 1994;12:17–22. doi: 10.1111/j.1365-2958.1994.tb00990.x. [DOI] [PubMed] [Google Scholar]
- 49.Kalogeraki V S, Winans S C. The octopine-type Ti plasmid pTiA6 of Agrobacterium tumefaciens contains a gene homologous to the chromosomal virulence gene acvB. J Bacteriol. 1995;177:892–897. doi: 10.1128/jb.177.4.892-897.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kalogeraki V S, Winans S C. Wound-released chemical signals may elicit multiple responses from an Agrobacterium tumefaciens strain containing an octopine-type Ti plasmid. J Bacteriol. 1998;180:5660–5667. doi: 10.1128/jb.180.21.5660-5667.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kalogeraki V S, Zhu J, Eberhard A, Madsen E L, Winans S C. The phenolic vir gene inducer ferulic acid is O-demethylated by the VirH2 protein of an Agrobacterium tumefaciens Ti plasmid. Mol Microbiol. 1999;34:512–522. doi: 10.1046/j.1365-2958.1999.01617.x. [DOI] [PubMed] [Google Scholar]
- 52.Kalogeraki V S, Zhu J, Stryker J L, Winans S C. The right end of the vir region of an octopine-type Ti plasmid contains four new members of the vir regulon that are not essential for pathogenesis. J Bacteriol. 2000;182:1774–1778. doi: 10.1128/jb.182.6.1774-1778.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim H, Farrand S K. Opine catabolic loci from Agrobacterium plasmids confer chemotaxis to their cognate substrates. Mol Plant-Microbe Interact. 1998;11:131–143. doi: 10.1094/MPMI.1998.11.2.131. [DOI] [PubMed] [Google Scholar]
- 54.Kim K S, Farrand S K. Ti plasmid-encoded genes responsible for catabolism of the crown gall opine mannopine by Agrobacterium tumefaciens are homologs of the T-region genes responsible for synthesis of this opine by the plant tumor. J Bacteriol. 1996;178:3275–3284. doi: 10.1128/jb.178.11.3275-3284.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klee H, Montoya A, Horodyski F, Lichtenstein C, Garfinkel D, Fuller S, Flores C, Peschon J, Nester E, Gordon M. Nucleotide sequence of the tms genes of the pTiA6NC octopine Ti plasmid: two gene products involved in plant tumorigenesis. Proc Natl Acad Sci USA. 1984;81:1728–1732. doi: 10.1073/pnas.81.6.1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koekman B P, Hooykaas P J, Schilperoort R A. A functional map of the replicator region of the octopine Ti plasmid. Plasmid. 1982;7:119–132. doi: 10.1016/0147-619x(82)90072-5. [DOI] [PubMed] [Google Scholar]
- 57.Korber H, Strizhov N, Staiger D, Feldwisch J, Olsson O, Sandberg G, Palme K, Schell J, Koncz C. T-DNA gene 5 of Agrobacterium modulates auxin response by autoregulated synthesis of a growth hormone antagonist in plants. EMBO J. 1991;10:3983–3991. doi: 10.1002/j.1460-2075.1991.tb04973.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lai E M, Kado C I. Processed VirB2 is the major subunit of the promiscuous pilus of Agrobacterium tumefaciens. J Bacteriol. 1998;180:2711–2717. doi: 10.1128/jb.180.10.2711-2717.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee K, Dudley M W, Hess K M, Lynn D G, Joerger R D, Binns A N. Mechanism of activation of Agrobacterium virulence genes: identification of phenol-binding proteins. Proc Natl Acad Sci USA. 1992;89:8666–8670. doi: 10.1073/pnas.89.18.8666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee Y W, Jin S, Sim W S, Nester E W. Genetic evidence for direct sensing of phenolic compounds by the VirA protein of Agrobacterium tumefaciens. Proc Natl Acad Sci USA. 1995;92:12245–12249. doi: 10.1073/pnas.92.26.12245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leroux B, Yanofsky M F, Winans S C, Ward J E, Ziegler S F, Nester E W. Characterization of the virA locus of Agrobacterium tumefaciens: a transcriptional regulator and host range determinant. EMBO J. 1987;6:849–856. doi: 10.1002/j.1460-2075.1987.tb04830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lessl M, Balzer D, Pansegrau W, Lanka E. Sequence similarities between the RP4 Tra2 and the Ti VirB region strongly support the conjugation model for T-DNA transfer. J Biol Chem. 1992;267:20471–20480. [PubMed] [Google Scholar]
