Significance
Bunyaviruses, like influenza viruses, possess a cap-dependent endonuclease (CEN) that mediates the critical cap-snatching step of viral RNA transcription. We identified the antibunyavirus compounds among influenza CEN inhibitors and identified specific structures involved in the inhibitory mechanism. These were 100- to 1,000-fold more active in vitro than ribavirin against bunyaviruses, including Lassa virus, lymphocytic choriomeningitis virus (LCMV), and Junin virus. In LCMV-infected mice, the compounds significantly decreased blood viral load, suppressed symptoms such as thrombocytopenia and hepatic dysfunction, and improved survival rates. Since effective compounds against other bunyaviruses including La Crosse virus, severe fever with thrombocytopenia syndrome virus, and hantavirus were also identified, we demonstrated the potential clinical utility of CEN inhibitors for the treatment of viral hemorrhagic diseases.
Keywords: antiviral compounds, cap-dependent endonuclease, bunyavirus
Abstract
Viral hemorrhagic fevers caused by members of the order Bunyavirales comprise endemic and emerging human infections that are significant public health concerns. Despite the disease severity, there are few therapeutic options available, and therefore effective antiviral drugs are urgently needed to reduce disease burdens. Bunyaviruses, like influenza viruses (IFVs), possess a cap-dependent endonuclease (CEN) that mediates the critical cap-snatching step of viral RNA transcription. We screened compounds from our CEN inhibitor (CENi) library and identified specific structural compounds that are 100 to 1,000 times more active in vitro than ribavirin against bunyaviruses, including Lassa virus, lymphocytic choriomeningitis virus (LCMV), and Junin virus. To investigate their inhibitory mechanism of action, drug-resistant viruses were selected in culture. Whole-genome sequencing revealed that amino acid substitutions in the CEN region of drug-resistant viruses were located in similar positions as those of the CEN α3-helix loop of IFVs derived under drug selection. Thus, our studies suggest that CENi compounds inhibit both bunyavirus and IFV replication in a mechanistically similar manner. Structural analysis revealed that the side chain of the carboxyl group at the seventh position of the main structure of the compound was essential for the high antiviral activity against bunyaviruses. In LCMV-infected mice, the compounds significantly decreased blood viral load, suppressed symptoms such as thrombocytopenia and hepatic dysfunction, and improved survival rates. These data suggest a potential broad-spectrum clinical utility of CENis for the treatment of both severe influenza and hemorrhagic diseases caused by bunyaviruses.
Human infections with certain members of the order Bunyavirales cause severe hemorrhagic fever syndromes which represent a significant threat to public health in endemic regions (1). Lassa virus (LASV) is the etiological agent associated with Lassa fever, and the number of cases has dramatically increased in Nigeria since 2016 (2). Junin virus (JUNV) causes Argentine hemorrhagic fever, which results in significant burdens of morbidity and mortality in affected regions in South America. The majority of bunyaviruses do not have effective therapeutic agents or vaccines, and the development of potent drugs against these viral diseases is a priority for global public health (3).
Cap-dependent endonucleases (CENs) represent an attractive target for antiviral drugs because they are specific to viruses and no CEN enzymes are encoded in the human genome. Since the CEN active site requires bivalent cations (Mg2+ or Mn2+) for enzymatic catalysis, metal-chelating molecules have been screened as potential inhibitors of viral metal-dependent enzymes. We previously discovered several influenza virus (IFV) CEN inhibitors (CENis) that potently inhibited viral replication (4, 5). The CEN active sites from different viral families, such as IFV, Hantaan virus, LASV, and La Crosse virus (LACV), exhibit structural homologies, which provides a rationale to develop broad-spectrum antiviral compounds that inhibit these enzymes (6). Recently, the antiviral activity of the CENi baloxavir marboxil (BXM) against hantavirus has been reported (7).
We have assembled a large number of metal-chelating molecules (ca. 40,000) including the HIV integrase inhibitors dolutegravir (8) and cabotegravir (9) and the IFV CENi baloxavir acid (BXA) (10). We have screened the antiviral compounds for several viruses, including members of the order Bunyavirales, lymphocytic choriomeningitis virus (LCMV), JUNV, LACV, severe fever with thrombocytopenia syndrome virus (SFTSV), Rift Valley fever virus (RVFV), and Thottapalayam virus (TPMV). After the screening of two-metal chelate compounds, we found highly effective antibunyavirus compounds structurally related to the IFV CENi but not to HIV integrase inhibitors. The CEN active center is also well-conserved across viruses in different families, suggesting fitness costs are associated with amino acid substitutions (11). Here, we report preclinical pharmacological data from the characterization of CENi antiviral activity against bunyaviruses in vitro and in vivo. These findings suggest that CENi could potentially serve as a selective, broad-spectrum antiviral not only for the treatment of severe influenza but also of hemorrhagic disease related to bunyavirus infections.
Results
Selection of Candidate Compounds for Broad-Spectrum Antiviral Activities.
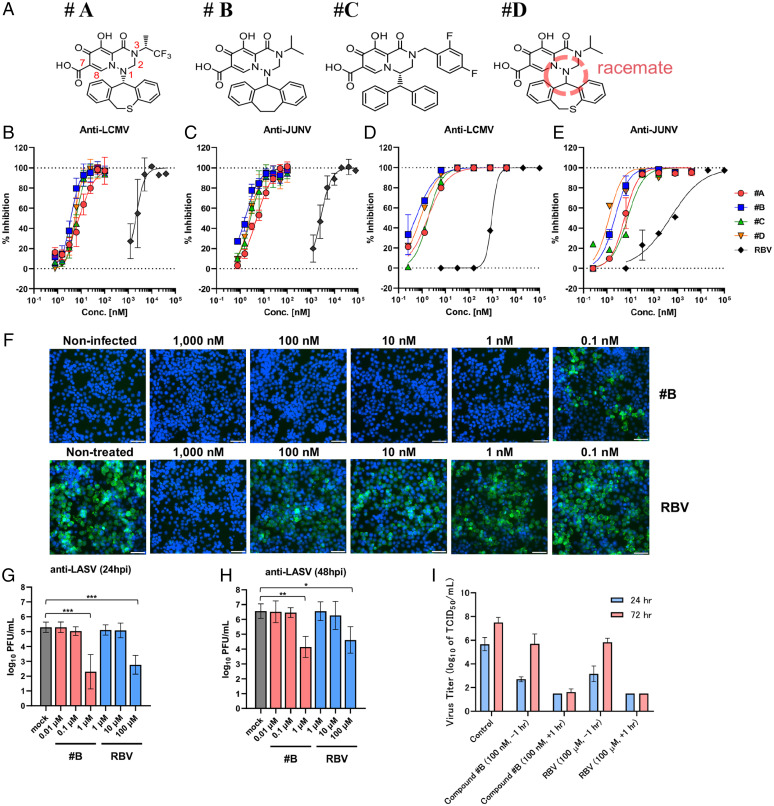
To conduct horizontal expansion and identification of antibunyavirus compounds from our antiviral library, we employed LCMV, which is closely related genetically to the risk group 4 pathogens LASV and JUNV, as a representative of South American hemorrhagic fever viruses, as screening target. To identify effective compounds, we constructed a 3-[4,5-dimethyl-2-thiazolyl]-2,5-diphenyl-2H-tetrazolium bromide (MTT) screening system, which we have used previously to identify dolutegravir (8) and BXA (10). We selected 6,077 candidates from our metal-chelating library (ca. 40,000). Of the 6,077 compounds screened in the LCMV and JUNV assay, 185 and 42 hits (3.04 and 0.69% hit rates) were identified, respectively. Each of these hits had 50% inhibition of cell viability (EC50) values at >2.5 μM concentrations and, strikingly, the chemical structures of the hits were related to the influenza CENi two-metal chelate compounds. Based on the structure of the compounds, antiviral activity was measured for about 1,000 compounds from our CENi library. Based on the concentration achieving EC50 values against LCMV and JUNV, we selected four compounds with different chemical structures: A through D (Fig. 1A). The antiviral activity against LCMV and JUNV was tested by the MTT assay. Ribavirin (RBV) was employed as a positive control for the antiviral compounds. Compounds A through D each showed >500- to 1,000-fold lower EC50 values when compared with RBV activity against LCMV (Fig. 1B) and JUNV (Fig. 1C). To assess the inhibitory activities of the compounds using another approach, viral replication assays examining LCMV infection in KB cells and JUNV infection in HEK293T cells were evaluated, and viral RNA in the supernatants was tested by qRT-PCR. The EC90 values of the four compounds were in the subnanomolar range of inhibitory activities against both LCMV (Fig. 1D) and JUNV (Fig. 1E). The antiviral data are presented in SI Appendix, Table 1. Furthermore, immunofluorescence assay revealed that LCMV nucleoprotein production was undetectable, with compound B at concentrations of 1 to 1,000 nM, and RBV inhibited only at 1,000 nM (Fig. 1F).
