Significance
Exosomes are a type of membranous extracellular vesicle and have been highlighted for their various roles in the transmission of microorganisms. Exportins are frequently found in exosomes. However, the function of exportins sorted in exosomes remains unknown. Here, we isolated exosomes from saliva of approximately 30,000 small brown planthoppers, which are vectors of rice stripe virus (RSV). We found that a fragmented exportin 6 acted as a vehicle for transporting RSV into exosomes to overcome the barrier of insect salivary glands for horizontal transmission. Interference with the expression of exportin 6 remarkably reduced viral transmission to rice plants. Therefore, exportin 6 would represent an ideal target for controlling the outbreak of insect-borne viruses.
Keywords: exosome, exportin, rice stripe virus, insect vector, salivary gland
Abstract
Exosomes play a key role in virus exocytosis and transmission. The exportin family is usually responsible for cargo nucleocytoplasmic trafficking, and they are frequently found in exosomes. However, the function of exportins sorted in exosomes remains unknown. Here, we successfully isolated “cup holder”-like exosomes from the saliva of ∼30,000 small brown planthoppers, which are vectors of rice stripe virus (RSV). RSV virions were packed in comparatively large exosomes. Four viral genomic RNAs at a certain ratio were identified in the saliva exosomes. The virions contained in the saliva exosomes were capable of replicating and causing disease in rice plants. Interference with each phase of the insect exosome system affected the transmission of RSV from the insect vectors to rice plants. Fragmented exportin 6 was coimmunoprecipitated with viral nucleocapsid protein in saliva and sorted to exosomes via interactions with the cargo sorting protein VPS37a. When the expression of exportin 6 was knocked down, the amounts of RSV secreted in saliva and rice plants were reduced by 60% and 74%, respectively. These results showed that exportin 6 acted as a vehicle for transporting RSV into exosomes to overcome the barrier of insect salivary glands for horizontal transmission. Exportin 6 would represent an ideal target that could be manipulated to control the outbreak of insect-borne viruses in the future.
Both arboviruses transmitted by blood-feeding insects and persistent plant viruses vectored by piercing–sucking agricultural insects are characterized by the systemic invasion of diverse organs inside the insects. These viruses initially invade the gut epithelial cells and are then released into the hemolymph, and they subsequently spread to salivary glands and other organs and are finally secreted from the salivary glands into host cells (1). Salivary glands are the last barrier for viruses to overcome, and this barrier includes two steps: entering and exiting salivary gland cells. Key factors that facilitate viral entry into salivary gland cells have been identified, such as an importin α protein that mediates the entry of rice stripe virus (RSV) into the salivary glands of the insect vector small brown planthopper (2); however, the key vector factors for virus release from salivary gland cells remain elusive. Exosomes may contain viruses to aid in their spread. For example, mosquito-borne dengue virus (3), tick-borne Langat virus (4), and leafhopper-borne rice dwarf virus (5) have been found in exosomes that were secreted in the culture medium of insect cell lines and infectious to the host cells. However, direct and solid evidence showing that exosomes exist in insect saliva is lacking.
Exosomes are a type of membranous extracellular vesicle with a diameter of 30 to 200 nm (6). They originate from intraluminal vesicles (ILVs) in multivesicular bodies (MVBs) (6). The formation of ILVs is involved in both endosomal sorting complexes required for transport (ESCRT)-dependent and ESCRT-independent machineries (7). MVBs are transported to and fuse with the plasma membrane to release ILVs as exosomes into the extracellular milieu. Proteins have been found in exosomes, and they include chaperone proteins (Hsc70 and Hsp90), ESCRT proteins (Tsg101 and Alix), tetraspanin proteins (CD9, CD63, CD81, and CD82), Rab proteins, and flotillins (6). Flotillins are frequently used as molecular markers to indicate exosomes because they bind cytosolic leaflets of endosome membranes in addition to cellular plasma membranes to facilitate the assembly of multiprotein complexes at the membrane–cytosol interface (7, 8). Exportins (exportin 1, exportin 2, exportin T, exportin 5, exportin 6, and exportin 7) have been identified in exosomes isolated from various mammal cell lines by mass spectrometry (9–12). However, the function of exportins sorted in exosomes is unknown.
RSV is a typical Tenuivirus that causes one of the most destructive rice stripe diseases in temperate and subtropical regions (13). The genome of RSV contains four single-stranded RNA segments and encodes a nucleocapsid protein (NP), an RNA-dependent RNA polymerase (RdRp), and five nonstructural proteins (NS2, NSvc2, NS3, SP, and NSvc4) (14–16). RSV is capable of proliferating in midgut epithelial cells and is efficiently transmitted by the small brown planthopper Laodelphax striatellus (17). In this study, we successfully isolated exosomes from the saliva of ∼30,000 small brown planthoppers and demonstrated that RSV was loaded in exosomes by binding with the exosome protein exportin 6 to overcome the barrier of salivary glands and facilitate viral horizontal transmission.
Results
Characteristics of Exosomes from Saliva and Hemolymph of Small Brown Planthoppers.
