Abstract
The metabolic consequences of two insertions, iscR1::MudJ and iscA2::MudJ, in the isc gene cluster of Salmonella enterica serovar Typhimurium were studied. Each of these insertions had polar effects and caused a nutritional requirement for the thiazole moiety of thiamine. Data showed that IscS was required for the synthesis of nicotinic acid and the thiazole moiety of thiamine and that one or more additional isc gene products were required for a distinct step in the thiazole biosynthetic pathway. Strains with isc lesions had reduced succinate dehydrogenase and aconitase activities. Furthermore, isc mutants accumulated increased levels of pyruvate in the growth medium in response to exogenously added iron (FeCl3), and this response required a functional ferric uptake regulator, Fur.
Historically, biosynthetic genes have been identified by mutations resulting in a predicted nutritional requirement. When the biosynthetic genes of several pathways were identified, it became clear that nutritional requirements were generated by lesions in genes that do not encode biosynthetic enzymes but rather have indirect roles in the respective pathways. The characterization of mutations that indirectly affect thiamine biosynthesis has provided insight into the synthesis of this essential vitamin and uncovered new features of metabolism in Salmonella enterica (1, 7, 8, 18, 19, 23, 25, 40).
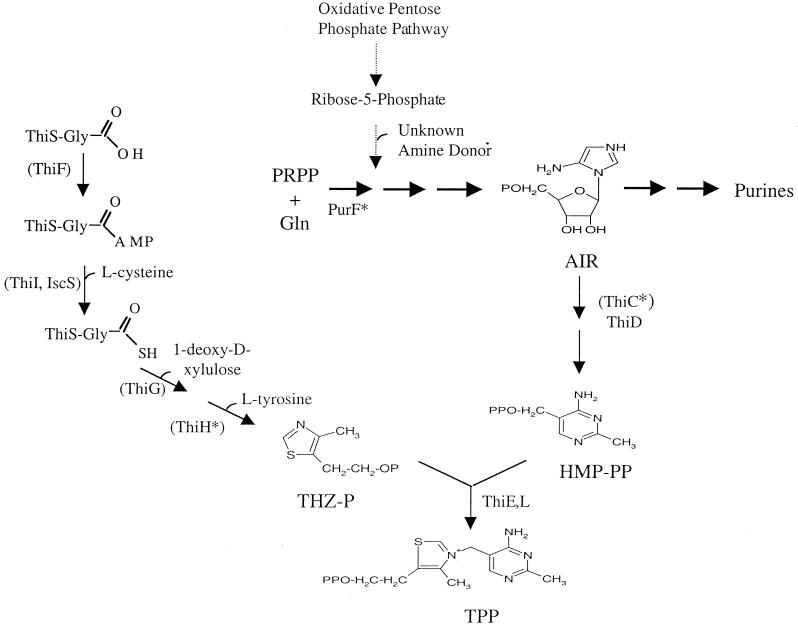
Thiamine pyrophosphate is an essential cofactor for several well-characterized enzymes, including pyruvate dehydrogenase, pyruvate decarboxylase, α-ketoglutarate dehydrogenase, and transketolase (57). Thiamine pyrophosphate is formed by the condensation and subsequent phosphorylation of 4-amino-5-hydroxymethyl pyrimidine pyrophosphate (HMP-PP) and 4-methyl-5-(β-hydroxyethyl)-thiazole monophosphate (THZ-P) (2). HMP-PP is derived from aminoimidazole ribotide, which is a shared intermediate with the purine pathway (20, 21, 36–38). Extensive labeling studies determined that cysteine, tyrosine, and 1-deoxy-d-xylulose contribute to the THZ-P moiety (10, 11, 15, 22, 52). Our present understanding of the thiamine biosynthetic pathway is depicted in Fig. 1.
FIG. 1.
Biosynthetic pathway for thiamine synthesis. Enzymes for the catalysis of each reaction are indicated above the relevant arrow. Enzymes that have not been demonstrated but are predicted to function at certain steps in thiamine biosynthesis are in parentheses. Putative [Fe-S]-containing enzymes are marked by asterisks. The dotted lines represent predicted reactions in the PurF-independent formation of phosphoribosyl amine (PRA). PRPP, phosphoribosylpyrophosphate; AIR, 5-aminoimidazole ribotide; TPP, thiamine pyrophosphate.
The strB genetic locus was defined genetically by mutations that resulted in low-level streptomycin resistance, affected histidine biosynthesis, and generated a thiamine and/or nicotinic acid auxotrophy (14, 44, 45). Mutations in this locus were reported to be resistant to 50 μg of streptomycin/ml in nutrient broth and 500 μg/ml in minimal salts medium, distinguishing them from the classical strA alleles of ribosomal protein S12 (rpsL), which are typically resistant to 2 mg of streptomycin/ml (12). Work by Zhu and Roth determined that the nicotinic acid (NA) requirement was due to a reduced activity of quinolinic acid synthetase in these strains (61). Here we mapped two independently isolated insertions in the strB locus and determined that it was allelic with the isc gene cluster in S. enterica.
The isc locus was originally described in Azotobacter vinlandii (59) and contained homologs of nif genes that are involved in formation of the iron-sulfur ([Fe-S]) center of nitrogenase (29, 58). Specifically, in a strain with nifS deleted (NifS is involved in the mobilization of sulfur for nitrogenase) (60), an enzyme with l-cysteine desulfurase activity was isolated and designated IscS. The complex locus containing the iscS gene was sequenced and found to be conserved in many non-nitrogen-fixing bacteria (59). The prevalence of this isc gene cluster and the presence of genes similar to those with a demonstrated role in [Fe-S] cluster assembly in nitrogenase suggested that genes in this locus had a general cellular role in [Fe-S] cluster assembly (59). Consistent with this proposal, Nakamura et al. recently demonstrated that coexpression of the isc gene cluster increased the production of five reporter ferredoxins (35). The authors attributed this increase in production to an increased ability to form stable [Fe-S] centers. A second study also showed that the specific isc gene products required for overproduction varied with each reporter [Fe-S] protein tested (50).
Recently, Kambampati and Lauhon (31) isolated IscS based on its ability to transfer sulfur to the 4-thiouridine modification of tRNA via a ThiI intermediate. ThiI has been shown to be required for the synthesis of both the thiazole (THZ) moiety of thiamine (55) and the 4-thiouridine modification of tRNA (34), causing the authors to suggest that IscS also has a role in the sulfur donation to THZ. The results presented in this study support this hypothesis.
