Significance
Human pregnancy relies on the formation of the transient organ placenta, and trophoblast cells are the major building blocks of the placenta. A defect in trophoblast progenitor self-renewal or its differentiation is associated with either pregnancy loss or pathological pregnancies, yet the underlying molecular mechanisms that regulate trophoblast differentiation are poorly understood. In this study, we discovered that WW domain containing transcription regulator 1 (WWTR1), a transcription cofactor and a component of the conserved Hippo signaling pathway, optimizes trophoblast progenitor self-renewal and is essential for its differentiation into the invasive extravillous trophoblast cell lineage. Our findings establish WWTR1 as a critical regulator for success in human placentation and progression of a healthy pregnancy.
Keywords: placenta, cytotrophoblast, human TSC, Hippo signaling, WWTR1
Abstract
Healthy progression of human pregnancy relies on cytotrophoblast (CTB) progenitor self-renewal and its differentiation toward multinucleated syncytiotrophoblasts (STBs) and invasive extravillous trophoblasts (EVTs). However, the underlying molecular mechanisms that fine-tune CTB self-renewal or direct its differentiation toward STBs or EVTs during human placentation are poorly defined. Here, we show that Hippo signaling cofactor WW domain containing transcription regulator 1 (WWTR1) is a master regulator of trophoblast fate choice during human placentation. Using human trophoblast stem cells (human TSCs), primary CTBs, and human placental explants, we demonstrate that WWTR1 promotes self-renewal in human CTBs and is essential for their differentiation to EVTs. In contrast, WWTR1 prevents induction of the STB fate in undifferentiated CTBs. Our single-cell RNA sequencing analyses in first-trimester human placenta, along with mechanistic analyses in human TSCs revealed that WWTR1 fine-tunes trophoblast fate by directly regulating WNT signaling components. Importantly, our analyses of placentae from pathological pregnancies show that extreme preterm births (gestational time ≤28 wk) are often associated with loss of WWTR1 expression in CTBs. In summary, our findings establish the critical importance of WWTR1 at the crossroads of human trophoblast progenitor self-renewal versus differentiation. It plays positive instructive roles in promoting CTB self-renewal and EVT differentiation and safeguards undifferentiated CTBs from attaining the STB fate.
Establishment of human pregnancy is associated with formation of an invasive primitive syncytium from cytotrophoblast (CTB) progenitors at the blastocyst implantation site (1–3). Subsequently, proliferation and differentiation of CTB progenitors result in formation of the functional villous placenta containing two types of matured villi: (i) floating villi, which float in the maternal blood and contain the syncytiotrophoblast (STB) population that establishes the maternal-fetal nutrient and gas exchange interface and secretes human chorionic gonadotropin (hCG) to maintain the corpus luteum (4, 5) and (ii) anchoring villi, which anchor to maternal tissue and contain the invasive extravillous trophoblast (EVT) population (Fig. 1A). In anchoring villi, CTB progenitors adapt a distinct differentiation pathway. At the base of the anchoring villi, CTB progenitors proliferate to form a CTB cell column. Eventually, cells at the distal ends of the CTB column differentiate to adapt a migratory phenotype, thereby establishing the invasive EVT lineage, which orchestrates the uterine environment, expresses nonclassical human leukocyte antigen (HLA)–G, and promotes immune tolerance to the fetus to secure progression of pregnancy (5–7). A subset of EVTs invades and remodels the uterine vasculature to establish enhanced maternal blood supply at the uterine–placental interface to fulfill the nutrient requirement of the growing fetus (8). Thus, self-renewal of CTB progenitors and their differentiation to STBs and EVTs in floating versus anchoring villi are the essential events for progression of human pregnancy. Failures in these processes are implicated in early pregnancy loss or pregnancy-associated diseases such as preeclampsia (PE), intrauterine growth restriction (IUGR), and preterm birth (9–14).
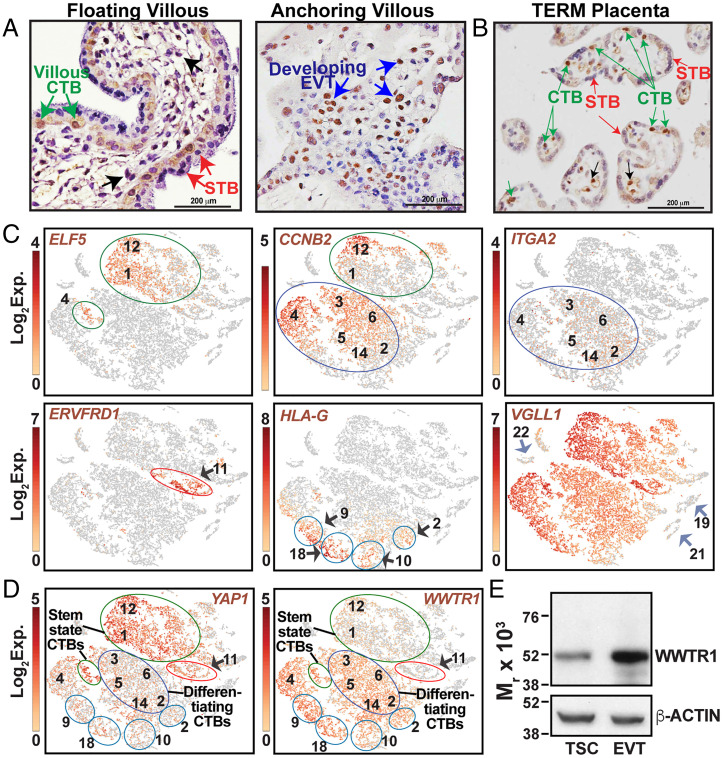
Fig. 1.
WWTR1 expression in human placentation site. (A) Immunostained images show WWTR1 protein expression in a first-trimester (week 8) human placental villous. WWTR1 expression was detected in CTBs within floating villi (green arrows) and in developing EVTs (blue arrows) in anchoring villi. However, WWTR1 protein expression was undetectable in STBs (red arrows) and in stromal cells (black arrows) within floating villi. (B) Immunostained image shows WWTR1 protein expression in a normal term human placenta. WWTR1 is predominantly expressed in CTBs (green arrows). However, STBs (red arrows) do not express WWTR1. Black arrows indicate a few endothelial cells in which WWTR1 expression was also detected. (C) t-SNE plots of the single-cell clusters of first-trimester human placental samples. [Two first-trimester placentae (one 6 wk and another 7 wk) were used for scRNA-seq analyses, and the data from two experiments were clustered together.] Expressions of specific genes were monitored to identify clusters of different CTB progenitors and developing EVTs. (D) t-SNE plots showing differential mRNA expression patterns of YAP1 and WWTR1 in single-cell clusters, obtained by scRNA-seq analyses in first-trimester human placentae. YAP1 is highly expressed in clusters of stem-state CTBs (green circles) and in CTBs of cluster 11 (red circle), which are mitotically arrested. In contrast, WWTR1 expression induced in mitotically active but differentiating CTBs and in cells of clusters 9,10, 18, and a part of clusters 2 (blue circles), which represent developing EVTs. (E) Western blots show induction of WWTR1 protein expression in human TSCs upon differentiation to EVTs. Black arrows, CTB clusters; Blue arrows, non-trophoblast cells; Green circles, stem state CTBs; Blue circles, differentiating CTBs.
Although CTBs establish the stem/progenitor compartment of a developing placenta, recent studies revealed that distinct populations of CTBs exist within a first-trimester placenta. Ex vivo developmental analyses of peri-implantation human embryos and global gene expression analyses, including single-cell RNA sequencing (scRNA-seq), revealed that during early stages of human placentation, along with undifferentiated CTBs, populations of mitotically active CTBs arise that are poised for either STB or EVT differentiation (15–17). In floating villi of the early first-trimester placenta (4 to 8 wk), expression of CDX2 and ELF5 marks the undifferentiated stem-state CTBs (18, 19), which are not committed to the differentiation pathway, whereas mitotically active but differentiating CTBs can be identified by the expression of genes that are linked to the interferon response, such as interferon gamma receptor 2 (IFNGR2), and cell cycle regulators such as CDK1 and CCNB2 (15, 16). In addition, a population of mitotically inactive CTBs, which are committed to STB differentiation, can be identified by the expression of retroviral protein ERVFRD1 (16). Unlike in floating villi, CDX2 expression is suppressed in column CTBs within anchoring villi, and ELF5 messenger RNA (mRNA) is expressed only in column CTBs near the base of the cell column (proximal column). Thus, it was proposed that ELF5 transcriptional activity regulates the trophoblast stem-state compartment of a developing human placenta (18). The undifferentiated column CTB subpopulation expresses integrin A2 (ITGA2) and NOTCH1 (20, 21). The transition of column CTBs to EVTs is associated with loss of ITGA2 expression and induction of specific genes, such as ITGA1, MMP2, and human leukocyte antigen-G (HLA-G) (22–24). Thus, CTB self-renewal and differentiation during human placentation is a highly dynamic process and relies on molecular mechanisms that fine-tune the gene expression programs in different CTB progenitor subpopulations.
Molecular mechanisms that regulate trophoblast development during early human placentation were poorly understood due to ethical restrictions and lack of appropriate model systems. However, successful derivation of human trophoblast stem cells (TSCs) from CTBs of first-trimester human placentae (25) and success in CTB-organoid culture (26, 27) have provided excellent models to identify regulatory pathways that are involved in CTB self-renewal and their differentiation to STBs and EVTs. Using human TSCs and CTB organoids, we identified conserved Hippo signaling components, transcription factor TEAD4, and cofactor YAP1 as important regulators to maintain the self-renewal ability of CTBs in a developing human placenta (11, 28). We showed that TEAD4 and YAP1 are selectively expressed in undifferentiated CTBs and loss of either TEAD4 or YAP1 in CTBs impairs their self-renewal ability (11, 28).