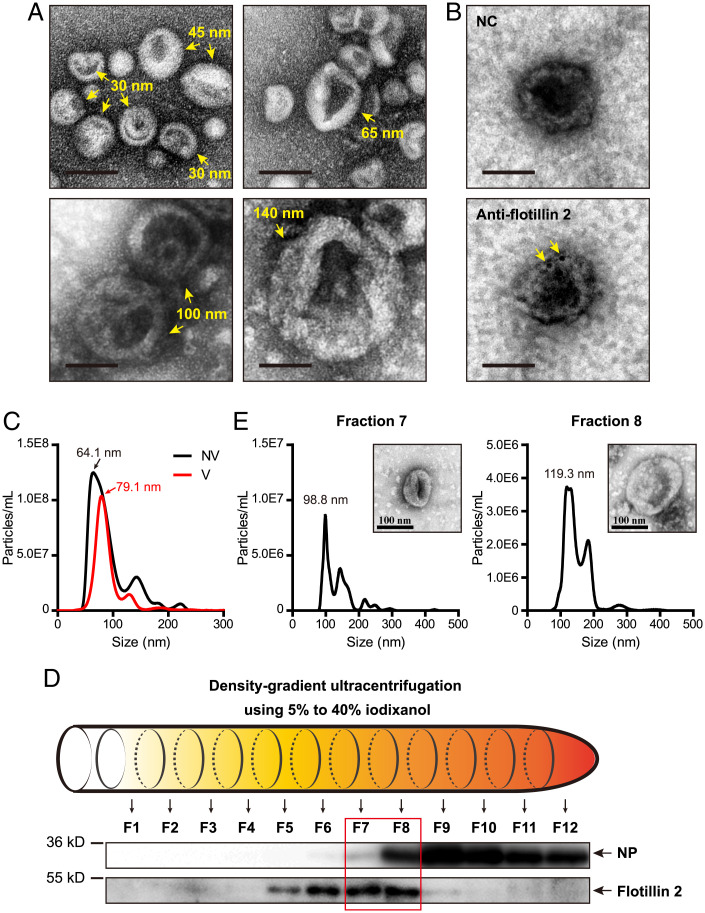
To characterize exosomes of small brown planthoppers, saliva from ∼30,000 nonviruliferous planthoppers was collected using saliva-collection glass bottles (SI Appendix, Fig. 1A) as described previously (18). Hemolymph from ∼3,000 nonviruliferous planthoppers was collected using a centrifugation method (19). After isolation from saliva and hemolymph samples, the exosomes resembled “cup holders” or “round cakes” with different sizes of ∼30, 45, 55, 60, 65, 100, and 140 nm under a transmission electron microscope (TEM) (Fig. 1A). Using a polyclonal antibody against planthopper flotillin 2 (20), which is regarded as a molecular marker associated with exosomes (7), we detected the existence of flotillin 2 in the cup holder-like exosomes under immunoelectron microscopy (Fig. 1B).
Fig. 1.
Characteristics of exosomes from saliva and hemolymph of small brown planthoppers. (A) Morphology of exosomes under a transmission electron microscope. (Scale bar, 50 nm.) (B) Immunoelectron microscopy showing exosomes and flotillin 2, which was labeled by gold-conjugated antibodies. Arrows indicate flotillin 2 molecules. (Scale bar, 50 nm.) (C) The size distributions of exosomes from the hemolymph of nonviruliferous (NV) and viruliferous (V) planthoppers analyzed by NTA. (D) Western blot assays for RSV NP and flotillin 2 in hemolymph exosomes distributed in twelve fractions (F1 to F12) after density-gradient ultracentrifugation. Anti-NP monoclonal and anti-flotillin 2 polyclonal antibodies were used. (E) NTA analysis of exosomes potentially containing RSV from fractions 7 and 8. The morphology of exosomes was shown using TEM.
The particle sizes of exosomes isolated from the hemolymph of nonviruliferous and viruliferous planthoppers were compared using nanoparticle tracking analysis (NTA). The mean sizes of the exosomes from nonviruliferous and viruliferous planthoppers were 92.8 ± 1.8 nm and 92.4 ± 2.1 nm, respectively. However, the peak diameter was 79.1 nm for viruliferous planthoppers, larger than the 64.1-nm peak diameter for nonviruliferous planthoppers (Fig. 1C), indicating that the particle diameter with the highest concentration of exosomes was larger in viruliferous insects. To further determine the sizes of the exosomes potentially containing RSV virions, density-gradient ultracentrifugation with a series of iodixanol concentrations was used to purify the exosomes. Twelve fractions were collected, among which flotillin 2 mainly accumulated in fractions 5 through 8, and RSV NPs were present in fractions 7 through 12 (Fig. 1D). Fractions 7 and 8, in which the exosomes potentially contained RSV virions, were isolated for NTA measurement. The results showed that the mean sizes of the exosomes in fractions 7 and 8 were 136.1 nm and 149.3 nm and that the major peaks were 98.8 nm and 119.3 nm (Fig. 1E). These results suggested that RSV was mostly loaded in comparatively large exosomes.
RSV Virions Were Packaged in Exosomes.