- 63.Lessl M, Lanka E. Common mechanisms in bacterial conjugation and Ti-mediated T-DNA transfer to plant cells. Cell. 1994;77:321–324. doi: 10.1016/0092-8674(94)90146-5. [DOI] [PubMed] [Google Scholar]
- 64.Li P L, Farrand S K. The replicator of the nopaline-type Ti plasmid pTiC58 is a member of the repABC family and is influenced by the TraR-dependent quorum-sensing regulatory system. J Bacteriol. 2000;182:179–188. doi: 10.1128/jb.182.1.179-188.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li P L, Hwang I, Miyagi H, True H, Farrand S K. Essential components of the Ti plasmid trb system, a type IV macromolecular transporter. J Bacteriol. 1999;181:5033–5041. doi: 10.1128/jb.181.16.5033-5041.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lichtenstein C, Klee H, Montoya A, Garfinkel D, Fuller S, Flores C, Nester E, Gordon M. Nucleotide sequence and transcript mapping of the tmr gene of the pTiA6NC octopine Ti-plasmid: a bacterial gene involved in plant tumorigenesis. J Mol Appl Genet. 1984;2:354–362. [PubMed] [Google Scholar]
- 67.Lin T S, Kado C I. The virD4 gene is required for virulence while virD3 and orf5 are not required for virulence of Agrobacterium tumefaciens. Mol Microbiol. 1993;9:803–812. doi: 10.1111/j.1365-2958.1993.tb01739.x. [DOI] [PubMed] [Google Scholar]
- 68.Luo Z Q, Farrand S K. Signal-dependent DNA binding and functional domains of the quorum-sensing activator TraR as identified by repressor activity. Proc Natl Acad Sci USA. 1999;96:9009–9014. doi: 10.1073/pnas.96.16.9009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Luo Z-Q, Qin Y, Farrand S K. The antiactivator TraM interferes with the autoinducer-dependent binding of TraR to DNA by interacting with the C-terminal region of the quorum-sensing activator. J Biol Chem. 2000;275:7713–7722. doi: 10.1074/jbc.275.11.7713. [DOI] [PubMed] [Google Scholar]
- 70.Lyi S, Jafri M S, Winans S C. Mannopinic acid and agropinic acid catabolism region of the octopine-type Ti plasmid pTi15955. Mol Microbiol. 1999;31:339–347. doi: 10.1046/j.1365-2958.1999.01178.x. [DOI] [PubMed] [Google Scholar]
- 71.Magrelli A, Langenkemper K, Dehio C, Schell J, Spena A. Splicing of the rolA transcript of Agrobacterium rhizogenes in Arabidopsis. Science. 1994;266:1986–1988. doi: 10.1126/science.7528444. [DOI] [PubMed] [Google Scholar]
- 72.Mayerhofer R, Koncz-Kalman Z, Nawrath C, Bakkeren G, Crameri A, Angelis K, Redei G P, Schell J, Hohn B, Koncz C. T-DNA integration: a mode of illegitimate recombination in plants. EMBO J. 1991;10:697–704. doi: 10.1002/j.1460-2075.1991.tb07999.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Melchers L S, Maroney M J, den Dulk-Ras A, Thompson D V, van Vuuren H A, Schilperoort R A, Hooykaas P J. Octopine and nopaline strains of Agrobacterium tumefaciens differ in virulence; molecular characterization of the virF locus. Plant Mol Biol. 1990;14:249–259. doi: 10.1007/BF00018565. [DOI] [PubMed] [Google Scholar]
