Significance
The lipolytic processing of triglyceride-rich lipoproteins by lipoprotein lipase (LPL) is crucial for delivering dietary lipids to tissues. For years, the mechanism by which LPL (which is secreted by myocytes and adipocytes) reaches its site of action within blood vessels was mysterious. We discovered that GPIHBP1, a protein of capillary endothelial cells, captures LPL within the interstitial spaces and shuttles it to the luminal surface of capillaries. GPIHBP1’s three-fingered LU (Ly6/uPAR) domain binds stably to LPL’s C-terminal domain. An intrinsically disordered acidic domain in GPIHBP1 accelerates the kinetics of LPL binding, stabilizes the conformational integrity of LPL’s N-terminal hydrolase domain (thereby preserving LPL activity), and plays a critical role in the transport of LPL to the capillary lumen.
Keywords: triglycerides, lipoprotein lipase, endothelial cells
Abstract
GPIHBP1, a protein of capillary endothelial cells (ECs), is a crucial partner for lipoprotein lipase (LPL) in the lipolytic processing of triglyceride-rich lipoproteins. GPIHBP1, which contains a three-fingered cysteine-rich LU (Ly6/uPAR) domain and an intrinsically disordered acidic domain (AD), captures LPL from within the interstitial spaces (where it is secreted by parenchymal cells) and shuttles it across ECs to the capillary lumen. Without GPIHBP1, LPL remains stranded within the interstitial spaces, causing severe hypertriglyceridemia (chylomicronemia). Biophysical studies revealed that GPIHBP1 stabilizes LPL structure and preserves LPL activity. That discovery was the key to crystallizing the GPIHBP1–LPL complex. The crystal structure revealed that GPIHBP1’s LU domain binds, largely by hydrophobic contacts, to LPL’s C-terminal lipid-binding domain and that the AD is positioned to project across and interact, by electrostatic forces, with a large basic patch spanning LPL’s lipid-binding and catalytic domains. We uncovered three functions for GPIHBP1’s AD. First, it accelerates the kinetics of LPL binding. Second, it preserves LPL activity by inhibiting unfolding of LPL’s catalytic domain. Third, by sheathing LPL’s basic patch, the AD makes it possible for LPL to move across ECs to the capillary lumen. Without the AD, GPIHBP1-bound LPL is trapped by persistent interactions between LPL and negatively charged heparan sulfate proteoglycans (HSPGs) on the abluminal surface of ECs. The AD interrupts the HSPG interactions, freeing LPL–GPIHBP1 complexes to move across ECs to the capillary lumen. GPIHBP1 is medically important; GPIHBP1 mutations cause lifelong chylomicronemia, and GPIHBP1 autoantibodies cause some acquired cases of chylomicronemia.
In this article, we review the roles of GPIHBP1 (glycosylphosphatidylinositol-anchored high-density lipoprotein (HDL)-binding protein 1) in plasma triglyceride (TG) metabolism. GPIHBP1, an endothelial cell (EC) protein, is a crucial partner for lipoprotein lipase (LPL) in the lipolytic processing of TG-rich lipoproteins (TRLs).
In 1955, Korn (1, 2) characterized LPL as an intravascular TG hydrolase that could be released into the plasma with a bolus of heparin. Five years later, Havel and Gordon (3) discovered that LPL deficiency impairs the processing of TRLs, resulting in severe hypertriglyceridemia (chylomicronemia). In affected subjects, the levels of TG hydrolase activity in the postheparin plasma were very low (3). In subsequent years, the biochemical properties of LPL, including its activation by apolipoprotein (apo) CII, were investigated in detail (4–8), but the mechanism for LPL binding to blood vessels received less attention. Nevertheless, the fact that LPL was released into the plasma by heparin suggested that LPL was attached to blood vessels by electrostatic interactions. That view was bolstered in the 1980s by the discovery that LPL, which is a positively charged protein, binds to negatively charged heparan sulfate proteoglycans (HSPGs) on the surface of cultured cells (9, 10), and by sequencing of the LPL complementary DNA (cDNA) (11, 12), which revealed that LPL contains several positively charged heparin-binding motifs (13–15). For the next three decades, models of plasma TG metabolism depicted LPL attached to HSPGs on the luminal surface of blood vessels (16). That model was widely accepted but incomplete. For example, it was unclear how LPL, which is produced by parenchymal cells (e.g., myocytes, adipocytes), reaches its site of action within blood vessels, nor did the model explain why lipoproteins would marginate along the luminal surface of blood vessels (thereby allowing LPL-mediated TG hydrolysis to proceed).
The discovery of GPIHBP1 transformed the prevailing model for plasma TG metabolism. GPIHBP1 is expressed by capillary ECs, binds LPL avidly, and is solely responsible for transporting LPL to the lumen of capillaries (17, 18). Also, the margination of TRLs along ECs depends on GPIHBP1-mediated transport of LPL into capillaries (19). In addition, GPIHBP1 stabilizes LPL activity—even in the face of regulatory proteins that function to inhibit LPL (20–23). The discovery of GPIHBP1 also increased our understanding of human disease. A deficiency of GPIHBP1 causes lifelong chylomicronemia (24–26), and some newly acquired cases of chylomicronemia are caused by GPIHBP1 autoantibodies (27, 28). The new insights into TG metabolism resulted from serendipity and new experimental approaches. Serendipity came with the discovery of chylomicronemia in Gpihbp1 knockout mice (Gpihbp1−/−) (18, 29) and by the discovery, while characterizing a GPIHBP1 immunoassay, of GPIHBP1 autoantibodies in patients with chylomicronemia (27). New experimental approaches included confocal microscopy (to localize GPIHBP1 and LPL in blood vessels) (17, 18, 30–32), hydrogen–deuterium exchange/mass spectrometry (MS) (to understand GPIHBP1–LPL interactions and LPL conformational stability) (20–23, 33), surface plasmon resonance (SPR) studies (to quantify GPIHBP1–LPL binding kinetics) (20, 21, 23, 33), X-ray crystallography (to define the structural basis for GPIHBP1–LPL interactions) (34, 35), and nanosecondary ion MS (NanoSIMS) imaging (to visualize intravascular TG metabolism) (36, 37). Progress was fueled by strong collaborations and a commitment to understanding disease.
Cloning of the GPIHBP1 cDNA and an Early Proposal Regarding GPIHBP1 Function
GPIHBP1 was first identified, by expression cloning, as a GPI-anchored protein that conferred upon LDL receptor–deficient Chinese hamster ovary (CHO) cells the ability to bind HDL (38). The expression cloning strategy also uncovered scavenger receptor, class B, type 1 (SRB1), a known HDL-binding protein (39). Incubating GPIHBP1-expressing CHO cells with phosphatidylinositol-specific phospholipase C (PIPLC), a bacterial enzyme that cleaves GPI anchors, reduced HDL binding (38). GPIHBP1 was noted to have an N-terminal acidic region rich in aspartates and glutamates, a three-fingered cysteine-rich LU (Ly6/uPAR) domain (40) resembling those in other “LU domain” proteins (e.g., CD59, uPAR), and a C-terminal hydrophobic region characteristic of GPI-anchored proteins. GPIHBP1 was reported, based on in situ hybridization studies, to be expressed widely, with high levels of expression in Kupffer cells, sinusoidal ECs of the liver, cardiomyocytes, bronchiolar epithelium, and pulmonary macrophages. Based on these observations, GPIHBP1 was proposed to function in the uptake of cholesterol by cells (38).
Our experimental findings differed from those in the expression cloning paper (38). We found that GPIHBP1 is expressed exclusively in capillary ECs (17, 18), and we did not detect binding of HDL to GPIHBP1-transfected cultured cells (41).
Uncovering a Role for GPIHBP1 in Plasma TG Metabolism
Phenotyping of knockout mice for 472 genes encoding secreted or membrane proteins uncovered very high plasma TG levels in Gpihbp1−/− mice (29). In our mouse colony, plasma TG levels in chow-fed Gpihbp1−/− mice are 2,500 mg/dL to 3,500 mg/dL, about 100-fold higher than in wild-type (WT) mice (Gpihbp1+/+). On a high-fat, low-cholesterol diet, Gpihbp1−/− mice have TG levels as high as 20,000 mg/dL (42). Plasma TG levels in Gpihbp1+/− mice are normal.
For us, the high TG levels in Gpihbp1−/− mice could mean only one thing—that GPIHBP1 is important for LPL-mediated TRL processing. We quickly made three observations that were consistent with that view. First, the high plasma TG levels in Gpihbp1−/− mice are accompanied by markedly reduced LPL levels in the postheparin plasma (18). Second, GPIHBP1 is expressed at particularly high levels in tissues where TRL processing is robust (e.g., heart, adipose tissue). Third, GPIHBP1-expressing CHO cells bind LPL avidly, and the LPL binding is inhibited by heparin (18). We also observed binding of TRLs to GPIHBP1-transfected CHO cells (18). CHO cells are known to produce hamster LPL (43). The mechanism for TRL binding to GPIHBP1-expressing cells was initially perplexing (18), but we are now confident that the TRL binding is mediated by GPIHBP1-bound LPL. LPL and TRL binding to GPIHBP1-expressing cells is abolished by incubating the cells with PIPLC (18).
GPIHBP1 and LPL are confined to capillary ECs, as judged by confocal immunofluorescence microscopy (17, 32). Indeed, as soon as capillaries double in size to become small venules, both GPIHBP1 expression and LPL binding disappear. This finding makes sense, given that the purpose of intravascular TRL processing is to deliver lipid nutrients to adjacent parenchymal cells. GPIHBP1 is not expressed in capillaries of the brain, which primarily uses glucose (rather than TGs) for fuel (17).
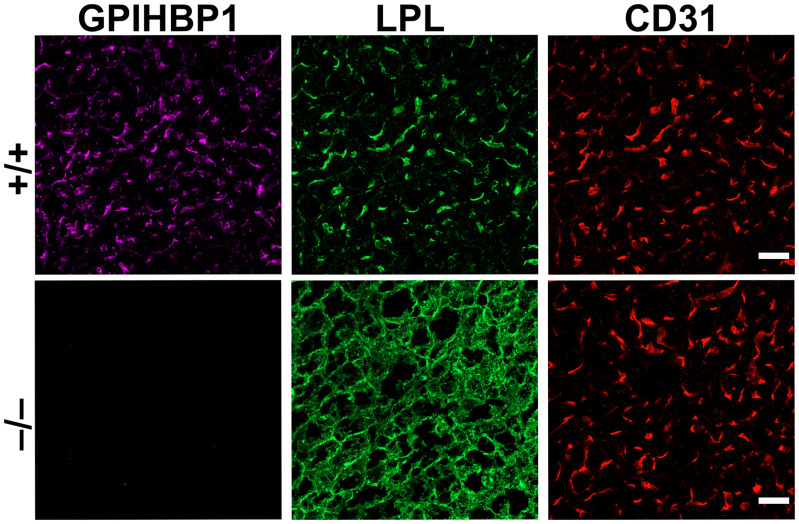
The fact that GPIHBP1 is expressed by capillary ECs and the observation that LPL binds to GPIHBP1 prompted us to propose that GPIHBP1 functions as a “platform for lipolysis” in capillaries (18). Soon thereafter, we discovered that GPIHBP1 is responsible for capturing LPL within the subendothelial spaces and transporting it across ECs to the capillary lumen (17). The crucial clues came from microscopy. In low-magnification images, most of the LPL in tissues of Gpihbp1+/+ mice was located on capillary ECs (colocalizing with GPIHBP1), whereas the LPL in tissues of Gpihbp1−/− mice was stranded within the interstitial spaces (bound to HSPGs on the surface of cells) (Fig. 1). With high-magnification images of capillary cross-sections, we were able to assess the distribution of LPL on the inner (luminal) and outer (abluminal) surfaces of ECs (17). In Gpihbp1+/+ mice, LPL and GPIHBP1 were abundant on the luminal surface of capillary ECs (Fig. 2). In Gpihbp1−/− mice, LPL was absent from the capillary lumen (Fig. 2). These findings left little doubt that GPIHBP1 functions as an LPL transporter.
