Significance
Splice-switching antisense oligonucleotides (ASOs) represent a unique class of drug molecules with the US Food and Drug Administration approval of Exondys 51, Vyondys 53, Amondys 45, and Viltepso for the treatment of Duchenne muscular dystrophy (DMD). Phosphorodiamidate morpholino oligomer (PMO) chemistry currently utilized for these drugs has significant limitations. PMOs show rapid kidney clearance and poor cellular uptake that leads to high and costly dosages. Therefore, it is crucial to develop next-generation splice-switching oligonucleotide chemistries with improved efficacy, safety, and biodistribution. The research outlined in this manuscript is highly significant as it demonstrates the impact of “Thiomorpholinos” as a robust and cost-effective splice-switching ASO platform that can be tested for enhanced biological activity and reduced toxicity relative to other chemistries.
Keywords: antisense oligonucleotide, thiomorpholino oligonucleotide, biological activity
Abstract
Recent advances in drug development have seen numerous successful clinical translations using synthetic antisense oligonucleotides (ASOs). However, major obstacles, such as challenging large-scale production, toxicity, localization of oligonucleotides in specific cellular compartments or tissues, and the high cost of treatment, need to be addressed. Thiomorpholino oligonucleotides (TMOs) are a recently developed novel nucleic acid analog that may potentially address these issues. TMOs are composed of a morpholino nucleoside joined by thiophosphoramidate internucleotide linkages. Unlike phosphorodiamidate morpholino oligomers (PMOs) that are currently used in various splice-switching ASO drugs, TMOs can be synthesized using solid-phase oligonucleotide synthesis methodologies. In this study, we synthesized various TMOs and evaluated their efficacy to induce exon skipping in a Duchenne muscular dystrophy (DMD) in vitro model using H2K mdx mouse myotubes. Our experiments demonstrated that TMOs can efficiently internalize and induce excellent exon 23 skipping potency compared with a conventional PMO control and other widely used nucleotide analogs, such as 2′-O-methyl and 2′-O-methoxyethyl ASOs. Notably, TMOs performed well at low concentrations (5–20 nM). Therefore, the dosages can be minimized, which may improve the drug safety profile. Based on the present study, we propose that TMOs represent a new, promising class of nucleic acid analogs for future oligonucleotide therapeutic development.
Khorana and colleagues laid the foundation for synthetic nucleic acid technology by demonstrating the first successful chemical synthesis of sequence-defined oligonucleotides in 1958 (1, 2). However, the process was inefficient due to its low yield and labor demands. A significant breakthrough was achieved in 1981 by Caruthers and coworkers who developed phosphoramidite chemistry for the synthesis of oligonucleotides, an approach that has become the most widely used method of modern oligonucleotide synthesis (3, 4). In the last 2 decades, improvements in synthesis, purification, and characterization techniques have allowed oligonucleotides, prepared by the phosphoramidite methodology, to be synthesized in large scale, with adequate yield and purity, for use as therapeutics. These advances have driven the remarkable progress of nucleic acid technologies in therapeutic and diagnostic applications—particularly antisense oligonucleotide (ASO) drugs. Following the approval of Vitravene by the US Food and Drug Administration in 1998 for the treatment of cytomegalovirus retinitis in immunocompromised patients (5), eight ASO drugs have been approved for clinical use, including Kynamro (2013) (6), Exondys 51 (2016) (7), Spinraza (2017) (8), Tegsedi (2018) (9), Waylivra (2019) (10), Vyondys 53 (2020) (11), Viltepso (2020) (12), and Amondys 45 (2021) (13). These approvals demonstrate the tremendous potential of ASOs for the treatment of various diseases.
Synthetic oligonucleotides, based upon using natural DNA and RNA, are not clinically relevant because of their vulnerability to nucleases and poor target binding affinities. In contrast, chemically modified nucleic acid analogs offer superior therapeutic properties, such as increased target binding affinity, improved resistance to nuclease degradation, and diminished immunogenicity. A comprehensive review of these analogs can be found elsewhere (14). Currently, there are three main chemically modified chemistries that have been utilized in the synthesis of approved oligonucleotide drugs: 2′-O-methyl (2′-OMe) and 2′-O-methoxyethyl (2′-MOE) nucleosides having a phosphorothioate (PS) backbone and also phosphorodiamidate morpholino oligomers (PMOs) (5–15) carrying a N,N-dimethylamino phosphorodiamidate backbone. However, there are significant limitations associated with these chemistries. 2′-OMe and 2′-MOE oligonucleotides can exhibit toxicity (16, 17), whereas PMOs, despite their excellent safety profile, are challenging to synthesize on a scale large enough for use as therapeutic drugs. Additionally, PMOs are rapidly excreted after administration in vivo and thereby require high dosages that subsequently contribute to the cost of treatment. Another limitation of PMOs is their inability to form complexes with commercial transfection reagents. This limitation challenges the rapid evaluation of any PMO drug in an in vitro system. Previously, we have demonstrated a novel chemistry leading to the synthesis of thiomorpholino oligonucleotides (TMOs) (18,19). In the present study, we utilize a well-known Duchenne muscular dystrophy (DMD) cellular model (H2Kb-tsA58 mdx [H2K mdx] mouse myotubes) (20, 21) to systematically evaluate these TMOs. Herein, we report the design, synthesis, and evaluation of TMOs for their exon 23 skipping activity using H2K mdx myotubes as an in vitro model system.
Results
Comparison of TMO and PMO ASO Efficiencies for Inducing DMD Exon 23 Skipping in the H-2Kb-tsA58 mdx Myotubes In Vitro Model.