Another transcriptional coactivator downstream of the Hippo signaling pathway is WW domain containing transcription regulator 1 (WWTR1). Similar to YAP1, WWTR1 also interacts with TEAD transcription factors to mediate overlapping gene expression programs in multiple cellular contexts. However, WWTR1 and YAP1 can also establish distinct transcriptional programs via interaction with specific transcriptional partners (29). Studies focusing on trophoblast lineage development in mouse and marsupials indicated that YAP1 and WWTR1 might have redundant or mutually distinct roles during trophoblast development. Studies with YAP1 and WWTR1 mutant mouse models revealed a redundant role in trophoblast lineage development. Although early trophoblast development was not impaired in either Wwtr1−/− or Yap1−/− embryos, the Yap1/Wwtr1 double knockout embryos failed to form blastocysts due to defective development of the trophectoderm lineage (30). In contrast to mouse, trophoblast cells of a developing marsupial embryo show distinct expression patterns of YAP1 and WWTR1. In marsupials, WWTR1 expression is strongly maintained within the nuclei of developing trophoblast lineage, whereas YAP1 expression is suppressed. Thus, it was predicted that WWTR1, but not YAP1, has a more important role in trophoblast maintenance in a developing marsupial embryo (31). Together, these studies indicated that WWTR1, either in conjunction with YAP1 or in an independent fashion, contributes to trophoblast lineage development during mammalian placentation. However, the importance of WWTR1 in human trophoblast development is yet to be defined.
Here, using human TSCs, primary CTBs, and placental explants, we focused our attention on the functional importance of WWTR1 in human trophoblast development, especially in the context of CTB self-renewal, and its differentiation to STBs and EVTs. In addition, we examined possible correlation of defective WWTR1 function in pathological pregnancies. We discovered that similar to YAP1, WWTR1 is required to maintain self-renewal of CTB progenitors. In addition, WWTR1 is important to prevent STB differentiation and to induce the EVT differentiation program in CTBs. We also found that pregnancies associated with extreme preterm birth as well as preterm birth along with IUGR/PE are often associated with loss of WWTR1 in CTBs. Collectively, our findings implicate WWTR1 as an important orchestrator of trophoblast development during human placentation.
Results
During Human Placentation, WWTR1 Is Expressed in CTB Progenitors and the Expression Is Induced during EVT Development.
To define the importance of WWTR1 in human trophoblast development, we tested WWTR1 protein expressions in first-trimester human placentae (6 to 8 wk of gestation). As mentioned earlier, floating villi in a first-trimester human placenta contain two different layers of trophoblast cells: (i) a layer of CTB progenitors and (ii) the postmitotic STB layer, overlaying the CTBs. In contrast, the anchoring villi contain the CTB column, and cells at the distal end of the CTB columns develop to the invasive EVTs. We found that WWTR1 was predominantly expressed in CTBs within floating villi (Fig. 1A, green arrows). We also noticed WWTR1 expression in emerging EVTs within anchoring villi (Fig. 1A, blue arrows). However, WWTR1 expression was suppressed in differentiated STBs (Fig. 1A, red arrows) and in stromal cells (Fig. 1A, black arrows) within floating villi. We also tested WWTR1 expression in term human placentae. Similar to first-trimester floating villi, WWTR1 was expressed only in CTBs (Fig. 1B, green arrows) within a normal term human placenta and was repressed in STBs (Fig. 1B, red arrows). Coimmunostaining analyses with TEAD4, which is selectively expressed in CTBs of a term placenta, further confirmed WWTR1 expression in CTBs and its absence in STBs (SI Appendix, Fig. S1A). However, we also noticed WWTR1 expression in a few endothelial cells within normal term placenta (Fig. 1B, black arrows).
scRNA-seq analyses with first-trimester human placentae revealed that a developing human placenta contains distinct CTB subpopulations that can be identified via expression of specific genes (16). We hypothesized that WWTR1 and YAP1 and another TEAD-interacting cofactor VGLL1, which is highly expressed in proliferative CTBs and is implicated in TEAD4-mediated gene expression programming during human trophoblast lineage development (19), might have distinct functions in different CTB subpopulations of a developing human placenta. Therefore, we compared expression of WWTR1, YAP1, and VGLL1 in different CTB subpopulations. To that end, we analyzed scRNA-seq data that we generated with first-trimester human placentae. Based on gene expression patterns, entire single-cell populations of 6- to 8-wk human placentae were distributed into 22 cell clusters (SI Appendix, Fig. S1B). Expressions of human cytokeratin 7 (KRT7) and human leukocyte antigen-A (HLA-A) distinguished the trophoblast cell clusters from nontrophoblast cells (SI Appendix, Fig. S1C). As GATA3 and TFAP2C are expressed in all mononuclear trophoblast cells of a first-trimester human placenta (32), we tested and confirmed expressions of GATA3 and TFAP2C mRNA in all single-cell clusters comprising trophoblast cells (SI Appendix, Fig. S1D).
To identify distinct CTB subpopulations in single-cell clusters, we compared expressions of ELF5, CCNB2, ITGA2, ERVFRD1, and HLA-G (Fig. 1C). ELF5, which is expressed in the CTB subpopulation that maintains an undifferentiated stem state, was detected mainly in cells of clusters 1 and 12 (Fig. 1 C, Upper Left) and within a few cells of cluster 4. A recent study showed that the stem-state CTBs also express Basal Cell Adhesion Molecule (BCAM) (17), and analyses of single-cell clusters showed that BCAM is predominantly expressed in ELF5-expressing cells of clusters 1, 12 and 4 (SI Appendix, Fig. S1E). Thus, we concluded that the ELF5/BCAM–expressing cells within clusters 1, 12 and 4 comprise the undifferentiated stem-state CTB subpopulation. Expression of CCNB2, which marks all mitotically active CTBs of a first-trimester placenta, was detected in all ELF5-expressing cells of clusters 1, 12 and 4. CCNB2 expression was also detected in a majority of cells within clusters 3 to 6 and 14 and in a few cells of cluster 2 (Fig. 1 C, Upper Middle), indicating that these cell clusters contain mitotically active but differentiating CTBs. Interestingly, mRNA expression of ITGA2, which marks the CTB progenitors at the proximal column of an anchoring villous, was not detected in ELF5-expressing CTBs within clusters 1 and 12. Rather, ITGA2-expressing cells were scattered within cells of clusters 2, 3, 4, 5, 6, and 14 (Fig. 1 C, Upper Right). We also noticed a similar expression pattern of NOTCH1 (SI Appendix, Fig. S1E). Thus, we concluded that the ITGA2-expressing cells of clusters 2, 3, 4, 5, 6, and 14 represent the proximal column CTB progenitors within anchoring villi.
ERVFRD1, an endogenous retroviral encoded cell-fusion gene, was shown to be expressed in mitotically inactive CTBs within a first-trimester human placenta (16). We detected ERVFRD1 mRNA expression only within single cells of cluster 11 (Fig. 1 C, Lower Left), indicating that cells in cluster 11 represent the mitotically inactive CTBs that are committed to STB differentiation. High-level expression of HLA-G, which is induced in developing EVTs, was detected within cells of clusters 9, 18, and 10 and a few cells of cluster 2 (Fig. 1 C, Lower Middle). Interestingly, the high-level HLA-G–expressing cells of clusters 9, 18, 10, and 2 did not express ITGA2. Thus, we concluded that these cells were committed to EVT differentiation.
Next, we tested expressions of Hippo signaling cofactors in single-cell clusters, representing different CTB subpopulations of a first-trimester human placenta. We found that VGLL1 was highly expressed in almost all single trophoblast cells within a first-trimester placenta (Fig. 1C). However, WWTR1 and YAP1 showed contrasting expression patterns in ELF5-expressing stem-state CTBs versus CCNB2-expressing differentiating CTB progenitors (Fig. 1D). WWTR1 and YAP1 were expressed in both stem-state and differentiating CTBs of clusters 1, 12 and 4, although YAP1 expression was higher in stem-state CTBs. In contrast, WWTR1 expression was induced in differentiating CTBs of cell clusters 2, 3, 4, 5, 6, and 14 (Fig. 1D). Cell clusters 2, 3, 5, 6, and 14 also contained ITGA2-expressing column CTB progenitors. We validated WWTR1 expression in nearly all ITGA2-expressing CTBs, whereas YAP1 expression was detected in a small fraction of ITGA2-expressing cells (SI Appendix, Fig. S2 A and B). Interestingly, a contrasting expression pattern of YAP1 and WWTR1 was also observed in mitotically inactive CTBs versus emerging EVTs. YAP1 expression was maintained in ERVFRD1-expressing, mitotically inactive CTBs of cluster 11 and was reduced in HLA-G–expressing cells of clusters 9, 18, 10, and 2 (Fig. 1D). In contrast, WWTR1 was highly expressed in HLA-G–expressing emerging EVTs and was repressed in mitotically arrested CTBs (Fig. 1D).
Next, we tested WWTR1 expression in human TSCs that were derived from first-trimester CTBs. Reverse transcription followed by quantitative PCR (RT-qPCR) showed that WWTR1 mRNA was expressed in undifferentiated human TSCs, and the expression was induced during EVT differentiation (SI Appendix, Fig. S3A). Western blot analyses confirmed induction of WWTR1 protein expression during EVT differentiation in human TSCs (Fig. 1E). Immunofluorescence (IF) analyses showed that WWTR1 was localized within nuclei in both undifferentiated TSCs and in TSC-derived EVTs (SI Appendix, Fig. S3B). Collectively, our expression analyses indicated that during human placentation WWTR1 might have important functional roles in CTB progenitors and might promote their differentiation to EVTs.
WWTR1 Regulates Self-Renewal of Human Trophoblast Progenitors.
WWTR1 is expressed in mitotically active CTBs within a first-trimester placenta, and expression is maintained in CTB-derived human TSCs. Therefore, we performed loss-of-function analyses to test the importance of WWTR1 in human trophoblast progenitor self-renewal. We depleted WWTR1 in human TSCs (WWTR1-KD human TSC) by RNA interference (RNAi) using lentiviral-mediated transduction of small hairpin RNAs (shRNAs) (Fig. 2 A and B). In a culture condition that promotes human TSC proliferation at stem state, WWTR1-KD human TSC showed loss of stem-state colony morphology and strong reduction in cell proliferation ability, which was confirmed via Bromodeoxyuridine (BrdU) incorporation assay (Fig. 2 C–E and SI Appendix, Fig. S4A). We also tested self-renewal of WWTR1-KD human TSC by assessing its ability to form self-renewing three-dimensional trophoblast organoids (TSC organoids). Unlike the control human TSCs, WWTR1-KD human TSCs showed severe impairment in organoid formation (Fig. 2F). Control human TSCs formed large organoids with prolonged culture (8 to 10 d) and could be dissociated and reorganized to form secondary organoids, indicating the self-renewing ability. In contrast, WWTR1-KD human TSCs formed much smaller organoids, which were not maintained upon passaging (SI Appendix, Fig. S4B).