Fig. 1.
Confocal immunofluorescence studies of LPL localization in BAT of Gpihbp1+/+ and Gpihbp1−/− mice. In Gpihbp1+/+ mice (+/+), most of the LPL is located on capillary ECs, bound to GPIHBP1 and colocalizing with CD31 (an EC marker). In Gpihbp1−/− mice (−/−), LPL is mislocalized within the interstitial spaces, bound to HSPGs on the surface of cells (adipocytes and ECs). The LPL in Gpihbp1−/− mice colocalizes with antibodies against collagen IV, a basal lamina protein of adipocytes (17). (Scale bar, 10 μm.)
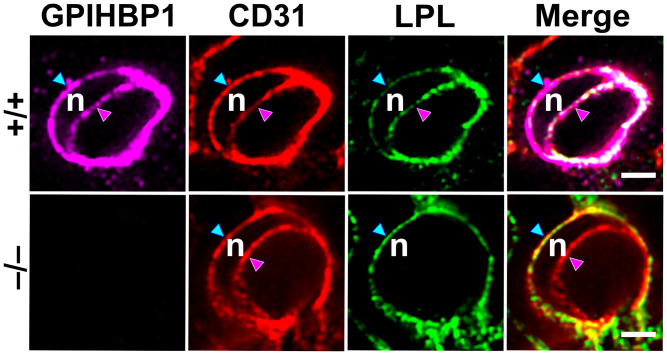
Fig. 2.
Immunofluorescence confocal microscopy images of capillary cross-sections in BAT of Gpihbp1+/+ and Gpihbp1−/− mice. Sections were stained with antibodies against GPIHBP1, LPL, and CD31. An EC nucleus (n) made it possible to examine distributions of GPIHBP1, CD31, and LPL along the abluminal surface of ECs (blue arrowhead) and the luminal plasma membrane (pink arrowhead). In Gpihbp1−/− mice, LPL was not transported to the capillary lumen. Different z slices of the capillary cross-sections were shown in a recent paper by Song et al. (30). (Scale bar, 2 µm.)
The notion that GPIHBP1 functions as a transporter was bolstered by experiments showing that GPIHBP1 moves LPL from the basal to the apical surface of cultured ECs and by studies showing that GPIHBP1 transports LPL across capillary ECs in living mice (17). When the GPIHBP1-specific monoclonal antibody (mAb) 11A12 was injected into a quadricep muscle bed of a Gpihbp1+/+ mouse, the antibody was rapidly captured by GPIHBP1 on the abluminal surface of capillary ECs. After 30 min, however, mAb 11A12 had been transported to the capillary lumen (17). In Gpihbp1−/− mice, there was no movement of mAb 11A12 into capillaries (17). We confirmed these observations in brown adipose tissue (BAT): mAb 11A12 was transported across capillary ECs in Gpihbp1+/+ mice but not in Gpihbp1−/− mice (31). GPIHBP1 movement across ECs is bidirectional (31). When Gpihbp1+/+ mice were given an intravenous injection of mAb 11A12, the antibody was rapidly captured by GPIHBP1 in the capillary lumen, but, within 120 min, it was transported to the abluminal surface of ECs. In cell culture studies, GPIHBP1-mediated LPL transport was blocked by dynasore, suggesting that GPIHBP1 moves across ECs by a vesicular mechanism (31). Also, GPIHBP1 was detected, by immunogold electron microscopy (EM), within vesicular structures of ECs (31).
GPIHBP1-mediated LPL movement to the capillary lumen is crucial in heart, skeletal muscle, and adipose tissue, but the transport function of GPIHBP1 is almost certainly less important in the liver, where ECs are fenestrated. Plasma TG levels are only modestly elevated (∼120 mg/dL) in Gpihbp1−/− mice during the suckling phase (when hepatic LPL expression is high), whereas Lpl−/− mice die soon after birth with severe chylomicronemia (44). We suspect that the milder phenotype of suckling Gpihbp1−/−mice relates to the ability of hepatic LPL in those mice to carry out TRL processing.
In the heart and BAT of Gpihbp1−/− mice, where intravascular LPL is absent, TRL margination along capillary ECs is negligible (19). In contrast, TRL margination is robust along heart and BAT capillaries in Gpihbp1+/+ mice. Interestingly, TRL margination was virtually undetectable along capillary ECs of the lung in Gpihbp1+/+ mice, a tissue with high levels of GPIHBP1 expression but extremely low levels of LPL. However, after an intravenous injection of purified LPL, LPL bound to GPIHBP1 on lung capillary ECs, and there was robust margination of TRLs. Thus, TRL margination along the luminal surface of capillaries requires LPL (19). Cell culture studies suggested that TRLs bind to a Trp-rich loop in the C-terminal domain(CTD) of LPL (residues 410 to 423) (19).
Early Insights into GPIHBP1–LPL Interactions
To define GPIHBP1 sequences required for LPL binding, we expressed mutant versions of GPIHBP1 in CHO cells and tested the capacity of transfected cells to bind LPL (45). These studies revealed that GPIHBP1’s LU domain is essential for LPL binding. Replacing GPIHBP1’s LU domain with CD59’s LU domain eliminated LPL binding (45), as did mutation of any of the 10 conserved cysteines in GPIHBP1’s LU domain (46). Alanine-scanning mutagenesis uncovered 12 other residues important for LPL binding, with the majority located in the second finger of the LU domain (residues 89 to 110) (47). Most of the mutations disrupted proper disulfide bond formation and led to the formation of disulfide-linked GPIHBP1 dimers and multimers (26, 46–48). However, mutating W109 abolished LPL binding without affecting disulfide bond formation, suggesting that it played a more direct role in LPL binding (48).
We identified several mAbs against human GPIHBP1 (e.g., RG3, RE3) that blocked the ability of GPIHBP1 to bind LPL (49). The epitopes for those mAbs were in the LU domain. A GPIHBP1-specific mAb (RF4) with an epitope upstream from the LU domain did not block LPL binding (33, 49).
The importance of the LU domain was nailed down by studying Gpihbp1 knock-in mice harboring a single amino acid substitution in the LU domain (p.C63Y) (50). The mutant GPIHBP1 was expressed in capillary ECs and was present along the luminal surface of capillary ECs, but it lacked the ability to bind LPL. Consequently, there was no LPL inside capillaries of the mutant mice, and they exhibited severe chylomicronemia (50).
To define LPL sequences required for GPIHBP1 binding, we examined the ability of hepatic lipase (HL)–LPL chimeric proteins to bind to GPIHBP1. HL (an LPL paralogue) had no ability to bind to GPIHBP1 (41), nor did an HL–LPL chimera containing the N terminus of LPL (residues 1 to 339) and the C terminus of HL (residues 351 to 499). In contrast, a chimera containing the N terminus of HL (residues 1 to 350) and the CTD of LPL (residues 340 to 475) bound to GPIHBP1 (51). Mutating C445 in LPL to Ser (C445 is disulfide-linked to C465) eliminated GPIHBP1 binding (49). Also, GPIHBP1 binding was markedly reduced by a E448K LPL mutation (52). While LPL’s C terminus is required for GPIHBP1 binding, we showed (with SPR studies) that a C-terminal fragment of bovine LPL (residues 343 to 478) binds to GPIHBP1 with a lower affinity than full-length LPL (20).
The conclusion that LPL binding to GPIHBP1 depends on its CTD was supported by studies with the LPL-specific mAb 88B8 (51). The 88B8 binds to LPL residues 357 to 465 (51) and blocks LPL binding to GPIHBP1. Also, the LPL residues that were found to be important for GPIHBP1 binding (e.g., C445, E448) are also important for 88B8 binding (51).
In early studies, we observed APOA5 binding to GPIHBP1-expressing CHO cells (41, 53). APOA5 binding appeared to be mediated by electrostatic interactions with GPIHBP1’s acidic domain. While APOA5 binding to GPIHBP1-expressing cells was reproducible, we caution that these studies were limited in scope and were subject to caveats. For example, the conformation and biological activity of the APOA5 were not well documented, and the specificity and affinity of APOA5 binding to GPIHBP1 were not investigated with purified proteins and cell-free binding assays.
Stoichiometry of GPIHBP1 Binding to LPL
LPL was proposed to be a homodimer in the mid-1970s (8), and that view gained widespread acceptance. LPL was thought to assume a head-to-tail homodimer conformation, with the lipid-binding sequences in the C terminus of one monomer in close proximity to the catalytic pocket in the N terminus of the partner monomer (54–56). Homodimer formation was thought to be crucial for LPL secretion (54, 57) and catalytic activity (15, 54, 55, 57–60). With the LPL homodimer model in mind, we predicted that two GPIHBP1 molecules would bind to LPL; however, we consistently failed to find support for this prediction. We incubated two GPIHBP1 preparations (each containing a different epitope tag) with LPL and then immunoprecipitated GPIHBP1–LPL complexes with an antibody against only one of the epitope tags. When we analyzed immunoprecipitates, we detected LPL but only one of the two GPIHBP1 proteins—we never observed both GPIHBP1 proteins. These observations led us to be skeptical of the LPL homodimer model. We went on to show, using density gradient ultracentrifugation and immunochemical studies, that freshly secreted and catalytically active LPL is monomeric. A recent study strongly supported this view (22). Native gel studies revealed similar electrophoretic migrations for GPIHBP1–LPL complexes and GPIHBP1 complexed to a Fab fragment of GPIHBP1-specific mAbs (RF3 or RF4); LPL and Fab fragments have comparable molecular weights. Also, GPIHBP1–LPL–Fab5D2 complexes and FabRF3–GPIHBP1–FabRF4 complexes had comparable electrophoretic migrations. Finally, the stoichiometry of GPIHBP1–LPL–Fab5D2 complexes was 1:1:1, as judged by mass determinations with SEC-MALS and SEC-SAXS (size-exclusion chromatography combined with either light or small-angle X-ray scattering) (22).