In order to address one of the main aims of our study, we envisioned a direct comparison between TMOs and PMOs primarily because PMOs are currently the most widely used analog relative to splice-switching applications. Toward this end, we designed and synthesized TMO1 ASO (Table 1) having the structure as shown in Fig. 1A. This sequence design was based upon the previously published best-performing ASO that targets exon 23 in the mouse dystrophin transcript (22–32). In parallel, an oligonucleotide having the PMO chemistry (Fig. 1A and Table 1) of the same sequence was used for direct comparison.
Table 1.
ASO names and sequences used in this study
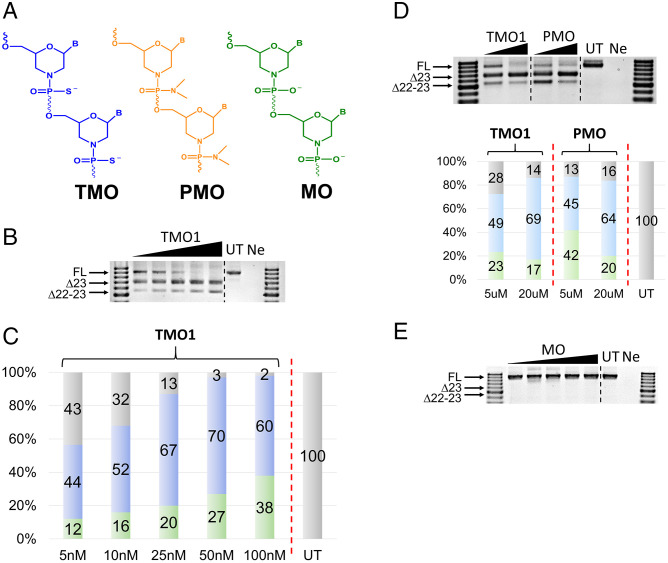
Fig. 1.
(A) Structural representation of TMO, PMO, and MO; (B) RT-PCR and (C) densitometry analysis of RNA prepared from H2K mdx mouse myotubes transfected with TMO1 ASO in the presence of lipofectin; (D) RT-PCR and densitometry analysis of RNA prepared from H2K mdx mouse myotubes transfected by nucleofection with TMO1 and PMO ASOs; (E) RT-PCR of RNA prepared from H2K mdx mouse myotubes transfected with MO ASO. The triangles above the gel images indicate increasing ASO concentrations (in B and E: 5, 10, 25, 50, and 100 nM; in D: 5 and 20 µM); FL (full-length) = 901 bp, Δ23 (exon-23 skipped) = 688 bp, Δ22–23 (exons-22 + 23 skipped) = 542 bp. In the bar graphs, FL is represented by gray, Δ23 by blue, and Δ22–23 by green. UT = untreated and Ne = negative control (RT-PCR sample with no RNA).
Toward assessing the splice-switching efficacy of these ASOs, a widely used DMD in vitro model—H2K mdx myotubes—was utilized. These myotubes contain a nonsense mutation in exon 23 that disrupts the dystrophin reading frame and leads to no dystrophin production (20, 21). Splice-switching oligonucleotides can target the dystrophin transcript, induce skipping of exon 23, and therefore restore the reading frame. For comparison purpose, TMO1 and PMO ASOs were transfected to H2K mdx mouse myotubes at doses ranging from 5 to 100 nM using the protocol as previously described (22–32). Briefly, the ASOs were complexed with lipofectin transfection reagent at a ratio of 2:1 (w:w) (lipofectin:ASO) and incubated with the cells for 24 h. Cells were then collected, the RNA extracted, and RT-PCR completed across exons 20–26 of the mouse dystrophin transcript. Remarkably, TMO1 showed efficient induction of exon 23 skipping at all concentrations in a dose-dependent manner (the 688 base pairs [bp] band, Fig. 1 B and C). On the other hand, the PMOs failed to induce any exon skipping (Fig. S1), which may not be surprising, as it does not complex with the lipid transfection reagent. It was also noted that TMO1 performed exceptionally well at lower concentrations (5 and 10 nM) to yield 44% and 52% of exon 23 skipped products, respectively (Fig. 1C). In addition, an unfavorable dual exon 22–23 skipped product was also detected at 542 bp (Fig. 1B) with the TMO. Overall, this 542-bp product increased proportionally with the concentration of TMO, ranging from 12 to 38% (Fig. 1 B and C).
In order to further explore TMO1 efficacy when compared with the PMO ASO, we employed nucleofection (electroporation) as an alternative transfection methodology—a procedure that is routinely used to transfect PMOs in cultured cells. Briefly, both ASOs were prepared at 5 and 20 µM concentrations (20-µL reactions) and subjected to the nucleofection protocol following the manufacturer’s recommendations. The cells were collected, and the RNA was extracted after 48 h of incubation. RT-PCR was performed to analyze the products as discussed above. The results at both concentrations demonstrated that TMO1 slightly outperformed (49% and 69%) the PMO (45% and 64%) relative to inducing exon 23 skipping (Fig. 1D). We also observed the unfavorable dual exon 22–23 skipped product (542 bp). Most notably, TMO1 limited this product to 23% and 17% compared with 42% and 20% for the PMO at these two concentrations (Fig. 1D).
Evaluation of Exon Skipping Efficiency Induced by the Phosphoroamidate Variant (MO).
After evaluating the TMO1, we examined the scope of the phosphoroamidate variant of TMO1 in our in vitro model. Therefore, we synthesized an MO analog having the same sequence as TMO1 (Table 1 and Fig. 1) and investigated its efficacy in inducing exon 23 skipping. Following the same transfection protocol as outlined for the results shown in Fig. 1B, the exon skipping efficiency of MO in H2K mdx myotubes was examined. The results demonstrated that the MO did not induce exon 23 skipping when compared with its TMO1 counterpart (Fig. 1E). We speculate that the MO might not have internalized into cells as efficiently as TMO1. Further evaluations are currently underway to assess the lack of MO potency.
Evaluation of Exon Skipping Efficiency Induced by Truncated Versions of TMO1.