Fig. 1.
Influenza CENi inhibits bunyavirus infection. (A) Chemical structures of compounds employed in the present studies. Red numbers in compound A show the position numbers of the main chelate structure. Compound D was a racemic mixture of the compound isomers E and F prepared by chromatographic resolution (SI Appendix, Fig. S1A). (B) Inhibitory effect of compounds A through D and RBV in LCMV-infected KB cells by MTT assay. (C) Inhibitory effect of compounds A through D and RBV in JUNV-infected HEK293T cells by MTT assay. (D) Inhibitory effect of compounds A through D and RBV in LCMV-infected KB cells by qRT-PCR assay. (E) Inhibitory effect of compounds A through D and RBV in JUNV-infected HEK293T cells by qRT-PCR assay. (F) Immunofluorescence staining of LCMV-infected cells. KB cells were infected with LCMV and then cultured in the presence of compound B for 72 h. Cells were stained with anti-LCMV antibody (green) and Hoechst 33342 (blue). (Scale bars, 100 μm.) (G and H) Viral infectivity (PFU) at 24 (G) and 48 hpi (H) in the cell-culture supernatant of LASV-infected VeroE6 cells. *P < 0.05, **P < 0.01, ***P < 0.001, one-way ANOVA (Tukey). (I) Time-of-addition assay for compound B. Error bars represent mean ± standard deviation (SD).
Next, we tested anti-LASV activity using compound B under biosafety level 4 (BSL-4) conditions. Viral replication in LASV-infected cells and viral infectivity in the supernatants of cultures were tested by plaque assay. At 24 h postinfection (hpi), compound B showed a 3 log10 viral reduction at 1 μM concentration, whereas RBV only showed a 2 log10 reduction at 100 μM concentration (Fig. 1G). At 48 hpi, compound B showed a 2.5 log10 viral reduction at 1 μM concentration, whereas RBV only showed a 2 log10 reduction at 100 μM concentration (Fig. 1H). These data demonstrated that compound B possessed antiviral activity against not only LCMV and JUNV but also LASV, and the efficacy was ∼100- to 1,000-fold higher than RBV.
To investigate the broad-spectrum inhibitory activity of these compounds, the antiviral activity of compounds A through D against LACV, SFTSV, RVFV, TPMV, and IFVs (A/H5N1 and A/H7N9) was tested using the MTT assay (SI Appendix, Fig. 1A). RBV and favipiravir were employed as positive controls of antiviral activity. BXA was employed as a positive control for antiinfluenza activity. Compounds A through D each showed more than 200-fold higher antiviral activities against LCMV, JUNV, and LACV, when compared with RBV and favipiravir. Compounds A, B, and D also showed antiviral activity against LACV, SFTSV, and influenza A/H5N1 and A/H7N9, and no detectable antiviral activity against RVFV and TPMV in vitro. BXA had strong antiviral efficacy against IFVs (H5N1 and H7N9) and weak activity against LACV, but was ineffective (>1,000 nM) against other viruses that were used in our studies.
Compound D was a racemic mixture of the compound isomers E and F prepared by chromatographic resolution (SI Appendix, Fig. 1A). Isomer E was more active than F, and was effective against LCMV, JUNV, LACV, SFTSV, and influenza A/H5N1 and A/H7N9. Compounds G and H had the same side chain (benzhydryl group) with different positions of the R1 side chain, and H with the R1 side chain in the second position had decreased activity against influenza A/H5N1. Compound I had no carboxylic acid moiety at the seventh position and showed decreased activity against LCMV and JUNV and increased activity for IFVs. Only F through H showed inhibitory activity against TPMV, which belongs to hantaviruses. All compounds, excluding E and F that were not tested, had an inhibitory activity on the influenza CEN enzyme.
We next investigated the toxicity of these compounds to cells and found that compounds A through D possessed less cytotoxicity and that the selectivity index (SI), which measures the difference between drug efficacy and toxicity, was high. Specifically, the SI of compound B was 1,000 times or greater in the LCMV and JUNV assays, and no cytotoxicity was observed in VeroE6 cells, employed in the LASV experiments, even at 10,000 nM (SI Appendix, Fig. 1B).
Furthermore, time-of-addition assays for compound B and RBV were performed to determine the effect on the viral life cycle. RBV is known to inhibit virus replication at the postentry stage by targeting the viral polymerase, resulting in the inhibition of viral propagation. While pretreatment with RBV or compound B decreased infectious viral particles in culture supernatants compared with a nontreatment control at 24 hpi, viral titers in supernatants following pretreatment with these compounds increased at 72 hpi. On the other hand, posttreatment with RBV or compound B significantly reduced viral propagation at both 24 and 72 hpi. This suggested that compound B mechanistically inhibited postentry steps in viral propagation in a comparable manner to RBV (Fig. 1I).
Identification of the Action Mechanism of the Compounds by Selection of Drug-Resistant Mutants.