Digging Deeper into GPIHBP1–LPL Interactions
To improve our understanding of GPIHBP1–LPL interactions, we used hydrogen–deuterium MS (HDX-MS) studies (61). Human GPIHBP1 (purified from the medium of Drosophila S2 cells) and purified bovine LPL were incubated in the presence of D2O. The binding of GPIHBP1 to LPL reduced deuterium exchange in GPIHBP1 residues 104 to 135 and LPL residues 429 to 446, suggesting that those sequences participate in the binding interface. Deuterium uptake was also reduced in LPL residues 306 to 320 (located in LPL’s hinge region and containing one of LPL’s heparin-binding motifs). A synthetic peptide corresponding to the acidic domain of human GPIHBP1 (residues 21 to 53) also reduced deuterium uptake in LPL residues 306 to 320, implying that the acidic domain binds, by electrostatic forces, to positively charged residues in LPL’s hinge region. Zero-length cross-linking studies with carbodiimide supported this conclusion (20).
Deuterium exchange in the acidic domain of full-length GPIHBP1 was extremely rapid, consistent with predictions that it is intrinsically disordered (20). The disordered nature of the acidic domain was corroborated by SEC-SAXS studies (33). Of note, the acidic domain had little or no effect on deuterium uptake in GPIHBP1’s LU domain, implying that it is not important for the conformation of the LU domain.
GPIHBP1 Stabilizes LPL Structure and Activity
Purified preparations of LPL, when incubated at 25 °C, lose enzymatic activity within minutes (62). To understand why LPL activity disappears so quickly, we used HDX-MS studies to monitor the conformational stability of LPL. At 25 °C, we observed a time-dependent increase in deuterium uptake in LPL’s N-terminal hydrolase domain with EX1 exchange kinetics (consistent with unfolding of secondary structure elements) but little or no unfolding in LPL’s C-terminal lipid-binding domain (20, 23, 33). Unfolding of the hydrolase domain was accompanied by loss of LPL catalytic activity. Both LPL unfolding and the loss of catalytic activity were markedly reduced by GPIHBP1 binding (20, 33).
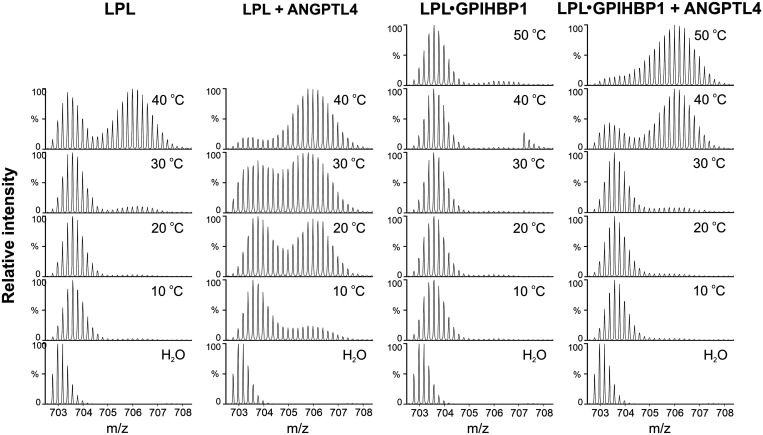
The spontaneous unfolding of purified preparations of LPL is not just a biochemical curiosity. A physiologic regulator of LPL, ANGPTL4, inhibits LPL activity in vivo by catalyzing the same LPL unfolding pathway (21, 23, 63). Even when the molar concentration of ANGPTL4 (relative to LPL) is low, ANGPTL4 fully inactivates LPL. When 10 µM LPL was incubated with 2 µM ANGPTL4 for 10 min, we observed, by HDX-MS, extensive unfolding of LPL’s hydrolase domain (accompanied by a 90% decrease in LPL activity) (21, 63). We found that ANGPTL4 reduces the conformational stability of LPL at a range of temperatures (Fig. 3). GPIHBP1 binding stabilizes the conformational stability of LPL, in both the presence and absence of ANGPTL4 (Fig. 3).
Fig. 3.
HDX-MS studies revealing temperature-dependent unfolding of the α/β-hydrolase (catalytic) domain of bovine LPL, either alone or in the presence of GPIHBP1 and/or ANGPTL4. Shown here are isotope envelopes of a peptic peptide from the hydrolase domain (residues 131 to 165 in the mature protein sequence of bovine LPL) containing LPL’s active-site serine after pulse labeling in deuterated solvent. LPL was preincubated in protiated solvent for 3 min at the indicated temperatures prior to pulse labeling for 10 s in the deuterated solvent at 25 °C. Emergence of bimodal isotope envelopes is a signature for irreversible protein unfolding. Adapted from Leth-Espensen et al. (23).
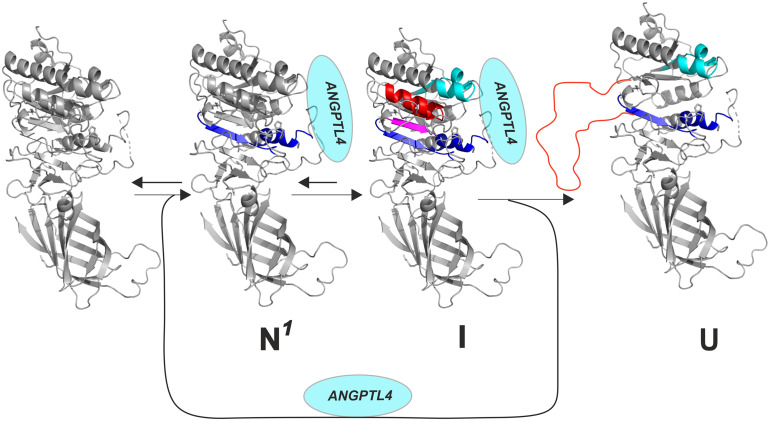
Recent HDX-MS studies yielded insights into mechanisms by which ANGPTL4 inhibits LPL (21, 23, 33, 63). ANGPTL4 binds initially to three segments of LPL’s hydrolase domain (residues 79 to 90, 112 to 130, 248 to 254), thereby protecting those sequences from deuterium uptake (23). However, this initial ANGPTL4 interaction triggers an allosteric increase in EX2-mediated deuterium uptake (i.e., increased dynamics) in sequences surrounding LPL’s catalytic pocket (residues 208 to 223 and 267 to 277) (Fig. 4). In the case of unbound LPL, the increased dynamics progress to irreversible unfolding of secondary structure elements forming LPL’s catalytic pocket (evident by EX1-mediated deuterium uptake kinetics). At 37 °C, the binding of GPIHBP1 to LPL markedly reduced but did not eliminate ANGPTL4-mediated LPL unfolding (23).
Fig. 4.
ANGPTL4-mediated inactivation of LPL triggers irreversible unfolding and collapse of the catalytic pocket in LPL’s N-terminal α/β-hydrolase (catalytic) domain. This figure illustrates the step-wise LPL unfolding elicited by ANGPTL4 binding. Upon binding to ANGPTL4 (N1), there is greater flexibility in the secondary structure elements of α5 and β6 (blue). The increased flexibility leads to an intermediate state (I) with more global but reversible unfolding of β5 (purple) and α3 (red). Ultimately, those changes trigger irreversible unfolding of structural elements (orange) forming LPL’s catalytic pocket (U). Adapted from Kristensen et al. (64), which is licensed under CC BY 4.0.
The capacity of GPIHBP1 to preserve the conformation of LPL’s hydrolase domain (Fig. 3) was supported by differential scanning fluorimetry (nano-DSF) measurements of LPL’s thermal stability (23). GPIHBP1 binding greatly augments the thermal stability of LPL’s hydrolase domain. The melting temperature (Tm) of the hydrolase domain is 34.8 °C with free (unbound) LPL but was 57.6 °C when the LPL was bound to GPIHBP1 (23). ANGPTL4 had opposite effects. In the presence of ANGPTL4, the melting temperature of the hydrolase domain in free LPL was reduced from 34.8 °C to ≤15 °C, and the melting temperature of GPIHBP1-bound LPL was reduced from 57.6 °C to 37.8 °C (23). The melting temperature of LPL’s CTD was 64.7 °C and was largely unaffected by ANGPTL4 or GPIHBP1 (23). The biophysics of the interactions between LPL, GPIHBP1, and its inhibitors (ANGPTL-3, ANGPTL-4, and ANGPTL-8) are discussed extensively in a recent review (64).
An Atomic Structure for the GPIHBP1–LPL Complex
The structure of pancreatic lipase (an LPL paralogue) was solved by X-ray crystallography more than 30 y ago (65, 66), but efforts to crystallize LPL were unsuccessful, likely because of LPL’s propensity to unfold and aggregate (34). Given GPIHBP1’s ability to mitigate LPL unfolding (20), we reasoned that it might be possible to crystallize LPL complexed to GPIHBP1 (34). Indeed, GPIHBP1–LPL complexes (purified by SEC) yielded crystals that diffracted at high resolution (2.8 Å) (34), and the structure of the GPIHBP1–LPL complex was solved by molecular replacement (34).
In the LPL–GPIHBP1 crystal structure, LPL was arranged in a head-to-tail homodimer conformation, resembling (at least superficially) old models for LPL homodimer structure (15, 54). At first glance, the LPL homodimers were disappointing, because we had evidence that LPL was catalytically active as a monomer (67). However, upon closer inspection, it was clear that the homodimers in the crystal structure were inconsistent with LPL catalytic activity. The Trp-rich lipid-binding loop in the C terminus of one monomer (crucial for TRL–LPL interactions) occluded the catalytic pocket of the partner monomer (obviating any possibility of TG hydrolysis) (34). The lid covering the catalytic pocket was in an open conformation (34). The formation of homodimers during protein crystallization presumably allowed hydrophobic sequences in LPL (i.e., the Trp-rich loop, the substrate-binding cavity) to be shielded from the aqueous environment. Unfortunately, the homodimer conformation limited our ability to define structures for portions of LPL’s lid region and Trp-rich loop. Subsequent crystal structures for the GPIHBP1–LPL complex also revealed a head-to-tail LPL conformation, and the authors of that study emphasized that LPL homodimers were not physiologic (68), referring to studies showing that catalytically active LPL is monomeric (67). In one of the crystal structures, LPL was crystallized with a small-molecule inhibitor in the catalytic pocket, which allowed for improved definition of the Trp-rich loop and lid domain (68). A cryo-EM structure of bovine LPL (3.8-Å resolution) also revealed a head-to-tail LPL homodimer conformation (69). The authors showed that catalytically inactive LPL homodimers at high concentration and low temperature self-associate and form helical fibrils, and they hypothesized that assembly of LPL fibrils in intracellular vesicles could be a physiologic mechanism for LPL storage (69).