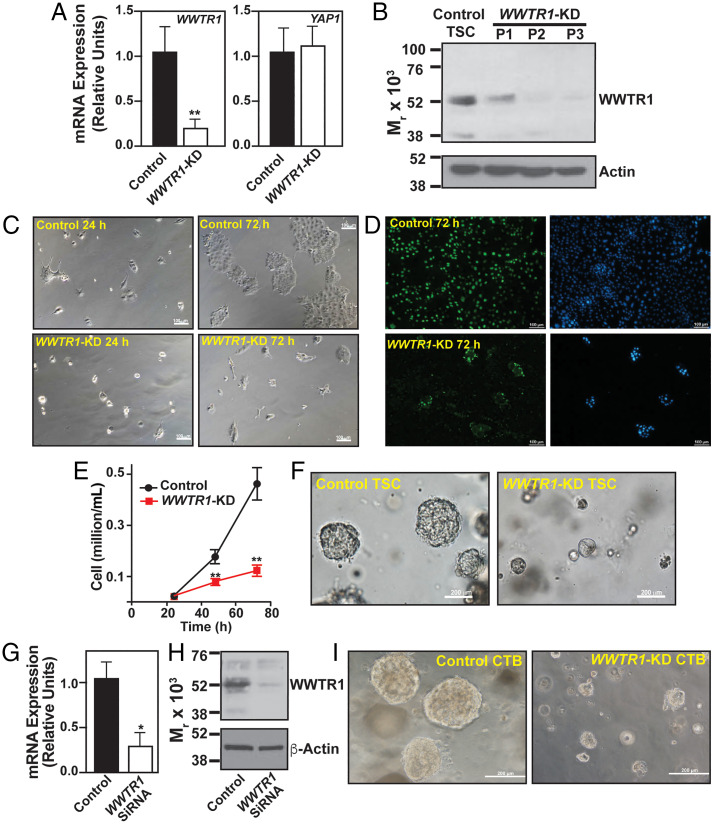
Fig. 2.
Loss-of WWTR1 impairs self-renewal in human TSCs and primary CTBs. (A) WWTR1 and YAP1 mRNA expressions were tested in human TSCs without (control) and with WWTR1 depletion by shRNA (WWTR1-KD). Plot shows that strong reduction of WWTR1-mRNA expression in WWTR1-KD human TSCs did not significantly alter YAP1 mRNA expression (error bars, mean ± SE; n = 3 independent experiments, **, P ≤ 0.005). (B) Western blot analyses showing depletion of WWTR1 protein expression in WWTR1-KD human TSCs over three passages. (C) Equal number of control and WWTR1-KD human TSCs were plated and cultured in stem-state culture condition. Micrographs confirm reduced cell proliferation in WWTR1-KD human TSCs. (D) Images show BrdU incorporation in control and WWTR1-depleted human TSCs when cultured in stem-state culture condition over 72 h. (E) Plot shows growth kinetics of human TSCs without and with WWTR1 depletion (error bars, mean ± SE; n = 3 independent experiments, **, P ≤ 0.005). (F) Micrographs show inefficient organoid formation by WWTR1-KD human TSCs. (G and H) RT-qPCR (error bars, mean ± SE; n = 3 independent experiments, *, P ≤ 0.005) and Western blot analysis, respectively, showing RNAi-mediated depletion of WWTR1 expression in primary CTBs isolated from human first-trimester placentae. (I) Micrographs show inefficient organoid formation by WWTR1-depleted (WWTR1-KD) primary CTBs. (Three individual experiments were performed, and representative images are shown.)
Next, we tested the impact of WWTR1 depletion on the self-renewal ability of primary CTBs that were isolated from first-trimester (6 to 10 wk) human placentae. We depleted WWTR1 expression in primary CTBs via small interfering RNA (siRNA) molecules (Fig. 2 G and H) and tested the ability to form self-renewing three-dimensional CTB-organoids. We found that similar to human TSCs, WWTR1 depletion in primary CTBs strongly inhibited organoid formation efficiency (Fig. 2I). Thus, loss-of-function studies in human TSCs and primary CTBs strongly indicated that WWTR1 plays an important role in maintaining the self-renewal ability within mitotically active CTB progenitors of developing human placenta.
WWTR1 Directly Regulates TP63 Expression in Human Trophoblast Progenitors.
To understand how WWTR1 regulates trophoblast progenitor self-renewal, we performed global gene expression (RNA sequencing [RNA-seq]) analyses in WWTR1-KD human TSCs. Depletion of WWTR1 in human TSCs significantly altered expression of 960 genes (216 down-regulated and 744 up-regulated; Dataset S1 and SI Appendix, Fig. S5A). RNA-seq analyses revealed that mRNA expression of TP63, which is implicated in the maintenance of CTB stem state (33), was strongly down-regulated in WWTR1-KD TSCs (Fig. 3A and SI Appendix, Fig. S5B). We confirmed TP63 down-regulation in WWTR1-KD TSCs via RT-qPCR and IF analyses (Fig. 3 B and C). Furthermore, using quantitative chromatin immunoprecipitation (ChIP-PCR) we detected WWTR1 occupancy at a conserved TEAD motif at the TP63 locus in undifferentiated TSCs (Fig. 3 D and E). These results indicate that WWTR1-mediated induction of TP63 expression might be one of the molecular mechanisms that maintains self-renewal in stem-state CTBs within a developing human placenta.
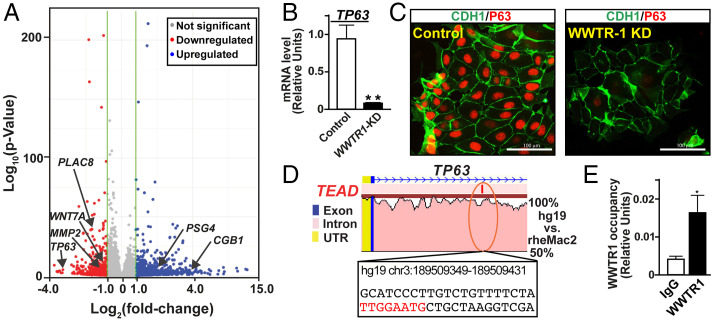
Fig. 3.
WWTR1 directly regulates TP63 expression in human TSCs. (A) Volcano plot showing global gene expression changes in WWTR1-KD human TSCs. Unbiased RNA-seq analyses were performed, and at least twofold gene-expression changes in WWTR1-KD human TSCs with a false discovery rate of P < 0.05 are indicated with colored dots (blue: up-regulated, red: down-regulated). Significant down-regulation in expression of TP63 (a marker of undifferentiated CTBs), PLAC8, MMP2 (important for EVT development), and WNT7A and up-regulation of CGB1 and PSG4 (STB-specific genes) are indicated. (B and C) RT-qPCR (mean ± SE; n = 3, **, P ≤ 0.001) and IF images, respectively, confirming down-regulation of TP63 expression in WWTR1-KD human TSCs. (D) rVISTA alignment plot of a conserved TEAD motif containing a region (orange oval) of human and rhesus macaque TP63 genes. The conserved TEAD motif (in red letters) along with adjacent base sequences and associated coordinates on the human TP63 gene are indicated. (E) The plot shows WWTR1 occupancy at the conserved TEAD motif of the TP63 locus (shown in D) in human TSCs (mean ± SE; n = 3, P ≤ 0.01). IgG, immunoglobulin G; UTR, untranslated region.
Discovery of a WWTR1–WNT Regulatory Axis in Human Trophoblast Progenitors.
Unbiased gene set enrichment analysis (GSEA) of RNA-seq data showed that loss of WWTR1 in human TSCs down-regulated transcription of various genes in the Wingless/Integrate (WNT) signaling pathway (Fig. 4A). A detailed look at the expression of WNT genes showed that six WNT genes, WNT3, WNT4, WNT5B, WNT7A, WNT8B, and WNT9A, are repressed in WWTR1-KD TSCs (Fig. 4B). The WNT signaling pathway has been implicated as a key regulator in maintaining CTBs at a progenitor state (26). Gene expression analyses in first-trimester CTBs showed that many of the WNT genes, including WNT3 and WNT7A, were expressed in undifferentiated CTBs (25), and activation of WNT signaling was key to successful derivation of human TSCs (25) and self-renewing CTB organoids (26, 27). Therefore, we looked at expressions of WNT genes in CTB progenitors of a first-trimester human placenta at single-cell resolution.
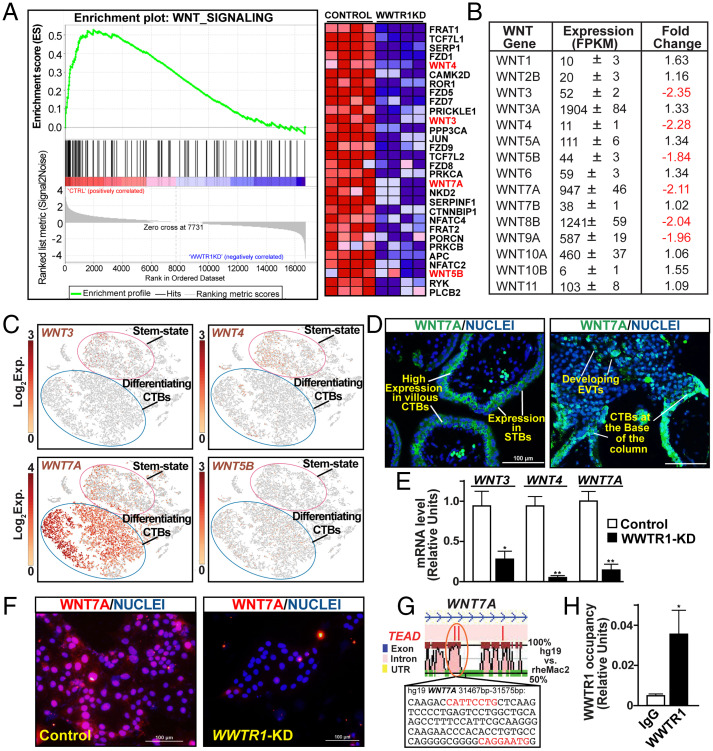
Fig. 4.