We have identified insertion mutations in two genes in the isc locus, iscR and iscA (Fig. 2). The insertions were polar on downstream genes, and complementation analyses showed that the iscR mutation affected two distinct steps in the synthesis of the THZ moiety of thiamine. Both insertion mutants in the isc gene cluster had decreased activities of at least two enzymes containing [Fe-S] centers and displayed novel metabolic phenotypes. In particular, spent medium from these mutants had increased levels of pyruvate when high levels of iron were present in the growth medium, and this increase was eliminated by a null mutation in fur, the gene that encodes the ferric uptake regulator (48).
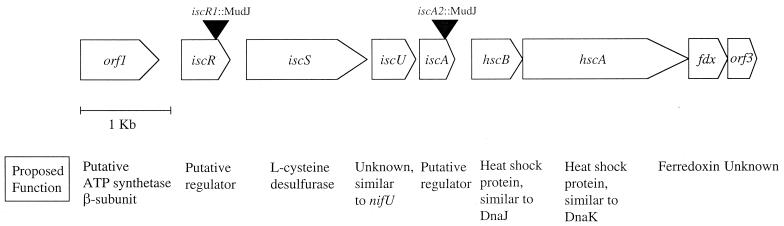
FIG. 2.
Organization of the isc gene cluster in S. enterica serovar Typhi. The relative locations of two MudJ insertions are indicated by solid triangles, and the assigned allele numbers are shown. The proposed functions of the products are listed below each gene. Note that orf2 in the annotated E. coli genome has been redesignated iscR (D. R. Dean, personal communication).
MATERIALS AND METHODS
Bacterial strains, media, and chemicals.
All strains used in this study are derived from S. enterica LT2 and are listed with their respective genotypes in Table 1. The NCE medium of Berkowitz et al. (3) supplemented with 1 mM MgSO4 was used as a minimal medium, and glucose or glycerol (11 mM) was added as a carbon source. When present in the culture media, these compounds were used at the following final concentrations: thiamine, 100 nM; NA, 20 μM; and sodium nitrate, 40 mM. When indicated, FeCl3 (Sigma Chemical Co., St. Louis, Mo.) was added to a final concentration of 50 nM or 50 μM. Luria broth and Difco nutrient broth (8 g/liter) with NaCl (5 g/liter) were used as rich media, with Difco BiTek agar added to a final concentration of 1.5% for solid medium. The final concentrations of antibiotics were as follows: tetracycline (Tc), 20 μg/ml; kanamycin (Km), 50 μg/ml; ampicillin (Ap), 30 μg/ml; and chloramphenicol (Cm), 20 μg/ml.
TABLE 1.
Strainsa
| Strain | Genotype | Source |
|---|---|---|
| LT2 | Wild type | Laboratory strain |
| TT16365 | strB1143::MudJb (iscR1::MudJ throughout) | J. Roth |
| TT16366 | strB1144::MudJ (iscA2::MudJ throughout) | J. Roth |
| TT20152 | fur-1 iroA1::MudJ zbj-5123::Tn10 (33% linked to fur-1) | J. Roth |
| TT20268 | entB::MudJ | J. Roth |
| DM2361 | sdhC203::Tn10dc(Tc) | Laboratory strain |
| DM5419 | iscR1::MudJ | |
| DM5420 | iscA2::MudJ | |
| DM5632 | zbj-5123::Tn10 (Tc) fur-1 | |
| DM5633 | zbj-5123::Tn10 (Tc) fur+ | |
| DM5634 | iscR1::MudJ zbj-5123::Tn10 (Tc) fur-1 | |
| DM5635 | iscR1::MudJ zbj-5123::Tn10 (Tc) fur+ | |
| DM5636 | iscA2::MudJ zbj-5123::Tn10 (Tc) fur-1 | |
| DM5637 | iscA2::MudJ zbj-5123::Tn10 (Tc) fur+ | |
| DM5694 | iscR1::MudJ(pSU19) | |
| DM5695 | iscR1::MudJ(piscS1) | |
| DM5708 | iscR1::MudJ(piscR1) | |
| DM5696 | iscA2::MudJ(pSU19) | |
| DM5710 | iscA2::MudJ(piscA1) | |
| DM5697 | iscA2::MudJ(piscS1) | |
| DM5719 | iscA2::MudJ(pfdx1) | |
| DM5720 | iscA2::MudJ(pfdx1, pT76) | |
| DM5721 | iscA2::MudJ(pfdx1, piscA3) |
Genetic techniques. (i) Transduction methods and strain construction.
The high-frequency general transducing mutant of bacteriophage P22 (HT105/1; int-201) (46) was used in all transductions. The methods for transduction and subsequent purification of transductants have been previously described (16). Point mutations in fur were cotransduced into strains LT2, DM5419, and DM5420 by selecting the tetracycline resistance associated with a linked Tn10 element. The presence of the fur mutation was confirmed by backcrossing into a strain containing a Mud-lacZ fusion (MudJ) insertion in entB (TT20268), a gene regulated by Fur. Constitutive expression of entB indicated the presence of a null allele of fur (53).
(ii) Phenotypic analysis.
Nutritional requirements were assessed on solid medium with soft agar overlays and by quantification of growth in liquid. Protocols for each have been described (1, 42). In growth curves, the starting A650 was routinely between 0.03 and 0.08, with a final A650 between 0.7 and 1.1.
(iii) Complementation analysis.
The plasmids used in this study are listed in Table 2. Orfmers (Genosys, The Woodlands, Tex.) were used to amplify the following genes from Escherichia coli K-12: iscR, iscA, iscS, and fdx. The corresponding plasmids, piscR1, piscA1, piscS1, and pfdx1, were generated by ligating the respective fragments into the SmaI site of pSU19 (33). Plasmid piscA3 was created by digesting piscA1 with PstI and EcoRI, purifying the fragment, and ligating it into appropriately digested plasmid (pT7-6) DNA (49). Plasmid inserts were confirmed by sequence analysis and/or restriction digest. The relevant plasmids were transformed into strains DM5419 and DM5420, and the nutritional requirements of the resulting strains were assessed.
TABLE 2.
Plasmidsa
| Plasmid | Vector (drug) | Insert |
|---|---|---|
| piscR1 | pSU19 (Cm) | iscR |
| piscA1 | pSU19 (Cm) | iscA |
| piscS1 | pSU19 (Cm) | iscS |
| pfdx1 | pSU19 (Cm) | fdx |
| piscA3 | pT76 (Ap) | iscA |
Inserts were obtained and plasmids were created as described in Materials and Methods.
(iv) Sequencing of isc::MudJ insertions.
The locations of the MudJ insertions were determined by sequencing with a PCR-based protocol (6, 56). A DNA product was amplified with degenerate primers and primers derived from the MudJ insertion sequence as described and sequenced by the University of Wisconsin Biotechnology Center-Nucleic Acid and Protein Facility.