We proposed that high concentrations of purified LPL drive homodimer formation (67). To create crystals for X-ray diffraction, GPIHBP1–LPL complexes were incubated at very high concentrations (∼10 mg/mL). In SAXS studies (where homodimers were also present), the concentration of GPIHBP1–LPL complexes was ∼0.7 mg/mL. In the medium of LPL-transfected CHO cells (where LPL is monomeric), the LPL concentration was very low (<1.0 μg/mL) (67).
The GPIHBP1–LPL crystal structure defined the molecular interface between LPL and GPIHBP1 (34). The concave surface of the LU domain covered one surface of LPL’s CTD; the binding interface (∼940 Å2) involved all three fingers of the LU domain and was mediated largely by hydrophobic contacts (34). Our prediction that GPIHBP1 residue W109 could play a direct role in the binding interface (48) was borne out by the crystal structure. The indole side chain of W109 lies flat within the interface and interacts with multiple residues in LPL’s C terminus (34). Our prediction, based on HDX-MS studies, that LPL residues 432 to 449 participate in the binding interface was also borne out by the crystal structure.
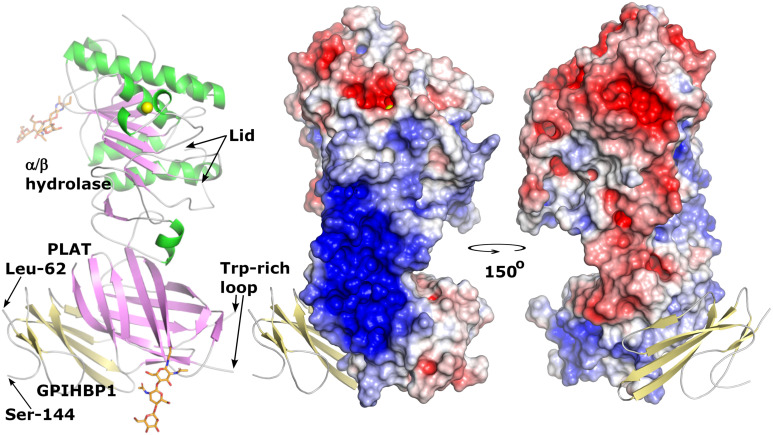
LPL contains multiple heparin-binding motifs (13–15). An electrostatic surface potential map of LPL, created from the crystal structure, revealed that LPL’s heparin-binding motifs converge to form a single large, flat, contiguous basic patch (∼2,400 Å2) (34) (Fig. 5). The basic patch extends across LPL’s hinge region and involves both C- and N-terminal domains (34). GPIHBP1’s disordered acidic domain (>60 Å in length) was not defined in the electron density map but was oriented to project across and interact with the entirety of LPL’s basic patch (34). Consistent with that view, we found, by HDX-MS studies, that GPIHBP1’s acidic domain protects LPL residues 279 to 293 from deuterium uptake (20). We also showed that the interactions between GPIHBP1’s acidic domain and LPL’s basic patch preserve LPL conformation and protect LPL from unfolding (34, 70).
Fig. 5.
Crystal structure of the human GPIHBP1–LPL complex. LPL assumed a head-to-tail homodimer conformation, with the lipid-binding Trp-rich loop in the C-terminal PLAT (polycystin-1, lipoxygenase, α-toxin) domain of one monomer buried in the catalytic pocket of the N-terminal α/β hydrolase domain of the partner monomer (34). (Left) A cartoon representation of the structure of a single LPL/GPIHBP1 complex. Both the α/β hydrolase and PLAT domains contain a single N-linked glycan (orange sticks) (34); the hydrolase domain contains a single calcium ion (yellow sphere). Portions of the Trp-rich loop and the lid covering the catalytic pocket were not defined in the electron density map. (Middle and Right) An electrostatic surface potential map of LPL with a ribbon diagram of human GPIHBP1 (bound to LPL’s PLAT domain). N-terminal sequences (residues 21 to 61) containing the disordered acidic domain were not defined in the electron density map. One surface of LPL, spanning both N-terminal domains and CTDs, contained a large basic patch (blue). Residues 21 to 61 of GPIHBP1 are predicted to extend from Leu-62 (the first residue defined in the crystal structure) and project over and form a fuzzy complex with LPL’s basic patch (34). The C terminus of GPIHBP1 (residues 145 to 151) was not defined; a GPI moiety attached to residue G151 anchors GPIHBP1 to the plasma membrane of ECs.
The GPIHBP1–LPL crystal structure made it possible for us to understand LPL mutations that cause chylomicronemia. For example, a p.M404R LPL mutation, identified in a Middle Eastern patient, had been reported to cause chylomicronemia by abolishing LPL secretion (71), but the crystal structure showed that M404 is located within the GPIHBP1–LPL binding interface (34). Follow-up studies revealed that the M404R substitution abolishes GPIHBP1 binding but has no effect on LPL secretion or catalytic activity (34). The crystal structure also made it possible to understand a p.D201V LPL mutation in Lebanese patients with chylomicronemia (72). D201 (along with A194, R197, S199, and D202) coordinates a calcium ion in LPL’s hydrolase domain (34). We suspected that the calcium ion might be crucial for the assembly of the hydrolase domain and that any defect in calcium coordination would interfere with LPL folding and secretion. Indeed, mutations in two of LPL’s calcium-coordinating residues abolished LPL secretion from cultured CHO cells (34).
Functional Relevance of GPIHBP1’s Acidic Domain
The GPIHBP1 in every mammalian species contains an N-terminal acidic domain. In human GPIHBP1, 21 of 26 consecutive residues are aspartate or glutamate. In the opossum, the acidic domain is more impressive (32 aspartates or glutamates in 39 residues, including 23 consecutive aspartates). The exact sequence of the acidic domain varies considerably in different mammals, and it appears that aspartates and glutamates are used interchangeably. In most mammals, the acidic domain contains a conserved tyrosine (Y38 in human GPIHBP1). Studies with a sulfotyrosine-specific antibody revealed that Y38 is sulfated (augmenting the negative charge of the acidic domain). Sulfation of Y38 was confirmed by MS (33). Mouse GPIHBP1 contains two tyrosines; MS revealed that one-half of mouse GPIHBP1 molecules contain two sulfate modifications, while the other half contain only one (30).
We uncovered roles for GPIHBP1’s acidic domain in binding LPL (20, 45), maintaining LPL conformation and catalytic activity (20, 23, 33), and transporting LPL to the luminal surface of capillaries (30).
A Role for the Acidic Domain in LPL Binding.
Soon after showing that LPL binds to GPIHBP1-transfected CHO cells, we tested LPL binding to cells that expressed mutant GPIHBP1 proteins with deletions in the acidic domain (45). The early experiments were not performed with optimized binding assays, but they suggested that the C-terminal half of the acidic domain (residues 38 to 48) is important for LPL binding (45). We also found that LPL binding to GPIHBP1 was inhibited by polyglutamate or polyaspartate (45). While the acidic domain appeared to contribute to LPL binding, it was clear that the acidic domain alone was insufficient for stable LPL interactions. For example, when the acidic domain was expressed in the context of CD59’s LU domain, there was no LPL binding (45), and single amino acid substitutions in the LU domain eliminated LPL binding—despite an intact acidic domain (46, 47). Based on those early observations, we speculated that the acidic domain bound positively charged proteins in a “promiscuous” fashion and that the “lock and key” specificity for LPL binding rested with the LU domain (46).
SPR studies clarified roles for GPIHBP1’s acidic domain and LU domain in LPL binding. In initial studies, the CTD of human LPL (residues 340 to 475) was immobilized on a sensor chip coated with the LPL-specific mAb 5D2 (20). WT-GPIHBP1 or a mutant GPIHBP1 lacking the acidic domain was injected over the sensor chip, making it possible for us to measure interactions of GPIHBP1 with the immobilized LPL. The affinity (Kd) of GPIHBP1–LPL CTD interactions was ∼10-fold higher for WT-GPIHBP1 than for a mutant GPIHBP1 lacking the acidic domain (0.12 μM vs. 1.4 μM) (20). The reduced affinity with the mutant GPIHBP1 was due to a lower association rate constant (kon). The dissociation rate constant (koff) was 0.1 s−1 for both the mutant and WT-GPIHBP1 proteins, implying that the acidic domain has little effect on the stability of GPIHBP1–LPL interactions (20). In these studies, a mutant GPIHBP1 with an LU domain mutation (p.W109S) had no ability to bind to LPL’s CTD, despite an intact acidic domain. Also, WT-GPIHBP1 failed to bind to an LPL CTD containing a p.C445Y mutation. We also used microscale thermophoresis to measure equilibrium binding constants for GPIHBP1–LPL interactions. In those studies, the affinity of GPIHBP1–LPL interactions was ∼25-fold higher for WT-GPIHBP1 than for a mutant GPIHBP1 lacking the acidic domain (20).
In subsequent SPR studies (33), performed with conditions that optimized the conformational integrity of the immobilized LPL, the kon for WT-GPIHBP1 binding to LPL was ∼2,500-fold higher than for a mutant GPIHBP1 lacking the acidic domain (kon 3 × 108 M−1⋅s−1 vs. 8.7 × 104 M−1⋅s−1). Furthermore, because the kon for WT-GPIHBP1 binding to LPL was markedly reduced at higher NaCl concentrations, we concluded that the binding kinetics were driven by electrostatic interactions between GPIHBP1’s acidic domain and LPL’s basic patch (33). Interestingly, the sulfation of Y38 contributed to the affinity of GPIHBP1–LPL interactions; replacing Y38 with phenylalanine reduced the kon for GPIHBP1 binding to LPL (33).
The ability of the acidic domain to accelerate the kinetics of GPIHBP1–LPL interactions is probably important in vivo. We suspect that GPIHBP1’s disordered acidic domain, which is predicted to extend >60 nm in every direction (20, 33), facilitates the capture of LPL from subendothelial HSPGs and promotes stable interactions of LPL with GPIHBP1’s LU domain.
The notion that the acidic domain augments the capture of LPL within the subendothelial spaces was inspired by SPR experiments in which LPL was bound to a sensor chip coated with heparin sulfate (33). When WT human GPIHBP1 was injected over the sensor chip, it rapidly detached the LPL from the heparin binding sites (releasing GPIHBP1–LPL complexes into the buffer flow). A mutant GPIHBP1 lacking the acidic domain failed to detach LPL and, instead, simply bound to the heparin-immobilized LPL (33).
A Role for the Acidic Domain in Stabilizing LPL Structure and Activity.
As noted earlier, HDX-MS studies revealed that full-length GPIHBP1 reduces the spontaneous unfolding of LPL’s hydrolase domain and preserves catalytic activity (20). GPIHBP1’s acidic domain protects LPL from spontaneous unfolding, but a mutant GPIHBP1 lacking the acidic domain has minimal ability to prevent unfolding (20). Moreover, an acidic domain peptide reduces spontaneous unfolding of LPL (although less efficiently than full-length GPIHBP1) (20).