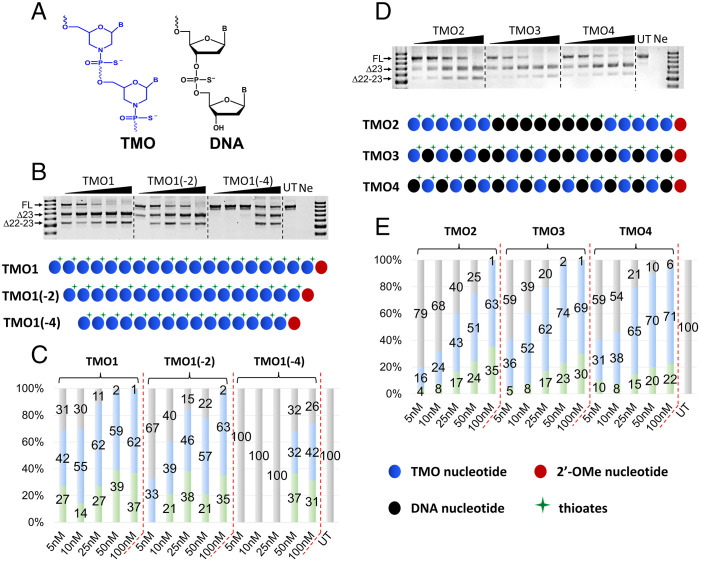
Because TMO1 generated excellent exon skipping activity, we decided to shorten this TMO ASO by eliminating two nucleotides [TMO1(–2)] and four nucleotides [TMO1(–4)] from the 5′ and 3′ ends of TMO1 (Table 1). These two truncated ASOs were synthesized and assessed for their exon skipping potency by transfecting with H2K mdx myotubes for 24 h at 5–100 nM. Not surprisingly, TMO1 remained the best TMO candidate for inducing exon 23 skipping (42–62%, 5–100 nM, respectively; Fig. 2 B and C). Remarkably, TMO1(–2) managed to yield 33% of exon 23 skipping at 5 nM (Fig. 2C). Although TMO1(–2) was not as efficient as TMO1 at the lower concentrations (5–50 nM), densitometry analysis showed that, at 100 nM, TMO1(–2) performed on par with the parent TMO1 ASO by yielding 63% of exon 23 skipped product when compared with 62% for TMO1 (Fig. 2C). We also observed exon 23 skipping with TMO1(–4) but only at 50 and 100 nM (32% and 42%, respectively). These results are encouraging and suggest that TMO1(–2) could be a promising candidate for additional evaluation in appropriate mouse models.
Fig. 2.
(A) Structural representation of TMO and DNA; (B) RT-PCR and (C) densitometry analysis of RNA prepared from H2K mdx mouse myotubes transfected with TMO1 and its truncated versions TMO1(–2) and TMO1(–4) in the presence of lipofectin; (D) RT-PCR and (E) densitometry analysis of RNA prepared from H2K mdx mouse myotubes transfected with TMO/DNA gapmer (TMO2) and mixmers (TMO3 and 4) ASOs in the presence of lipofectin. The triangles above the gel images indicate increasing ASO concentrations 5, 10, 25, 50, and 100 nM (B and D); FL = 901 bp, Δ23 (exon-23 skipped) = 688 bp, Δ22–23 (exons-22 + 23 skipped) = 542 bp. In the bar graphs, FL is represented by gray, Δ23 by blue, and Δ22–23 by green.
Evaluation of Exon Skipping Efficiency Induced by TMO/DNA Mixmer and Gapmer ASOs.
In order to further explore the scope of utilizing TMOs in various ASO design modalities for splice-switching, we incorporated DNA nucleotides (Fig. 2A) into the TMO1 sequence at various positions and therefore designed three additional TMOs, including TMO2, TMO3, and TMO4 (Table 1). Briefly, TMO2 was designed as a gapmer ASO with 6-8-6 structure, where eight nucleotides in the middle are 2′-deoxynucleoside 3′-thiophosphosphate. On the other hand, TMO3 and TMO4 were designed as mixmers of TMO and 2′-deoxynucleoside 3′-thiophosphate throughout the sequence. To evaluate the exon skipping efficiency, transfection of these TMOs was carried out as discussed above in H2K mdx myotubes using lipofectin. The cells were collected after 24 h of incubation and subjected to RNA extraction and RT-PCR as previously described (21–31). The results showed that all three TMO/DNA mixmers induced exon 23 skipping in a dose-dependent manner, except for TMO3 at 100 nM (Fig. 2 D and E). Remarkably, at lower concentrations (5 and 10 nM), these mixmer and gapmer ASOs performed well, as they yielded 16% and 24% (TMO2), 36% and 52% (TMO3), and 31% and 38% (TMO4) of exon 23 skipped product (Fig. 2 D and E). At 25 nM, TMO4 induced more exon 23 skipping than the other two ASOs, with 65% compared with 62% and 43% as observed for TMO3 and TMO2, respectively. Notably, at 50 nM, TMO3 yielded higher skipping efficacy (74%) when compared with 51% and 70% for TMO2 and TMO4 (Fig. 2E), respectively. Finally, at 100 nM, while we recorded an increase in the dual exon skipped product (542 bp) in the case of TMO2, the other two ASOs still resulted in higher amount of exon 23 skipped product (71% for TMO4 and 69% for TMO3). Dual skipped product also increased proportionally with concentration, ranging from 4 to 35% (TMO2), 5 to 30% (TMO3), and 10 to 22% (TMO4), except for TMO2 and TMO4 at 100 nM (Fig. 2 D and E). In summary, although there is variation in the proportion of induced exon 23 and dual exon 22–23 skipped mRNA, TMOs 1, 2, 3, and 4 are capable of inducing exon skipping in the dystrophin transcript of H2K mdx myotubes.