- 74.Melchers L S, Regensburg T T J, Bourret R B, Sedee N J, Schiperoort R A, Hooykaas P J. Membrane topology and functional analysis of the sensory protein VirA of Agrobacterium tumefaciens. EMBO J. 1989;8:1919–1925. doi: 10.1002/j.1460-2075.1989.tb03595.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Messens E, Lenaerts A, VanMontagu M, Hedges R W. Genetic basis for opine secretion from crown gall tumor cells. Mol Gen Genet. 1985;199:344–348. [Google Scholar]
- 76.Miranda A, Janssen G, Hodges L, Peralta E G, Ream W. Agrobacterium tumefaciens transfers extremely long T-DNAs by a unidirectional mechanism. J Bacteriol. 1992;174:2288–2297. doi: 10.1128/jb.174.7.2288-2297.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Moré M I, Finger L D, Stryker J L, Fuqua C, Eberhard A, Winans S C. Enzymatic synthesis of a quorum-sensing autoinducer through use of defined substrates. Science. 1996;272:1655–1658. doi: 10.1126/science.272.5268.1655. [DOI] [PubMed] [Google Scholar]
- 78.Mushegian A R, Fullner K J, Koonin E V, Nester E W. A family of lysozyme-like virulence factors in bacterial pathogens of plants and animals. Proc Natl Acad Sci USA. 1996;93:7321–7326. doi: 10.1073/pnas.93.14.7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nishiguchi R, Takanami M, Oka A. Characterization and sequence determination of the replicator region in the hairy-root-inducing plasmid pRiA4b. Mol Gen Genet. 1987;206:1–8. [Google Scholar]
- 80.Oger P, Kim K-S, Sackett R L, Piper K R, Farrand S K. Octopine-type Ti plasmids code for a mannopine-inducible dominant-negative allele of traR, the quorum-sensing activator that regulates Ti plasmid conjugal transfer. Mol Microbiol. 1998;27:277–288. doi: 10.1046/j.1365-2958.1998.00671.x. [DOI] [PubMed] [Google Scholar]
- 81.Oger P, Petit A, Dessaux Y. Genetically engineered plants producing opines alter their biological environment. Nat Biotechnol. 1997;15:369–372. doi: 10.1038/nbt0497-369. [DOI] [PubMed] [Google Scholar]
- 82.Okamoto S, Toyoda-Yamamoto A, Ito K, Takebe I, Machida Y. Localization and orientation of the VirD4 protein of Agrobacterium tumefaciens in the cell membrane. Mol Gen Genet. 1991;228:24–32. doi: 10.1007/BF00282443. [DOI] [PubMed] [Google Scholar]
- 83.Otten L, Canaday J, Gerard J C, Fournier P, Crouzet P, Paulus F. Evolution of agrobacteria and their Ti plasmids—a review. Mol Plant-Microbe Interact. 1992;5:279–287. doi: 10.1094/mpmi-5-279. [DOI] [PubMed] [Google Scholar]
- 84.Otten L, De Greve H, Leemans J, Hain R, Hooykaas P, Schell J. Restoration of virulence of vir region mutants of Agrobacterium tumefaciens strain B6S3 by coinfection with normal and mutant Agrobacterium strains. Mol Gen Genet. 1984;195:159–163. [Google Scholar]
- 85.Pan S Q, Charles T, Jin S, Wu Z L, Nester E W. Preformed dimeric state of the sensor protein VirA is involved in plant-Agrobacterium signal transduction. Proc Natl Acad Sci USA. 1993;90:9939–9943. doi: 10.1073/pnas.90.21.9939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pansegrau W, Schoumacher F, Hohn B, Lanka E. Site-specific cleavage and joining of single-stranded DNA by VirD2 protein of Agrobacterium tumefaciens Ti plasmids: analogy to bacterial conjugation. Proc Natl Acad Sci USA. 1993;90:11538–11542. doi: 10.1073/pnas.90.24.11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Regensburg-Tuink A J, Hooykaas P J. Transgenic N. glauca plants expressing bacterial virulence gene virF are converted into hosts for nopaline strains of A. tumefaciens. Nature. 1993;363:69–71. doi: 10.1038/363069a0. [DOI] [PubMed] [Google Scholar]