Full-length GPIHBP1 also protects against ANGPTL4-catalyzed LPL unfolding (Fig. 4). Again, that protection requires GPIHBP1’s acidic domain. A mutant GPIHBP1 lacking the acidic domain had little effect on ANGPTL4-mediated unfolding (21), and protection against ANGPTL4-mediated LPL unfolding was reduced when the sulfated tyrosine in the acidic domain (Y38) was replaced with a phenylalanine (33). Furthermore, the ability of a GPIHBP1 acidic domain peptide to inhibit the equilibrium binding between LPL and full-length GPIHBP1 was enhanced when Y38 was sulfated (33).
Thermostability measurements by nano-DSF underscored the role of GPIHBP1’s acidic domain in stabilizing LPL conformation (23). WT-GPIHBP1 augmented the thermal stability of LPL’s hydrolase domain (increasing the Tm from 34.8 °C to 57.6 °C), whereas a mutant GPIHBP1 lacking the acidic domain increased the Tm to only 42.2 °C (23).
A Role for the Acidic Domain in Transporting LPL across ECs.
SPR, HDX-MS, microscale thermophoresis, and nano-DSF experiments—all involving purified proteins—revealed that GPIHBP1’s acidic domain preserves LPL conformation, but we wanted to define the biological importance of the acidic domain for plasma TG metabolism. For this reason, we created mice with a mutant allele, Gpihbp1S, in which 17 residues in the acidic domain (including the sulfated tyrosine and the long stretch of acidic residues) were replaced with an S-protein tag. Expression of the Gpihbp1S allele in ECs was robust, and the mutant protein (S-GPIHBP1) reached the luminal surface of capillaries (30). The LU domain in S-GPIHBP1 was intact; thus, it retained the capacity to bind LPL (30). However, SPR studies revealed that the kon for S-GPIHBP1 binding to LPL was significantly lower than that for WT-GPIHBP1. In contrast, the koff for S-GPIHBP1 binding to LPL (reflecting stability of GPIHBP1–LPL interactions) was similar to the koff for WT-GPIHBP1 (30).
Gpihbp1S/S mice had elevated plasma TG levels (∼500 mg/dL), and the TG levels increased to >1,000 mg/dL after an intragastric oil gavage (30). After an intravenous bolus of heparin, plasma LPL activity levels were ∼70% lower in Gpihbp1S/S mice than in Gpihbp1+/+ mice. In the heart and BAT, we observed, by immunofluorescence microscopy, a modest increase in LPL within the interstitial spaces, suggesting a reduced capacity of S-GPIHBP1 to capture interstitial LPL. However, the most remarkable microscopy finding was increased amounts of LPL on the capillary ECs in Gpihbp1S/S mice (30). That observation posed an obvious question: With greater-than-normal amounts of LPL on capillaries, why did Gpihbp1S/S mice have hypertriglyceridemia? We imagined two possibilities. The first was that LPL on capillaries in Gpihbp1S/S mice was distributed disproportionately to the abluminal surface of capillaries (where it would be useless for plasma TG metabolism). The second was that the catalytic activity of LPL in Gpihbp1S/S mice was reduced (reflecting a reduced capacity of S-GPIHBP1 to protect LPL from unfolding).
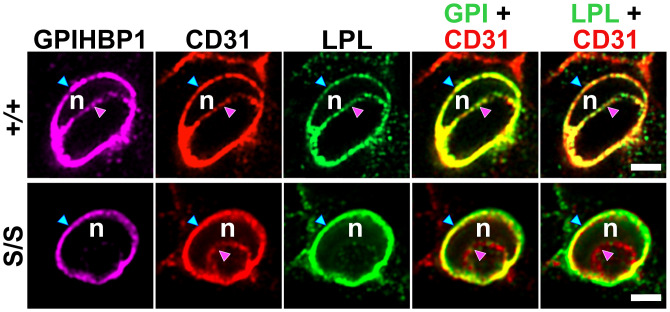
To define the distribution of LPL along capillary ECs in Gpihbp1S/S mice, we imaged capillary cross-sections (30). In BAT and the heart of Gpihbp1+/+ mice, LPL and GPIHBP1 were distributed equally along the abluminal and luminal surfaces of ECs. In Gpihbp1S/S mice, we found markedly reduced amounts of GPIHBP1 and LPL on the luminal surface of ECs but increased amounts on the abluminal surface (30) (Fig. 6). The reduced amounts of LPL inside the capillaries of Gpihbp1S/S mice were confirmed by experiments in which we gave mice intravenous injections of LPL- and CD31-specific antibodies. The amount of LPL antibody binding to blood vessels (relative to the binding of the CD31 antibody) was significantly lower in tissues of Gpihbp1S/S mice than in tissues of Gpihbp1+/+ mice (30).
Fig. 6.
Immunofluorescence confocal microscopy images of capillary cross-sections in BAT from a WT mouse (Gpihbp1+/+) and a mutant mouse (Gpihbp1S/S) in which the acidic domain was replaced with an S-protein tag. Sections were stained with antibodies against GPIHBP1 (GPI), LPL, and CD31. An EC nucleus (n) made it possible to examine distributions of GPIHBP1, CD31, and LPL along the abluminal surface of ECs (blue arrowhead) and the luminal plasma membrane (pink arrowhead). In Gpihbp1+/+ mice (+/+), LPL was distributed roughly equally between the abluminal and luminal surfaces of ECs. In Gpihbp1S/S mice (−/−), LPL was distributed asymmetrically, with only trace amounts in the capillary lumen. (Scale bar, 2 µm.)
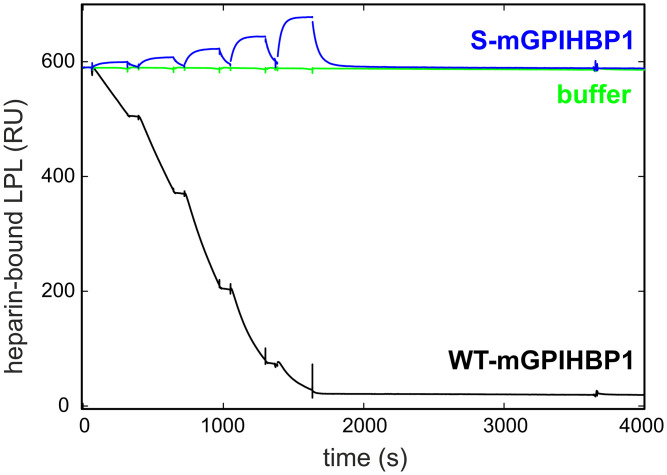
We suspected that the disproportionate distribution of LPL and S-GPIHBP1 to the abluminal surface of blood vessels in Gpihbp1S/S mice was due to persistent binding of GPIHBP1-bound LPL to abluminal HSPGs (30). In Gpihbp1S/S mice, we reasoned that LPL’s basic patch would be exposed and free to bind to abluminal HSPGs. In Gpihbp1+/+ mice, we suspected that GPIHBP1’s acidic domain would sheath LPL’s basic patch, abrogating HSPG interactions and freeing GPIHBP1–LPL complexes to move to the capillary lumen. That interpretation was supported by SPR studies that examined the ability of WT- and S-GPIHBP1 to bind to LPL captured on heparin-coated sensor chips (30). When S-GPIHBP1 was flowed over the sensor chip, it bound (via its LU domain) to the LPL (Fig. 7). In contrast, when WT-GPIHBP1 was flowed over the sensor chip, it detached the LPL, releasing GPIHBP1–LPL complexes into the buffer flow (Fig. 7). Thus, WT-GPIHBP1, but not S-GPIHBP1, was able to disrupt LPL binding to heparin sulfate chains.
Fig. 7.
SPR study demonstrating that WT mouse GPIHBP1 (WT-mGPIHBP1) detaches LPL from a heparin sulfate–coated sensor chip, whereas a mutant mouse GPIHBP1 protein lacking the acidic domain (S-mGPIHBP1) does not. When buffer was injected over the sensor chip, LPL remained stably bound to heparin sulfate chains (green). When WT-mGPIHBP1 was injected over the sensor chip (in five consecutive serial dilutions, 12.5 nM to 200 nM), it dislodged LPL from the heparin sulfate–coated sensor chip (black). When S-mGPIHBP1 was injected over the sensor chip, it simply bound to the LPL and did not dislodge it. Adapted from Song et al. (30), which is licensed under CC BY 4.0.
The idea that the GPIHBP1–LPL complexes in the heart and BAT of Gpihbp1S/S mice were trapped on the abluminal surface of capillaries was supported by studies of GPIHBP1 localization in lung capillaries (30). GPIHBP1 is abundant in lung capillary ECs, whereas LPL expression in the lung is extremely low. Because the lung contains such low levels of LPL, we suspected that abluminal trapping of S-GPIHBP1 in lung capillaries would be absent in Gpihbp1S/S mice. Indeed, S-GPIHBP1 was distributed evenly along the abluminal and luminal surfaces of lung capillary ECs in Gpihbp1S/S mice (30).
The notion that the GPIHBP1–LPL complexes in the heart and BAT of Gpihbp1S/S mice were trapped by binding to HSPGs on the abluminal surface of capillaries was also supported by in vivo studies of GPIHBP1 movement across capillary ECs (30). We injected the fluorescently labeled GPIHBP1-specific mAb 11A12 into interscapular BAT pads of Gpihbp1+/+ and Gpihbp1S/S mice. In Gpihbp1+/+ mice, mAb 11A12 was rapidly captured by GPIHBP1 on the abluminal surface of capillaries. After 90 min, however, a large fraction of the antibody had been transported to the luminal surface of capillary ECs. In Gpihbp1S/S mice, mAb 11A12 did not move across ECs and, instead, remained at the abluminal surface of ECs (reflecting trapping of GPIHBP1-bound LPL by abluminal HSPGs) (30). In follow-up studies, we coinjected mAb 11A12 into BAT along with heparin or a high concentration of a GPIHBP1 acidic domain peptide (30). We reasoned that disrupting electrostatic interactions between LPL’s basic patch and abluminal HSPGs would make it possible for S-GPIHBP1 to reach the capillary lumen. Indeed, both heparin and the acidic domain peptide normalized the movement of mAb 11A12 across BAT capillary ECs in Gpihbp1S/S mice (30).
Abluminal trapping of S-GPIHBP1–LPL complexes undoubtedly contributed to the hypertriglyceridemia in Gpihbp1S/S mice, but we also identified a second mechanism. The specific activity of LPL in the postheparin plasma was ∼50% lower in Gpihbp1S/S mice than in Gpihbp1+/+ mice (30). The lower specific activity in Gpihbp1S/S mice is consistent with the properties of S-GPIHBP1. The binding of WT mouse GPIHBP1 to mouse LPL increased the Tm of LPL’s hydrolase domain from 34.5 ± 0.5 °C to 52.5 ± 0.2 °C, whereas the binding of S-GPIHBP1 increased the Tm only to 39.3 ± 0.8 °C (30). Also, S-GPIHBP1 was less effective than WT mouse GPIHBP1 in preserving the catalytic activity of purified preparations of mouse LPL (30).