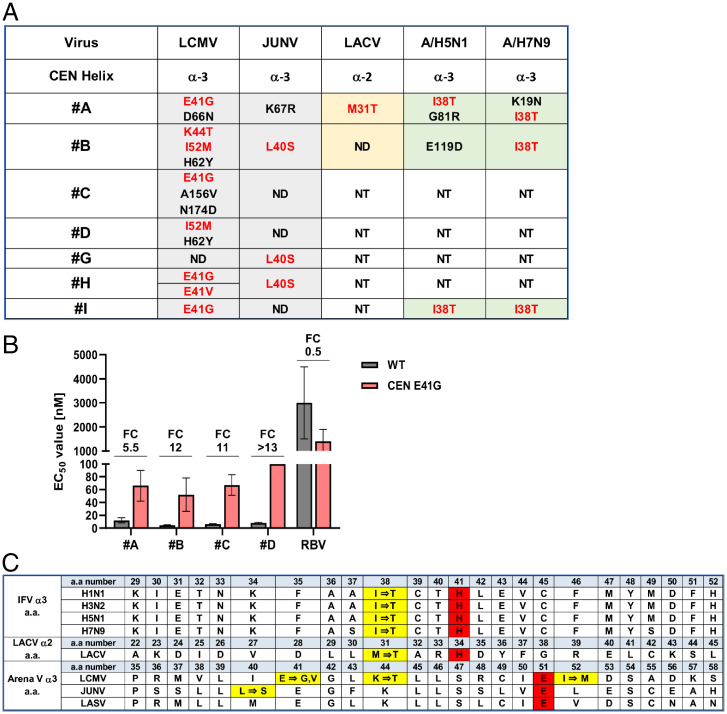
Notably, all of the hit compounds were analogs of IFV CENi; however, the mechanism of action of these compounds against bunyaviruses remained unknown. To address this, we isolated drug-resistant mutants derived from LCMV and JUNV propagated in cultured cells under drug selection with antibunyavirus compounds A through D and G through I. Whole-genome sequences of the isolated viruses were analyzed by next-generation sequencing (SI Appendix, Table 2). A summary of the drug-resistant mutant data on the CEN is shown in Fig. 2A. The adjacent mutations E41G, E41V, K44T, and I52M for LCMV and the L40S and K67R mutations in JUNV were detected in the CEN α3-helix following long-term passage under drug selection.
Fig. 2.
Identification of target viral proteins of CENi compounds by isolation of drug-escape mutants. (A) Amino acid substitutions of drug-escape mutants of LCMV, JUNV, LACV, and IFVs A/H5N1 and A/H7N9. Red letters are corresponding mutations in the CEN α3-region of LCMV, JUNV, and IFVs A/H5N1 and A/H7N9 or the α2-region of LACV. ND, not detected; NT, not tested. (B) Inhibitory effect of compounds A through D in LCMV-infected KB cells by MTT assay. Fold change (FC) is the EC50 of the LCMV E41V mutant divided by that of the WT virus. Raw data are shown in SI Appendix, Table 3. Error bars represent mean ± SD. (C) Amino acid (a.a.) sequence alignment of the α3- or α2-region of CEN. The alignment was based on prior research (6). Yellow columns are the amino acid substitutions in drug-resistant mutants. Red columns are catalytic sites essential for CEN activity.
To confirm the drug susceptibility of drug-resistant mutants, the viral replication kinetics of the selected clones of LCMV were tested. All viruses had decreased replicative capacity with the exception of LCMV clone 1 (E41G), which possessed sufficient replication for evaluation by the MTT assay. Fig. 2B shows the EC50 values of compounds for LCMV clone 1 (E41G) compared with the wild-type (WT) virus. LCMV with the E41G mutation showed an ∼10-fold or greater antiviral resistance in the presence of compounds B through D compared with the parental WT virus. These findings suggest that the LCMV E41G mutation in the CEN α3-helix is the residue related to the development of CENi drug resistance (SI Appendix, Table 3). To investigate the virological characteristics of the E41G mutant, we compared the growth curves of the LCMV WT and the E41G mutant. In KB cells, propagation of the E41G mutant was significantly reduced compared with that of WT, and the decreased propagation of the LCMV E41G was independent of the viral infection dose employed (multiplicity of infection [MOI] 0.1 or 0.01) (SI Appendix, Fig. 2). These results suggested that the LCMV mutant with the E41G substitution in the L protein possessed a replication deficiency compared with the WT virus. This is consistent with the strategy that antiviral targeting of the virally encoded CEN activity of bunyaviruses could be of benefit.
Some of the compounds had antiviral activity against LACV, A/H5N1, and A/H7N9, and their drug-resistant mutants were derived by propagation in cultured cells under drug selection. For IFV A/H5N1 and A/H7N9 mutants, amino acid substitutions, including I38T in the CEN α3-helix of the polymerase acidic subunit (PA), were detected in all tested compounds (A, B, and I). These results were consistent with prior findings that IFV A/H1N1 and A/H3N2 CENi drug-resistant mutants harbored the adjacent amino acid substitution I38T in the CEN α3-helix of the PA (12). For LACV, the M31T mutation was detected in the CEN α2-helix following selection for drug-resistant mutants to compound A. Fig. 2C shows amino acid sequence alignment of the α3- or α2-region of CEN. The alignment was based on prior research and red columns are the catalytic sites essential for CEN activity (6). Yellow columns are the amino acid substitutions in drug-resistant mutants, and amino acid substitutions were found at approximately the same location in all viruses. SI Appendix, Table 3 shows the EC50 values of compounds for the A/H5N1 clone (I38T) compared with the WT virus. A/H5N1 with the I38T mutation showed an ∼10-fold or greater antiviral resistance in the presence of compounds A through D. These findings suggest that the A/H5N1 I38T mutation in the CEN α3-helix is the residue responsible for the development of CENi drug resistance.
Effect of CENi Chelate Compounds on Survival in a Murine Model of LCMV.