Enzyme assays. (i) AC assays.
Aconitase (AC) activity was assayed by modifying a published protocol (27). Briefly, strains were grown in nutrient broth, and 3 ml was harvested in early stationary phase (A650, ca. 0.9). The cells were pelleted and washed with Tris-citrate buffer (20 mM; pH 8) at 0°C. The washed cells were resuspended in 300 μl of cold Tris-citrate buffer (20 mM; pH 8). The cells were sonicated with a Sonic Dismembrator 550 (Fisher Scientific, Pittsburgh, Pa.). Two rounds of sonication were performed, with a 30-s cooling period in between; a round consisted of sonication for 10 s with 0.5-s pauses between bursts. The cell extract was clarified by centrifugation at 10,000 × g for 20 s in a Marathon microcentrifuge (Fisher Scientific). Assays were performed immediately and were initiated by adding cell extract (0 to 100 μg of protein) to 20 mM isocitrate in a 200-μl final volume. AC activity was assayed at room temperature by following the formation of cis-aconitate by monitoring the increase in absorbance at 240 nm in a quartz microtiter plate using a Spectramax Plus (Molecular Devices, Sunnyvale, Calif.) plate reader. The linear range of the assay was determined for each strain, and specific activities were calculated as ΔA240/minute/milligram of protein. The protein concentration was determined by the method of Bradford (5).
(ii) SDH.
Cells were grown in minimal glucose medium (supplemented with NA and thiamine) to late log phase (ca. 100 Klett units [red filter]). Cells from a 3-ml sample were pelleted and washed with cold 50 mM potassium phosphate buffer (pH 7.5). The cells were resuspended in 300 μl of the same buffer and sonicated and clarified as described above. Succinate dehydrogenase (SDH) activity was measured by the method of Spencer and Guest (47) except that a final volume of 200 μl was used and absorbance at 600 nm was measured in a microtiter plate with a Spectramax Plus plate reader. Activities were measured in crude extracts at protein concentrations between 0 and 100 μg. The linear range was determined for each strain, and specific activities were calculated as ΔA600/minute/milligram of protein. The activity contributed by boiled cell extracts was subtracted from the activity obtained in each assay.
Measurement of pyruvate accumulation. (i) Derivatization with DNPH.
The derivatization procedure for α-keto acids was modified from the method of Kadner et al. (30). Strains were grown overnight in minimal glucose medium supplemented with thiamine and NA. A 200-μl sample of this culture was subcultured into iron-free tubes (Fisher Scientific) containing 6 ml of minimal glucose medium supplemented with thiamine, NA, and 50 nM or 50 μM FeCl3. The concentration of 50 nM FeCl3 was sufficiently low that the growth rate of a wild-type strain was noticeably impaired. Spent culture medium (50 μl) was derivatized by adding 66 μl of 2,4-dintrophenylhydrazine (DNPH) (0.25 mg/ml in 2 N HCl) and incubating the mixture for 15 min at room temperature. In general, spent medium was derivatized after 3 h of growth and every 1.5 h thereafter. Color was visible 15 min after neutralization with 83 μl of 10% NaOH, and a UV-visible scan showed a major absorbence peak at 443 nm.
(ii) Lactate dehydrogenase assays.
The lactate dehydrogenase assay was modified from the method of Oeschger (39). The reaction mixture contained pyruvate-specific d-lactic acid dehydrogenase (catalog no. L3888; Sigma Chemical Co.) (8.7 U), 10 mM Tris-HCl (pH 8), 240 mM KCl, 20 mM MgCl2, and 1 mM pyruvate or 10 to 100 μl of spent growth medium. The preassay mixture was incubated at 30°C for 2 min before the reaction was initiated with NADH (final concentration, 25 mM). The oxidation of NADH was monitored as a decrease in the absorbance at 340 nM in a quartz microtiter plate using a Spectramax Plus plate reader.
Determination of glucose.
The concentration of glucose in the medium was monitored using the Glucose LiquiColor kit (Stan Bio, San Antonio, Tex.) according to the manufacturer's instructions.
RESULTS
Mutations in the strB genetic locus map to the isc gene cluster in S. enterica.
Our interest in the strB genetic locus was prompted by reports of thiamine auxotrophy associated with several strB mutants. To pursue this observation, two strains carrying independent MudJ insertions in the strB locus were analyzed (strB1143::MudJ and strB1144::MudJ) (61). The MudJ insertions were transduced into our laboratory LT2 strain, generating DM5419 and DM5420, respectively. Sequence analysis of these two strains determined that both MudJ insertions were located in the isc gene cluster in the 57 centisome region of the S. enterica chromosome. This locus has been implicated in the formation of [Fe-S] clusters in E. coli and other organisms (35, 50, 59). The physical organization of this locus (as found in the annotated E. coli genome [4]) is shown in Fig. 2 with the positions of the strB1143::MudJ and strB1144::MudJ insertions at the ends of iscR and iscA, respectively. To reflect these positions, the insertions were renamed iscR1 and iscA2. Data produced by the S. typhi Sequencing Group at the Sanger Center indicate that a similar physical organization exists in S. enterica serovar Typhi (http://www.sanger.ac.uk/Projects/S_typhi) and is likely to exist in S. enterica serovar Typhimurium.
Lesions in isc genes result in varied nutritional requirements.
The nutritional requirements for the iscR1 and iscA2 mutant strains were determined on solid growth media. Data from three independent experiments were averaged and are presented in Table 3. As reported previously, the iscR1::MudJ insertion generated a nutritional requirement for NA and thiamine (61). Although the iscA2::MudJ insertion mutant was reported to be prototrophic, under the conditions reflected in Table 3, strain DM5420 required thiamine for growth. The thiamine auxotrophy of both the iscR1 and iscA2 insertion mutants on solid growth medium was satisfied by the addition of the THZ moiety of thiamine, indicating a block in the biosynthetic pathway for THZ. Notably, the THZ requirements of the two isc strains could be distinguished by the addition of tyrosine. As shown in Table 3, tyrosine satisfied the THZ requirement of DM5420 but failed to correct the phenotype of strain DM5419. We have previously isolated mutations that result in a requirement for THZ or tyrosine and hypothesized that this phenotype reflected a defect in ThiH activity (1, 54; J. Gralnick, E. Webb, B. Beck, and D. M. Downs, submitted for publication).