GPIHBP1 expression is unique to mammals, whereas LPL is expressed in all vertebrates. We propose (with a teleological argument) that LPL’s basic patch, by binding to negatively charged HSPGs within the interstitial spaces (including subendothelial HSPGs), functions in vertebrates to keep LPL in the local environment, thereby facilitating delivery of lipoprotein-derived nutrients to nearby parenchymal cells (33). We suspect that GPIHBP1 (complete with an N-terminal disordered acidic domain) appeared in mammals to promote capture of LPL from HSPGs (20, 33) and to provide an “electrostatic sheath” for LPL’s basic patch. Sheathing of the basic patch limits LPL interactions with HSPGs on the abluminal surface of capillaries and frees GPIHBP1-bound LPL to move across ECs to the capillary lumen (30).
GPIHBP1 Expression Is Important for the Uptake of TRL-Derived Lipids by Tissues
Aside from causing elevated plasma TG levels, GPIHBP1 deficiency impairs the delivery of lipid nutrients to peripheral tissues (36, 37, 73). To examine uptake of TRL-derived lipids in tissues, we administered 13C-labeled fatty acids to Gpihbp1−/− and Gpihbp1+/+ mice by gastric gavage. After 4 d, NanoSIMS was used to visualize (and quantify) 13C enrichment in tissues (36). The 13C enrichment in the heart and BAT of Gpihbp1−/− mice was reduced, but the level of 13C enrichment was increased in the liver (36). Consistent with that observation, we found higher levels of 16:1 fatty acids—and lower levels of 18:2 and 18:3 fatty acids—in adipose tissue TGs of Gpihbp1−/− mice (73). In contrast, the 18:2,18:3/16:1 fatty acid ratio in liver TGs was higher in Gpihbp1−/− mice than in Gpihbp1+/+ mice (73).
In another series of experiments, we produced TRLs that were highly enriched in 2H-labeled TGs (37). The [2H]TRLs were then administered intravenously to Gpihbp1−/− and Gpihbp1+/+ mice. After 2 min, the mice were euthanized, and tissue sections were prepared for NanoSIMS imaging. In the hearts of Gpihbp1+/+ mice, [2H]TRLs had marginated along the luminal surface of capillaries (37). In contrast, [2H]TRLs margination was absent in heart capillaries of Gpihbp1−/− mice. Also, the levels of 2H enrichment in heart capillary ECs, cardiomyocytes, and cardiomyocyte cytosolic lipid droplets were significantly reduced in Gpihbp1−/− mice (37).
In Gpihbp1+/+ mice, [2H]TRL margination was absent in brain capillaries (consistent with the absence of GPIHBP1 in ECs in the brain), and there was no 2H enrichment in brain parenchymal cells. Interestingly, GPIHBP1 and LPL were easily detectable by immunohistochemistry in capillaries of glioma tumors in the cerebral cortex (74). After administering [2H]TRLs to mice with glioma tumors in the cerebral cortex, we observed both [2H]TRL margination along glioma capillaries and 2H enrichment in glioma cells (74).
GPIHBP1 and Clinical Medicine
GPIHBP1 Deficiency.
Given the phenotype of Gpihbp1−/− mice, we suspected that GPIHBP1 mutations would eventually be uncovered in patients with chylomicronemia. The first such case was a 33-y-old male who was homozygous for a p.Q115P mutation (24). Plasma TG levels were >3,000 mg/dL but fell to 744 mg/dL on a fat-free diet. LPL mass and activity levels in the postheparin plasma were ∼10% of those in normal subjects. The Q115P substitution, located next to a cysteine in the LU domain, impaired proper disulfide bond formation and abolished GPIHBP1’s capacity to bind LPL (48).
GPIHBP1 deficiency causes lifelong chylomicronemia. Multiple missense mutations have now been reported, with many involving a cysteine within the LU domain. All of these mutations prevent LPL binding (24–26, 75). One patient had a missense mutation in GPIHBP1’s C-terminal signal peptide (downstream from the LU domain), which likely interfered with GPI anchoring (76). A 17.5-kb deletion spanning the entire GPIHBP1 gene was identified in a 2-mo-old boy with failure to thrive and plasma TG levels of >30,000 mg/dL (77). A point mutation in intron 2, uncovered in chylomicronemia patients from Brazil, was proposed to cause skipping of exon 3 (78). There have been no reports of exon 2 acidic domain deletions, but we strongly suspect, based on our mouse studies (30), that any such deletion would cause severe hypertriglyceridemia.
Most patients with GPIHBP1 deficiency have plasma TG levels of >1,500 mg/dL, although several subjects ascertained through family studies had TG levels less than 1,000 mg/dL (26, 77). A Chinese woman with GPIHBP1 deficiency had baseline plasma TG levels of 234 mg/dL, but the TG levels increased to >1,500 mg/dL during pregnancy. Several affected patients have had a history of acute pancreatitis. A biochemical hallmark of GPIHBP1 deficiency (apart from the high plasma TG levels) is a low level of LPL in the preheparin and postheparin plasma (very likely due to absent LPL transport into capillaries) (24–26, 75). Plasma TG levels in heterozygotes appear to be normal (26, 75).
Chylomicronemia is also caused by LPL missense mutations that abolish the ability of LPL to bind to GPIHBP1 (e.g., p.M404R, p.C445Y, p.E448K) (71, 79, 80). Because these mutations are distant from LPL’s catalytic domain, the mechanism for the chylomicronemia was initially puzzling (79, 80). Follow-up studies revealed that the mutations have no effect on LPL catalytic activity but cause disease by abolishing LPL binding to GPIHBP1 (34, 52).
GPIHBP1 Autoantibodies.
We discovered that some acquired cases of chylomicronemia are caused by GPIHBP1 autoantibodies (“GPIHBP1 autoantibody syndrome”) (27, 28). GPIHBP1 autoantibodies were uncovered while developing a mAb-based enzyme-linked immunosorbent assay (ELISA) to measure plasma levels of GPIHBP1 (27). As part of efforts to validate the ELISA, 40 human plasma samples were spiked with recombinant human GPIHBP1 (27). In 38 of 40 samples, the “recovery” of the spiked GPIHBP1 (as judged by the ELISA) averaged ∼98%. In the other two samples, both from patients with chylomicronemia, the recovery was <10% (27). In those cases, the “immunoassay interference” (i.e., the failure to detect GPIHBP1) was caused by GPIHBP1 autoantibodies (27).
GPIHBP1 autoantibodies bind to the LU domain (33) and abolish LPL binding (27). We have identified ∼25 chylomicronemia patients with GPIHBP1 autoantibodies. Nearly all have had plasma TG levels of >1,500 mg/dL, and many had a history of acute pancreatitis (27). Most but not all carried a diagnosis of an autoimmune disease (e.g., systemic lupus erythematosus [SLE], Sjögren’s syndrome, thryroiditis) or had serological evidence of autoimmune disease (e.g., antinuclear antibodies) (27, 28). A female SLE patient with chylomicronemia and GPIHBP1 autoantibodies became pregnant and delivered a baby girl; the infant was born with severe but transient chylomicronemia (caused by maternal transfer of GPIHBP1 autoantibodies) (27). In another case, chylomicronemia and GPIHBP1 autoantibodies appeared after initiating interferon (IFN) β1α therapy for multiple sclerosis (81). In that case, the autoantibodies disappeared, plasma LPL levels increased, and plasma TG levels normalized after discontinuing the IFN β1α treatment (81). In some cases, GPIHBP1 autoantibodies have been intermittent, resulting in intermittent chylomicronemia (82).
Patients with GPIHBP1 autoantibodies have extremely low levels of LPL in the preheparin and postheparin plasma. In our initial report, all of the patients had extremely low plasma levels of GPIHBP1 (a result of immunoassay interference) (28). Subsequently, we encountered patients in which immunoassay interference was absent and plasma levels of GPIHBP1 were extremely high (likely due to an accumulation of GPIHBP1 immune complexes in the plasma) (28). Thus, the properties of GPIHBP1 autoantibodies in individual patients can vary (28). Most patients with GPIHBP1 autoantibodies have a preponderance of IgG4 and IgA autoantibodies (28). IgG4 autoantibodies cannot activate the classic complement cascade and typically cause disease by disrupting protein–protein interactions rather than by inducing tissue injury (83).
We have not yet encountered GPIHBP1 autoantibodies in cases of mild to moderate hypertriglyceridemia. When GPIHBP1 autoantibodies exceed the threshold of detection by our immunoassay, the plasma TG levels have been greater than 1,500 mg/dL.
Experience in treating the GPIHBP1 autoantibody syndrome patients is limited, but several patients have been treated successfully with rituximab (28, 84). In those cases, GPIHBP1 autoantibodies disappeared, and the plasma LPL and TG levels normalized.
We suspect that many cases of GPIHBP1 autoantibody syndrome go undiagnosed. In a recent review of causes of chylomicronemia (85), the GPIHBP1 autoantibody syndrome was not mentioned in the differential diagnosis.
Perspectives
We now recognize that GPIHBP1 is responsible for transporting LPL to the capillary lumen (17), that LPL transport into capillaries is required for TRL margination along capillaries (19), and that GPIHBP1 stabilizes LPL conformation and enzymatic activity—even in the face of inhibitory proteins (20–23, 33). GPIHBP1 was the key to determining the atomic structure of LPL (34), and it led to the realization that freshly secreted, catalytically active LPL is monomeric (22, 67). GPIHBP1 also expanded the differential diagnosis of genetic and acquired forms of chylomicronemia (24, 28). Collectively, these findings expanded our understanding of GPIHBP1 and LPL physiology, but, at the same time, they highlighted persistent lacunae in the field of plasma TG metabolism.
At the level of basic physiology, we still have little understanding of LPL’s transit from the interstitium to the capillary lumen. The machinery for GPIHBP1-mediated LPL transport across ECs has received little attention. Also, we have little understanding of LPL turnover and the extent to which turnover is affected by fasting and refeeding. There have been suggestions that GPIHBP1 is a long-lived protein (86), but the half-life of GPIHBP1 has never been investigated.
The location of LPL along the luminal surface of capillary ECs needs more study. We know that GPIHBP1 moves LPL into capillaries (17) and that GPIHBP1-bound LPL is catalytically active (67), but whether the GPIHBP1-bound LPL on the luminal plasma membrane of ECs is solely responsible for TRL processing is not known. SPR studies revealed that GPIHBP1–LPL complexes have a half-life of 55 s (33). If LPL were to detach from GPIHBP1 on the luminal plasma membrane of capillary ECs, it seems possible that some of that LPL might be captured by the HSPG-rich glycocalyx lining of ECs. A glycocalyx-bound pool of LPL—if it exists—could play a role in TRL margination and intravascular TG hydrolysis. High-resolution imaging studies are needed to better define the location of LPL in capillaries.