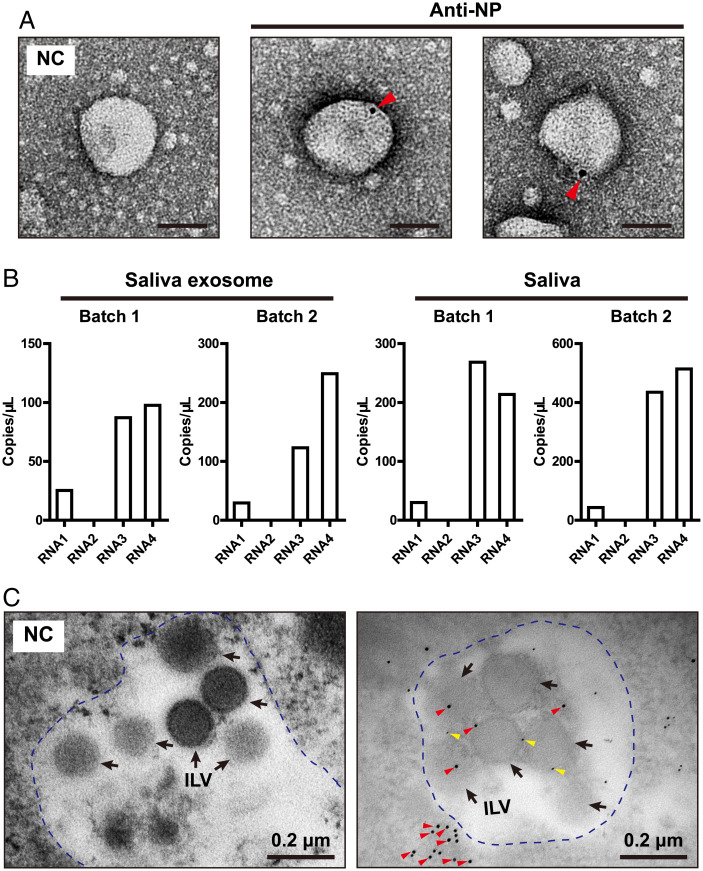
To verify whether RSV virions were contained in exosomes, we used a colloidal gold-conjugated anti-NP antibody to label virions, and the four genomic segments (RNA1 to RNA4) of RSV were quantified using absolute real-time quantification PCR (qPCR) based on standard curves (SI Appendix, Fig. 2). Virions were observed inside of the exosomes isolated from saliva under immunoelectron microscopy (Fig. 2A). The copy numbers of RNA3 and RNA4 in the saliva exosomes were much higher than those of the other two segments, and RNA2 was nearly undetectable due to the low quantity of collected exosomes (Fig. 2B). The ratios of RNA3 and RNA4 were also the highest in the whole saliva (Fig. 2B). Considering that exosomes are derived from ILVs in MVBs before being released out of the cells, we examined MVBs in the primary salivary gland cells of viruliferous planthoppers. Multiple ILVs were contained in one MVB. Flotillin 2 labeled by 5 nm gold-conjugated anti-flotillin 2 antibodies was found on the plasma membrane of ILVs (Fig. 2C). RSV virions labeled by 10 nm gold-conjugated anti-NP antibodies existed inside of ILVs and on the plasma membrane of ILVs (Fig. 2C). Interestingly, RSV virions were frequently found to accumulate near MVBs (Fig. 2C).
Fig. 2.
RSV virions were packaged in exosomes. (A) Immunoelectron microscopy showing RSV virions in saliva exosomes labeled with gold-conjugated anti-NP monoclonal antibodies. The negative control (NC) was not treated with the monoclonal antibody. Arrowheads indicate RSV NP molecules. (Scale bar, 50 nm.) (B) Copy numbers of four genomic RNAs of RSV in saliva exosomes and whole saliva measured by absolute real-time qPCR. Two batches of samples are shown. (C) Double immunogold labeling of RSV NPs and flotillin 2 in MVBs of primary salivary gland cells of planthoppers. MVB is outlined by blue-dotted line. Red arrowheads indicate the NPs labeled by 10 nm gold-conjugated anti-NP monoclonal antibodies. Yellow arrowheads indicate flotillin 2 labeled by 5 nm gold-conjugated anti-flotillin 2 polyclonal antibodies. Black arrows indicate ILVs in MVBs. The NC was not treated with antibodies.
Saliva Exosomes Containing RSV Were Infectious to Rice Plants.
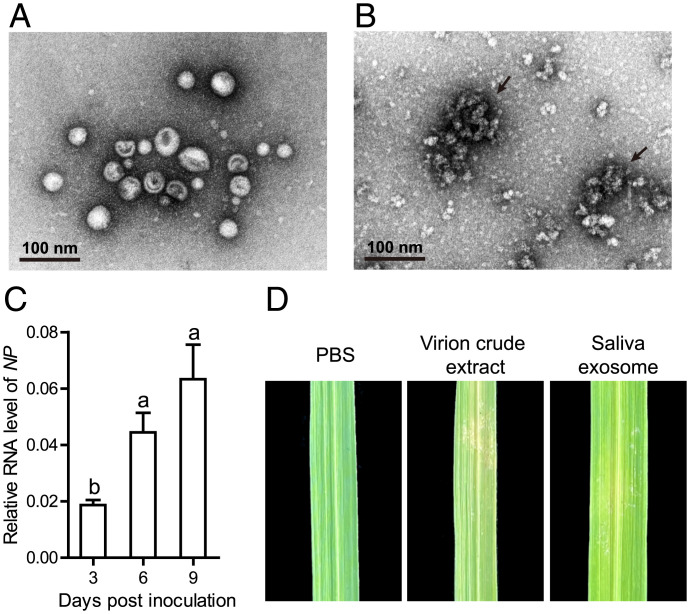
To clarify whether RSV virions contained in the saliva exosomes were infectious to rice plants, we isolated exosomes from the saliva of viruliferous planthoppers with a high purity without free virions (Fig. 3A). Filamentous viral particles were clearly observed in virion crude extracts from viruliferous planthoppers under a TEM (Fig. 3B). When the saliva exosomes were injected into the midrib of rice leaves, the viral load in the local leaves increased significantly over time from 3 to 9 d postinoculation (dpi) in terms of the RNA level of RSV NP (Fig. 3C). A typical stripe symptom appeared in the leaves after 4 wk, similar to the positive control, which was inoculated with virion crude extracts from viruliferous planthoppers (Fig. 3D). Thus, RSV virions contained in the saliva exosomes were capable of replicating and causing diseases in rice plants.
Fig. 3.
Saliva exosomes containing RSV were infectious to rice plants. (A) Purity of saliva exosomes isolated from viruliferous planthoppers and checked under a TEM. (B) Filamentous viral particles in virion crude extracts from viruliferous planthoppers under a TEM. Arrows indicate viral particles. (C) Relative RNA levels of RSV NP in rice leaves at different times after inoculation with saliva exosomes isolated from viruliferous planthoppers measured by real-time qPCR. The RNA level of NP was normalized to the transcript level of tubulin. Different letters indicate a statistically significant difference. (D) Disease symptoms in rice leaves 4 wk after inoculation with saliva exosomes or virion crude extracts from viruliferous planthoppers. The negative control was inoculated with PBS.