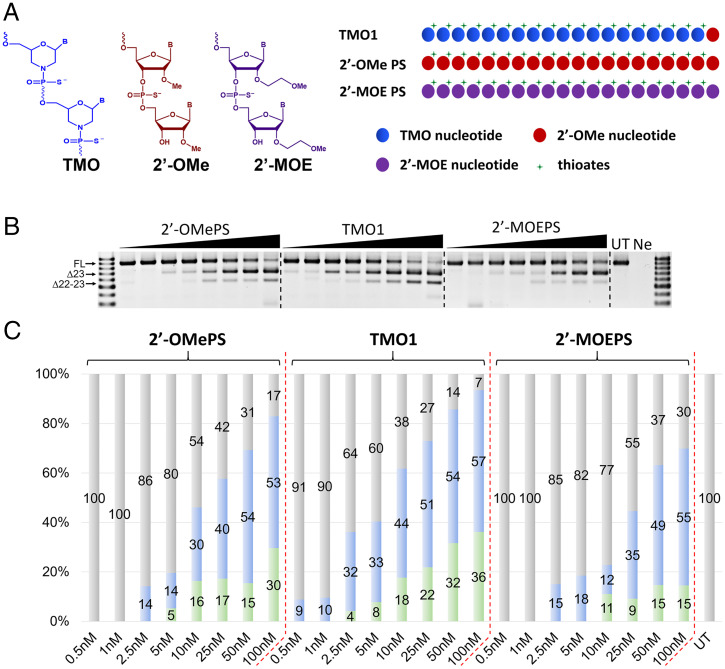
Comparison of Exon 23 Skipping Efficiency of TMO1, 2′-OMePS, and 2′-MOEPS ASOs.
Currently for various antisense experiments, 2′-OMePS and 2′-MOEPS (Fig. 3A) are the most widely used ASO platform chemistries other than PMOs. In order to evaluate TMO1 against these two chemistries, we synthesized the fully modified 2′-OMePS and 2′-MOEPS ASOs having the same sequence as TMO1 (Table 1) and evaluated these three oligonucleotides for their exon skipping potency. From previous experiments as outlined in Fig. 1 and Fig. S1, we have shown that TMO1 outperformed the PMO analog at lower concentrations. Therefore, in order to further explore the efficacy of TMOs, we tested these three ASOs from 0.5 to 100 nM. The results demonstrated excellent efficiency for TMO1, with 9% and 10% exon 23 skipping induced at 0.5 and 1 nM, respectively (Fig. 3 B and C). This result compares with no skipping at both concentrations for the other two ASOs (Fig. 3 B and C). Additionally, from 2.5 to 25 nM, TMO1 surpassed the other two ASOs at all concentrations relative to exon 23 skipping, yielding 32–51%. A dual exon 22–23 skipped product was also recorded at various levels (Fig. 3 B and C). In general, this dual exon 22–23 skipped product increased proportionally with the concentrations, except for the 2′-OMePS and 2′-MOEPS at 50 nM. Surprisingly, 2′-MOEPS ASO yielded lesser amount of this dual skipping product at all concentrations in comparison to the other two ASOs (Fig. 3 B and C).
Fig. 3.
(A) Structural representation of TMO, 2′-OMe, and 2′-MOE (left) and schematic illustration of the ASOs (right); (B) RT-PCR and (C) densitometry analysis of RNA prepared from H2K mdx mouse myotubes transfected with TMO1, 2′-OMePS and 2′-MOEPS ASOs at 0.5–100 nM in the presence of lipofectin. The triangles above the gel images indicate increasing ASO concentrations: 0.5, 1, 2.5, 5, 10, 25, 50, and 100 nM; FL = 901 bp, Δ23 (exon-23 skipped) = 688 bp, Δ22–23 (exons-22 + 23 skipped) = 542 bp. In the bar graphs, FL is represented by gray, Δ23 by blue, and Δ22–23 by green.
Internalization of TMO1 in H2K mdx Myotubes.
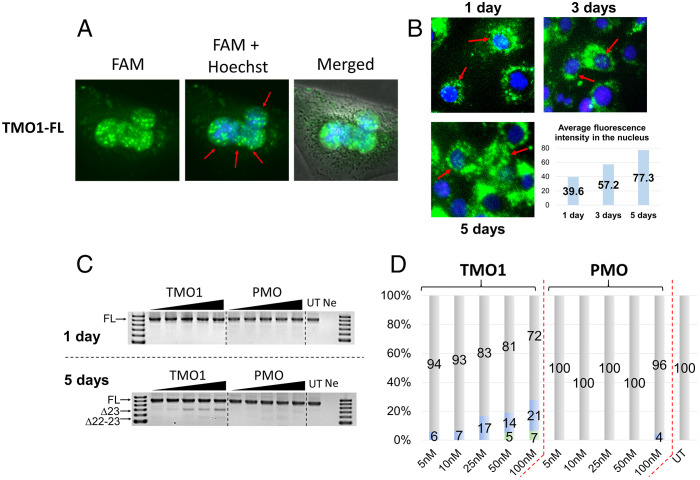
To demonstrate nucleus internalization of TMOs, we performed an imaging experiment using 6-carboxyfluorescein dye-labeled TMO (TMO1-FL) and PMO (PMO-FL) (Table 1). Dye-labeled ASOs (the dye was linked to the oligonucleotides via a PS linkage) were transfected to H2K mdx cells using lipofectin transfection reagent at 100 nM. Cells were incubated with the ASOs for 24 h before staining with Hoechst, washed, and observed under the fluorescence microscope. The captured images showed that TMO1-FL efficiently localized in myotube nuclei (Fig. 4A). On the other hand, PMO-FL failed to internalize into the cell nucleus, with no observed fluorescence signal (Fig. S2A).
Fig. 4.