At the level of molecular biology, we have little understanding of GPIHBP1 gene expression. We know that an upstream enhancer element influences gene expression (87), but we have no understanding of why GPIHBP1 is expressed in capillary ECs but not in ECs of venules or larger blood vessels. We also do not understand why GPIHBP1 is absent in capillaries of the brain.
At the level of biochemistry, we have little understanding of how APOC2 interacts with GPIHBP1-bound LPL (or free LPL), nor do we understand the structural features of LPL that confer lipoprotein and glycerolipid substrate specificity. We know that LPL’s C-terminal Trp-rich cluster is important for TRL binding, but whether those tryptophans are directly involved in mobilizing core TGs for hydrolysis requires more study (35). Also, while the ability of ANGPTL4 to catalyze LPL unfolding is well documented (21–23, 33, 64), the activity of the ANGPTL3/ANGPTL8 complex needs more study. The recent proposal that APOA5 suppresses the activity of the ANGPTL3/8 complex is an exciting development (88). It will now be important to determine whether APOA5 affects LPL levels on the luminal surface of capillaries.
At the level of human genetics, we still have little understanding of why a two–amino acid truncation of LPL (S474X), present in ∼10% of the population, increases plasma LPL levels and lowers plasma TG levels (89). Based on the crystal structure of the GPIHBP1–LPL complex, it is likely that the S474X mutation changes LPL’s basic patch (34). One possibility is that GPIHBP1 is particularly effective in preserving the structure and activity of S474X-LPL.
At the level of comparative biology, we have been intrigued by the fact that LPL is conserved throughout vertebrate evolution, whereas GPIHBP1 is present only in mammals. Because TRL processing is important for milk production by the mammary gland, we suspect that GPIHBP1 may have appeared in mammals to support “nursing the young.” In any case, the mechanisms for intravascular TRL processing in “lower vertebrates” need further study. We have presented evidence that LPL is present in heart capillaries of chickens (90), but the mechanism for the entry of chicken LPL into capillaries is unclear. New antibody reagents are needed to define the distribution of LPL in tissues of reptiles, amphibians, fish, and birds. Thanks to a new method for purifying LPL (91), creating the required antibodies should be straightforward.
Acknowledgments
This work was supported by National Heart, Lung, and Blood Institute Grants HL087228, HL146358, and HL139725; Fondation Leducq Transatlantic Network Grant 19CVD04; NOVO Nordisk Foundation Grant NNF20OC0063444; and The John and Birthe Meyer Foundation. We thank Kazuya Miyashita and Katsuyuki Nakajima for collaborative studies on GPIHBP1 autoantibodies.
Footnotes
Reviewers: D.G., University of Michigan; and R.Z., Karl-Franzens-Universitat Graz.
The authors declare no competing interest.
Data, Materials, and Software Availability
All study data are included in the article.
References
- 1.Korn E. D., Clearing factor, a heparin-activated lipoprotein lipase. I. Isolation and characterization of the enzyme from normal rat heart. J. Biol. Chem. 215, 1–14 (1955). [PubMed] [Google Scholar]
- 2.Korn E. D., Clearing factor, a heparin-activated lipoprotein lipase. II. Substrate specificity and activation of coconut oil. J. Biol. Chem. 215, 15–26 (1955). [PubMed] [Google Scholar]
- 3.Havel R. J., Gordon R. S. Jr., Idiopathic hyperlipemia: Metabolic studies in an affected family. J. Clin. Invest. 39, 1777–1790 (1960). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fielding C. J., Human lipoprotein lipase. I. Purification and substrate specificity. Biochim. Biophys. Acta 206, 109–117 (1970). [DOI] [PubMed] [Google Scholar]
- 5.LaRosa J. C., Levy R. I., Herbert P., Lux S. E., Fredrickson D. S., A specific apoprotein activator for lipoprotein lipase. Biochem. Biophys. Res. Commun. 41, 57–62 (1970). [DOI] [PubMed] [Google Scholar]
- 6.Morley N., Kuksis A., Positional specificity of lipoprotein lipase. J. Biol. Chem. 247, 6389–6393 (1972). [PubMed] [Google Scholar]
- 7.Bengtsson G., Olivecrona T., Apolipoprotein CII enhances hydrolysis of monoglycerides by lipoprotein lipase, but the effect is abolished by fatty acids. FEBS Lett. 106, 345–348 (1979). [DOI] [PubMed] [Google Scholar]
- 8.Iverius P. H., Ostlund-Lindqvist A. M., Lipoprotein lipase from bovine milk. Isolation procedure, chemical characterization, and molecular weight analysis. J. Biol. Chem. 251, 7791–7795 (1976). [PubMed] [Google Scholar]
- 9.Cisar L. A., Hoogewerf A. J., Cupp M., Rapport C. A., Bensadoun A., Secretion and degradation of lipoprotein lipase in cultured adipocytes. Binding of lipoprotein lipase to membrane heparan sulfate proteoglycans is necessary for degradation. J. Biol. Chem. 264, 1767–1774 (1989). [PubMed] [Google Scholar]
- 10.Shimada K., Gill P. J., Silbert J. E., Douglas W. H., Fanburg B. L., Involvement of cell surface heparin sulfate in the binding of lipoprotein lipase to cultured bovine endothelial cells. J. Clin. Invest. 68, 995–1002 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirchgessner T. G., Svenson K. L., Lusis A. J., Schotz M. C., The sequence of cDNA encoding lipoprotein lipase. A member of a lipase gene family. J. Biol. Chem. 262, 8463–8466 (1987). [PubMed] [Google Scholar]
- 12.Senda M., Oka K., Brown W. V., Qasba P. K., Furuichi Y., Molecular cloning and sequence of a cDNA coding for bovine lipoprotein lipase. Proc. Natl. Acad. Sci. U.S.A. 84, 4369–4373 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma Y., et al. , Mutagenesis in four candidate heparin binding regions (residues 279-282, 291-304, 390-393, and 439-448) and identification of residues affecting heparin binding of human lipoprotein lipase. J. Lipid Res. 35, 2049–2059 (1994). [PubMed] [Google Scholar]
- 14.Sendak R. A., Melford K., Kao A., Bensadoun A., Identification of the epitope of a monoclonal antibody that inhibits heparin binding of lipoprotein lipase: New evidence for a carboxyl-terminal heparin-binding domain. J. Lipid Res. 39, 633–646 (1998). [PubMed] [Google Scholar]
- 15.van Tilbeurgh H., Roussel A., Lalouel J.-M., Cambillau C., Lipoprotein lipase. Molecular model based on the pancreatic lipase x-ray structure: Consequences for heparin binding and catalysis. J. Biol. Chem. 269, 4626–4633 (1994). [PubMed] [Google Scholar]
- 16.Olivecrona T., Bengtsson-Olivecrona G., Lipoprotein lipase and hepatic lipase. Curr. Opin. Lipidol. 1, 222–230 (1990). [Google Scholar]
- 17.Davies B. S. J., et al. , GPIHBP1 is responsible for the entry of lipoprotein lipase into capillaries. Cell Metab. 12, 42–52 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beigneux A. P., et al. , Glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1 plays a critical role in the lipolytic processing of chylomicrons. Cell Metab. 5, 279–291 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goulbourne C. N., et al. , The GPIHBP1-LPL complex is responsible for the margination of triglyceride-rich lipoproteins in capillaries. Cell Metab. 19, 849–860 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mysling S., et al. , The acidic domain of the endothelial membrane protein GPIHBP1 stabilizes lipoprotein lipase activity by preventing unfolding of its catalytic domain. eLife 5, e12095 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mysling S., et al. , The angiopoietin-like protein ANGPTL4 catalyzes unfolding of the hydrolase domain in lipoprotein lipase and the endothelial membrane protein GPIHBP1 counteracts this unfolding. eLife 5, e20958 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kristensen K. K., et al. , Unfolding of monomeric lipoprotein lipase by ANGPTL4: Insight into the regulation of plasma triglyceride metabolism. Proc. Natl. Acad. Sci. U.S.A. 117, 4337–4346 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leth-Espensen K. Z., et al. , The intrinsic instability of the hydrolase domain of lipoprotein lipase facilitates its inactivation by ANGPTL4-catalyzed unfolding. Proc. Natl. Acad. Sci. U.S.A. 118, e2026650118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beigneux A. P., et al. , Chylomicronemia with a mutant GPIHBP1 (Q115P) that cannot bind lipoprotein lipase. Arterioscler. Thromb. Vasc. Biol. 29, 956–962 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franssen R., et al. , Chylomicronemia with low postheparin lipoprotein lipase levels in the setting of GPIHBP1 defects. Circ. Cardiovasc. Genet. 3, 169–178 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plengpanich W., et al. , Multimerization of glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1 (GPIHBP1) and familial chylomicronemia from a serine-to-cysteine substitution in GPIHBP1 Ly6 domain. J. Biol. Chem. 289, 19491–19499 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beigneux A. P., et al. , Autoantibodies against GPIHBP1 as a cause of hypertriglyceridemia. N. Engl. J. Med. 376, 1647–1658 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyashita K., et al. , Chylomicronemia from GPIHBP1 autoantibodies. J. Lipid Res. 61, 1365–1376 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang T., et al. , A mouse knockout library for secreted and transmembrane proteins. Nat. Biotechnol. 28, 749–755 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Song W., et al. , Electrostatic sheathing of lipoprotein lipase is essential for its movement across capillary endothelial cells. J. Clin. Invest. 132, e157500 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davies B. S., et al. , Assessing mechanisms of GPIHBP1 and lipoprotein lipase movement across endothelial cells. J. Lipid Res. 53, 2690–2697 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meng X., Zeng W., Young S. G., Fong L. G., GPIHBP1, a partner protein for lipoprotein lipase, is expressed only in capillary endothelial cells. J. Lipid Res. 61, 591 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kristensen K. K., et al. , A disordered acidic domain in GPIHBP1 harboring a sulfated tyrosine regulates lipoprotein lipase. Proc. Natl. Acad. Sci. U.S.A. 115, E6020–E6029 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Birrane G., et al. , Structure of the lipoprotein lipase-GPIHBP1 complex that mediates plasma triglyceride hydrolysis. Proc. Natl. Acad. Sci. U.S.A. 116, 1723–1732 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luz J. G., et al. , The structural basis for monoclonal antibody 5D2 binding to the tryptophan-rich loop of lipoprotein lipase. J. Lipid Res. 61, 1347–1359 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang H., et al. , High-resolution imaging of dietary lipids in cells and tissues by NanoSIMS analysis. J. Lipid Res. 55, 2156–2166 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He C., et al. , NanoSIMS analysis of intravascular lipolysis and lipid movement across capillaries and into cardiomyocytes. Cell Metab. 27, 1055–1066.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ioka R. X., et al. , Expression cloning and characterization of a novel glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein, GPI-HBP1. J. Biol. Chem. 278, 7344–7349 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Acton S. L., Scherer P. E., Lodish H. F., Krieger M., Expression cloning of SR-BI, a CD36-related class B scavenger receptor. J. Biol. Chem. 269, 21003–21009 (1994). [PubMed] [Google Scholar]