Interference with Insect Exosome Systems Reduced RSV Transmission to Plants.
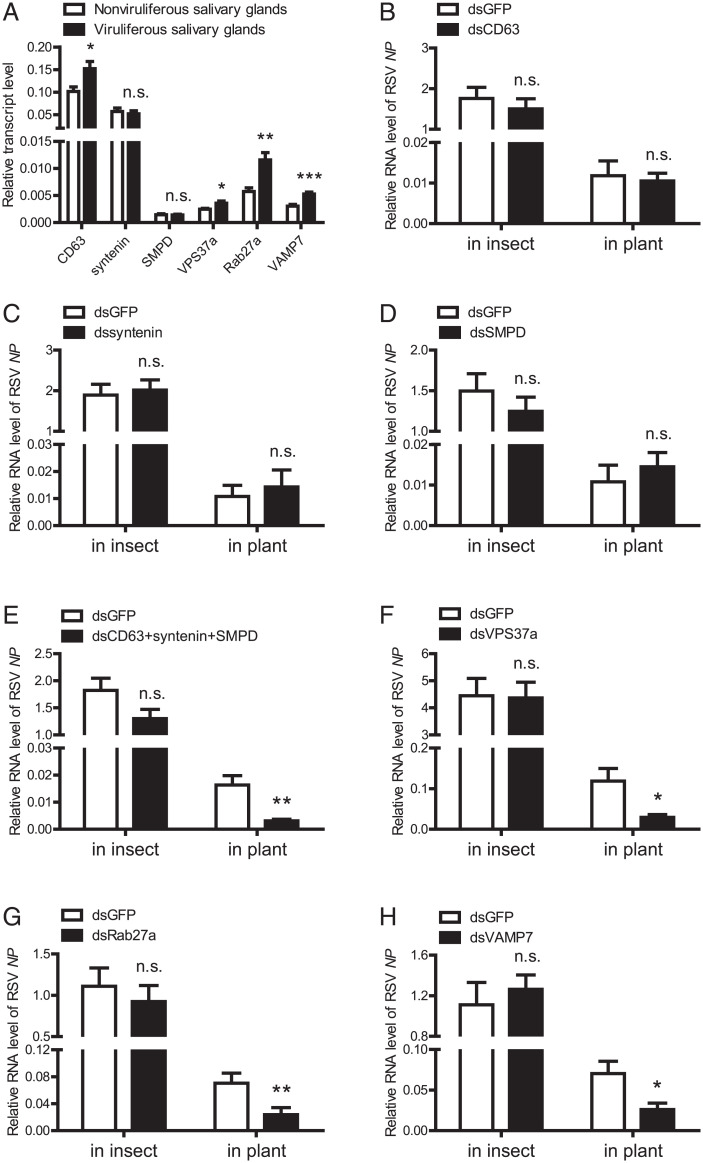
The effects of several key genes of insect exosome systems on RSV transmission were explored, including tetraspanin CD63, syntenin, and SMPD, involved in exosome formation (21–23); VPS37a, involved in cargo sorting (24); Rab27a, involved in MVB transport (25); and VAMP7, involved in exosome secretion (26). The expression of the six genes did not change in the insect whole body between viruliferous and nonviruliferous planthoppers (SI Appendix, Fig. 3A). However, the transcript levels of CD63, VPS37a, Rab27a, and VAMP7 were up-regulated in viruliferous salivary glands (Fig. 4A), while CD63 was up-regulated and SMPD and VPS37a were down-regulated in viruliferous guts (SI Appendix, Fig. 3B). These results indicated that RSV probably affects insect exosome systems with organ specificity.
Fig. 4.
Interference with the insect exosome systems reduced RSV transmission to plants. (A) Comparison of relative transcript levels of CD63, syntenin, SMPD, VPS37a, Rab27a, or VAMP7 in the salivary glands of viruliferous and nonviruliferous adult planthoppers as measured by real-time qPCR. The transcript level of each gene was normalized to that of EF2. (B–H) Relative RNA levels of RSV NP in the viruliferous planthoppers at 7 d after the injection of dsRNAs for each gene (dsCD63, dssyntenin, dsSMPD, dsVPS37a, dsRab27a, and dsVAMP7) or the three genes together (dsCD63+syntenin+SMPD) and in the rice plants fed on by the dsRNA-injected planthoppers for 3 d, as measured by qPCR. The transcript levels of planthopper EF2 and rice tubulin were quantified to normalize the RNA level of NP. The control group was injected with GFP dsRNA (dsGFP). *P < 0.05. **P < 0.01. ***P < 0.001. n.s., no significant difference.
When the expression of each gene was interfered with an efficiency of over 70% after injection of double-stranded RNAs (dsRNAs) targeting each gene in viruliferous third-instar planthoppers (SI Appendix, Fig. 4), the RSV load did not change in planthoppers at 7 dpi compared to the control groups that were injected with GFP dsRNA, and even the simultaneous interference of the expression of three exosome formation genes, i.e., CD63, syntenin, and SMPD, did not affect viral load in planthoppers (Fig. 4 B–H). However, the amounts of RSV secreted to rice plants in terms of the RNA level of NP were reduced by 75%, 66%, and 63% after being fed on by the planthoppers for 3 d with a VPS37a, Rab27a, or VAMP7 knockdown, respectively, or reduced by 81% with the simultaneous knockdown of the three exosome formation genes (Fig. 4 E–H). These results indicated that each phase of the insect exosome systems affected the transmission of RSV from insect vectors to rice plants.