(A) Fluorescence images showing nuclear uptake of TMO1-FL at 100 nM when transfected with lipofectin (24 h time point); (B) fluorescence images showing nuclear uptake of TMO1-FL at 200 nM without a transfection reagent (1-, 3-, and 5-d time points). The graphical illustration shows average fluorescence intensity of the corresponding images inside the nucleus. Red arrows indicate representative cells with internalized ASOs (green dots). (C) RT-PCR and (D) densitometry analysis of RNA prepared from H2K mdx mouse myotubes incubated with the “naked” form of TMO1 and PMO for 1 and 5 d. The triangles above the gel images indicate increasing ASO concentrations: 5, 10, 25, 50, and 100 nM. FL = 901 bp, Δ23 (exon-23 skipped) = 688 bp, Δ22–23 (exons-22 + 23 skipped) = 542 bp. In the bar graphs, FL is represented by gray, Δ23 by blue, and Δ22–23 by green.
In order to further examine the nuclear uptake ability of TMO1, we investigated whether TMO1 can internalize into cells without a transfection reagent. Therefore, we performed fluorescence imaging experiments where TMO1-FL was incubated with H2K mdx mouse myotubes for 1, 3, and 5 d at 200 nM in the absence of lipofectin or nucleofection. The captured images demonstrate that TMO1-FL could efficiently localize inside the cells at all time points (Fig. 4B). However, unlike the previous case (Fig. 4A), where we observed high intensity of TMO1-FL localized in the cell nuclei when transfected with lipofectin, in the absence of lipofectin or nucleofection, TMO1-FL mostly localized in the cell cytoplasm as a “ring” surrounding the cell nucleus. This is not surprising, as the presence of lipofectin helps facilitate ASO nuclear uptake when compared with transfection in the absence of lipofectin. Furthermore, we also conducted quantification of the average fluorescence intensity in the cell nuclei using Image J software (33). The resulting nos. of 39.6, 57.2, and 77.3 for 1, 3, and 5 d, respectively, represent the average fluorescence signal present inside each nucleus detected by Image J software. These values, therefore, are also correlated with the amount of TMO1-FL localized inside the cell nuclei. This is remarkable as, even in the absence of a transfection reagent, TMO1-FL ASO internalizes into the cell nuclei and the fluorescence intensity increased proportionally from day 1 to day 5 (Fig. 4B). In addition, we also performed the same experiment with the PMO-FL in the absence of lipofectin. The average fluorescence intensity was recorded as 30.44, 34.81 and 50.82 for 1, 3, and 5 d, respectively (Fig. S2B showing significantly lower uptake in the cell nuclei in comparison to the TMO1-FL.
TMO1 was also shown to induce exon skipping when transfected without lipofectin or nucleofection. This was demonstrated by incubating TMO1 and PMO with H2K mdx myotubes for 1 and 5 d. The results showed that, at the 1-d time point, there was no exon skipping observed by either ASO (Fig. 4C). Remarkably, extended incubation to 5 d yielded 21% of the exon 23 skipped product with the TMO at 100 nM, while the PMO induced a negligible amount of exon 23 skipped product (4%, Fig. 4 C and D). These results validate our observations from the above imaging experiments (Fig. 4B).
Discussion
Nucleic-acid-based therapeutics using ASO-mediated splice modulation was first described by Dominski and Kole (34, 35). This approach has attracted considerable attention in recent years, especially with the approval of Exondys 51, Vyondys 53, Amondys 45, and Viltepso for the treatment of DMD and Spinraza for the treatment of spinal muscular atrophy. Despite the advances in nucleic acid chemical synthesis, large-scale production of oligonucleotides remains a challenge; in particular, PMO (Fig. 1) synthesis (the chemistry used for the DMD drugs) requires intensive time and labor. In addition, another main drawback of the PMO chemistry is the rapid clearance of the administered drug. This means that a significantly high dosage is required, which leads to a very costly treatment. PMOs are also known to be very poorly complexed with commercial transfection reagents, and this significantly affects the choice of assays available for in vitro evaluation. Clinical trial data using other chemistries, such as 2′-OMePS and 2′-MOEPS, have also shown major limitations, such as possible toxicity issues, including inflammation at the injection site, as in the case of Drisapersen (16), or systematic toxicity that could affect vital organs (17). In order to potentially address these issues, we developed a new class of oligonucleotide analogs called TMOs (Fig. 1) and previously reported a comprehensive protocol for their synthesis (18). We also characterized the basic biochemical properties of TMOs, including melting temperature, binding affinity, catalysis with RNase H, stability toward nucleases, etc. In the present study, we explore the ability of TMOs to induce exon skipping in an in vitro DMD model—H2K mdx myotubes.
We initially synthesized TMO1 as a fully TMO. When TMO1 was compared with a PMO of the same sequence using lipofectin transfection in H2K mdx myotubes, TMO1 showed excellent exon skipping efficiency. On the other hand, the PMO failed to induce exon skipping at any concentration (Fig. S1). Having shown promising data, TMO1 was selected for subsequent studies. An additional experiment was conducted to see rather the phosphodiester (PO) form of TMO1 could also be useful for exon skipping, since ASOs with PO backbones possess higher target binding affinity than their PS counterparts. Surprisingly, the MO did not exhibit any exon skipping with lipofectin transfection (Fig. 1E). One possible explanation could be the poor internalization of MO into cells. Nevertheless, further assessment of the MO is currently underway in order to address this issue.
Additionally, we also tested alternative strategies for designing TMO1. One was to truncate the 20-mer TMO1 to 18- and 16-mer ASOs and to evaluate their exon skipping ability. Notably, TMO1(–2) showed on-par exon skipping efficacy with the TMO1 parent sequence (Fig. 2). This is not surprising, as we observed similar results in previous studies where we systematically truncated a 2′-OMePS ASO and found that the 18-mer ASO performed similarly to the 20-mer (27).