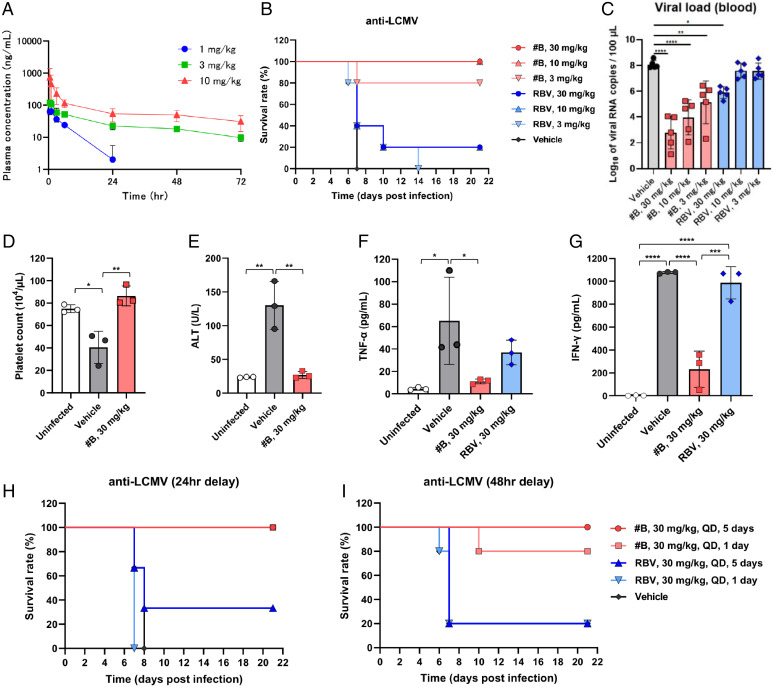
To confirm whether the compound also shows potent in vivo efficacy, a mouse model of lethal LCMV infection was first established. We selected compound B using this model, which had the most potent antiviral activity and favorable pharmacokinetics among compounds A through D in a rat study (SI Appendix, Table 4). For compound B, plasma concentrations after intramuscular administration were also measured (Fig. 3A). The survival rate of ICR mice infected with the LCMV WE strain was 0% at 14 d postinfection. Using this lethal mouse model, the in vivo efficacy of compound B was evaluated (Fig. 3B). Following intraperitoneal infection with 3.0 × 103 50% tissue culture infectious dose (TCID50) LCMV, groups of mice (n = 5) were treated by single daily (QD) administrations of either 3, 10, or 30 mg/kg dose of compound B, RBV, or vehicle on the same day of virus challenge. Intramuscular treatment with 10 or 30 mg/kg or 3 mg/kg of compound B showed 100 or 80% survival rates, respectively. In contrast, the administration of 30 mg/kg of RBV resulted in a 20% survival rate. Moreover, compound B reduced the viral load in blood compared with vehicle-treated mice at 1 d after infection in a dose-dependent manner. Compound B–treated mice maintained similar alanine aminotransferase (ALT) and platelet counts as uninfected mice (Fig. 3 C–E).
Fig. 3.
Compound B improved survival rates in a lethal murine model with LCMV infection. (A) Concentrations of compound B in plasma after a single intramuscular administration in mouse. (B) Survival rates in a lethal murine model with LCMV infection (n = 5 per group). Compound B (3 to 30 mg/kg), RBV (3 to 30 mg/kg), or vehicle control (0 mg/kg) was administered intramuscularly for 1 d. Dose-dependent effects of compound B and RBV were compared on survival rates. Dosing regimens are shown (Right). (C) Viral load in blood of compound B– and RBV-treated groups was compared with vehicle-treated groups at 1 d after infection. (D and E) The platelet counts (D) and ALT levels (E) in the blood of the compound B–treated group were compared with those in the vehicle-treated group at 5 d after infection. (F and G) The serum TNF-α (F) and IFN-γ levels (G) in the compound B– and RBV-treated groups were compared with those in the vehicle-treated group at 5 d after infection. Survival rates in a lethal murine model of LCMV infection with delayed treatment. (H and I) Compound B (30 mg/kg), RBV (30 mg/kg), or vehicle control (0 mg/kg) was administered intramuscularly at 24 (H) or 48 hpi (I). The survival rates of compound B (treatment for 1 or 5 d) and RBV (treatment for 1 or 5 d) were compared with that in the vehicle-treated group. Dosing regimens are shown (Right). Error bars represent mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, one-way ANOVA (Tukey).
Additionally, pathology and immunohistochemistry (IHC) analyses were conducted. In the vehicle-treated and RBV-treated mice, inflammatory foci were observed, which were absent in the uninfected and compound B–treated mice. Moreover, IHC showed almost no viral antigen was detected in the compound B–treated mice. In contrast, viral antigens were detected in hepatocytes of both vehicle- and RBV-treated mice (SI Appendix, Fig. 3). Next, serum interferon γ (IFN-γ) and tumor necrosis factor α (TNF-α) levels were evaluated. In the compound B–treated mice, IFN-γ levels were fivefold lower than that in vehicle-treated mice and TNF-α levels were comparable to that of the uninfected mice. On the other hand, in the RBV-treated mice, IFN-γ was at an equivalent level to the vehicle-treated mice and TNF-α was higher than that in compound B–treated mice (Fig. 3 F and G). These results demonstrated that compound B had much more potent anti-LCMV efficacy than RBV in vivo.
Finally, the delayed-treatment efficacy of compound B was evaluated. LCMV-infected mice (n = 3) were treated QD by intramuscular administration for 1 or 5 d, with a 30 mg/kg dose of compound B, RBV, or vehicle at 24 or 48 hpi. At 24 hpi, compound B–treated mice in a 1- or 5-d group showed a 100% survival rate (Fig. 3H). And, at 48 hpi, compound B–treated mice in a 1- or 5-d group showed an 80 or 100% survival rate, respectively (Fig. 3I). These results suggest that compound B remains effective even after the virus has propagated in vivo.
Discussion
Our drug discovery goal is the identification of compounds that are effective against a broad spectrum of viral families, and at present we are specifically targeting the discovery of drugs with efficacy against members of the order Bunyavirales. These viral infections have high mortality rates, and no currently available licensed antivirals or vaccines have been proven to be effective in the treatment of their respective associated viral hemorrhagic diseases. In this study, we constructed an MTT assay system for different bunyaviruses to identify antiviral candidate compounds from an influenza CENi library. We identified broad-spectrum antiviral compounds with inhibitory effects in vitro and in vivo against numerous members of the family Arenaviridae within the order Bunyavirales.
Arenavirus infections are important zoonotic diseases transmitted from rodents with high morbidity and mortality and are divided into two groups, Old and New World viruses, with at least eight pathogens known to cause hemorrhagic fevers (13). Thus, the development of antiviral therapies and prophylactic vaccines is a major unmet global public health need that could significantly reduce disease burdens. LCMV was the first described arenavirus and causes aseptic meningitis, a severe human disease involving inflammation of the brain and spinal cord, while JUNV, LASV, Guanarito virus, Lujo virus, Machupo virus, Sabia virus, and Whitewater Arroyo virus cause viral hemorrhagic fever syndromes (3). Among them, LASV, belonging to the family Arenaviridae, was scored as the top-ranked virus for the risk of animal-to-human spillover for newly discovered viruses (14). We have succeeded in the identification of a compound with nanomolar levels of antiviral activity against LCMV and JUNV and, importantly, which were also proven to possess antiviral efficacy, as assessed by the decreased symptoms and suppressed lethality, in an in vivo LCMV mouse model. Notably, this compound also has CEN enzyme-inhibitory activity against IFVs, and we infer from the arenavirus drug selection experiments that the mechanism of action was inhibition of the virally encoded CEN enzyme. Since this compound also had antiviral effects on LASV, LACV, and IFV, it seems reasonable to conclude that it may be effective against all arenaviruses and thus represent the long-sought-after broad-spectrum antiviral for treatment of arenavirus infections.