WWTR1 regulates WNT signaling components in human TSCs. (A) GSEA of differentially expressed genes showing down-regulation of WNT-signaling genes in WWTR1-KD human TSCs. The heat map shows specific genes that were down-regulated in WWTR1-KD human TSCs. Various WNT family members are highlighted. CTRL, control. (B) The table shows average expression levels of all WNT genes that were expressed in human TSCs. WNT genes that were down-regulated in WWTR1-KD human TSCs are highlighted. FPKM, Fragments Per Kilobase of transcript per Million mapped reads. (C) t-SNE plots showing differential mRNA expression patterns of WNT genes in single-cell clusters representing stem state and differentiating CTBs of first-trimester human placentae. Exp., mRNA expression. (D) IF images show WNT7A protein expression in trophoblast cells of first-trimester placental villi. Representative images of floating and anchoring villi are shown. (E) RT-qPCR analyses confirming WNT3, WNT4, and WNT7A mRNA expressions in WWTR1-KD human TSCs (mean ± SE; n = 4, *, P < 0.01; **, P ≤ 0.001). (F) IF images confirm loss of and WNT7A protein expression in WWTR1-KD human TSCs. (G) rVISTA alignment plot of a conserved TEAD motif containing a region of human and rhesus macaque WNT7A genes. Conserved TEAD motif (in red letters) along with adjacent base sequences and associated coordinates on the human WNT7A gene, where WWTR1 occupancy was detected in human TSCs, are indicated (orange oval). (H) Quantitative ChIP analysis identified WWTR1 occupancy at the region with highlighted conserved TEAD motifs of the WNT7A locus (shown in G) in human TSCs (mean ± SE; n = 3, *, P < 0.01). UTR, untranslated region.
scRNA-seq analyses showed that among six WNT genes, namely WNT3, WNT4, WNT5B, WNT7A, WNT8B, and WNT9A, that are regulated by WWTR1 in human TSCs, WNT7A was most abundantly and widely expressed in mitotically active CTBs (Fig. 4C). WNT3 and WNT4 were also expressed in undifferentiated stem-state CTBs. However, compared to WNT7A, WNT3- and WNT4-expressing CTBs were less abundant in a first-trimester placenta (Fig. 4C). WNT5B mRNA was not expressed in undifferentiated CTBs; rather, WNT5B mRNA expression was detected in a small number of differentiating, mitotically active CTBs (Fig. 4C), and WNT8B and WNT9A were not expressed in primary CTBs (SI Appendix, Fig. S6A).
As scRNA-seq analyses identified WNT7A as the most abundantly expressed WNT gene in first-trimester CTBs, we tested WNT7A protein expression in human first-trimester placenta. We found that WNT7A was highly expressed in CTBs within floating villi, and expression was reduced but maintained in STBs (Fig. 4 D, Left). In anchoring villi, WNT7A was also expressed at the base of the CTB column and in the emerging EVTs at the distal cell column (Fig. 4 D, Right).
Our expression analyses showed that among WWTR1-regulated WNT genes in human TSCs, WNT3, WNT4, and WNT7A were expressed in primary CTBs and may contribute to the CTB self-renewal process. Therefore, we validated down-regulation of WNT3, WNT4, and WNT7A mRNA expression in WWTR1-KD TSCs via RT-qPCR (Fig. 4E). We also validated loss of WNT7A protein expression in WWTR1-KD TSCs (Fig. 4F). Furthermore, using ChIP-PCR we found that WNT7A (Fig. 4 G and H) as well as WNT3 and WNT4 are direct WWTR1 target genes in human TSCs (SI Appendix, Fig. S6B).
The WNT signaling pathway is pivotal for maintenance of self-renewal in CTB progenitors [reviewed in (34)]. Along with WNT molecules, other components of the WNT-signaling pathway, including Frizzled 5,6, beta catenin, and several T cell factor (TCF) family members, are highly expressed in CTBs and CTB-derived human TSCs (25, 34). Thus, our experiments showing direct regulation of WNT7A, WNT3, and WNT4 in human TSCs indicate a WWTR1–WNT regulatory axis that could orchestrate gene expression programs in human CTB progenitors.
WWTR1 Prevents Induction of STB Fate and Promotes EVT Differentiation in Human Trophoblast Progenitors.
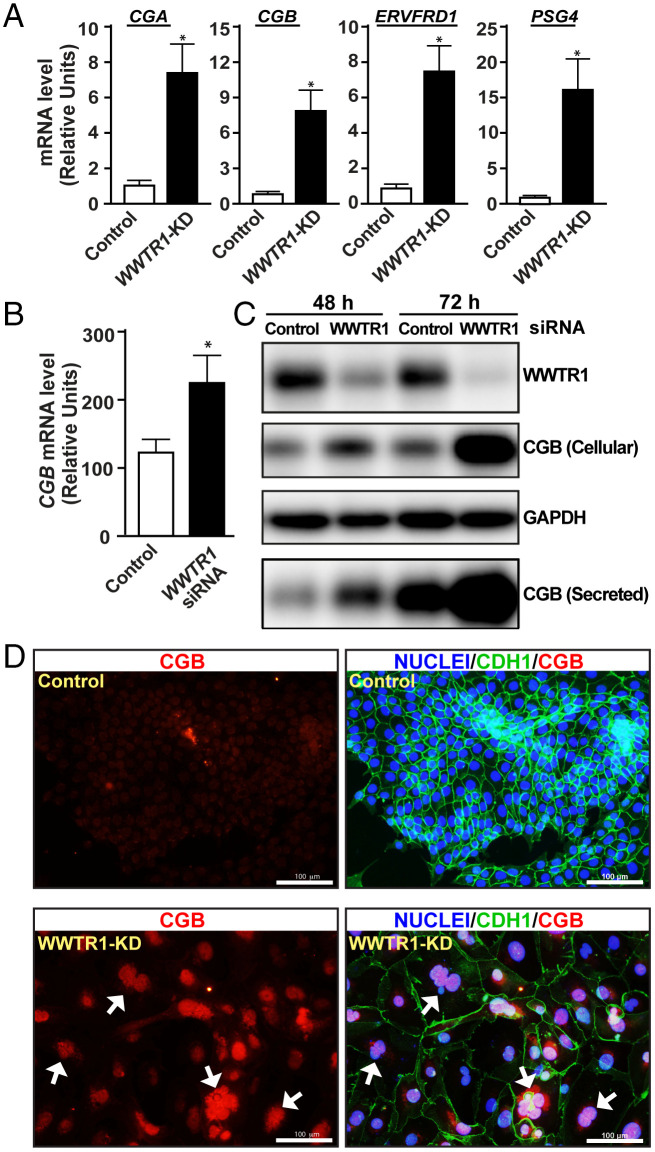
Global gene expression analyses in WWTR1-KD human TSCs revealed strong up-regulation of many STB-specific genes, such as chorionic gonadotropin A (CGA), chorionic gonadotropin B isoforms (CGBs), and pregnancy-specific β-1-glycoproteins (PSGs), in a culture condition that maintains human TSC stem state (Fig. 3A and Dataset S1). We confirmed induction of STB-specific gene transcripts in WWTR1-KD human TSCs via RT-qPCR (Fig. 5A). We also found that siRNA-mediated depletion of WWTR1 in primary CTBs of first-trimester human placentae strongly induced CGB protein expression and secretion (Fig. 5 B and C). Furthermore, at human TSC stem-state culture condition, extended culture of WWTR1-KD human TSCs often resulted in spontaneous cell fusion and formation of multinucleated syncytium (Fig. 5D). The nuclei of these multinucleated syncytium expressed high levels of CGB, confirming induction of STB-differentiation fate in WWTR1-KD human TSCs in a culture condition that should maintain the TSC stem state. Taken together, our studies strongly indicated that during human placentation, WWTR1 function in CTB progenitors prevents induction of the STB differentiation program by suppressing expression of STB-specific genes.
Fig. 5.