TABLE 3.
isc mutations generate nutritional requirementsa
| Strain | Genotype | Diameter of growth zone (mm)b
|
||||
|---|---|---|---|---|---|---|
| THI | NA | NA-THI | NA-THZ | NA-TYR | ||
| DM5419 | iscR1::MudJ | ND | ND | 24 | 46 | ND |
| DM5420 | iscA2::MudJ | 32 | ND | 32 | 41 | 40 |
| DM5694 | iscR1::MudJ(pSU19) | ND | ND | 33 | 52 | ND |
| DM5708 | iscR1::MudJ(piscR1) | ND | ND | 33 | 52 | ND |
| DM5695 | iscR1::MudJ(piscS1) | 42 | ND | 41 | 52 | 48 |
| DM5696 | iscA2::MudJ(pSU19) | 35 | ND | 35 | 50 | 44 |
| DM5710 | iscA2::MudJ(piscA1) | 35 | ND | 36 | 49 | 44 |
Soft agar overlays of the indicated strains were plated on minimal glucose medium. The indicated compounds were spotted as follows: thiamine (THI), 0.2 nmol in 2 μl; NA, 40 nmol in 2 μl; THZ, 0.2 nmol in 2 μl; tyrosine (TYR), 1 μmol in 10 μl.
Results are the means of three independent experiments with variation of less than 4 mm. ND, not detectable.
To test whether the phenotypes associated with the isc::MudJ insertions were generated by polar effects on downstream genes, clones containing iscR, iscA, iscS, or fdx in trans were constructed and utilized in complementation analyses (see Materials and Methods). The results of some of these experiments (Table 3) indicated that in both strains, one or more genes downstream of the MudJ insertion site were involved in generating the observed phenotype. In the case of the iscA2 mutant (DM5420), providing iscA in trans failed to alter the nutritional requirements of the strain (Table 3, lines 6 and 7). Additional experiments showed that neither the presence of fdx nor both iscA and fdx in trans altered the requirements (data not shown). Although the relevant constructs were confirmed by sequence analyses, it is formally possible that these genes were not being expressed. From these results we concluded that one or more of the downstream genes were involved in the thiamine requirement generated by the iscA2 mutation. In the case of the iscR1 mutant, the presence of iscR in trans did not change the nutritional requirements of the strain. However, the presence of iscS in trans had two detectable effects on the nutritional requirement caused by the iscR1::MudJ insertion: (i) the NA requirement was eliminated, and (ii) the remaining THZ requirement was satisfied by tyrosine. Our interpretation of these results was that two distinct steps in THZ synthesis were affected by mutations in the isc genes. The data in Table 3 indicated that a lack of the iscS gene resulted in a requirement for NA and generated a THZ requirement that was not satisfied by tyrosine. We suggest that this defect is at the ThiI step (31). Consistent with these results, nonpolar insertions in iscS have recently been generated in E. coli and found to cause a requirement for NA and thiamine (C. Lauhon, personal communication).
isc cluster mutants have phenotypes consistent with [Fe-S] center defects.
Based on the predicted functions of the genes in the isc locus, it was likely that several metabolic processes, specifically, those involving [Fe-S] centers, would be disrupted in the mutant strains. Although not demonstrably linked to a lack of [Fe-S] centers, the ratio of growth rates on glycerol to those on glucose of both isc mutant strains under anaerobic conditions was reduced compared to that of a wild-type strain. The specific growth rates (μ) of LT2, DM5419, and DM5420 were 0.257, 0.123, and 0.216 in glucose and 0.269, 0.082, and 0.191 in glycerol, respectively. Together with the final A650s of the same strains in glucose (0.652, 0.426, and 0.62) and glycerol (0.588, 0.178, and 0.432), these results suggested that the isc mutant strains had a decreased ability to grow under conditions requiring anaerobic respiration. Anaerobic conditions were used to minimize the negative effects that lack of the isc gene products might have on repairing oxidative damage to known enzymes containing [Fe-S] centers. The finding that DM5419 (iscR1) had a more severe defect than DM5420 (iscA2) was consistent with a role for multiple isc gene products in this phenotype.
The specific activities of [Fe-S]-containing proteins, SDH, and AC were determined in strains DM5419, DM5420, and LT2. The results from these experiments (Table 4) demonstrated that the isc mutants had decreased SDH activity (∼3-fold) and decreased AC activity (∼4-fold). Together, the results in this section were consistent with a defect in [Fe-S] center formation and/or maintenance in these strains that was anticipated based on the sequence analysis of the disrupted genes.
TABLE 4.
SDH and AC activities are reduced in isc mutants
| Strain | Relevant genotype | SDH activitya | AC activitya |
|---|---|---|---|
| LT2 | Wild type | 1.58 ± 0.03 | 3.73 ± 0.16 |
| DM5419 | iscR1::MudJ | 0.48 ± 0.06 | 0.93 ± 0.07 |
| DM5420 | iscA2::MudJ | 0.56 ± 0.09 | 1.04 ± 0.01 |
| DM2361 | sdhC203::Tn10d (Tc) | 0.06 ± 0.01 | ND |
Activities of SDH and AC are expressed as the change in relative absorbance/minute/milligram of protein (± standard deviation) as described in Materials and Methods. ND, not determined.
Lesions in the isc genes cause accumulation of large amounts of pyruvate in the growth medium.
When isc mutants were grown in Luria broth supplemented with glucose, the cultures failed to reach full density. This growth arrest correlated with the accumulation of acid in the growth media of the mutants (pH 4.5; pH 8.5 for the wild type) (data not shown). Derivatization of the culture medium with DNPH determined that α-keto acids were accumulating. When a buffered minimal medium was used, growth of the isc mutants was not arrested and DNPH derivatizable α-keto acid(s) accumulated and was depleted in a growth-dependent manner (Fig. 3).
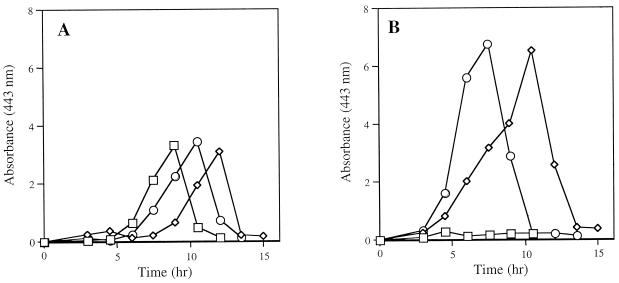
FIG. 3.
isc mutants are altered in pyruvate accumulation. Pyruvate was monitored over time through the A443 after derivatization with DNPH. Strains were grown at 37°C in minimal glucose medium supplemented with 100 nM thiamine and 20 μM NA with 50 nM (A) or 50 μM (B) FeCl3 as described in Materials and Methods. Medium samples were taken every 1.5 h during growth. The strains tested were LT2 (□), DM5419 (◊), and DM5420 (○).