- 88.Roitsch T, Wang H, Jin S G, Nester E W. Mutational analysis of the VirG protein, a transcriptional activator of Agrobacterium tumefaciens virulence genes. J Bacteriol. 1990;172:6054–6060. doi: 10.1128/jb.172.10.6054-6060.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Savka M A, Farrand S K. Modification of rhizobacterial populations by engineering bacterium utilization of a novel plant-produced resource. Nat Biotechnol. 1997;15:363–368. doi: 10.1038/nbt0497-363. [DOI] [PubMed] [Google Scholar]
- 90.Scheiffele P, Pansegrau W, Lanka E. Initiation of Agrobacterium tumefaciens T-DNA processing. Purified proteins VirD1 and VirD2 catalyze site- and strand-specific cleavage of superhelical T-border DNA in vitro. J Biol Chem. 1995;270:1269–1276. doi: 10.1074/jbc.270.3.1269. [DOI] [PubMed] [Google Scholar]
- 91.Sheng J, Citovsky V. Agrobacterium-plant cell DNA transport: have virulence proteins, will travel. Plant Cell. 1996;8:1699–1710. doi: 10.1105/tpc.8.10.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shimoda N, Toyoda-Yamamoto A, Aoki S, Machida Y. Genetic evidence for an interaction between the VirA sensor protein and the ChvE sugar-binding protein of Agrobacterium. J Biol Chem. 1993;268:26552–26558. [PubMed] [Google Scholar]
- 93.Shirasu K, Koukolikova-Nicola Z, Hohn B, Kado C I. An inner-membrane-associated virulence protein essential for T-DNA transfer from Agrobacterium tumefaciens to plants exhibits ATPase activity and similarities to conjugative transfer genes. Mol Microbiol. 1994;11:581–588. doi: 10.1111/j.1365-2958.1994.tb00338.x. [DOI] [PubMed] [Google Scholar]
- 94.Shurvington C E, Ream W. Stimulation of Agrobacterium tumefaciens T-DNA transfer by overdrive depends on a flanking sequence but not on helical position with respect to the border repeat. J Bacteriol. 1991;173:5558–5563. doi: 10.1128/jb.173.17.5558-5563.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Smith E F, Townsend C O. A plant-tumor of bacterial origin. Science. 1907;25:671–673. doi: 10.1126/science.25.643.671. [DOI] [PubMed] [Google Scholar]
- 96.Spudich G M, Fernandez D, Zhou X R, Christie P J. Intermolecular disulfide bonds stabilize VirB7 homodimers and VirB7/VirB9 heterodimers during biogenesis of the Agrobacterium tumefaciens T-complex transport apparatus. Proc Natl Acad Sci USA. 1996;93:7512–7517. doi: 10.1073/pnas.93.15.7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stachel S E, Timmerman B, Zambryski P. Generation of single-stranded T-DNA molecules during the initial stages of T-DNA transfer from Agrobacterium tumefaciens to plant cells. Nature. 1986;322:706–712. [Google Scholar]
- 98.Stachel S E, Timmerman B, Zambryski P. Activation of Agrobacterium tumefaciens vir gene expression generates multiple single-stranded T-strand molecules from the pTiA6 T-region: requirement for 5′ virD gene products. EMBO J. 1987;6:857–863. doi: 10.1002/j.1460-2075.1987.tb04831.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Stachel S E, Zambryski P C. Agrobacterium tumefaciens and the susceptible plant cell: a novel adaptation of extracellular recognition and DNA conjugation. Cell. 1986;47:155–157. doi: 10.1016/0092-8674(86)90437-x. [DOI] [PubMed] [Google Scholar]
- 100.Stachel S E, Zambryski P C. virA and virG control the plant-induced activation of the T-DNA transfer process of A. tumefaciens. Cell. 1986;46:325–333. doi: 10.1016/0092-8674(86)90653-7. [DOI] [PubMed] [Google Scholar]
- 101.Sundberg C, Meek L, Carroll K, Das A, Ream W. VirE1 protein mediates export of the single-stranded DNA-binding protein VirE2 from Agrobacterium tumefaciens into plant cells. J Bacteriol. 1996;178:1207–1212. doi: 10.1128/jb.178.4.1207-1212.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Suzuki K, Hattori Y, Uraji M, Ohta N, Iwata K, Murata K, Kato A, Yoshida K. Complete nucleotide sequence of a plant tumor-inducing Ti plasmid. Gene. 2000;242:331–336. doi: 10.1016/s0378-1119(99)00502-8. [DOI] [PubMed] [Google Scholar]