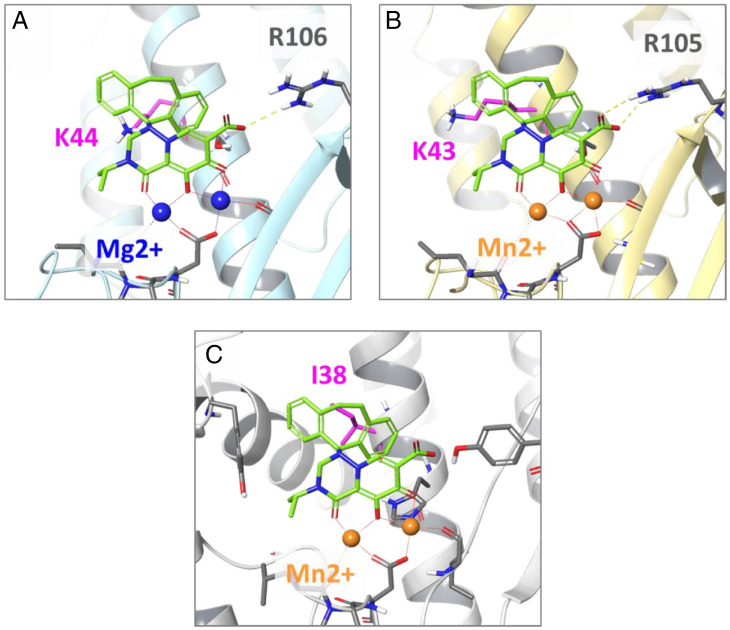
CEN is conserved in the viral families Orthomyxoviridae and order Bunyavirales and mediates cap snatching during viral RNA transcriptional initiation, which is essential for viral replication. Furthermore, CEN enzymes are not present in humans and therefore represent ideal broad-spectrum antiviral drug targets. Based on the present findings using drug-resistant mutants selected in the presence of the antivirals, the examined agents were associated with the clustering of nonsynonymous mutations in the CEN α3-regions of both IFVs and bunyaviruses. CEN coordinates two divalent cations, which are absolutely required for enzyme function, with catalytic amino acids constituting the active site. The cocrystal structures of influenza A and B virus CENs with BXA have been solved, and the I38T substitution in the α-chain affects the interaction with the hydrophobic aromatic ring of the compound (4). Based on the crystal structures of LCMV and LASV, we speculate that compound B has an active center capable of binding tightly to the CEN active site (Fig. 4). The model suggests that the seventh carboxylic acid moiety of the compounds is also required for their antiviral activity against bunyavirus when bound to CEN R105 (or R106).
Fig. 4.
Binding model of compound B to LASV, LCMV, and influenza A virus. Superimposed structures of compound B bound to the endonuclease of (A) LASV (PDB ID code 4miw), (B) LCMV (PDB ID code 5t2t), and (C) influenza A (PDB ID code 6fs6). Residues where amino acid substitutions in the CEN active center were identified following drug selection and the predicted location of the divalent metal cation are indicated. The 44th amino acid in the CEN region of the LCMV Armstrong strain was K44, and the genetic information of LCMV used in this model is K43.
To test this hypothesis, antiviral activities were measured for the 7-position transformants for compound B and BXA. As a result, for LCMV and JUNV, the compound with the 7-position or carboxylic acid showed strong antiviral activities whereas, in contrast, in IFV assays, the compound without the 7-position–modifying group also showed strong antiviral activity. Furthermore, we investigated other side chains in this 7-position carboxylic acid and found that the carboxylic acid was the most active against both LCMV and JUNV (SI Appendix, Fig. 4).
Strikingly, a carboxylic acid moiety at the seventh position of the main structure was necessary for the antiviral activity. We examined ∼1,000 compounds and investigated the correlation between the chemical structures and antiviral activities against LCMV and JUNV. The EC50 values of the 116 compounds against LCMV and JUNV were plotted, and a significant correlation was observed between the LCMV and JUNV EC50 values (SI Appendix, Fig. 5), confirming the requirement of a carboxylic acid at the seventh position of the main structure for nanomolar-level anti-LCMV and anti-JUNV activity.
As two-metal chelators bind divalent cations, which are essential for CEN catalytic activity, highly active hit compounds can be identified at the early stages of drug discovery. We demonstrated that these compounds had nanomolar-level activity against LCMV, JUNV, LASV, LACV, and IFV, but no or lowered activity against SFTSV, RVFV, and TPMV. Recently, Wang et al. (15) reported that BXA inhibited the replication of bunyaviruses (SFTSV and Heartland virus) and the EC50 value for SFTSV was 263 nM; however, BXA did not inhibit the replication of SFTSV at 1,000 nM in our experiment. We believe that the reason for this was the difference in assay methods. Some articles have previously reported mechanistic differences among cap-snatching endonucleases from segmented negative-strand RNA viruses, including LCMV, RVFV, and hantavirus (16, 17). We speculate the structure of the target region for the compounds could be different, particularly for RVFV and TPMV. Thus, it will be necessary to screen new chemical compound libraries in the future to expand the number of potential antiviral candidate structures. In addition, various chemical side chains can be introduced onto different two-metal chelate skeletons, and compounds with various desirable physicochemical properties (e.g., solubility, pharmacokinetics) can be designed in combination with the second pharmacophore, making it possible to design inhibitors for specific pathogen targets or compounds that are conceivably more broadly active against a number of viral families. Thus, two-metal chelators have been demonstrated to have the potential to inhibit the replication of a broad range of viral families, and could potentially increase the development of new therapeutic agents for the treatment of viral hemorrhagic fevers.
Materials and Methods
Cell Culture and Viruses.
Baby hamster kidney 21 (BHK-21) cells, human A549 cells, Madin–Darby canine kidney (MDCK) cells, Madin–Darby bovine kidney (MDBK) cells, and African green monkey VeroE6 cells were maintained in high-glucose Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) at 37 °C under 5% CO2. HEK293T cells and KB cells were maintained in minimum essential medium (MEM) supplemented with 5% FBS. For IFV A/Anhui/1/2013 (A/H7N9), the cleavage of hemagglutinin by host cell proteases is essential for virus infectivity and propagation. MA104 cells stably expressing the transmembrane protease TMPRSS2 (18, 19) (MA104-TP2 cells) were used for propagation of A/H7N9. MA104-TP2 was maintained in high-glucose DMEM supplemented with 10% FBS. LCMV (Armstrong and WE strains), JUNV (Candid 1 strain), LACV (ATCC VR-1834), and TPMV (VRC-66412 strain) were grown in BHK-21 cells. Influenza A/H1N1 (A/WSN/33 strain) and SFTSV (ArtLN/2017 strain) (20) were grown in MDCK cells. IFV A/H5N1 (A/Hong Kong/483/97) and A/H7N9 (A/Anhui/1/2013) were propagated in embryonated chicken eggs and harvested from virus-containing allantoic fluids. Recombinant RVFV (MP-12 strain) was rescued from the plasmids (21) using BSR-T7/5 cells (22) and grown in BHK-21 cells. LASV (Josiah strain) (23) was grown in VeroE6 cells.
Compounds.
All screening compounds were supplied by Shionogi & Co., Ltd. RBV was purchased from Wako Pure Chemical Industries. Favipiravir (T-705) was purchased from PharmaBlock Sciences. For in vitro experiments, all compounds were dissolved in dimethyl sulfoxide (DMSO) and diluted in 2% FBS-MEM for antiviral assays. For in vivo experiments, compounds were dissolved in phosphate-buffered saline (PBS) with 5% DMSO to prepare 3 mg/mL solutions and each solution was diluted with the same vehicle for dosing.