A UV-visible spectrum of the DNPH-derivatized culture supernatant showed a single peak of absorbance at 443 nm, suggesting the predominant acid was pyruvate (13). The presence of pyruvate was confirmed by using spent culture medium as a substrate for d-lactic acid dehydrogenase (39). When spent minimal glucose medium (supplemented with NA and thiamine) from cultures of isc mutants was used, activity in the lactate dehydrogenase assay indicated the presence of pyruvate. Control experiments determined that the activity in this assay correlated with the accumulation, over the growth of the cultures, of a DNPH-derivatized species that absorbed at 443 nm (data not shown). In addition, aliquots of spent medium samples were derivatized before (A443, 2.1 and 1.3) and after (A443, 0.48 and 0.33) the sample was depleted of pyruvate by incubation with d-lactic acid dehydrogenase. This decrease in the A443 was similar to that found when a control pyruvate solution (1 mM) was subjected to the same treatment. In that case, an A443 of 1.53 was found upon DNPH derivatization, and when derivatization occurred after pyruvate depletion, an A443 of 0.24 was measured. Taken together, these results demonstrated that the major species resulting in the A443 peak after DNPH derivatization was pyruvate, and subsequent experiments addressing this phenomenon were performed by monitoring the absorbance of the DNPH-derivatized culture medium at 443 nM. As the data in Fig. 3 and 4 show, pyruvate accumulated in a time-dependent manner followed by a rapid decrease in concentration after 10 to 15 h of growth. This decrease correlated with the disappearance of glucose from the medium, suggesting that after the depletion of glucose, pyruvate was utilized as a carbon source (data not shown).
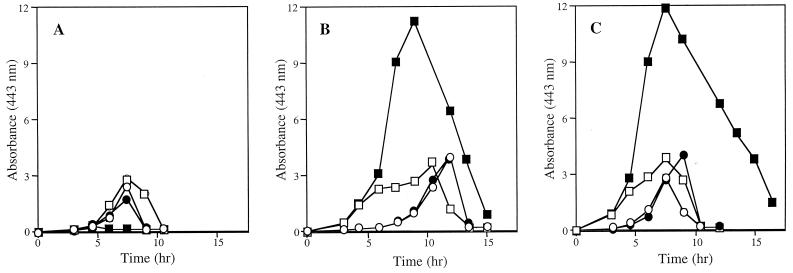
FIG. 4.
Regulation of pyruvate accumulation involves Fur. Pyruvate accumulation in the growth medium was monitored every 1.5 h as described in Materials and Methods. Strains were grown at 37°C in minimal medium supplemented with 100 nM thiamine and 20 μM NA with 50 nM (open symbols) or 50 μM (solid symbols) FeCl3. The circles represent strains containing the fur-1 null mutation, and the squares represent the isogenic fur+ strains. The strains tested were the wild type (A), iscR1::MudJ (B), and iscA2::MudJ (C).
Under the growth conditions described here, no absorbance peaks that would indicate a significant presence of other keto acids in the spent supernatant were visible after derivatization with DNPH. However, the presence of other metabolites (i.e., lactate) that would not have been derivatized was not determined.
Exogenous iron affects the level of pyruvate in the spent culture medium.
During the initial experiments described above, no efforts were made to determine or alter the levels of iron in the medium. Subsequent experiments found that the level of pyruvate detected in the spent medium was affected by the concentration of iron (Fig. 3). As shown in Fig. 3A, when the growth medium contained a low concentration of iron (50 nM FeCl3), spent culture media of isc mutant strains had levels of pyruvate similar to those of the wild-type strain. When the concentration of iron in the growth medium was increased to 50 μM FeCl3, pyruvate was undetectable in the wild-type strain culture medium. In contrast, under high-iron conditions, levels of pyruvate in the spent media from the isc mutants increased twofold relative to the low-iron medium. Titration experiments determined that as little as 1 μM FeCl3 increased pyruvate in the cultures of isc mutants while decreasing pyruvate in a wild-type culture. The pyruvate levels in cultures of isc mutants reached a plateau when 10 μM FeCl3 was added (data not shown).
Null mutations in fur eliminate the effect of iron on pyruvate levels.
Because the ferric uptake repressor protein, Fur, controls many of the processes in the cell that are responsive to iron (9), null alleles of fur were introduced into strains DM5419, DM5420, and LT2. The profile of pyruvate accumulation was determined in each of the resulting three isogenic pairs. As shown by the data presented in Fig. 4, the presence of a fur mutation eliminated the effect of exogenous iron on pyruvate levels in all cases. The three strains containing the mutant fur allele had approximately the same level of pyruvate in the medium as the isogenic fur+ strains in the presence of low iron (50 nM FeCl3). However, the addition of 50 μM FeCl3 altered the level of pyruvate in the growth medium of the fur+ strains, yet had no effect on the pyruvate levels in the culture medium of the fur mutants. Thus, the fur mutation eliminated the change in pyruvate levels in response to iron in both the wild-type and isc mutant strains.
DISCUSSION
The goal of this study was to physically define the genetic strB locus and understand the thiamine requirement resulting from mutations in this locus. We have shown that IscS is required for NA biosynthesis. In addition, we found that mutations in the isc gene cluster in S. enterica affected thiamine synthesis in at least two steps in addition to generating other significant metabolic phenotypes.
IscS function is required for THZ synthesis.
With respect to thiamine synthesis, the simplest interpretation of our results is that IscS is involved in at least one step in the biosynthesis of THZ and NA, and one or more genes downstream of iscS are also required for an additional step in THZ synthesis. A role for IscS in THZ biosynthesis is not unexpected based on the recent finding that this protein is a sulfurtransferase that can transfer sulfur to the 4-thiouridine modification of tRNA via a ThiI intermediate (31). Since ThiI is involved in the donation of sulfur to the THZ moiety of thiamine (51, 55) in addition to its role in the formation of the 4-thiouridine modification (34), it is anticipated that IscS is involved in transferring the sulfur to THZ via a ThiI intermediate.
isc functions are required for ThiH activity.