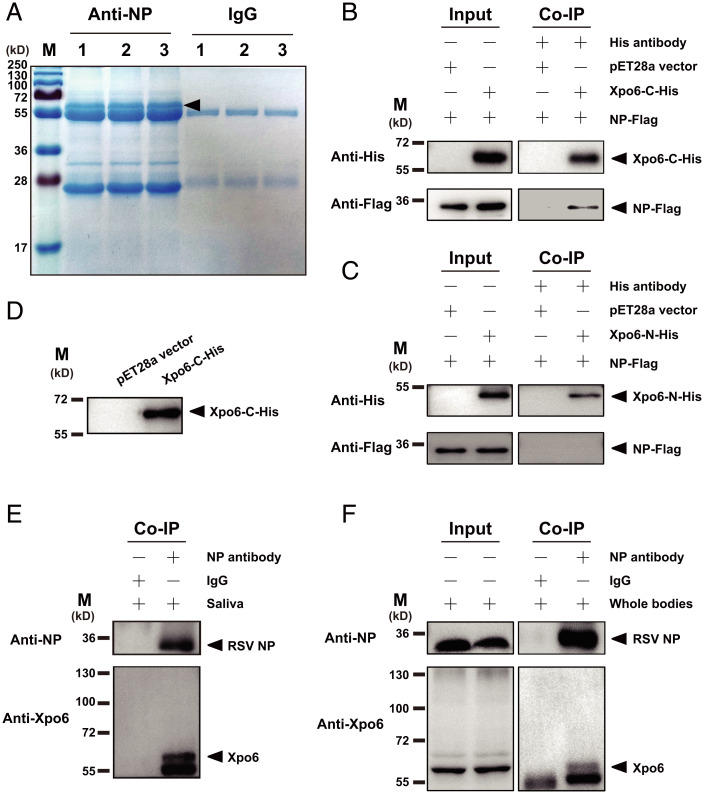
RSV NP Bound Fragmented Exportin 6 in Saliva.
The molecular mechanism of RSV loaded in exosomes was further investigated using RSV NP monoclonal antibody as a bait to pull down NP-binding proteins in the saliva of viruliferous planthoppers through coimmunoprecipitation (Co-IP) assays. A predominant band appeared between 55 kDa and 72 kDa in three Co-IP assay replicates (Fig. 5A). Liquid chromatography-tandem mass spectrometry (LC-MS/MS) identified 10 NP-binding saliva proteins with a peptide score higher than 21, including exportin 6, coiled-coil domain-containing protein 40, endoplasmin, armadillo protein, alanine-glyoxylate aminotransferase 2-like, elongator complex protein 1, and four unknown proteins. Exportin 6 and coiled-coil domain-containing protein 40 were detected in two replicates and found in exosomes (11, 27). Surprisingly, exportin 6 was also identified in the predominant band between 55 kDa and 72 kDa by mass spectrometry (Fig. 5A).
Fig. 5.
RSV NP bound fragmented exportin 6 in saliva. (A) SDS-PAGE of the NP-binding proteins pulled down using RSV NP monoclonal antibody from the saliva of viruliferous planthoppers in three replicates of Co-IP assays. Mouse IgG was used as a negative control. The arrowhead indicates the predominant band between 55 kDa and 72 kDa. (B and C) Interactions between recombinantly expressed NP-Flag and His-tagged C fragment (Xpo6-C-His) or N fragment (Xpo6-N-His) of exportin 6 in the Co-IP and Western blot assays using anti-His and anti-Flag monoclonal antibodies. The expression products from the pET28a vector were used as a negative control. (D) Verification of the anti-exportin 6 polyclonal antibody using Xpo6-C-His in Western blot assays. (E and F) In vivo Co-IP assays on the binding of RSV NP to exportin 6 (Xpo6) in saliva and whole bodies of planthoppers using the anti-NP monoclonal antibody and anti-Xpo6 polyclonal antibody. Mouse IgG was used as a negative control.
Based on the genome of the small brown planthopper (28), the full-length open reading frame (ORF) of exportin 6 was 2,949 bp, and it putatively encoded a protein of 982 amino acid residues (aa) with a molecular weight of 113.5 kDa (GenBank accession no. RZF34382.1). Phylogenetic analyses showed that exportin 6 of the small brown planthopper was clustered with exportin 6 from mammals and other insects (SI Appendix, Fig. 5). The exportin 6 bound to NP in saliva seemed to be fragmented. Considering that the peptide from amino acids 433 to 442 of exportin 6 was retrieved by mass spectrometry and that the possible protease cleavage sites were predicted in the region from 417 to 426 aa in the PeptideCutter website, we expressed two fragments of exportin 6, the N fragment from 1 to 416 aa (native molecular weight of 48.2 kDa) and the C fragment from 417 to 982 aa (native molecular weight of 65.3 kDa), with His-tags in Escherichia coli. Co-IP assays showed that the C-fragment of exportin 6, but not the N fragment, bound the recombinantly expressed NP-Flag (Fig. 5 B and C).
To clarify the occurrence of fragmented exportin 6 in the small brown planthopper, a polyclonal antibody against exportin 6 was produced using a synthesized polypeptide from 937 to 961 aa of exportin 6. This antibody was first proven to recognize the recombinantly expressed C fragment of exportin 6 (Fig. 5D). In the NP-binding proteins pulled down using the NP monoclonal antibody from saliva or whole bodies of viruliferous planthoppers, the exportin 6 antibody showed only one band with a size between 55 kDa and 72 kDa (Fig. 5 E and F), in which exportin 6 was identified by mass spectrometry (Fig. 5A). These results indicated that RSV virions bound fragmented exportin 6, a putative exosome protein, in planthopper saliva.