WWTR1 prevents STB differentiation in human TSCs and primary CTBs. (A) RT-qPCR analyses confirming induction of mRNA expressions of STB-specific genes CGA, CGB, ERVFRD1, and PSG4 in WWTR1-KD human TSCs (mean ± SE; n = 4, *, P ≤ 0.005). (B) RT-qPCR analyses confirming CGB mRNA expressions in CTBs isolated from first-trimester human placentae upon siRNA-mediated depletion of WWTR1 (mean ± SE; n = 3, *, P ≤ 0.01). (C) Western blot analyses confirming induction of CGB protein expression in WWTR1-depleted CTBs. (D) Fluorescence images show propensity of STB differentiation in WWTR1-KD human TSCs when cultured in stem-state culture conditions for three passages. WWTR1-KD human TSCs show enhanced propensity of cell fusion with multinucleated cells and loss of E-Cadherin expression (white arrows). The nuclei of fused cells also show a higher level of CGB protein expression. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
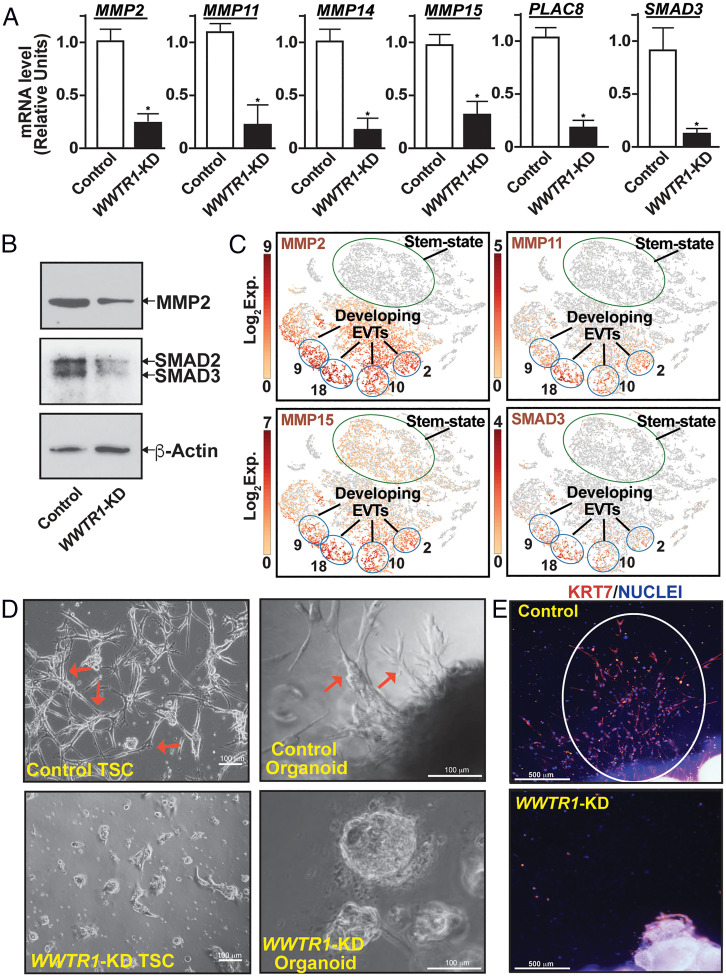
The global gene expression analyses also revealed that mRNA expressions of matrix metalloproteinase 2 (MMP2), Placenta Associated 8 (PLAC8), and SMAD Family Member 3 (SMAD3), which are implicated in EVT development (35–37), were down-regulated in WWTR1-KD human TSCs (Dataset S1). MMP2 and other MMPs have been implicated in EVT development and invasion (35, 38). Thus, we tested mRNA expressions of MMP family members along with PLAC8 and SMAD3 in WWTR1-KD TSCs using RT-qPCR. We found that along with PLAC8 and SMAD3, mRNA expressions of four MMP genes, MMP2, MMP11, MMP14, and MMP15, were significantly down-regulated in WWTR1-KD human TSCs (Fig. 6A). We also confirmed loss of MMP2 and SMAD3 protein expressions in WWTR1-KD human TSCs (Fig. 6B). Our single-cell gene expression analyses in first-trimester human placenta showed that all of these genes were either highly expressed (MMP2, MMP14, and PLAC8) or highly induced (MMP11, MMP15, and SMAD3) in developing EVTs (cell clusters 9, 18, 10, and 2; Fig. 6C and SI Appendix Fig. S7A), in which HLA-G expression was also induced (shown in Fig. 1C). Therefore, we next tested the importance of WWTR1 in EVT development.
Fig. 6.
WWTR1 regulates EVT development. (A) RT-qPCR analyses confirming down-regulation of mRNA expressions of MMP2, MMP11, MMP14, MMP15, PLAC8, and SMAD3 in WWTR1-KD human TSCs (mean ± SE; n = 4, *, P < 0.01). (B) Western blot analyses confirming loss of MMP2 and SMAD3 protein expressions in WWTR1-KD human TSCs. (C) t-SNE plots showing mRNA induction of MMP2, MMP11, MMP15, and SMAD3 in single-cell clusters representing developing EVTs of first-trimester human placentae. Note that the cell clusters representing stem-state CTBs mostly lack mRNA expressions of MMP2, MMP11, and SMAD3 and have much less mRNA expression of MMP15. Exp., mRNA expression. (D) Representative phase contrast images show inefficient EVT development from WWTR1-KD human TSCs (Left) and first-trimester CTB-organoids (Right). In a culture condition that promotes EVT differentiation, control TSCs and CTB-organoids readily developed EVTs with characteristic elongated spindle-shaped cell protrusions (shown in red arrows). However, EVT development was strongly impaired from WWTR1-KD human TSCs and WWTR1-KD CTB organoids. (E) IF images show impairment of EVT emergence from human first-trimester placental explants upon WWTR1 depletion. Invasive EVTs were readily developed (highlighted with white ellipse) when first-trimester placental explants were cultured on Matrigel in a culture condition that promotes EVT differentiation. EVT emergence was strongly inhibited from placental explants, in which WWTR1 expression was depleted.
We performed three different experiments to test the importance of WWTR1 in EVT development. First, we tested EVT differentiation efficiency of WWTR1-KD human TSCs and found that loss of WWTR1 in human TSC strongly inhibited the efficiency of EVT differentiation (Fig. 6 D, Left). Next, we studied first-trimester CTB-derived organoids, which has been successfully utilized to test EVT development from primary CTBs (26, 27). EVT development was readily noticed when control CTB-organoids were cultured on Matrigel. However, RNAi-mediated silencing of WWTR1 expression nearly abrogated EVT emergence from CTB-organoid (Fig. 6 D, Right). Finally, we tested EVT emergence from human first-trimester placental explants after depleting WWTR1 expression via RNAi (SI Appendix, Fig. S7 B and C). Similar to our findings with human TSCs and primary CTBs, EVT emergence from first-trimester placental explants was strongly inhibited upon depletion of WWTR1 expression (Fig. 6E and SI Appendix, Fig. S7D). Collectively, our studies in human TSCs, primary CTBs, and placental explants identified WWTR1 as an important regulator of EVT development.
Extreme Preterm Birth Is Associated with Loss of WWTR1 Expression in CTBs.
Defective trophoblast development has been implicated as a major cause of pregnancy-associated diseases, including preterm birth, IUGR, and PE. It has been shown that extreme preterm birth is often associated with premature differentiation of villous CTBs (39). Pregnancies associated with severe PE or severe IUGR often demonstrate depletion of proliferating CTBs (40), and severe PE is also associated with increased syncytial knot formation as well (41). This indicates that these pregnancies are associated with an imbalance in either CTB self-renewal or the differentiation process. After discovering WWTR1 as an important regulator to maintain self-renewal capacity in CTBs, we tested whether WWTR1 mRNA expression was altered in placentae from pregnancies associated with preterm birth, IUGR, and PE (Fig. 7 A and B).
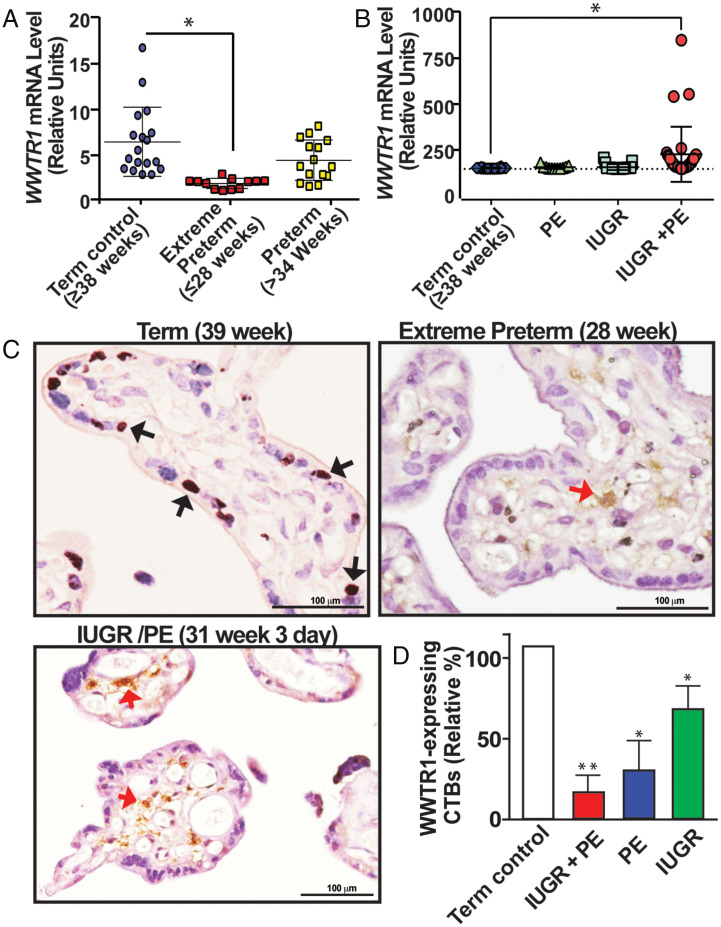
Fig. 7.
WWTR1 expression is impaired in CTBs in pathological pregnancies. (A) RT-qPCR analyses of WWTR1 mRNA expression from mRNAs isolated from whole placental tissues from pregnancies with gestational age ≥38 wk (Term control), preterm-birth (≥34 wk), or extreme preterm birth (≤28 wk). [Error bars, mean ± SE, * Indicates significant change (P < 0.01) in WWTR1 mRNA expression in placentae from extreme preterm birth]. (B) RT-qPCR analyses of WWTR1 mRNA expression from mRNAs isolated from whole placentae that were collected from pregnancies with preterm birth along with IUGR, PE, or both IUGR and PE (IUGR/PE). [* Indicates significant change (P < 0.01) in WWTR1 mRNA expression in placentae with preterm birth along with IUGR/PE]. (C) Representative immunostained images show lack of WWTR1-expressing CTBs (black arrows) in placentae from pregnancies that were associated with extreme preterm birth or preterm birth in association with IUGR/PE. Red arrows indicate WWTR1-expressing nontrophoblast cells. (D) WWTR1-expressing CTBs were quantitated from 10 different placental sections from normal term pregnancy or from preterm pregnancies with IUGR, PE, or IUGR/PE. The plot shows significant (*, P < 0.01) reduction in WWTR1-expressing CTBs in pathological pregnancies.