Providing iscS in trans did not eliminate the thiamine requirement of the iscR1::MudJ insertion mutant but rather resulted in a requirement that was satisfied by either THZ or tyrosine, similar to the one displayed by the iscA2::MudJ insertion. The ability of tyrosine to satisfy the THZ requirement had been noted for other mutants indirectly affected in thiamine synthesis and was subsequently shown for specific missense alleles of the thiH gene (1, 54; Gralnick et al., submitted). We have suggested that this phenotype is diagnostic of a defect in ThiH activity. The ThiH sequence contains the motif CXXXCXXCXnC and thus has the potential to contain an [Fe-S] center. In our working model, this putative [Fe-S] center is essential for the efficient function of ThiH, either because it is involved in catalysis or because it increases the stability of the protein. We suggest that tyrosine is either serving to stabilize the protein or providing increased substrate to facilitate turnover of the limited number of active ThiH molecules present. Glutathione has been shown to function in the formation and repair of [Fe-S] centers (26); thus, our recent demonstration that mutations in gshA, a gene required for the synthesis of glutathione, result in a similar tyrosine-correctable defect in THZ synthesis is consistent with this model (Gralnick et al., submitted).
isc functions are also required for the synthesis of the HMP moiety of thiamine.
The phenotypic results presented here indicated that while the THZ biosynthetic pathway is impaired in the isc mutants, the synthesis of the hydroxymethyl pyrimidine (HMP) moiety remains functional. However, in a purF mutant, isc mutations caused an additional requirement for HMP (data not shown). This finding placed isc mutations in a class of lesions that affect HMP synthesis only when flux through the purine biosynthetic pathway is reduced (1, 24, 41). The potential target for the defect in HMP synthesis caused by isc mutants was not addressed here.
Lack of isc functions disrupts iron metabolism.
While it was formally possible that the increased pyruvate in the culture media of isc mutants was simply the result of a metabolic imbalance caused by inactive enzymes (i.e., those containing [Fe-S] centers), the data suggested that this was a regulated response. Pyruvate, as well as other α-keto acids, has been associated with iron chelation in many bacterial systems, including S. enterica (17, 28, 32, 43). This was consistent with our observation that pyruvate levels in the medium were altered in response to iron concentration and that this response required the Fur protein. Reissbrodt et al. previously suggested that the intracellular iron status of the cell was important in determining the level of α-keto acids excreted (43). While the isc mutant phenotype suggested a possible role (direct or indirect) for these genes in responding to exogenous iron, a detailed model to account for the diverse metabolic phenotypes is premature.
Conclusions.
The results presented here have increased our understanding of several aspects of metabolism. The isc mutants belong to a class, also including gshA mutants, of THZ auxotrophs that can be corrected by tyrosine (Gralnick et al., submitted). The similarity of the defect in thiamine synthesis caused by these two lesions is consistent with a model involving compromised formation and repair of an [Fe-S] center in a critical enzyme, suggested to be ThiH (Gralnick et al., submitted). It is somewhat surprising that if isc mutants are defective in the formation and repair of [Fe-S] centers their phenotypes are not more severe. In the scenario we propose, the THZ requirement of these mutants indicates the cell does not tolerate a three- to fourfold decrease in the activity of ThiH, whereas a similar decrease in AC and some of the other [Fe-S] enzymes can be accommodated. The presence of significant activity of [Fe-S] proteins in isc mutants suggests there are redundant mechanisms for formation of the clusters in these cells. The identification of a Fur-related metabolic phenomenon in the isc mutants uncovered exciting questions about [Fe-S] centers, genes regulated in response to iron, and potentially new features of bacterial metabolism.
ACKNOWLEDGMENTS
We are grateful to J. Roth for providing us with strains and directing us toward their respective thiamine requirements. We appreciate the technical assistance of Shara Weber, who first sequenced the isc::MudJ insertions. We thank T. Kiley for providing results prior to publication and for helpful discussion and C. Lauhon for providing results prior to publication.
This work was supported by NIH grant GM47296 and the Shaw Scientist Program of the Milwaukee Foundation.
ADDENDUM IN PROOF
Lauhon and Kambampati have recently demonstrated that in vitro IscS mobilizes the sulfur for transfer to the ThiS thiocarboxylate, which is the ultimate sulfur donor in thiazole synthesis. These authors also show that ThiI stimulates this transfer by sevenfold. (C. T. Lauhon and R. Kambampati, J. Biol. Chem., in press).
REFERENCES
- 1.Beck B J, Downs D M. The apbE gene encodes a lipoprotein involved in thiamine synthesis in Salmonella typhimurium. J Bacteriol. 1998;180:885–891. doi: 10.1128/jb.180.4.885-891.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Begley T P, Downs D M, Ealick S, McLafferty F, van Loon D, Taylor S, Chiu H, Kinsland C, Reddick J, Xi J, Campobasso N. Thiamine synthesis in prokaryotes. Arch Microbiol. 1999;171:293–300. doi: 10.1007/s002030050713. [DOI] [PubMed] [Google Scholar]
- 3.Berkowitz D, Hushon J M, Whitfield H J, Roth J, Ames B N. Procedure for identifying nonsense mutations. J Bacteriol. 1968;96:215–220. doi: 10.1128/jb.96.1.215-220.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Caetano-Annoles G. Amplifying DNA with arbitrary oligonucleotide primers. PCR Methods Appl. 1993;3:85–92. doi: 10.1101/gr.3.2.85. [DOI] [PubMed] [Google Scholar]
- 6a.Castilho B A, Olfson P, Casadaban M J. Plasmid insertion mutagenesis and lac gene fusion with mini Mu bacteriophage transposons. J Bacteriol. 1984;158:488–495. doi: 10.1128/jb.158.2.488-495.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christian T, Downs D M. Defects in pyruvate kinase cause a conditional increase of thiamine synthesis in Salmonella typhimurium. Can J Microbiol. 1999;45:565–572. [PubMed] [Google Scholar]