Inhibition of In Vitro Cap-Dependent Endonuclease Activity.
The methods for the inhibition of CEN activity were previously described (4, 5). In brief, viral ribonucleoproteins (vRNPs) of the A/WSN/33 (H1N1) strain were purified from viruses propagated in embryonated chicken eggs. The allantoic fluids were recovered and virus particles were purified by ultracentrifugation using 20% sucrose and solubilized using Triton X-100 and lysolecithin. The vRNP fractions were collected by ultracentrifugation and used as sources of CEN activity.
The oligonucleotide RNA (sequence: 5′-cap-8AA UA5 GCA UCA CUA GUA AGC UUU GCU CUA-BHQ2-3′) was used as a substrate, where “8” indicates 2′-O-methyl-G, and “5” indicates cyanine 3′-labeled U. The enzymatic reaction was performed in reaction buffer (1 mM MgCl2, 50 mM NaCl, 1 mM dithiothreitol, 0.05% Tween 20, 3 nM capped RNA substrate, and 50 mM Tris⋅HCl, pH 7.8). Each reaction mixture was incubated at 37 °C for 1 h followed by adding excess ethylenediaminetetraacetic acid (EDTA) to stop the reaction. The products were examined using the 3130xL Genetic Analyzer and relatively quantitated using GeneMapper version 4.1 software (Life Technologies). The drug concentration achieving 50% inhibition of the enzymatic production (IC50) was calculated by dividing the peak area of the Cy3-labeled enzymatic product by the peak area of the internal standard (15-nt-sized marker in GeneScan-120 LIZ).
In Vitro Antivirus Activity and Cell Viability.
The MTT assay was performed to evaluate cell viability following viral infection according to methods previously described (24). For experiments evaluating drug inhibition of viral replication, each sample was serially diluted in twofold increments in duplicates and plated on 96-well microplates. Cells used for each virus were infected with 4 to 10 TCID50 and added to the plates. Plates were incubated for 3 to 4 d, and cytopathic effect (CPE) was determined by calculation of 50% end points using MTT. The detailed assay conditions for each virus are shown in SI Appendix, Fig. 6. The EC50 was defined in Prism version 8.4.3 (GraphPad Software) with a variable slope (four parameters). Noninfected cells were used as a control for 100% inhibition whereas, for infected cells, DMSO-treated cells were used as a control for 0% inhibition. The concentration achieving 50% cytotoxicity (CC50) was also measured for each cell line using the same method. Cell-free samples were used as a 100% cytotoxicity control, and DMSO-treated cells were used as a 0% cytotoxicity control. The selectivity index was calculated as mean EC50/mean CC50.
Virus Yield Reduction Assay.
The qRT-PCR assay was performed to calculate the levels of viral RNA in culture supernatants following viral infection. KB cells were infected with LCMV and HEK293T cells were infected with JUNV in 96-well plates with 10 TCID50 per well. The infected cells were treated with serial fivefold dilutions of inhibitors and cultured at 37 °C in a 5% CO2 incubator. At 72 h after infection, the culture supernatant was collected, and RNA was extracted. The expression levels of vRNA from CEN regions were quantified as described. qRT-PCR was performed using the Brilliant III Ultra-Fast SYBR Green QRT PCR Master Mix, 1-Step Kit (Agilent Technologies). The primers for the qRT-PCR are listed in SI Appendix, Table 5. The EC90 value was defined in GraphPad Prism version 8.4.3 with a variable slope (Find ECanything, F = 90).
For LASV, the plaque assay was performed to calculate the viral titer in culture supernatants following viral infection. VeroE6 cells were infected with LASV in 24-well plates at an MOI of 0.01, and incubated for 1 h at 37 °C. Followed by washing with DMEM containing 2% FBS three times, infected cells were cultured in DMEM, 2% FBS and various doses of the compound at 37 °C. Samples were collected at 24 and 48 h after infection and titrated by the plaque assay to calculate plaque-forming units (PFUs) as described elsewhere (25). All work with LASV was performed in the BSL-4 facility of the Galveston National Laboratory at the University of Texas Medical Branch in accordance with institutional guidelines.
Virus Antigen Reduction Assay.
For immunofluorescence staining, KB cells were infected with LCMV in 24-well plates with 10 TCID50 per well. The infected cells were treated with serial 10-fold dilutions of inhibitors and cultured in a 5% CO2 incubator. At 72 h after infection, infected cells were fixed with 100% methanol at −20 °C and stained with anti-LCMV nucleoprotein rabbit monoclonal antibody (Bio X Cell) in 25% Block Ace (KAC) in PBS for 1 h. Alexa Fluor 555–conjugated anti-rabbit immunoglobulin G antibody (Invitrogen, Thermo Fisher Scientific) was used as the secondary antibody. Nuclei were stained with Hoechst 33342 (Invitrogen). Fluorescent images were captured using an IX73 fluorescence microscope (Olympus).
Time-of-Addition Assay.
LCMV Armstrong strain was incubated in the presence or absence of compound B (100 nM) or RBV (100 μM) for 1 h at 37 °C, and then KB cells were inoculated with LCMV incubated with or without inhibitors at an MOI of 0.1 for 1 h. After washing, KB cells were cultured with fresh media for 24 or 72 h. As a posttreatment condition, KB cells were infected with LCMV without pretreatment with inhibitors and were cultured in the presence of inhibitors (compound B [100 nM] or RBV [100 μM]) for 24 or 72 h after cell washing. Culture supernatants were collected at 24 or 72 hpi, and infectious viral titers in supernatants were measured by TCID50 assay using MDCK cells. In brief, collected culture supernatants including LCMV were serially diluted 10-fold with MEM supplemented with 2% FBS. MDCK cells were inoculated with serially diluted supernatants in a 96-well plate and cultured with MEM supplemented with 2% FBS for 72 h. At 72 hpi, viral titers were determined by microscopy of CPE.
Isolation and Characterization of Drug-Resistant Mutants.
To isolate drug-resistant LCMV mutants, LCMV-infected and noninfected KB cells were seeded into the same 12-well plate in the presence of the compounds. Three drug concentrations, specifically 2× EC50, 6× EC50, and 20× EC50, were used. Infected cells were passaged every 2 to 3 d. When a CPE was observed by microscopy, LCMV-infected culture supernatant was transferred to noninfected KB cells in the presence of the compounds. After continuous culture of the viruses for ∼26 to 30 d for LCMV, the viral genome was then examined by sequencing the CEN region. To isolate drug-resistant JUNV mutants, JUNV-infected and noninfected HEK293T cells were seeded into the same 12-well plate in the presence of the compounds, and passage was performed as described for the isolation of drug-escape LCMV variants. To isolate drug-escape LACV mutants, BHK-21 cells were infected with LACV, passaged twice (7 d), used to infect NIH 3T3 cells, and passaged six times every other day (12 and 19 d in total).