First, we tested WWTR1 expression in placentae associated with preterm birth without reported complications of IUGR and PE and compared it with its expression in normal term pregnancy placentae. We analyzed 37 placentae from preterm pregnancies without IUGR and PE, 12 of which were associated with extreme preterm birth (babies born at or prior to 28 wk of gestation). Notably, we found that WWTR1 mRNA expression levels were significantly reduced in placentae from pregnancies associated with extreme preterm birth (Fig. 7A). As WWTR1 is predominantly expressed in CTBs within a term placenta, we tested WWTR1 protein expressions in placental sections associated with extreme preterm birth and found that the number of WWTR1-expressing CTBs was drastically reduced within these placentae (Fig. 7C). Additionally, we analyzed 10 preterm placentae from babies born between 30 and 34 wk of gestation and 15 placentae from babies born between 34 and 36 wk of gestation. These preterm pregnancies were not associated with IUGR or PE. We observed variable levels of WWTR1-expressing CTBs in the placentae from the preterm pregnancies between 30 and 34 wk of gestation (SI Appendix, Fig. S8). However, we did not notice any significant change in WWTR1 mRNA expression or the number of WWTR1-expressing CTBs in placentae from preterm births after 34 wk of pregnancy (Fig. 7A and SI Appendix, Fig. S8).
As IUGR and PE are also often associated with preterm birth, we tested WWTR1 mRNA expression in placentae from preterm pregnancies with IUGR or PE or both (IUGR/PE), where gestation was less than 34 wk. We analyzed 44 IUGR placentae, 11 PE placentae, and 31 IUGR/PE placentae with average gestation of 33 wk, 32 wk, and 30 wk, respectively. qRT-PCR analyses with whole placental extracts showed no significant induction of WWTR1 mRNA expressions in pregnancies associated with either IUGR or PE (Fig. 7B). In contrast, RT-PCR analyses with whole placental extracts from pregnancies associated with both IUGR and PE showed significant induction of WWTR1 mRNA expression (Fig. 7B). However, when we tested WWTR1 protein expressions via immunostaining, there was no noticeable presence of WWTR1-expressing CTBs in preterm birth placentae associated with IUGR/PE (Fig. 7 C and D and SI Appendix, Fig. S9). We also noticed a significant reduction in WWTR1-expressing CTBs in placentae from preterm pregnancies with IUGR or with PE (Fig. 7D and SI Appendix, Fig. S8). Rather, we observed an increased number of WWTR1-expressing cells within the stroma of these placentae (red arrows, Fig. 7C). Incidentally, while placentae associated with IUGR/PE from pregnancies of more than 36 wk contained WWTR1-expressing CTBs comparable to the amount in normal term pregnancies (SI Appendix, Fig. S10), these placentae also showed extensive presence of WWTR1-expressing nontrophoblast cells within the stroma (SI Appendix, Fig. S10). These results suggest that the increased WWTR1 mRNA expression that we observed with whole placental tissues from IUGR/PE were not due to increased WWTR1 expression in CTBs or STBs. Rather, preterm birth with IUGR/PE was often associated with loss of WWTR1-expressing CTB population in placental villi. Collectively, our findings imply that impaired WWTR1 function in CTB progenitors could be a trophoblast-associated molecular cause contributing to pathological pregnancies, including extreme preterm birth.
Discussion
Our findings in this study establish the Hippo signaling cofactor WWTR1 as an essential regulator in the orchestration of the gene expression program and the balance of self-renewal versus differentiation in CTB progenitors. Our experimental findings indicate a bimodal function of WWTR1 in human trophoblast progenitors. In floating villi, it promotes CTB self-renewal and suppresses premature instigation of the STB differentiation fate, whereas in anchoring villi, WWTR1’s function in CTB progenitors is important in instigating EVT differentiation. We also discovered that pregnancies associated with extreme preterm birth as well as IUGR/PE are often characterized with loss of WWTR1 expression in CTBs. Collectively, our findings implicate defective WWTR1 expression/function in CTBs as one of the molecular causes of adverse pregnancies.
The observation that WWTR1 promotes CTB self-renewal, along with our earlier reports showing essential roles of TEAD4 and YAP1 in maintenance of CTB self-renewal (11, 28), establishes the critical importance of the Hippo signaling pathway in human trophoblast development. These findings also indicate a functional redundancy of WWTR1 and YAP1 in CTB progenitors. However, our single-cell resolution gene expression analyses indicated that YAP1 and WWTR1 are differentially expressed in distinct CTB subpopulations. A high level of YAP1 expression was confined within the ELF5-expressing undifferentiated/stem-state CTB progenitors. Interestingly, expression of TEAD4 was also predominantly detected within the ELF5-expressing CTB subpopulations (11). In contrast, WWTR1 expression was induced in the CTB subpopulation, which is mitotically active but poised for differentiation. Thus, we propose that a TEAD4/YAP1 transcriptional complex is important in maintaining a ground level of stemness within undifferentiated CTB progenitors, whereas WWTR1 can interact with other TEAD family members to maintain the self-renewal ability in CTB progenitors, which are priming for differentiation, including the column CTBs of anchoring villi. Future studies involving identification of global targets in CTBs along with spatial single-cell genomics of the developing human placenta will be instrumental in gaining insights into transcriptional programs that are established by YAP1 and WWTR1 in distinct CTB subpopulations.
The WNT signaling pathway has also been implicated in the CTB self-renewal process (26), as well as the differentiation to EVTs (42). Gene expression analyses reported by earlier studies (25, 34) showed that along with various WNT molecules, several other components of the WNT signaling pathway are highly expressed in trophoblast cells of a developing placenta. The WNT receptors FZD5 and FZD6 are highly expressed in CTBs and EVTs, and β-catenin is abundantly expressed in all three trophoblast cell types of a developing human placenta (25, 34). The WNT processing enzyme PORCN is also expressed, albeit at a low level, in CTBs and EVTs, and its expression is induced in STBs. Furthermore, repressors of the WNT signaling pathway, including GSK3β, TCF7, and TCF7L2, are highly expressed in CTBs and EVTs of a developing human placenta (25, 34). These findings indicate a major role of the WNT signaling pathway in the dynamics of human trophoblast development. However, the roles of individual WNT molecules in maintaining CTB self-renewal versus differentiation are not well understood.
Our findings in this study indicate a prominent role of WNT7A in human trophoblast development. We discovered WNT7A as the most abundantly expressed WNT molecule in CTBs of a first-trimester placenta, and its expression pattern is similar to that of WWTR1 in subpopulations of mitotically active CTBs and is a direct target of WWTR1 in CTB-derived human TSCs. WNT7A is highly expressed in CTBs within floating villi and at the base of the CTB column in anchoring villi. WNT7A expression is also detected in emerging EVTs at the distal columns of anchoring villi along with WNT7B, indicating that the WWTR1–WNT7A signaling axis could be important for the CTB self-renewal process within the floating villi and EVT development in anchoring villi. WWTR1 also directly regulates expressions of WNT3 and WNT4 in human TSCs. Interestingly, our scRNA-seq analyses showed that in a first-trimester human placenta, WNT3 and WNT4 expressions, albeit at low level, are confined to stem-state CTBs, which also express high levels of YAP1 and TEAD4. Thus, crosstalk among distinct WNT molecules and Hippo signaling components might regulate gene expressions in different CTB subpopulations within a developing human placenta.
We found that WWTR1 is essential for EVT differentiation in human TSCs and emergence of EVT cells from first-trimester placental explants. Interestingly, WWTR1 function in EVT development appears to be distinct from that of YAP1, which is suppressed in EVTs. We have shown that during EVT differentiation of human TSCs, WWTR1 is required for optimal expression of MMP2, MMP11, MMP14, and MMP15. MMP2 has been shown to regulate EVT invasion (35, 38). Expressions of MMP11, MMP14, and MMP15 were earlier detected in human trophoblast cells, including EVTs within maternal decidua (38, 43). An earlier study showed that PLAC8 is selectively induced during EVT development and induces the formation of filopodia in migratory trophoblast cells (36). SMAD3 has also been implicated in EVT development. It was shown that depletion of SMAD3 but not SMAD2 suppressed EVT emergence from first-trimester human placental explants (37), indicating a specific role of SMAD3 during EVT development.
Interestingly, our scRNA-seq analyses with first-trimester human placentae showed that all of these MMPs, as well as PLAC8 and SMAD3, are induced in developing EVTs. Thus, our findings indicate that WWTR1 might mediate multipronged roles during EVT development by regulating expressions of MMPs, PLAC8, and SMAD3. Given the dynamic nature of EVT development and the essential role of EVTs in human placentation, it is important to institute future studies to better understand the role of WWTR1 in EVT development and function.
The loss of WWTR1 expression in CTBs from pregnancies with extreme preterm birth and IUGR/PE indicates a direct correlation of WWTR1 with adverse pregnancies. We also showed that loss of WWTR1 in human TSCs and primary CTBs promotes a premature differentiation to STB lineage. Inductions of STB-specific gene expression were also noticed when TEAD4 and YAP1 were depleted in human TSCs and CTBs, respectively. Intriguingly, elevated maternal serum levels of hCG and Inhibin-A, measured at 15 to 20 wk of gestation, increase the subsequent risk of IUGR/PE and extreme preterm birth (39). Since both hCG and Inhibin-A are produced by STBs and extreme preterm births are often associated with loss of proliferating CTBs (39), it was proposed that elevated levels of hCG and/or Inhibin-A may result from premature differentiation of the CTBs to adopt STB fate (35). Thus, our findings from this study and prior studies with TEAD4 and YAP1 (11, 28) support the hypothesis that during human placental development, loss of Hippo signaling components such as WWTR1, YAP1, and TEAD4 may result in premature accelerated differentiation of CTBs to STBs, which subsequently contributes to adverse pregnancies, such as extreme preterm birth and IUGR/PE.
Experimental Procedures
Human Placental Sample Analysis.