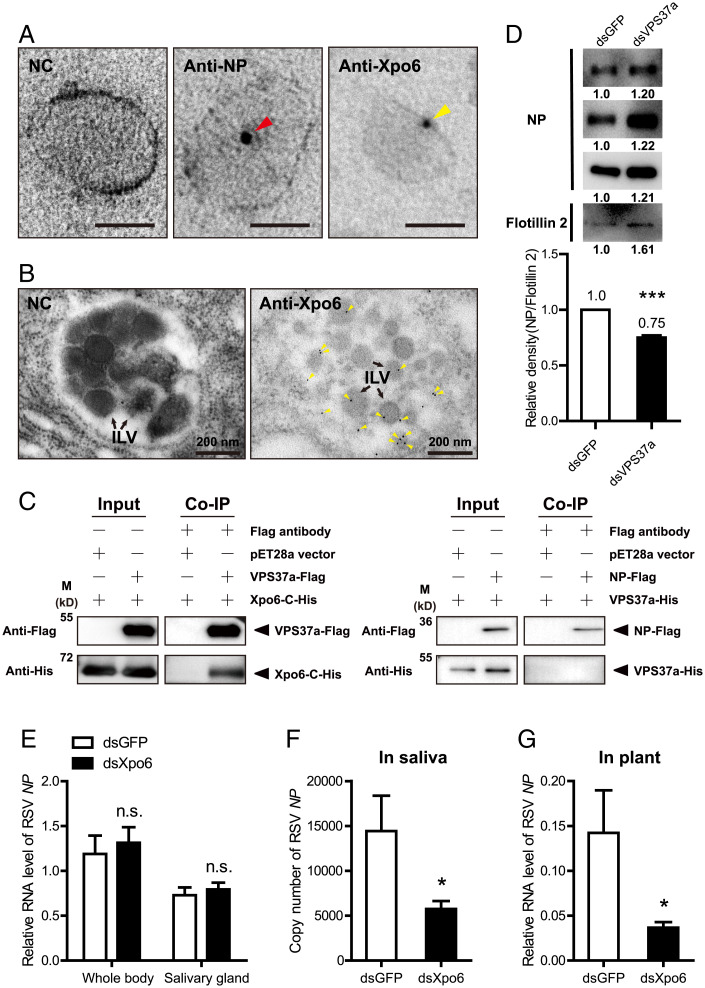
Exportin 6 Was Sorted into Exosomes by VPS37a and Facilitated RSV Transmission.
This study further explored whether the fragmented exportin 6 resides in exosomes. In the ultrathin sections of hemolymph exosomes from viruliferous planthoppers analyzed under immunoelectron microscopy, exportin 6 was found to be labeled by colloidal gold-conjugated exportin 6 polyclonal antibody as well as RSV NP, which was also labeled by NP monoclonal antibody (Fig. 6A). Exportin 6 was also observed in the ILVs of MVBs in the primary salivary gland cells of viruliferous planthoppers (Fig. 6B). How exportin 6 and RSV were loaded in exosomes was further explored. In the six key genes of insect exosome systems, VPS37a, Rab27a, and VAMP7 affected the transmission of RSV from insect vectors to rice plants (Fig. 4 F–H). Considering that only VPS37a is responsible for sorting cargoes into the ILVs while Rab27a and VAMP7 play a role in MVB transport and docking at cell membrane (24–26), we chose VPS37a to test its binding with fragmented exportin 6 and the NP of RSV through Co-IP assays. The recombinantly expressed VPS37a pulled down the C fragment of exportin 6, whereas it did not pull down the NP of RSV (Fig. 6C). When the expression of VPS37a was knocked down with injection of dsRNAs in viruliferous planthoppers, we purified hemolymph exosomes in fractions 7 and 8 of density-gradient ultracentrifugation at 7 dpi (Fig. 1D) and found that the amounts of RSV in terms of NP protein levels in the exosomes were reduced by one quarter (Fig. 6D). These results indicated that the load of RSV in exosomes was dependent on the sorting of exportin 6 in exosomes by VPS37a.
Fig. 6.
Exportin 6 was sorted into exosomes by VPS37a and facilitated RSV transmission. (A) Immunoelectron microscopy showing RSV virions and exportin 6 (Xpo6) in ultrathin cryosections of hemolymph exosomes labeled by gold-conjugated anti-NP monoclonal antibody and anti-Xpo6 polyclonal antibody. The NC was not treated with antibodies. Arrowheads indicate RSV NP and Xpo6 molecules. (Scale bar, 50 nm.) (B) Immunogold labeling of Xpo6 in ultrathin sections of primary salivary gland cells of planthoppers. Yellow arrowheads indicate Xpo6 labeled with gold-conjugated anti-Xpo6 polyclonal antibody. Black arrows indicate ILVs. NC was not treated with antibodies. (C) Interactions between recombinantly expressed VPS37a-Flag and the His-tagged C fragment of exportin 6 (Xpo6-C-His) or between NP-Flag and VPS37a-His in the Co-IP and Western blot assays using anti-His and anti-Flag monoclonal antibodies. The expression products from the pET28a vector were used as a negative control. (D) Western blot assays for RSV NP and flotillin 2 in hemolymph exosomes distributed in fractions 7 and 8 of density-gradient ultracentrifugation at 7 d after the injection of double-stranded RNAs for VPS37a (dsVPS37a) or GFP (dsGFP). Anti-NP monoclonal and anti-flotillin 2 polyclonal antibodies were used. Three repeats are shown with hemolymph exosomes isolated from 1,000 planthoppers. The raw gray values are indicated below each band and the relative gray value of NP to that of flotillin 2 was compared between groups. ***P < 0.001. (E) Relative RNA levels of RSV NP to that of planthopper EF2 in the whole bodies and salivary glands of viruliferous planthoppers at 8 d after injection of dsRNAs for exportin 6 (dsXpo6). The control group was injected with GFP dsRNA (dsGFP). n.s., no significant difference. (F) Copy number of RSV NP in planthopper saliva secreted in PBS buffer after injection of dsXpo6 or dsGFP measured by absolute real-time qPCR. (G) Relative RNA levels of RSV NP to that of rice tubulin in rice plants fed by dsXpo6- or dsGFP-injected planthoppers for 1 d. *P < 0.05.