Another alternative strategy was to introduce DNA thiophosphate nucleotides into this oligonucleotide using different ASO design modalities, including a gapmer (TMO2) and two mixmers (TMO3 and TMO4). Similar strategy utilizing 2′-OMe and 2′-MOE chemistries as mixmer or gapmer constructs to induce exon skipping has been previously reported (24, 26–32, 36). Upon lipid transfection, all three TMOs induced efficient exon 23 skipping at concentrations from 5 to 100 nM. However, the mixmers (TMO3 and TMO4) performed better than the gapmer (TMO2) at every concentration. This is an interesting observation, as TMO3 and TMO4 have fewer TMO nucleotides than TMO2. Remarkably, TMO4 yielded the highest amount of exon 23 skipped product at 100 nM while limiting the unfavorable dual exon 22–23 skipped product in comparison to TMO1, 2, and 3.
Toward clinical translation of the TMO, we endeavored to compare the efficacy among the TMO and ASOs of the same sequence having two widely used chemistries, 2′-OMePS and 2′-MOEPS. In this experiment (Fig. 3), a lower range of concentrations was used (0.5 nM to 25 nM) in order to assess the behavior of these three chemistries at the lower limits of expected activity. The results demonstrated the superiority of the TMO1 at lower concentrations when compared with the other two chemistries. This experiment once again confirms the relevance of TMOs relative to potential clinical settings. This is because lower dosages of TMOs may generate biological activity and thus perhaps have lower toxicity than other chemistries where higher dosages of the ASO would be required. However, we have no evidence at this time that supports this possibility.
In order to confirm the localization of TMO1 in the cell nucleus, we performed an imaging experiment where we incubated the FAM conjugated TMO1-FL with H2K mdx myotubes with and without lipofectin (Fig. 4). In both cases, we found that the fluorescein-labeled ASO (TMO1-FL) localizing in the nucleus was consistent with the excellent exon skipping activity of TMO1. This is because exon skipping is a nuclear event (37). The captured images demonstrated that TMO1-FL was efficiently internalized into cell nuclei in both cases, while the PMO of the same sequence failed to internalize in any cell nuclei, even when transfected using lipofectin (Fig. S2A). Remarkably, TMO1-FL was observed to internalize into the cell nuclei proportionally from day 1 to day 5 of incubation in the absence of transfection reagent. In line with this observation, we demonstrated that TMO1 can induce exon 23 skipping after 5 d of incubation without any transfection reagent, while the PMO of the same sequence did not induce any skipping.
One limitation of the present study is to demonstrate the efficacy of TMO at the protein level. The major challenge comes from the H-2Kb-tsA58 mdx (H2K mdx) cells that we used in our experiments. This is a well-known in vitro model system for DMD; however, this cell line does not express any trace of dystrophin protein (due to a nonsense mutation in exon 23 that leads to a complete dystrophin protein depletion). Therefore, detection of dystrophin protein after treatment with an ASO at the cellular level is challenging, if not impossible. This is not surprising, as these protein assays are only feasible with in vivo studies using mdx mice under a high dose of drug and prolonged treatment periods (38, 39). In a comprehensive study by Fletcher et al. (39), the detection of dystrophin, using a western blot and immunofluorescence assay, was observed when the ASO was injected into mdx mice using doses from 2 to 10 μg per mouse over a 2- to 4-wk period. In future research with this system, we will inject TMO1 into mdx mice and hence the dystrophin protein can be quantified.
In conclusion, our work has demonstrated the ability of TMO1 to induce exon skipping in an in vitro model. The results from various experiments demonstrated the superiority of TMO1 in comparison to other chemistries, especially at the lower concentrations that were studied, making it an exceptionally promising candidate for subsequent studies with mdx mice. We therefore propose that these results in the H2K mdx myotubes demonstrates an opportunity for using TMOs as therapeutic ASO in the treatment of various genetic diseases.
Materials and Methods
Design and Synthesis of ASOs Used in This Study.
The TMOs were synthesized using the procedures as described previously (18). All other ASOs were synthesized in house on GE AKTA Oligopilot 10 (GE Healthcare Life Sciences, Parramatta, NSW, Australia) oligonucleotide synthesizer using standard phosphoramidite chemistry at 1 µmol scale unless specified (24). All synthesis reagents were purchased from Merck Millipore (Bayswater, VIC, Australia). The appropriately protected morpholino 3′-phosphorodiamidite synthons were prepared according to a published procedure (18) or purchased from ChemGenes. Synthesized oligonucleotides were deprotected and cleaved from the solid support by treatment with NH4OH (Merck Millipore) at 55 °C overnight, and the crude oligonucleotides were then purified by desalting through Illustra NAP-10 Columns (GE Healthcare Life Sciences). PMO was purchased from Gene Tools (Oregon, USA).
Cell Culture and Transfection.
H-2Kb-tsA58 mdx myoblasts (H2K mdx cells) were cultured and differentiated as described previously (22–32). Briefly, 60–80% confluent, myoblast cultures were treated with trypsin (Thermo Fisher Scientific, Scoresby, VIC, Australia) and seeded on 24-well plates pretreated with 50 μg/mL poly-D-lysine (Merck Millipore, Bayswater, VIC, Australia), followed by 100 μg/mL Matrigel (Corning, supplied through In Vitro Technologies, Noble Park North, VIC, Australia) at a density of 2.5 × 104 cells/well. Cells were differentiated into myotubes in low-glucose Dulbecco's Modified Eagle Medium (DMEM) (Thermo Fisher Scientific) containing 5% horse serum (Thermo Fisher Scientific) by incubating at 37 °C, 5% CO2 for 24 h.
ASOs were complexed with lipofectin transfection reagent (Thermo Fisher Scientific) at the ratio of 2:1 (w:w) (lipofectin:ASO) and used in a final transfection volume of 500 μL/well in a 24-well plate as per the manufacturer's instructions, except that the solution was not removed after 3 h. For “naked” transfection, the ASOs were mixed directly with Optimem reduced serum medium and added to the cells. For long-term incubations, the cells were collected at 1-, 3-, and 5-d intervals. All experiments were repeated independently at least three times unless specified.