- 8.Claas K, Weber S, Downs D M. Lesions in the nuo operon, encoding NADH dehydrogenase complex I, prevent PurF-independent thiamine synthesis and reduce flux through the oxidative pentose phosphate pathway in Salmonella enterica serovar Typhimurium. J Bacteriol. 2000;182:228–232. doi: 10.1128/jb.182.1.228-232.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crosa J H. Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiol Mol Biol Rev. 1997;61:319–336. doi: 10.1128/mmbr.61.3.319-336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.David S, Estramareix B, Fischer J C, Therisod M. The biosynthesis of thiamine. Synthesis of [1,1,1,5-2H4]-1-deoxy-d-threo-2-pentulose and incorporation of this sugar in biosynthesis of thiazole by Escherichia coli cells. J Chem Soc [Perkin 1] 1982;1:2131–2137. [Google Scholar]
- 11.David S, Estramareix J, Therisod M. 1-Deoxy-d-threo-2-pentulose; the precursor of the five-carbon chain of the thiazole of thiamine. J Am Chem Soc. 1981;103:7341–7342. [Google Scholar]
- 12.Davies J, Anderson P, Davis B. Inhibition of protein synthesis by spectinomycin. Science. 1965;149:1096–1098. doi: 10.1126/science.149.3688.1096. [DOI] [PubMed] [Google Scholar]
- 13.Dawson R M, Elliott D, Elliott W, Jones K M. Data for biochemical research. 3rd ed. Oxford, United Kingdom: Clarendon Press; 1986. [Google Scholar]
- 14.Demerec M, Lahr E L, Balbinder E, Miyake T, Ishidsu J, Mizobuchi K, Mahler B. Bacterial genetics. Annual report of the director of the department of genetics. Washington, D.C.: Carnegie Institution of Washington; 1960. [Google Scholar]
- 15.DeMoll E, Shive W. Determination of the metabolic origin of the sulfur atom in thiamine of Escherichia coli by mass spectrometry. Biochem Biophys Res Commun. 1985;132:217–222. doi: 10.1016/0006-291x(85)91010-1. [DOI] [PubMed] [Google Scholar]
- 16.Downs D M, Petersen L. apbA, a new genetic locus involved in thiamine biosynthesis in Salmonella typhimurium. J Bacteriol. 1994;176:4858–4864. doi: 10.1128/jb.176.16.4858-4864.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drechsel H, Thieken A, Reissbrodt R, Jung G, Winkelmann G. Alpha-keto acids are novel siderophores in the genera Proteus, Providencia, and Morganella and are produced by amino acid deaminases. J Bacteriol. 1993;175:2727–2733. doi: 10.1128/jb.175.9.2727-2733.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enos-Berlage J L, Downs D M. Involvement of the oxidative pentose phosphate pathway in thiamine biosynthesis in Salmonella typhimurium. J Bacteriol. 1996;178:1476–1479. doi: 10.1128/jb.178.5.1476-1479.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Enos-Berlage J L, Langendorf M J, Downs D M. Complex metabolic phenotypes caused by a mutation in yjgF, encoding a member of the highly conserved YER057c/YjgF family of proteins. J Bacteriol. 1998;180:6519–6528. doi: 10.1128/jb.180.24.6519-6528.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Estramareix B, David S. Conversion of 5-aminoimidazole ribotide to the pyrimidine of thiamin in enterobacteria: study of the pathway with specifically labeled samples of riboside. Biochim Biophys Acta. 1990;1035:154–160. doi: 10.1016/0304-4165(90)90110-i. [DOI] [PubMed] [Google Scholar]
- 21.Estramareix B, Therisod M. Biosynthesis of thiamine: 5-aminoimidazole ribotide as the precursor of all the carbon atoms of the pyrimidine moiety. J Am Chem Soc. 1984;106:3857–3860. [Google Scholar]
- 22.Estramareix B, Therisod M. Tyrosine as a factor in biosynthesis of the thiazole moiety of thiamine in Escherichia coli. Biochim Biophys Acta. 1972;273:275–282. [PubMed] [Google Scholar]
- 23.Frodyma M, Downs D M. The panE gene, encoding ketopantoate reductase, maps at 10 minutes and is allelic to apbA in Salmonella typhimurium. J Bacteriol. 1998;180:4757–4759. doi: 10.1128/jb.180.17.4757-4759.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frodyma M, Rubio A, Downs D M. Reduced flux through the purine biosynthetic pathway results in an increased requirement for coenzyme A in thiamine synthesis in Salmonella enterica serovar Typhimurium. J Bacteriol. 2000;182:236–240. doi: 10.1128/jb.182.1.236-240.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frodyma M E, Downs D M. ApbA, the ketopantoate reductase enzyme of Salmonella typhimurium, is required for the synthesis of thiamine via the alternative pyrimidine biosynthetic pathway. J Biol Chem. 1998;273:5572–5576. doi: 10.1074/jbc.273.10.5572. [DOI] [PubMed] [Google Scholar]
- 26.Gardner P R, Fridovich I. Effect of glutathione on aconitase in Escherichia coli. Arch Biochem Biophys. 1993;301:98–102. doi: 10.1006/abbi.1993.1120. [DOI] [PubMed] [Google Scholar]
- 27.Gruer M J, Guest J R. Two genetically-distinct and differentially-regulated aconitases (AcnA and AcnB) in Escherichia coli. Microbiology. 1994;140:2531–2541. doi: 10.1099/00221287-140-10-2531. [DOI] [PubMed] [Google Scholar]
- 28.Heuck D, Beer W, Reissbrodt R. Iron supply of staphylococci and of micrococci by alpha-ketoacids. J Med Microbiol. 1995;43:26–32. doi: 10.1099/00222615-43-1-26. [DOI] [PubMed] [Google Scholar]
- 29.Jacobson M R, Cash V L, Weiss M C, Laird N F, Newton W E, Dean D R. Biochemical and genetic analysis of the nifUSVWZM cluster from Azotobacter vinelandii. Mol Gen Genet. 1989;219:49–57. doi: 10.1007/BF00261156. [DOI] [PubMed] [Google Scholar]
- 30.Kadner R J, Murphy G P, Stephens C M. Two mechanisms for growth inhibition by elevated transport of sugar phosphates in Escherichia coli. J Gen Microbiol. 1992;138:2007–2014. doi: 10.1099/00221287-138-10-2007. [DOI] [PubMed] [Google Scholar]
- 31.Kambampati R, Lauhon C T. IscS is a sulfurtransferase for the in vitro biosynthesis of 4-thiouridine in Escherichia coli tRNA. Biochemistry. 1999;38:16561–16568. doi: 10.1021/bi991119r. [DOI] [PubMed] [Google Scholar]
- 32.Kingsley R, Rabsch W, Roberts M, Reissbrodt R, Williams P H. TonB-dependent iron supply in Salmonella by alpha-ketoacids and alpha-hydroxyacids. FEMS Microbiol Lett. 1996;140:65–70. doi: 10.1111/j.1574-6968.1996.tb08316.x. [DOI] [PubMed] [Google Scholar]
- 33.Martinez E, Bartolome B, de la Cruz F. pACYC184-derived cloning vectors containing the multiple cloning site and lacAa reporter gene of pUC8/9 and pUC18/19 plasmids. Gene. 1988;68:159–162. doi: 10.1016/0378-1119(88)90608-7. [DOI] [PubMed] [Google Scholar]