When the expression of exportin 6 was knocked down by the injection of dsRNAs for exportin 6 (SI Appendix, Fig. 6A), the survival rate and feeding behavior of viruliferous planthoppers were not affected (SI Appendix, Fig. 6 B and C); moreover, the RSV amounts in the whole bodies and salivary glands of planthoppers did not change at 8 dpi (Fig. 6E). Instead, the amounts of RSV were reduced by 60% in planthopper saliva secreted in phosphate buffer saline (PBS) buffer and 74% in rice plants fed by planthoppers compared to the control groups, which were injected with GFP dsRNA (Fig. 6 F and G and SI Appendix, Fig. 2). Therefore, exportin 6 did not affect RSV accumulation in insect vectors; instead, it facilitated RSV release from insect vectors to plants.
Discussion
As mediators of molecular transmission between cells or organs, exosomes have been highlighted for their biological significance based on the discovery of their various specific roles, including their function as vehicles of transmission for a variety of microorganisms. Here we isolated exosomes from tiny amounts of saliva from an insect vector with a size of less than 4 mm and identified viral proteins and genomes of RSV in the exosomes. The entry of RSV into exosomes relied on fragmented exportin 6, which was sorted into exosomes by interaction with VPS37a. Interference with any phase of the exosome systems or exportin 6 greatly reduced the amount of RSV secreted to rice plants. This study provides explicit evidence to show the function of exportin 6 in transporting viruses to exosomes for viral horizontal transmission.
This study found that exportin transports viruses into exosomes. The exportin family is usually responsible for trafficking proteins with a nuclear export signal from the nucleus to the cytoplasm in eukaryotes (29). We found that exportin 6 acted as a VPS37a-recognized vehicle to transport RSV virions into exosomes, while RSV could not bind VPS37a directly. This virus-transporter function was fulfilled by fragmented exportin 6. Although we did not determine the accurate cleavage site of the C fragment of exportin 6, fragmented exportin 6 did not contain the two conserved domains of the IBN_N and Xpo1 superfamilies and seemed to be the predominant form in the small brown planthopper. Homologs of exportin 6 in humans (30), Xenopus oocytes (31), and Drosophila (32) have a full protein length of over 100 kDa. Interestingly, a 72-kDa protein recognized by the Drosophila exportin 6 antibody was previously revealed as an untargeted protein due to the cross-reaction (32). Thus, fragmented exportin 6 may also occur in these species.
The main mechanism by which RSV is released from salivary glands could be by hijacking exosomes. We found that the ratios of viral RNA3 and RNA4 were higher than those of the other two genomic RNAs in saliva exosomes and in the whole saliva but were different from those in the whole insect body, where RNA1 was the most dominant segment (33). Coincidently, the ratios of RNA3 and RNA4 were triple that of RNA1 or RNA2 when RSV was secreted from planthoppers to rice leaves for 1 d (33). The amounts of RSV secreted to rice plants were reduced by at least 63% when insect exosome systems were inhibited. This evidence suggests that hijacking exosomes is probably the dominant pathway for RSV transmission from salivary glands to plants. The dominance of RNA3 and RNA4 in saliva exosomes has biological significance because the NP (structural protein) (34), NS3 (gene silencing suppressor) (35), SP (disease-specific protein) (36), and NSvc4 (movement protein) (14) encoded by the two segments are indispensable for RSV in establishing the initial infection in rice plants. Rice dwarf virus (RDV) also predominantly relies on exosomes for transmission, as virions were observed to be enclosed in the exosomes and the transmission ratio by leafhoppers was reduced by ∼75% when Rab27a was knocked down (5). Other pathways may take part in viral release. Rice gall dwarf virus induces the formation of virus-associated filaments within the salivary glands of leafhoppers, and such filaments attach to the apical plasmalemma and induce an exocytosis-like process for viral release (37).
The sizes of the exosomes are affected by viral infection. Exosomes usually range from 30 to 200 nm in diameter (6). RSV is a nonenveloped and 3 nm- to 8 nm-wide filament of different lengths that folds to form pleomorphic or branched configurations (38). We found that the mean size of the exosomes containing RSV was between 136.1 nm and 149.3 nm, which were larger than that of exosomes from nonviruliferous planthoppers (92.8 nm). RDV is an icosahedral and nonenveloped virus with an average diameter of ∼70 nm (39). In the salivary cavities of leafhoppers, the packaging of RDV enlarged exosomes to 110 to 302 nm in diameter, while the virus-free exosomes were 53 to 135 nm in diameter (5). Langat virions are small round icosahedral enveloped particles with a diameter of 50 nm, similar to tick-borne encephalitis virus (40). The ratio of 150- to 200-nm exosomes is much higher in Langat virus-infected tick cells (18%) than in uninfected cells (6.5%) (4). Regardless of the size, configuration, and presence or absence of an envelope, viruses always take advantage of exosomes with comparatively large sizes.
In conclusion, we identified a key role of exportins in exosome-mediated horizontal transmission of RSV from insect vectors to host plants. Exportin 6 as well as genes in each phase of the insect exosome systems represent potential targets for controlling future RSV outbreaks.
Materials and Methods
Small Brown Planthoppers.