RNA Extraction and RT-PCR.
RNA was extracted from transfected cells using ISOLATE II RNA Mini Kit (Bioline, Eveleigh, NSW, Australia) as per the manufacturer’s instructions. The mouse dystrophin transcripts were amplified by nested RT-PCR using SuperScript III Reverse Transcriptase III and AmpliTaq Gold 360 DNA Polymerase (Thermo Fisher Scientific) across exons 20–26 as described previously (22–32). The amount of RNA used for the primary amplification (primer set Ex20Fo and Ex26Ro (Table 2); 55 °C for 30 min, 94 °C for 2 min before entering 31 cycles of 94 °C for 30 s, 55 °C for 30 s, and 68 °C for 90 s) was 50 ng/µL, and 1 µL of this primary PCR product was subjected to the secondary PCR (primer set Ex20Fi and Ex26Ri (Table 2); 94 °C for 6 min before entering 33 cycles of 94 °C for 30 s, 55 °C for 1 min, and 72 °C for 2 min). The secondary PCR products were separated on 2% agarose gels in Tris-acetate-EDTA buffer, and the images were captured on a Fusion Fx gel documentation system (Vilber Lourmat, Marne-la-Vallee, France). Densitometry was performed by Image J software (33). The actual exon skipping efficiency was determined by expressing the amount of the skipped exon RT-PCR products as a percentage of total dystrophin transcript products.
Table 2.
Primers for amplifying mouse dystrophin transcript
| Primer name | Sequence (5′–3′) |
|---|---|
| Ex20Fo | CAGAATTCTGCCAATTGCTGAGa |
| Ex26Ro | TTCTTCAGCTTGTGTCATCC |
| Ex20Fi | CCCAGTCTACCACCCTATCAGAGC |
| Ex26Ri | CCTGCCTTTAAGGCTTCCTT |
aAll primers were synthesized as DNA on a phosphodiester backbone.
Internalization of Fluorescein-Labeled TMOs in H2K mdx Myotubes.
H-2Kb-tsA58 mdx myoblasts (H2K mdx cells) were cultured and differentiated as described previously (22–32). For internalization experiment, fluorescein-(FAM)-labeled thiomorpholino ASO (TMO1-FL and PMO-FL, Table 1) was transfected at 100 nM when complexed with lipofectin transfection reagent (Thermo Fisher Scientific) at the ratio of 2:1 (w:w) (lipofectin:ASO) and used in a final transfection volume of 500 μL/well in a 24-well plate as per the manufacturer's instructions, except that the solution was not removed after 3 h. In addition, to assess the ability of TMO to internalize into cells without transfection reagent, TMO1-FL and PMO-FL were incubated with cells at 200 nM for 1, 3, and 5 d. After indicated time points, cell nuclei were stained with Hoechst (Sigma Aldrich) for 10 min and washed five times with PBS containing 10% fetal bovine serum before the images were captured using Olympus TS-100 inverted fluorescence microscopy system with the NIS-Elements software (Nikon Instruments Inc., Hilton, South Australia, Australia). Quantitation of average fluorescence intensity was performed using Image J software.
Supplementary Material
Acknowledgments
R.N.V. acknowledges funding from the Department of Health Western Australia Merit Award Scheme (project no. 17993). R.N.V. also acknowledges funding from The McCusker Charitable Foundation and the Perron Institute for Neurological and Translational Science (project no. 19008). B.T.L. and R.N.V. acknowledge funding from the Caruthers Family Foundation and Perron Institute for Neurological and Translational Science (project no. 18873). S.P., H.L., and K.J. acknowledge funding by The University of Colorado (project no. 11704). We thank Molecular Therapy Laboratory, Murdoch University for providing H2K mdx cells.
Footnotes
Reviewers: S.D., University of California San Diego; and M.E., Vanderbilt University.
Competing interest statement: The authors declare a competing interest. The authors are included in the following patent: US Patent Application: Patent Application (CU ref. no. CU4506B-US1:) Thiomorpholino Oligonucleotides for the Treatment of Muscular Dystrophy.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2207956119/-/DCSupplemental.
Data, Materials, and Software Availability
All study data are included in the main text and the Supplementary Information.
References
- 1.Khorana H. G., Synthesis in the study of nucleic acids. The Fourth Jubilee Lecture. Biochem. J. 109, 709–725 (1968). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khorana H. G., Nucleic acid synthesis in the study of the genetic code. https://www.nobelprize.org/prizes/medicine/1968/khorana/lecture/. Accessed 10 August 2022.