- 103.Suzuki K, Ohta N, Hattori Y, Uraji M, Kato A, Yoshida K. Novel structural difference between nopaline- and octopine-type trbJ genes: construction of genetic and physical map and sequencing of trb/traI and rep gene clusters of a new Ti plasmid pTi-SAKURA. Biochim Biophys Acta. 1998;1396:1–7. doi: 10.1016/s0167-4781(97)00182-6. [DOI] [PubMed] [Google Scholar]
- 104.Tabata S, Hooykaas P J, Oka A. Sequence determination and characterization of the replicator region in the tumor-inducing plasmid pTiB6S3. J Bacteriol. 1989;171:1665–1672. doi: 10.1128/jb.171.3.1665-1672.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Thomashow M F, Nutter R, Montoya A L, Gordon M P, Nester E W. Integration and organization of Ti plasmid sequences in crown gall tumors. Cell. 1980;19:729–739. doi: 10.1016/s0092-8674(80)80049-3. [DOI] [PubMed] [Google Scholar]
- 106.Thompson D V, Melchers L S, Idler K B, Schilperoort R A, Hooykaas P J. Analysis of the complete nucleotide sequence of the Agrobacterium tumefaciens virB operon. Nucleic Acids Res. 1988;16:4621–4636. doi: 10.1093/nar/16.10.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Thorstenson Y R, Kuldau G A, Zambryski P C. Subcellular localization of seven VirB proteins of Agrobacterium tumefaciens: implications for the formation of a T-DNA transport structure. J Bacteriol. 1993;175:5233–5241. doi: 10.1128/jb.175.16.5233-5241.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tinland B, Fournier P, Heckel T, Otten L. Expression of a chimaeric heat-shock-inducible Agrobacterium 6b oncogene in Nicotiana rustica. Plant Mol Biol. 1992;18:921–930. doi: 10.1007/BF00019206. [DOI] [PubMed] [Google Scholar]
- 109.Tinland B, Schoumacher F, Gloeckler V, Bravo-Angel A M, Hohn B. The Agrobacterium tumefaciens virulence D2 protein is responsible for precise integration of T-DNA into the plant genome. EMBO J. 1995;14:3585–3595. doi: 10.1002/j.1460-2075.1995.tb07364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Toro N, Datta A, Yanofsky M, Nester E. Role of the overdrive sequence in T-DNA border cleavage in Agrobacterium. Proc Natl Acad Sci USA. 1988;85:8558–8562. doi: 10.1073/pnas.85.22.8558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Toro N, Datta A, Carmi O A, Young C, Prusti R K, Nester E W. The Agrobacterium tumefaciens virC1 gene product binds to overdrive, a T-DNA transfer enhancer. J Bacteriol. 1989;171:6845–6849. doi: 10.1128/jb.171.12.6845-6849.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Valdivia R H, Wang L, Winans S C. Characterization of a putative periplasmic transport system for octopine accumulation encoded by Agrobacterium tumefaciens Ti plasmid pTiA6. J Bacteriol. 1991;173:6398–6405. doi: 10.1128/jb.173.20.6398-6405.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.van Haaren M J, Sedee N J, Schilperoort R A, Hooykaas P J. Overdrive is a T-region transfer enhancer which stimulates T-strand production in Agrobacterium tumefaciens. Nucleic Acids Res. 1987;15:8983–8997. doi: 10.1093/nar/15.21.8983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.von Lintig J, Kreusch D, Schroder J. Opine-regulated promoters and LysR-type regulators in the nopaline (noc) and octopine (occ) catabolic regions of Ti plasmids of Agrobacterium tumefaciens. J Bacteriol. 1994;176:495–503. doi: 10.1128/jb.176.2.495-503.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang L, Helmann J D, Winans S C. The A. tumefaciens transcriptional activator OccR causes a bend at a target promoter, which is partially relaxed by a plant tumor metabolite. Cell. 1992;69:659–667. doi: 10.1016/0092-8674(92)90229-6. [DOI] [PubMed] [Google Scholar]