A/H5N1-infected BHK-21 cells and A/H7N9-infected MA104-TP2 cells were also used to isolate drug-resistant variants. Three drug concentrations, namely 10× EC50, 30× EC50, and 100× EC50, were used. In dose-escalating and dose-constant conditions, the drug-escape mutants were detected following 3 wk in culture. However, the replication of A/H5N1 and A/H7N9 was rapid, and it was difficult to suppress viral replication using drugs. Taking advantage of the fact that CENis have strong ability to suppress viruses, we performed a method to isolate drug-escape viruses by changing the drug concentration in two steps, namely in the virus growth phase in the presence of drugs and in the virus selection phase. We employed modified isolation methods (two-step method) and isolated CEN variants carrying I38T in the PA (∼7 to 8 d).
Genome Sequence Analysis.
To analyze mutations, viral RNA was extracted from culture supernatant or infected cells using a PureLink Viral RNA/DNA Mini Kit (Thermo Fisher Scientific), Wizard SV 96 Binding Plates (Promega), or Direct-zol RNA Mini Prep Kit (Zymo Research). The CEN region of each virus was amplified by qRT-PCR using a PrimeScript II High Fidelity One Step RT-PCR Kit (Takara Bio) and specific primers (SI Appendix, Table 5). Sequencing of the products was performed using a 3500xL Genetic Analyzer (Life Technologies). The nucleotide sequence of the CEN region was compared with that of the WT virus, and amino acid substitutions were identified. Whole-genome sequencing was performed using the Ion Torrent PGM System (Life Technologies) as previously described (26). Briefly, viral RNA was extracted using a PureLink Viral RNA/DNA Mini Kit, and double-stranded complementary DNA (cDNA) was synthesized using a PrimeScript Double Strand cDNA Synthesis Kit (Takara Bio). Double-stranded cDNA was fragmented using a Covaris S2 focused ultrasonicator and used to prepare a 400-base-read library using an Ion Plus Fragment Library Kit (Life Technologies). After emulsion PCR, sequencing was performed using an Ion PGM sequencer with an Ion PGM Sequencing 400 Kit and Ion 318 Chip V2 (Life Technologies). Data analysis was performed using CLC Genomics Workbench version 7.5.1 software (CLC bio, Qiagen).
Pharmacokinetic Parameters of Compounds in Rat Plasma.
The animal study protocol was approved by the director of the institute after review by the Institutional Animal Care and Use Committee of Shionogi & Co., Ltd. relative to the 3R (replacement/reduction/refinement) principle. Male Crl:CD (SD) rats (8 wk old) were purchased from Japan SLC. The oral dosing vehicle was 25% DMSO, 0.5% (weight/volume) methylcellulose. Compounds were orally administered at 2 μmol⋅5 mL−1⋅kg−1 (n = 2) under the nonfasting condition. Blood samples were collected with disposable syringes containing anticoagulants (EDTA 2K and heparin) at 0.25, 0.5, 1, 2, 4, 6, 8, and 24 h after dosing. The intravenous dosing vehicle was 50% dimethylacetamide/propylene glycol. Compounds were intravenously administered at 1 μmol⋅1 mL−1⋅kg−1 (n = 2) under nonfasting conditions. Blood samples were collected with disposable syringes containing anticoagulants (EDTA 2K and heparin) at 0.033, 0.083, 0.25, 0.5, 1, 2, 4, and 6 h after dosing. Blood samples were centrifuged (3,000 rpm, 4 °C, 10 min) to obtain plasma samples, which were transferred to individual tubes and stored in a freezer until analysis. Plasma concentrations were determined using a liquid chromatography–tandem mass spectrometry system following protein precipitation with acetonitrile. The pharmacokinetic parameters of compounds in plasma were calculated using WinNonlin (version 8.3, Certara) based on noncompartment analysis with uniform weighting. For intramuscular injection of compound B, five female ICR mice (6 wk old) were purchased from CLEA Japan. Compound B was dissolved in PBS with 5% DMSO to prepare 0.3, 1, or 3 mg/mL solutions. The solutions were intramuscularly administered under nonfasting conditions. Blood samples were collected with disposable capillaries containing heparin at 0.5, 1, 3, 6, 24, 48, and 72 h after administration. Blood samples were centrifuged (12,000 rpm, 4 °C, 2 min) to obtain plasma samples, which were transferred to individual tubes and stored at −20 °C until analysis. Plasma concentrations were determined and the pharmacokinetic parameters of compounds in plasma were calculated by the same methodology as shown above.
In Vivo Efficacy of CENis in a Lethal Murine Model of LCMV Infection.
Six-week-old specific pathogen-free female ICR mice (CLEA Japan) were used for all experiments. All mouse studies were conducted under applicable laws and guidelines and with the approval of the Animal Care and Use Committee of the Graduate School of Veterinary Medicine of Hokkaido University. To compare the in vivo efficacy of compound B with RBV, mice were intraperitoneally inoculated with 3.0 × 103 TCID50 of the LCMV WE strain. Immediately after infection, mice were intramuscularly administered compound B (3, 10, or 30 mg/kg), RBV (3, 10, or 30 mg/kg), or Ctrl (vehicle) QD for 1 d. Survival was monitored daily for up to 21 d after infection. For delayed treatment, mice were intramuscularly administered compound B (30 mg/kg), RBV, or only the empty vehicle QD for 1 or 5 d starting 24 hpi. Survival was monitored daily up to day 21 after infection.
Viral Load in Blood.
To quantify viral RNA levels in blood, mice were intraperitoneally inoculated with 3.0 × 103 TCID50 of the LCMV WE strain. Immediately after infection, mice were intramuscularly administered compound B (3, 10, or 30 mg/kg), RBV (3, 10, or 30 mg/kg), or vehicle QD for 1 d. At 1 d after infection, whole blood was collected from each mouse, mixed with TRIzol (Invitrogen, Thermo Fisher Scientific), and subjected to RNA extraction using a Direct-zol RNA Mini Prep Kit. The viral RNA levels were analyzed by qRT-PCR using Brilliant III Ultra-Fast qRT-PCR Master Mix. qRT-PCR was performed using a StepOne Plus qPCR System (Applied Biosystems).
Blood Biochemistry Profile.
To investigate the blood biochemistry profiles, mice were intraperitoneally inoculated with 3.0 × 103 TCID50 of the LCMV WE strain. Immediately after infection, mice were intramuscularly administered compound B (30 mg/kg) or vehicle QD for 1 d. At 5 d after infection, whole blood was collected from each mouse and analyzed for platelet counts and ALT levels using a Celltac Alpha MEK-6450 (Nihon Kohden) and VETSCAN VS2 (Abaxis), respectively.
Cytokine Quantification.
To evaluate the cytokine levels in serum, mice were intraperitoneally inoculated with 3.0 × 103 TCID50 of the LCMV WE strain. Immediately after infection, mice were intramuscularly administered compound B (30 mg/kg), RBV (30 mg/kg), or vehicle QD for 1 d. At 5 d after infection, serum was collected from each mouse, and cytokine levels were quantified using a Quantikine ELISA Kit (R&D Systems).
Histopathological Examination.
Liver tissues at 6 d after infection were fixed in 3.7% formaldehyde in PBS. Histopathological analysis was performed on snap-frozen tissue sections stained with rat monoclonal antibodies targeting LCMV nucleoprotein (clone VL4) using Histofine Simple Stain Mouse Max-PO (Rat) and a diaminobenzidine substrate kit (Nichirei Biosciences).