- 34.Mueller E G, Buck C J, Palenchar P M, Barnhart L E, Paulson J L. Identification of a gene involved in the generation of 4-thiouridine in tRNA. Nucleic Acids Res. 1998;26:2606–2610. doi: 10.1093/nar/26.11.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakamura M, Saeki K, Takahashi Y. Hyperproduction of recombinant ferredoxins in Escherichia coli by coexpression of the ORF1-ORF2-iscS-iscU-iscA-hscB-hscA-fdx-ORF3 gene cluster. J Biochem (Tokyo) 1999;126:10–18. doi: 10.1093/oxfordjournals.jbchem.a022409. [DOI] [PubMed] [Google Scholar]
- 36.Newell P C, Tucker R G. Biosynthesis of the pyrimidine moiety of thiamine. Biochem J. 1968;106:279–287. doi: 10.1042/bj1060279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Newell P C, Tucker R G. New pyrimidine pathway involved in the biosynthesis of the pyrimidine of thiamine. Nature (London) 1967;215:1384–1385. doi: 10.1038/2151384a0. [DOI] [PubMed] [Google Scholar]
- 38.Newell P C, Tucker R G. Precursors of the pyrimidine moiety of thiamine. Biochem J. 1968;106:271–277. doi: 10.1042/bj1060271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oeschger M P. Guanylate kinase from Escherichia coli B. Methods Enzymol. 1978;51:473–482. doi: 10.1016/s0076-6879(78)51065-3. [DOI] [PubMed] [Google Scholar]
- 40.Petersen L, Downs D M. Mutations in apbC (mrp) prevent function of the alternative pyrimidine biosynthetic pathway in Salmonella typhimurium. J Bacteriol. 1996;178:5676–5682. doi: 10.1128/jb.178.19.5676-5682.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petersen L A, Downs D M. Identification and characterization of an operon in Salmonella typhimurium involved in thiamine biosynthesis. J Bacteriol. 1997;179:4894–4900. doi: 10.1128/jb.179.15.4894-4900.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petersen L A, Enos-Berlage J E, Downs D M. Genetic analysis of metabolic crosstalk and its impact on thiamine synthesis in Salmonella typhimurium. Genetics. 1996;143:37–44. doi: 10.1093/genetics/143.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reissbrodt R, Kingsley R, Rabsch W, Beer W, Roberts M, Williams P H. Iron-regulated excretion of alpha-keto acids by Salmonella typhimurium. J Bacteriol. 1997;179:4538–4544. doi: 10.1128/jb.179.14.4538-4544.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roberts L M, Reeve E C R. Two mutations giving low-level streptomycin resistance in Escherichia coli K12. Genet Res. 1970;16:359–365. doi: 10.1017/s0016672300002640. [DOI] [PubMed] [Google Scholar]
- 45.Sanderson K E, Roth J R. Linkage map of Salmonella typhimurium, edition VII. Microbiol Rev. 1988;52:485–532. doi: 10.1128/mr.52.4.485-532.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmieger H. Phage P22 mutants with increased or decreased transduction abilities. Mol Gen Genet. 1972;119:75–88. doi: 10.1007/BF00270447. [DOI] [PubMed] [Google Scholar]
- 47.Spencer M E, Guest J R. Isolation and properties of fumarate reductase mutants of Escherichia coli. J Bacteriol. 1973;114:563–570. doi: 10.1128/jb.114.2.563-570.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stojiljkovic I, Baumler A J, Hantke K. Fur regulon in gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a fur titration assay. J Mol Biol. 1994;236:531–545. doi: 10.1006/jmbi.1994.1163. [DOI] [PubMed] [Google Scholar]
- 49.Tabor S. Expression using the T7 RNA polymerase/promoter system. In: Ausubel P A, Brent R, Kingston R E, Moore D D, Seidman J C, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Wiley; 1990. pp. 16.2.1–16.2.11. [Google Scholar]
- 50.Takahashi Y, Nakamura M. Functional assignment of the ORF2-iscS-iscU-iscA-hscB-hscA-fdx-ORF3 gene cluster involved in the assembly of Fe-S clusters in Escherichia coli. J Biochem (Tokyo) 1999;126:917–926. doi: 10.1093/oxfordjournals.jbchem.a022535. [DOI] [PubMed] [Google Scholar]
- 51.Taylor S V, Kelleher N L, Kinsland C, Chiu H-J, Costello C A, Backstrom A D, McLafferty F W, Begley T P. Thiamin biosynthesis in Escherichia coli. Identification of ThiS thiocarboxylate as the immediate sulfur donor in the thiazole formation. J Biol Chem. 1998;273:16555–16560. doi: 10.1074/jbc.273.26.16555. [DOI] [PubMed] [Google Scholar]
- 52.Taylor S V, Vu L D, Begley T P, Sprenger G A, Gringer-Meyer S, Sahm H. Chemical and enzymatic synthesis of 1-deoxy-d-xylulose-5-phosphate. J Org Chem. 1998;63:2375–2377. [Google Scholar]
- 53.Tsolis R M, Baumler A J, Stojiljkovic I, Heffron F. Fur regulon of Salmonella typhimurium: identification of new iron-regulated genes. J Bacteriol. 1995;177:4628–4637. doi: 10.1128/jb.177.16.4628-4637.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53a.Way J C, Davis M C, Morisato D, Roberts D E, Kleckner N. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene. 1984;32:369–379. doi: 10.1016/0378-1119(84)90012-x. [DOI] [PubMed] [Google Scholar]
- 54.Webb E. Ph.D. thesis. University of Wisconsin, Madison; 1999. [Google Scholar]
- 55.Webb E, Claas K, Downs D. Characterization of thiI, a new gene involved in thiazole biosynthesis in Salmonella typhimurium. J Bacteriol. 1997;179:4399–4402. doi: 10.1128/jb.179.13.4399-4402.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Webb E, Claas K, Downs D. thiBPQ encodes an ABC transporter required for transport of thiamine and thiamine pyrophosphate in Salmonella typhimurium. J Biol Chem. 1998;273:8946–8950. doi: 10.1074/jbc.273.15.8946. [DOI] [PubMed] [Google Scholar]
- 57.White R C, Spenser I D. Biosynthesis of thiamin. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: Cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 680–686. [Google Scholar]
- 58.Yuvaniyama P, Agar J N, Cash V L, Johnson M K, Dean D R. NifS-directed assembly of a transient [2Fe-2S] cluster within the NifU protein. Proc Natl Acad Sci USA. 2000;97:599–604. doi: 10.1073/pnas.97.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zheng L, Cash V L, Flint D H, Dean D R. Assembly of iron-sulfur clusters. Identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J Biol Chem. 1998;273:13264–13272. doi: 10.1074/jbc.273.21.13264. [DOI] [PubMed] [Google Scholar]
- 60.Zheng L, White R H, Cash V L, Jack R F, Dean D R. Cysteine desulfurase activity indicates a role for NIFS in metallocluster biosynthesis. Proc Natl Acad Sci USA. 1993;90:2754–2758. doi: 10.1073/pnas.90.7.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu N. Ph.D. thesis. Salt Lake City: University of Utah; 1990. [Google Scholar]