- 40.Leth J. M., et al. , Evolution and medical significance of LU domain–containing proteins. Int. J. Mol. Sci. 20, 2760 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gin P., et al. , Binding preferences for GPIHBP1, a glycosylphosphatidylinositol-anchored protein of capillary endothelial cells. Arterioscler. Thromb. Vasc. Biol. 31, 176–182 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weinstein M. M., et al. , Cholesterol intake modulates plasma triglyceride levels in glycosylphosphatidylinositol HDL-binding protein 1-deficient mice. Arterioscler. Thromb. Vasc. Biol. 30, 2106–2113 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olivecrona G., Vilaró S., Esko J. D., Olivecrona T., Synthesis and secretion of lipoprotein lipase in heparan sulfate-deficient Chinese hamster ovary cells. Isr. J. Med. Sci. 32, 430–444 (1996). [PubMed] [Google Scholar]
- 44.Weinstock P. H., et al. , Severe hypertriglyceridemia, reduced high density lipoprotein, and neonatal death in lipoprotein lipase knockout mice. Mild hypertriglyceridemia with impaired very low density lipoprotein clearance in heterozygotes. J. Clin. Invest. 96, 2555–2568 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gin P., et al. , The acidic domain of GPIHBP1 is important for the binding of lipoprotein lipase and chylomicrons. J. Biol. Chem. 283, 29554–29562 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beigneux A. P., et al. , Highly conserved cysteines within the Ly6 domain of GPIHBP1 are crucial for the binding of lipoprotein lipase. J. Biol. Chem. 284, 30240–30247 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beigneux A. P., et al. , Assessing the role of the glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1 (GPIHBP1) three-finger domain in binding lipoprotein lipase. J. Biol. Chem. 286, 19735–19743 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beigneux A. P., et al. , GPIHBP1 missense mutations often cause multimerization of GPIHBP1 and thereby prevent lipoprotein lipase binding. Circ. Res. 116, 624–632 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu X., et al. , Monoclonal antibodies that bind to the Ly6 domain of GPIHBP1 abolish the binding of LPL. J. Lipid Res. 58, 208–215 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Allan C. M., et al. , Mutating a conserved cysteine in GPIHBP1 reduces amounts of GPIHBP1 in capillaries and abolishes LPL binding. J. Lipid Res. 58, 1453–1461 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Allan C. M., et al. , An LPL-specific monoclonal antibody, 88B8, that abolishes the binding of LPL to GPIHBP1. J. Lipid Res. 57, 1889–1898 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Voss C. V., et al. , Mutations in lipoprotein lipase that block binding to the endothelial cell transporter GPIHBP1. Proc. Natl. Acad. Sci. U.S.A. 108, 7980–7984 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gin P., et al. , Normal binding of lipoprotein lipase, chylomicrons, and apo-AV to GPIHBP1 containing a G56R amino acid substitution. Biochim. Biophys. Acta 1771, 1464–1468 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kobayashi Y., Nakajima T., Inoue I., Molecular modeling of the dimeric structure of human lipoprotein lipase and functional studies of the carboxyl-terminal domain. Eur. J. Biochem. 269, 4701–4710 (2002). [DOI] [PubMed] [Google Scholar]
- 55.Wong H., et al. , A molecular biology-based approach to resolve the subunit orientation of lipoprotein lipase. Proc. Natl. Acad. Sci. U.S.A. 94, 5594–5598 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wong H., et al. , Lipoprotein lipase domain function. J. Biol. Chem. 269, 10319–10323 (1994). [PubMed] [Google Scholar]
- 57.Doolittle M. H., Ehrhardt N., Péterfy M., Lipase maturation factor 1: Structure and role in lipase folding and assembly. Curr. Opin. Lipidol. 21, 198–203 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zambon A., Schmidt I., Beisiegel U., Brunzell J. D., Dimeric lipoprotein lipase is bound to triglyceride-rich plasma lipoproteins. J. Lipid Res. 37, 2394–2404 (1996). [PubMed] [Google Scholar]
- 59.Zhang L., Lookene A., Wu G., Olivecrona G., Calcium triggers folding of lipoprotein lipase into active dimers. J. Biol. Chem. 280, 42580–42591 (2005). [DOI] [PubMed] [Google Scholar]
- 60.Doolittle M. H., Péterfy M., Mechanisms of lipase maturation. Clin. Lipidol. 5, 71–85 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Masson G. R., et al. , Recommendations for performing, interpreting and reporting hydrogen deuterium exchange mass spectrometry (HDX-MS) experiments. Nat. Methods 16, 595–602 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Twu J. S., Garfinkel A. S., Schotz M. C., Rat heart lipoprotein lipase. Atherosclerosis 22, 463–472 (1975). [DOI] [PubMed] [Google Scholar]
- 63.Kristensen K. K., Leth-Espensen K. Z., Young S. G., Ploug M., ANGPTL4 inactivates lipoprotein lipase by catalyzing the irreversible unfolding of LPL’s hydrolase domain. J. Lipid Res. 61, 1253 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kristensen K. K., et al. , GPIHBP1 and ANGPTL4 utilize protein disorder to orchestrate order in plasma triglyceride metabolism and regulate compartmentalization of LPL activity. Front. Cell Dev. Biol. 9, 702508 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Winkler F. K., D’Arcy A., Hunziker W., Structure of human pancreatic lipase. Nature 343, 771–774 (1990). [DOI] [PubMed] [Google Scholar]
- 66.van Tilbeurgh H., Sarda L., Verger R., Cambillau C., Structure of the pancreatic lipase-procolipase complex. Nature 359, 159–162 (1992). [DOI] [PubMed] [Google Scholar]
- 67.Beigneux A. P., et al. , Lipoprotein lipase is active as a monomer. Proc. Natl. Acad. Sci. U.S.A. 116, 6319–6328 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arora R., et al. , Structure of lipoprotein lipase in complex with GPIHBP1. Proc. Natl. Acad. Sci. U.S.A. 116, 10360–10365 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gunn K. H., et al. , The structure of helical lipoprotein lipase reveals an unexpected twist in lipase storage. Proc. Natl. Acad. Sci. U.S.A. 117, 10254–10264 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Young S. G., et al. , GPIHBP1 and lipoprotein lipase, partners in plasma triglyceride metabolism. Cell Metab. 30, 51–65 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pingitore P., et al. , Identification and characterization of two novel mutations in the LPL gene causing type I hyperlipoproteinemia. J. Clin. Lipidol. 10, 816–823 (2016). [DOI] [PubMed] [Google Scholar]
- 72.Abifadel M., et al. , Identification of the first Lebanese mutation in the LPL gene and description of a rapid detection method. Clin. Genet. 65, 158–161 (2004). [DOI] [PubMed] [Google Scholar]
- 73.Weinstein M. M., et al. , Reciprocal metabolic perturbations in the adipose tissue and liver of GPIHBP1-deficient mice. Arterioscler. Thromb. Vasc. Biol. 32, 230–235 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hu X., et al. , GPIHBP1 expression in gliomas promotes utilization of lipoprotein-derived nutrients. eLife 8, e47178 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Olivecrona G., et al. , Mutation of conserved cysteines in the Ly6 domain of GPIHBP1 in familial chylomicronemia. J. Lipid Res. 51, 1535–1545 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Charrière S., et al. , GPIHBP1 C89F neomutation and hydrophobic C-terminal domain G175R mutation in two pedigrees with severe hyperchylomicronemia. J. Clin. Endocrinol. Metab. 96, E1675–E1679 (2011). [DOI] [PubMed] [Google Scholar]
- 77.Rios J. J., et al. , Deletion of GPIHBP1 causing severe chylomicronemia. J. Inherit. Metab. Dis. 35, 531–540 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lima J. G., et al. , A novel GPIHBP1 mutation related to familial chylomicronemia syndrome: A series of cases. Atherosclerosis 322, 31–38 (2021). [DOI] [PubMed] [Google Scholar]
- 79.Henderson H. E., Hassan F., Marais D., Hayden M. R., A new mutation destroying disulphide bridging in the C-terminal domain of lipoprotein lipase. Biochem. Biophys. Res. Commun. 227, 189–194 (1996). [DOI] [PubMed] [Google Scholar]
- 80.Henderson H., Leisegang F., Hassan F., Hayden M., Marais D., A novel Glu421Lys substitution in the lipoprotein lipase gene in pregnancy-induced hypertriglyceridemic pancreatitis. Clin. Chim. Acta 269, 1–12 (1998). [DOI] [PubMed] [Google Scholar]
- 81.Eguchi J., et al. , GPIHBP1 autoantibody syndrome during interferon β1a treatment. J. Clin. Lipidol. 13, 62–69 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ashraf A. P., et al. , Intermittent chylomicronemia caused by intermittent GPIHBP1 autoantibodies. J. Clin. Lipidol. 14, 197–200 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huijbers M. G., Plomp J. J., van der Maarel S. M., Verschuuren J. J., IgG4-mediated autoimmune diseases: A niche of antibody-mediated disorders. Ann. N. Y. Acad. Sci. 1413, 92–103 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lutz J., et al. , Chylomicronemia from GPIHBP1 autoantibodies successfully treated with rituximab: A case report. Ann. Intern. Med. 173, 764–765 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chait A., Eckel R. H., The chylomicronemia syndrome is most often multifactorial: A narrative review of causes and treatment. Ann. Intern. Med. 170, 626–634 (2019). [DOI] [PubMed] [Google Scholar]
- 86.Olafsen T., et al. , Unexpected expression pattern for glycosylphosphatidylinositol-anchored HDL-binding protein 1 (GPIHBP1) in mouse tissues revealed by positron emission tomography scanning. J. Biol. Chem. 285, 39239–39248 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Allan C. M., et al. , An upstream enhancer regulates Gpihbp1 expression in a tissue-specific manner. J. Lipid Res. 60, 869–879 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen Y. Q., et al. , ApoA5 lowers triglyceride levels via suppression of ANGPTL3/8-mediated LPL inhibition. J. Lipid Res. 62, 100068 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rip J., et al. , Lipoprotein lipase S447X: A naturally occurring gain-of-function mutation. Arterioscler. Thromb. Vasc. Biol. 26, 1236–1245 (2006). [DOI] [PubMed] [Google Scholar]
- 90.He C., et al. , Lipoprotein lipase reaches the capillary lumen in chickens despite an apparent absence of GPIHBP1. JCI Insight 2, e96783 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lund Winther A. M., Kristensen K. K., Kumari A., Ploug M., Expression and one-step purification of active LPL contemplated by biophysical considerations. J. Lipid Res. 62, 100149 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All study data are included in the article.