- 3.Beaucage S. L., Caruthers M. H., Deoxynucleoside phosphoramidites—A new class of key intermediates for deoxypolynucleotide synthesis. Tetrahedron Lett. 22, 1859–1862 (1981). [Google Scholar]
- 4.Matteucci M. D., Caruthers M. H., Synthesis of deoxyoligonucleotides on a polymer support. J. Am. Chem. Soc. 103, 3185–3191 (1981). [PubMed] [Google Scholar]
- 5.Roehr B., Fomivirsen approved for CMV retinitis. J. Int. Assoc. Physicians AIDS Care 4, 14–16 (1998). [PubMed] [Google Scholar]
- 6.Hair P., Cameron F., McKeage K., Mipomersen sodium: First global approval. Drugs 73, 487–493 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Syed Y. Y., Eteplirsen: First global approval. Drugs 76, 1699–1704 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Hoy S. M., Nusinersen: First global approval. Drugs 77, 473–479 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Keam S. J., Inotersen: First global approval. Drugs 78, 1371–1376 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Paik J., Duggan S., Volanesorsen: First global approval. Drugs 79, 1349–1354 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Heo Y. A., Golodirsen: First approval. Drugs 80, 329–333 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Dhillon S., Viltolarsen: First approval. Drugs 80, 1027–1031 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Shirley M., Casimersen: First approval. Drugs 81, 875–879 (2021). [DOI] [PubMed] [Google Scholar]
- 14.Wan W. B., Seth P. P., The medicinal chemistry of therapeutic oligonucleotides. J. Med. Chem. 59, 9645–9667 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Stein C. A., Castanotto D., FDA-approved oligonucleotide therapies in 2017. Mol. Ther. 25, 1069–1075 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDonald C. M., et al. ; DEMAND V study group, Placebo-controlled phase 2 trial of drisapersen for duchenne muscular dystrophy. Ann. Clin. Transl. Neurol. 5, 913–926 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dirin M., Winkler J., Influence of diverse chemical modifications on the ADME characteristics and toxicology of antisense oligonucleotides. Expert Opin. Biol. Ther. 13, 875–888 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Langner H. K., Jastrzebska K., Caruthers M. H., Synthesis and Characterization of thiophosphoramidate Morpholino Oligonucleotides and chimeras. J. Am. Chem. Soc. 142, 16240–16253 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Paul S., Caruthers M., Synthesis of Backbone Modified Morpholino Oligonucleotides and Chimeras Using Phosphoramidite Chemistry, US Patent 11,230,565 B2, 2022.
- 20.Bulfield G., Siller W. G., Wight P. A., Moore K. J., X chromosome-linked muscular dystrophy (mdx) in the mouse. Proc. Natl. Acad. Sci. U.S.A. 81, 1189–1192 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rando T. A., Blau H. M., Primary mouse myoblast purification, characterization, and transplantation for cell-mediated gene therapy. J. Cell Biol. 125, 1275–1287 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilton S. D., et al. , Specific removal of the nonsense mutation from the mdx dystrophin mRNA using antisense oligonucleotides. Neuromuscul. Disord. 9, 330–338 (1999). [DOI] [PubMed] [Google Scholar]
- 23.Mann C. J., et al. , Antisense-induced exon skipping and synthesis of dystrophin in the mdx mouse. Proc. Natl. Acad. Sci. U.S.A. 98, 42–47 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le B. T., Agarwal S., Veedu R. N., Evaluation of DNA segments in 2′-modified RNA sequences in designing efficient splice switching antisense oligonucleotides. RSC Advances 11, 14029–14035 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le B. T., Kosbar T. R., Veedu R. N., Novel disulfide-bridged bioresponsive antisense oligonucleotide induces efficient splice modulation in muscle myotubes in vitro. ACS Omega 5, 18035–18039 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen S., Le B. T., Chakravarthy M., Kosbar T. R., Veedu R. N., Systematic evaluation of 2′-Fluoro modified chimeric antisense oligonucleotide-mediated exon skipping in vitro. Sci. Rep. 9, 6078 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le B. T., Adams A. M., Fletcher S., Wilton S. D., Veedu R. N., Rational design of short locked nucleic acid-modified 2′-o-methyl antisense oligonucleotides for efficient exon-skipping in vitro. Mol. Ther. Nucleic Acids 9, 155–161 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le B. T., Murayama K., Shabanpoor F., Asanuma H., Veedu R. N., Antisense oligonucleotide modified with serinol nucleic acid (SNA) induces exon skipping in mdx myotubes. RSC Advances 7, 34049–34052 (2017). [Google Scholar]
- 29.Le B. T., Hornum M., Sharma P. K., Nielsen P., Veedu R. N., Nucleobase-modified antisense oligonucleotides containing 5-(phenyltriazol)-2′-deoxyuridine nucleotides induce exon-skipping in vitro. RSC Advances 7, 54542–54545 (2017). [Google Scholar]
- 30.Le B. T., Filichev V. V., Veedu R. N., Investigation of twisted intercalating nucleic acid (TINA)-modified anti-sense oligonucleotides for splice modulation by induced exon-skipping in vitro. RSC Advances 6, 95169–95172 (2016). [Google Scholar]
- 31.Le B. T., Chen S., Abramov M., Herdewijn P., Veedu R. N., Evaluation of anhydrohexitol nucleic acid, cyclohexenyl nucleic acid and D-altritol nucleic acid-modified 2′-O-methyl RNA mixmer antisense oligonucleotides for exon skipping in vitro. Chem. Commun. (Camb.) 52, 13467–13470 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Chen S., et al. , Synthesis of a morpholino nucleic acid (MNA)-uridine phosphoramidite, and exon skipping using MNA/2′-O-methyl mixmer antisense oligonucleotide. Molecules 21, 1582 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schneider C. A., Rasband W. S., Eliceiri K. W., NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dominski Z., Kole R., Restoration of correct splicing in thalassemic pre-mRNA by antisense oligonucleotides. Proc. Natl. Acad. Sci. U.S.A. 90, 8673–8677 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sierakowska H., Sambade M. J., Agrawal S., Kole R., Repair of thalassemic human beta-globin mRNA in mammalian cells by antisense oligonucleotides. Proc. Natl. Acad. Sci. U.S.A. 93, 12840–12844 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scharner J., et al. , Hybridization-mediated off-target effects of splice-switching antisense oligonucleotides. Nucleic Acids Res. 48, 802–816 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee Y., Rio D. C., Mechanisms and regulation of alternative pre-mRNA splicing. Annu. Rev. Biochem. 84, 291–323 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fletcher S., et al. , Morpholino oligomer-mediated exon skipping averts the onset of dystrophic pathology in the mdx mouse. Mol. Ther. 15, 1587–1592 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Fletcher S., et al. , Dystrophin expression in the mdx mouse after localised and systemic administration of a morpholino antisense oligonucleotide. J. Gene Med. 8, 207–216 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the main text and the Supplementary Information.