De-identified and discarded first-trimester placental tissues and term placental samples from normal and pathological pregnancies were obtained from Mount Sinai Hospital, Toronto, ON, Canada, or collected at the University of Kansas Medical Center. The Institutional Review Board (IRB) at Mount Sinai Hospital and the University of Kansas IRB approved all collections and studies. Fresh first-trimester placental tissues were embedded in optimal cutting temperature compound and cryo-sectioned or used for scRNA-seq analyses. To test EVT development, first-trimester placental explants were cultured on Matrigel for 6 to 8 d in medium that promoted EVT differentiation in human TSCs (see below).
scRNA-Seq Analysis.
scRNA-seq analysis with first-trimester placenta was performed, and details were reported earlier (11). Briefly, single-cell suspensions from two first-trimester placentae were generated, and transcriptomic profiles were obtained using the 10x Genomics Chromium Single Cell Gene Expression Solution (https://www.10xgenomics.com). The primary analysis of the scRNA-seq data was performed using the 10x Genomics Cell Ranger pipeline (version 3.1.0). This pipeline performs sample de-multiplexing, barcode processing, and single-cell 3′ gene counting. The quality of the sequenced data was assessed using FastQC software. Sequenced reads were mapped to the human reference genome (GRCh38) using STAR software. Individual samples were aggregated using the “cellranger aggr” tool in Cell Ranger to produce a single-feature barcode matrix containing all the sample data. Cell Ranger software was used to perform t-distributed stochastic neighbor embedding (t-SNE) projections of cells and k-means clustering. The 10x Genomics Loupe Cell Browser software was used to find significant genes, cell types, and substructure within the single-cell data. The raw data for scRNA-seq analyses have been submitted to the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/gds), with accession No. GSE145036.
Human TSC Culture.
Human TSC lines, derived from first-trimester CTBs, were described earlier (11, 25). To maintain stem-state culture, human TSCs were cultured on a Collagen IV-coated (5 µg/mL) plate in Dulbecco’s modified Eagle’s medium (DMEM)/F12 medium supplemented with 0.1 mM 2-mercaptoethanol, 0.2% fetal bovine serum (FBS), 0.5% penicillin-streptomycin, 0.3% bovine serum albumin (BSA), 1% ITS-X supplement, 1.5 μg/mL L-ascorbic acid, 50 ng/mL epidermal growth factor, 2 μM CHIR99021, 0.5 μM A83-01, 1 μM SB431542, 0.8 mM Valproic acid, and 5 μM Y27632. For EVT differentiation, TSCs were resuspended in 2% Matrigel (Corning, NY) and media containing DMEM/F12 supplemented with 0.3% BSA, 1% ITS-X, 0.5% penicillin-streptomycin, 100 µM β-Mercaptoethanol, 2.5 µM Y27632, 7.5 µM A83-01, 100 ng/mL hNRG1, and 4% Knockout serum (KSR). EVT differentiation medium without hNRG1 was replaced on day 3. On day 6, the medium, lacking hNRG1 and KSR, was again replaced and finally analyzed on day 8.
RNAi in Human TSCs.
Lentiviral shRNAs were used to knock down WWTR1 (target sequence: GCGATGAATCAGCCTCTGAAT) in human TSCs. A scramble shRNA (Addgene-1864; CCTAAGGTTAAGTCGCCCTCGC) was used as control. Lentiviral particles were generated by transfecting plasmids into HEK-293T cells. Virus-containing supernatant was collected, and virus particles were concentrated by Lenti-X concentrator (Clontech Laboratories) according to the manufacturer’s protocol. Human TSCs were transduced using viral particles at 60 to 70% confluency. Transduced cells were selected in the presence of puromycin (1.5 to 2 µg/mL). Selected cells were tested for knockdown efficiency and used for further experimental analyses.
CTB Isolation from First-Trimester Placenta.
CTBs were isolated from 8th- to 10th-week first-trimester pooled placentae (n = 8) as described (28). Briefly, placentae were kept overnight in DMEM HAM’s F12 (Gibco; 31331–28)/0.05 mg/mL gentamicin (Gibco; 15710–049), and 0.5 µg/mL fungizone (Gibco; 15290026). Next-day placental villi were scraped in 1x Hank’s Balanced Salt Solution (HBSS; Sigma-Aldrich; H4641), collected by centrifugation, and incubated for two consecutive digestions with 1x HBSS containing 0.125% trypsin (Gibco; 15090–046) and 0.125 mg/mL DNase I (Sigma-Aldrich; DN25) at 37 °C in the incubator. Cells were purified by Percoll (Cytiva; 17089101) gradient centrifugation. Contaminating erythrocytes were lysed by incubation with erythrocyte lysis buffer (155 mM NH4Cl, 10 mM KHCO3, and 0.1 mM ethylenediaminetetraacetic acid (EDTA), pH 7.3) for 5 min at room temperature (RT). The cell suspension was seeded onto cell culture dishes for 45 min to allow contaminating stromal cells to adhere to the plastic. Trophoblasts were collected from the supernatants by centrifugation. HLAG+ EVTs were depleted from the cell suspension by immune-purification using HLA-G Phycoerythrin-labeled antibodies (Exbio; Clone MEM-G/9, 1P-292-C100), PE MACS beads (Miltenyi Biotec; 130–048-801), and MACS MS columns (Miltenyi Biotec; 130–042-201). Purified Trophoblasts were seeded in DMEM-Ham’s F12 (Gibco; 31331–28)/10% FBS (Sigma-Aldrich; S0615-500ML), 0.05 mg/mL gentamicin (Gibco; 15710–049), and 0.5 µg/mL fungizone (Gibco; 15290026) onto fibronectin-coated cell culture dishes (2 µg/cm2; Merck; FC010). For siRNA-mediated gene silencing, 1 h later a proportion of the cell culture medium was replaced by siRNA/RNAiMax containing media prepared as described (44) by using nontargeting (D-001810-10-20) or TAZ (L-016083-00-0005) ON-TARGETplus SMARTpools and Lipofectamine RNAimax (Invitrogen; 13778–075).
IF and Immunohistochemistry (IHC).
IF was performed with human TSCs for expression analyses of distinct proteins as described in the respective figures. In addition, IF was performed with normal term human placental sections to test coexpression of WWTR1 and TEAD4 in CTBs. Cells and placental sections were fixed using 4% paraformaldehyde in 1X phosphate buffer saline (PBS) (Sigma-Aldrich; D8537), permeabilized using 0.25% Triton X-100 (Sigma-Aldrich; ×100) in 1X PBS and blocked for 1 h using blocking buffer (10% FBS and 0.1% TritonX100 in PBS). Sections were incubated with primary antibodies against WWTR1 and TEAD4 (1:100, overnight at 4 °C), washed with 0.1% Triton X-100 in PBS. After incubation with conjugated secondary antibodies (1:400, 1 h, RT), sections were washed and mounted using anti-fade mounting medium (Thermo Fisher Scientific) containing DAPI. For all IF experiments, four or more individual experiments were performed, and representative images are shown in the figures.
IHC was performed using human placenta paraffinized sections. The slides were deparaffinized by Histo-clear (National Diagnostics; HS-200), followed by rehydration with 100%, 90%, 80%, and 70% ethanol. Antigen retrieval was done using a decloaking chamber at 80 °C for 15 min. The endogenous peroxidase was inactivated by treating with 3% H2O2 (Sigma-Aldrich; 216763) followed by wash with 1X PBS. Nonspecific immunoglobulin binding was blocked with 10% goat serum (Thermo Fisher Scientific; 50062Z) for 10 min at RT followed by 1-h incubation with 1:100 dilution of anti-WWTR1 antibody or immunoglobulin G at RT. The slides were washed three times with 1X PBS and incubated with biotin conjugated secondary antibody (1:200 dilution) for 30 min at RT. The slides were washed with 1X PBS followed by treatment with streptavidin-conjugated horseradish peroxidase (Vector Laboratories; SA-5704) for 10 min at RT. Reactivity was detected using DAB+ Substrate Chromogen System (Dako; TA-125-QHDX). The slides were counterstained with Mayer’s hematoxylin (Sigma-Aldrich; MH516). The slides were then dehydrated by sequential treatment using 70%, 80%, 90%, and 100% ethanol and xylene. The sections were dried, mounted with Toluene, and imaged using a Nikon TE2000 microscope. At least five different placentae for each experimental condition were analyzed by IHC for WWTR1 expression. Representative images are included in the figures. Antibodies that were used for the study are mentioned in SI Appendix.
Placental Explants Culture.
First-trimester placental explants were submerged in DMEM/F12 (Gibco) media and divided into smaller pieces under a dissection microscope under sterile conditions. Pieces containing branching villous-like structures were washed in PBS supplemented with 10% FBS and then subjected to lentiviral treatment. The explants were divided into two groups; one group was incubated with scrambled lentiviral particles, while the other group was incubated with lentiviral particles carrying shRNA for WWTR1 gene knockdown. Both groups were incubated with the respective lentiviral particles for 6 h at 37 °C in a humidified chamber in a 5% CO2/95% air gas mixture. After 6 h, the explant pieces were rinsed and encapsulated in gel for further culture. For the encapsulation, growth factor reduced Matrigel (Corning) was mixed 1:1 with DMEM/F12 on ice to make a Matrigel suspension; 200 μL of the Matrigel suspension was added to each well of a 24-well plate, and explants were placed centrally and covered with another 200 μL of Matrigel suspension. The plate was then incubated at 37 °C in a humidified chamber in 5% CO2 for the gel suspension to solidify, thereby encapsulating the explant. Finally, 300 μL EVT medium was added to each well and allowed to culture. EVT medium was changed on days 3 and 5 as mentioned earlier.
RNA-Seq Analysis.
RNA-seq analysis was performed according to published protocol (45, 46). Total RNA from the control human TSCs as well as WWTR1-KD human TSCs were isolated using RNeasy Mini Kit (Qiagen; 74104) per the manufacturer’s protocol with on-column deoxyribonuclease digestion. RNA concentrations were quantified using a NanoDrop Spectrophotometer at a wavelength of 260 nm. Integrity of the total RNA samples was evaluated using an Agilent Technologies 2100 Bioanalyzer). The total RNA fraction was processed by oligo dT bead capture of mRNA, fragmentation, and reverse transcription into cDNA. After ligation with the appropriate Unique Dual Index (UDI) adaptors, the cDNA library was prepared using the Universal Plus mRNA-seq +UDI library preparation kit (NuGEN; 0508–08, 0508–32).
Statistical Significance.
Statistical significance was determined for quantitative RT-PCR analyses for mRNA expression and for cell proliferation analyses. We performed at least n = 3 experimental replicates for all of these experiments. For statistical significance of generated data, statistical comparisons between two means were determined with Student’s t test, and significantly altered values (P ≤ 0.01) are highlighted in the figures by an asterisk. RNA-seq data were generated with n = 4 experimental replicates per group. The statistical significance of altered gene expression (absolute fold change ≥2.0 and false discovery rate (FDR) q-value ≤0.05) was initially confirmed with right-tailed Fisher’s exact test. For WWTR1 mRNA expression analyses in pathological placentae, one-way ANOVA was used to determine statistically significant differences in mean WWTR1 expression in a specific pathological condition, such as extreme preterm birth, with term control placentae.