- 116.Ward J E, Akiyoshi D E, Regier D, Datta A, Gordon M P, Nester E W. Characterization of the virB operon from an Agrobacterium tumefaciens Ti plasmid. J Biol Chem. 1988;263:5804–5814. [PubMed] [Google Scholar]
- 117.Waters V L, Hirata K H, Pansegrau W, Lanka E, Guiney D G. Sequence identity in the nick regions of IncP plasmid transfer origins and T-DNA borders of Agrobacterium Ti plasmids. Proc Natl Acad Sci USA. 1991;88:1456–1460. doi: 10.1073/pnas.88.4.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Winans S C, Jin S, Komari T, Johnson K M, Nester E W. The role of virulence regulatory loci in determining Agrobacterium host range. In: von Wettstein D, Chua N-H, editors. Plant molecular biology. New York, N.Y: Plenum Press; 1987. pp. 573–582. [Google Scholar]
- 119.Winans S C, Kerstetter R A, Nester E W. Transcriptional regulation of the virA and virG genes of Agrobacterium tumefaciens. J Bacteriol. 1988;170:4047–4054. doi: 10.1128/jb.170.9.4047-4054.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Winans S C, Zhu J, Moré M I. Cell density-dependent gene expression by Agrobacterium tumefaciens during colonization of crown gall tumors. In: Dunny G M, Winans S C, editors. Cell-cell communication in bacteria. Washington, D.C.: ASM Press; 1999. pp. 117–128. [Google Scholar]
- 121.Yadav N S, Vanderlayden J, Bennett D R, Barnes W M, Chilton M-D. Short direct repeats flank the T-DNA on a nopaline Ti plasmid. Proc Natl Acad Sci USA. 1982;79:6322–6326. doi: 10.1073/pnas.79.20.6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yanofsky M F, Nester E W. Molecular characterization of a host-range-determining locus from Agrobacterium tumefaciens. J Bacteriol. 1986;168:244–250. doi: 10.1128/jb.168.1.244-250.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yanofsky M F, Porter S G, Young C, Albright L M, Gordon M P, Nester E W. The virD operon of Agrobacterium tumefaciens encodes a site-specific endonuclease. Cell. 1986;7:471–477. doi: 10.1016/0092-8674(86)90604-5. [DOI] [PubMed] [Google Scholar]
- 124.Yusibov V M, Steck T R, Gupta V, Gelvin S B. Association of single-stranded transferred DNA from Agrobacterium tumefaciens with tobacco cells. Proc Natl Acad Sci USA. 1994;91:2994–2998. doi: 10.1073/pnas.91.8.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zambryski P. Agrobacterium-plant cell DNA transfer. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C.: American Society for Microbiology; 1989. pp. 309–333. [Google Scholar]
- 126.Zhang L, Murphy P J, Kerr A, Tate M E. Agrobacterium conjugation and gene regulation by N-acyl-l-homoserine lactones. Nature. 1993;362:446–448. doi: 10.1038/362446a0. [DOI] [PubMed] [Google Scholar]
- 127.Zhu J, Winans S C. Activity of the quorum-sensing regulator TraR of Agrobacterium tumefaciens is inhibited by a truncated, dominant defective TraR-like protein. Mol Microbiol. 1998;27:289–297. doi: 10.1046/j.1365-2958.1998.00672.x. [DOI] [PubMed] [Google Scholar]
- 128.Zhu J, Winans S C. Autoinducer binding by the quorum-sensing regulator TraR increases affinity for target promoters in vitro and decreases TraR turnover rates in whole cells. Proc Natl Acad Sci USA. 1999;96:4832–4837. doi: 10.1073/pnas.96.9.4832. [DOI] [PMC free article] [PubMed] [Google Scholar]