Molecular Modeling.
Models of compound B bound to CEN of LASV, LCMV, and influenza A virus were constructed via superimposition on a crystal structure of BXA and CEN of influenza A complex (Protein Data Bank [PDB] ID code 6fs6). First, the CEN structures for LASV (PDB ID code 4miw) and LCMV (PDB ID code 5t2t) were aligned to that of influenza A using three active-site atoms, including two atoms involved in metal chelation and a backbone Ca atom on the same position of the α3-helix (residues H41, S47, and S46 for influenza A, LASV, and LCMV, respectively). Because the sequence similarity of CENs between bunyaviruses and influenza A was relatively low (22%), the superposition of two-metal chelators and the α3-helix resulted in more feasible active-site alignment than the protein backbone alignment. Second, a three-dimensional structure of B was built using the LigPrep program and superimposed on BXA using the Flex align program of Schrödinger suite 2019-1. Consequently, we obtained three binding models that agree with the experimental results (Fig. 4). We also attempted molecular docking of B to the CENs, but we could not obtain a plausible binding mode for LASV and LCMV. We considered that some small conformational changes around the active sites of CENs may be needed to reveal actual binding modes.
Supplementary Material
Acknowledgments
We thank Dr. Shinji Makino (University of Texas Medical Branch) for providing the RVFV plasmid, Dr. Karl-Klaus Conzelmann (Max von Pettenkofer Institute, Germany) for providing BSR-T7/5 cells, and Dr. Masayuki Saijo (National Institute of Infectious Diseases, Japan) for providing LCMV. We also thank Shinya Omoto, Keiko Baba, Kayo Ishida, Haruka Maeda, and Etsuko Hayashi for their excellent assistance. This study was partly supported by the Japan Agency for Medical Research and Development (AMED) under Grants JP21wm0125008 and JP21jk0210037; Japan Science and Technology Agency (JST) Moonshot R&D under Grant JPMJMS2025; and the World-Leading Innovative and Smart Education (WISE) Program from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT), Japan under Grant 1801.
Footnotes
Competing interest statement: S.T., A.S., M. Kawai, Y.T., Y.U., S.K., H.N., S.U., K.U., K.T., T.N., R.Y., A.N., and T.S. are employees of Shionogi & Co., Ltd. and have filed an application with the Japanese patent office.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2206104119/-/DCSupplemental.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Paessler S., Walker D. H., Pathogenesis of the viral hemorrhagic fevers. Annu. Rev. Pathol. 8, 411–440 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Bagcchi S., Lassa fever outbreak continues across Nigeria. Lancet Infect. Dis. 20, 543 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Regional Office for the Eastern Mediterranean, World Health Organization, Haemorrhagic fevers, viral. http://www.emro.who.int/health-topics/haemorrhagic-fevers-viral/index.html. Accessed 19 August 2022.
- 4.Omoto S., et al. , Characterization of influenza virus variants induced by treatment with the endonuclease inhibitor baloxavir marboxil. Sci. Rep. 8, 9633 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyagawa M., et al. , Synthesis and SAR study of carbamoyl pyridone bicycle derivatives as potent inhibitors of influenza cap-dependent endonuclease. J. Med. Chem. 62, 8101–8114 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Reguera J., et al. , Comparative structure and functional analysis of bunyavirus and arenavirus cap-snatching endonucleases. PLoS Pathog. 12, e1005636 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ye C., et al. , An improved enzyme-linked focus formation assay revealed baloxavir acid as a potential antiviral therapeutic against hantavirus infection. Front. Pharmacol. 10, 1203 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kobayashi M., et al. , In vitro antiretroviral properties of S/GSK1349572, a next-generation HIV integrase inhibitor. Antimicrob. Agents Chemother. 55, 813–821 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshinaga T., et al. , Antiviral characteristics of GSK1265744, an HIV integrase inhibitor dosed orally or by long-acting injection. Antimicrob. Agents Chemother. 59, 397–406 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noshi T., et al. , In vitro characterization of baloxavir acid, a first-in-class cap-dependent endonuclease inhibitor of the influenza virus polymerase PA subunit. Antiviral Res. 160, 109–117 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Tomassini J., et al. , Inhibition of cap (m7GpppXm)-dependent endonuclease of influenza virus by 4-substituted 2,4-dioxobutanoic acid compounds. Antimicrob. Agents Chemother. 38, 2827–2837 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayden F. G., et al. ; Baloxavir Marboxil Investigators Group, Baloxavir marboxil for uncomplicated influenza in adults and adolescents. N. Engl. J. Med. 379, 913–923 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Brisse M. E., Ly H., Hemorrhagic fever-causing arenaviruses: Lethal pathogens and potent immune suppressors. Front. Immunol. 10, 372 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grange Z. L., et al. ; Expert Panel; PREDICT Consortium, Ranking the risk of animal-to-human spillover for newly discovered viruses. Proc. Natl. Acad. Sci. U.S.A. 118, e2002324118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang W., et al. , The cap-snatching SFTSV endonuclease domain is an antiviral target. Cell Rep. 30, 153–163.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holm T., et al. , Biochemical and structural studies reveal differences and commonalities among cap-snatching endonucleases from segmented negative-strand RNA viruses. J. Biol. Chem. 293, 19686–19698 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X., et al. , Structure of Rift Valley fever virus RNA-dependent RNA polymerase. J. Virol. 96, e0171321 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zmora P., Moldenhauer A. S., Hofmann-Winkler H., Pöhlmann S., TMPRSS2 isoform 1 activates respiratory viruses and is expressed in viral target cells. PLoS One 10, e0138380 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Limburg H., et al. , TMPRSS2 is the major activating protease of influenza A virus in primary human airway cells and influenza B virus in human type II pneumocytes. J. Virol. 93, e00649-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsuno K., et al. , Fatal tickborne phlebovirus infection in captive cheetahs, Japan. Emerg. Infect. Dis. 24, 1726–1729 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikegami T., Won S., Peters C. J., Makino S., Rescue of infectious Rift Valley fever virus entirely from cDNA, analysis of virus lacking the NSs gene, and expression of a foreign gene. J. Virol. 80, 2933–2940 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buchholz U. J., Finke S., Conzelmann K. K., Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73, 251–259 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yun N. E., et al. , Mice lacking functional STAT1 are highly susceptible to lethal infection with Lassa virus. J. Virol. 87, 10908–10911 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pauwels R., et al. , Rapid and automated tetrazolium-based colorimetric assay for the detection of anti-HIV compounds. J. Virol. Methods 20, 309–321 (1988). [DOI] [PubMed] [Google Scholar]
- 25.Maruyama J., et al. , Lethal infection of Lassa virus isolated from a human clinical sample in outbred guinea pigs without adaptation. mSphere 4, e00428-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sasaki M., et al. , Metagenomic analysis of the shrew enteric virome reveals novel viruses related to human stool-associated viruses. J. Gen. Virol. 96, 440–452 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.