Supplementary Material
Acknowledgments
This research was supported by NIH Grants HD101319, HD062546, HD0098880, HD103161, and HD102188 and by a pilot grant under the NIH Center of Biomedical Research Program (P30GM122731) to S.P. We acknowledge the Genomics Core, the Imaging and Histology Core, and the Bioinformatics Core of the University of Kansas Medical Center. We thank Drs. Hiroaki Okae and Takahiro Arima of Tohoku University Graduate School of Medicine, Japan, for sharing human TSC lines. We thank Ms. Brandi Miller for critical comments on the manuscript.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2204069119/-/DCSupplemental.
Data, Materials, and Software Availability
The raw data for RNA-seq analyses are available at the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/gds) with Accession No. GSE188738 (47). The raw data for single-cell RNA-seq in the first-trimester human placenta are also available in the GEO database (Accession No. GSE145036 (48)). Additional details of experimental procedures are mentioned in SI Appendix.
References
- 1.Knofler M., et al. , Human placenta and trophoblast development: Key molecular mechanisms and model systems. Cell. Mol. Life Sci. 76, 3479–3496 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.James J. L., Carter A. M., Chamley L. W., Human placentation from nidation to 5 weeks of gestation. Part I: What do we know about formative placental development following implantation? Placenta 33, 327–334 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Boss A. L., Chamley L. W., James J. L., Placental formation in early pregnancy: How is the centre of the placenta made? Hum. Reprod. Update 24, 750–760 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Chang C. W., Wakeland A. K., Parast M. M., Trophoblast lineage specification, differentiation and their regulation by oxygen tension. J. Endocrinol. 236, R43–R56 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costa M. A., The endocrine function of human placenta: An overview. Reprod. Biomed. Online 32, 14–43 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Beer A. E., Sio J. O., Placenta as an immunological barrier. Biol. Reprod. 26, 15–27 (1982). [DOI] [PubMed] [Google Scholar]
- 7.PrabhuDas M., et al. , Immune mechanisms at the maternal-fetal interface: Perspectives and challenges. Nat. Immunol. 16, 328–334 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soares M. J., Varberg K. M., Iqbal K., Hemochorial placentation: Development, function, and adaptations. Biol. Reprod. 99, 196–211 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burton G. J., Fowden A. L., Thornburg K. L., Placental origins of chronic disease. Physiol. Rev. 96, 1509–1565 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brosens I., Pijnenborg R., Vercruysse L., Romero R., The “Great Obstetrical Syndromes” are associated with disorders of deep placentation. Am. J. Obstet. Gynecol. 204, 193–201 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saha B., et al. , TEAD4 ensures postimplantation development by promoting trophoblast self-renewal: An implication in early human pregnancy loss. Proc. Natl. Acad. Sci. U.S.A. 117, 17864–17875 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim K. H., et al. , Human cytotrophoblast differentiation/invasion is abnormal in pre-eclampsia. Am. J. Pathol. 151, 1809–1818 (1997). [PMC free article] [PubMed] [Google Scholar]
- 13.Hustin J., Jauniaux E., Schaaps J. P., Histological study of the materno-embryonic interface in spontaneous abortion. Placenta 11, 477–486 (1990). [DOI] [PubMed] [Google Scholar]
- 14.Romero R., Kusanovic J. P., Chaiworapongsa T., Hassan S. S., Placental bed disorders in preterm labor, preterm PROM, spontaneous abortion and abruptio placentae. Best Pract. Res. Clin. Obstet. Gynaecol. 25, 313–327 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.West R. C., et al. , Dynamics of trophoblast differentiation in peri-implantation-stage human embryos. Proc. Natl. Acad. Sci. U.S.A. 116, 22635–22644 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y., et al. , Single-cell RNA-seq reveals the diversity of trophoblast subtypes and patterns of differentiation in the human placenta. Cell Res. 28, 819–832 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shannon M. J., et al. , Cell trajectory modeling identifies a primitive trophoblast state defined by BCAM enrichment. Development 149, dev199840 (2022). [DOI] [PubMed] [Google Scholar]
- 18.Hemberger M., Udayashankar R., Tesar P., Moore H., Burton G. J., ELF5-enforced transcriptional networks define an epigenetically regulated trophoblast stem cell compartment in the human placenta. Hum. Mol. Genet. 19, 2456–2467 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Soncin F., et al. , Comparative analysis of mouse and human placentae across gestation reveals species-specific regulators of placental development. Development 145, dev156273 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee C. Q. E., et al. , Integrin α2 marks a niche of trophoblast progenitor cells in first trimester human placenta. Development 145, dev162305 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haider S., et al. , Notch1 controls development of the extravillous trophoblast lineage in the human placenta. Proc. Natl. Acad. Sci. U.S.A. 113, E7710–E7719 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Damsky C. H., Fitzgerald M. L., Fisher S. J., Distribution patterns of extracellular matrix components and adhesion receptors are intricately modulated during first trimester cytotrophoblast differentiation along the invasive pathway, in vivo. J. Clin. Invest. 89, 210–222 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Isaka K., et al. , Expression and activity of matrix metalloproteinase 2 and 9 in human trophoblasts. Placenta 24, 53–64 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Kovats S., et al. , A class I antigen, HLA-G, expressed in human trophoblasts. Science 248, 220–223 (1990). [DOI] [PubMed] [Google Scholar]
- 25.Okae H., et al. , Derivation of human trophoblast stem cells. Cell Stem Cell 22, 50–63.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Haider S., et al. , Self-renewing trophoblast organoids recapitulate the developmental program of the early human placenta. Stem Cell Reports 11, 537–551 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turco M. Y., et al. , Trophoblast organoids as a model for maternal-fetal interactions during human placentation. Nature 564, 263–267 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meinhardt G., et al. , Pivotal role of the transcriptional co-activator YAP in trophoblast stemness of the developing human placenta. Proc. Natl. Acad. Sci. U.S.A. 117, 13562–13570 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reggiani F., Gobbi G., Ciarrocchi A., Sancisi V., YAP and TAZ are not identical twins. Trends Biochem. Sci. 46, 154–168 (2021). [DOI] [PubMed] [Google Scholar]
- 30.Nishioka N., et al. , The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev. Cell 16, 398–410 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Frankenberg S., Shaw G., Freyer C., Pask A. J., Renfree M. B., Early cell lineage specification in a marsupial: A case for diverse mechanisms among mammals. Development 140, 965–975 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Lee C. Q., et al. , What is trophoblast? A combination of criteria define human first-trimester trophoblast. Stem Cell Reports 6, 257–272 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y., Moretto-Zita M., Leon-Garcia S., Parast M. M., p63 inhibits extravillous trophoblast migration and maintains cells in a cytotrophoblast stem cell-like state. Am. J. Pathol. 184, 3332–3343 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dietrich B., Haider S., Meinhardt G., Pollheimer J., Knöfler M., WNT and NOTCH signaling in human trophoblast development and differentiation. Cell. Mol. Life Sci. 79, 292 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Staun-Ram E., Goldman S., Gabarin D., Shalev E., Expression and importance of matrix metalloproteinase 2 and 9 (MMP-2 and -9) in human trophoblast invasion. Reprod. Biol. Endocrinol. 2, 59 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang W. L., et al. , PLAC8, a new marker for human interstitial extravillous trophoblast cells, promotes their invasion and migration. Development 145, dev148932 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brkić J., et al. , Differential role of Smad2 and Smad3 in the acquisition of an endovascular trophoblast-like phenotype and preeclampsia. Front. Endocrinol. (Lausanne) 11, 436 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohen M., Meisser A., Bischof P., Metalloproteinases and human placental invasiveness. Placenta 27, 783–793 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Fitzgerald B., et al. , Villous trophoblast abnormalities in extremely preterm deliveries with elevated second trimester maternal serum hCG or inhibin-A. Placenta 32, 339–345 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Macara L., et al. , Structural analysis of placental terminal villi from growth-restricted pregnancies with abnormal umbilical artery Doppler waveforms. Placenta 17, 37–48 (1996). [DOI] [PubMed] [Google Scholar]
- 41.Huppertz B., Kadyrov M., Kingdom J. C., Apoptosis and its role in the trophoblast. Am. J. Obstet. Gynecol. 195, 29–39 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Knöfler M., Pollheimer J., Human placental trophoblast invasion and differentiation: A particular focus on Wnt signaling. Front. Genet. 4, 190 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anacker J., et al. , Human decidua and invasive trophoblasts are rich sources of nearly all human matrix metalloproteinases. Mol. Hum. Reprod. 17, 637–652 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Rosner M., et al. , Efficient siRNA-mediated prolonged gene silencing in human amniotic fluid stem cells. Nat. Protoc. 5, 1081–1095 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Home P., et al. , Genetic redundancy of GATA factors in the extraembryonic trophoblast lineage ensures the progression of preimplantation and postimplantation mammalian development. Development 144, 876–888 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bhattacharya B., et al. , Atypical protein kinase C iota (PKCλ/ι) ensures mammalian development by establishing the maternal-fetal exchange interface. Proc. Natl. Acad. Sci. U.S.A. 117, 14280–14291 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paul S., Kumar R., RNA-Seq analysis in human trophoblast stem cells (CT27) upon depletion of Hippo signaling Component TAZ (WWTR1). NCBI GEO. http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE188738. Deposited 12 November 2021.
- 48.Soumen P., Avishek G., Pratik H., Ananya G., Sumedha G., Single cell Transcriptome profiling of Human first trimester placenta. NCBI GEO. http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE145036. Deposited 10 February 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data for RNA-seq analyses are available at the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/gds) with Accession No. GSE188738 (47). The raw data for single-cell RNA-seq in the first-trimester human placenta are also available in the GEO database (Accession No. GSE145036 (48)). Additional details of experimental procedures are mentioned in SI Appendix.