The small brown planthoppers used in this study were derived from a field population collected in Nanjing, Jiangsu Province, China in 2019. The nonviruliferous and viruliferous populations were separately reared on fresh rice seedlings, Oryza sativa Wuyujing, in glass beakers at 25 °C with 16 h of light daily. The RSV-carrying frequency of the viruliferous population was maintained above 90% through a purification selection every 3 mo via a dot enzyme-linked immunosorbent assay (dot-ELISA) with a homemade anti-NP monoclonal antibody (41).
Isolation and Purification of Hemolymph Exosomes.
Each hemolymph sample was collected from ∼3,000 nonviruliferous or viruliferous adult planthoppers using a centrifugation method (18). The exosomes were isolated by serial centrifugation as described in SI Appendix, Fig. 1B and used directly for electron microscopy or immunoelectron microscopy assays or further purified in a density-gradient ultracentrifugation with a series of iodixanol concentrations. Discontinuous iodixanol gradients from 40%, 20%, and 10 to 5% were made by diluting a stock solution of 60% iodixanol (OptiPrep, Axis-Shield) with 0.25 M sucrose in 1 mM ethylenediaminetetraacetic acid (EDTA) and 10 mM Tris⋅HCl (pH 7.4). The exosome sample was layered on top of an iodixanol gradient and centrifuged at 100,000 × g for 18 h at 4 °C. Twelve fractions with 400 μL in each fraction were collected for subsequent Western blot analysis using a homemade anti-flotillin 2 polyclonal antibody and an anti-NP monoclonal antibody. Exosomes from fractions 7 and 8 were used for NTA analysis and electron microscopy.
Isolation of Saliva Exosomes.
Each saliva sample was collected from ∼30,000 viruliferous fourth-instar nymphs. Fifty nymphs were transferred to a saliva-collection glass bottle (SI Appendix, Fig. 1A). One end of the bottle was covered with two layers of parafilm membranes that sandwiched 400 μL of PBS (10 mM PBS, pH 7.4). The other end was sealed with a thin layer of gauze. After feeding for 24 h at 23 °C in an incubator, the saliva contained in PBS was collected as described previously (18). Exosomes were isolated using the exosome precipitation solution ExoQuick-TC (System Biosciences Inc.) following a procedure described in detail in SI Appendix, Fig. 1B.
Nanoparticle Tracking Analysis.
NTA was performed in a NanoSight NS300 particle size analyzer (Malvern Panalytical) following the manufacturer’s instructions. Exosome samples were diluted with PBS to concentrations falling into the range between 1 × 108 and 20 × 108 particles/mL. Each sample was measured in triplicate. The recorded movie data were analyzed with the NTA 3.4 program to calculate the sizes and numbers of particles included.
Electron Microscopy of the Exosomes.
Hemolymph or saliva exosomes were loaded onto 200 mesh copper grids, which were coated with a formvar/carbon film and glow charged for 45 s. Exosomes were stained with 1% uranyl acetate for 1 min in the dark and then examined by a JEM-1400 transmission electron microscope (JEOL) at an accelerating voltage of 80 kV.
Immunoelectron Microscopy of the Exosomes.
Saliva or hemolymph exosomes were fixed with 2% paraformaldehyde in phosphate buffer (PB) (0.1 M phosphate, pH 7.4) and loaded onto 200 mesh copper grids, which were coated with a formvar/carbon film and glow charged for 45 s. After permeation with 0.1% Triton X-100 for 20 min, the exosomes were immunolabeled with a homemade anti-NP monoclonal antibody or an anti-flotillin 2 polyclonal antibody, followed by a gold-conjugated goat anti-mouse or goat anti-rabbit secondary antibody (Merck). The negative control was not treated with primary antibodies. The samples were negatively stained with 1% uranyl acetate in the dark and observed using a JEM-1400 transmission electron microscope (JEOL) at an accelerating voltage of 80 kV.
Hemolymph exosomes were also prepared for ultrathin cryosections. After fixation with a mixture of 4% paraformaldehyde and 0.5% glutaraldehyde in PB (pH 7.4), the exosomes were embedded in 2% gelatin and infused in 2.3 M sucrose. The mounted gelatin blocks were frozen in liquid nitrogen, and 80-nm ultrathin sections were prepared with a Leica EM FC7 μLtramicrotome (Leica Microsystems). The sections were adhered to 50 mesh copper grids using 2% methylcellulose in 2.3 M sucrose, blocked with 1% bovine serum albumin (BSA), and immunolabeled with an anti-NP monoclonal antibody or anti-exportin 6 polyclonal antibody, followed by a gold-conjugated goat anti-mouse or goat anti-rabbit secondary antibody. The samples were embedded in a mixture of 4% methylcellulose and 2% uranyl acetate (9:1) on ice and viewed with a Tecnai Spirit transmission electron microscope (FEI) at 120 kV.
Immunoelectron Microscopy of the Salivary Glands.
The salivary glands were dissected and fixed with 4% paraformaldehyde and 0.5% glutaraldehyde in PB (pH 7.4) at 4 °C overnight. After dehydration in 30%, 50%, 70%, 85%, 95%, and 100% alcohol, the salivary glands were embedded in LR White Resin (Merck) and cut into 70-nm ultrathin sections using a Leica EM UC6 μLtramicrotome (Leica Microsystems). The sections were placed onto 50 mesh copper grids before being blocked for 1 h with 1% BSA. After immunolabeling with the anti-NP monoclonal antibody and anti-flotillin 2 polyclonal antibody, or the anti-exportin 6 polyclonal antibody, 10 nm gold-conjugated goat anti-mouse and 5 nm gold-conjugated goat anti-rabbit secondary antibodies were added. The negative control was not treated with primary antibodies. The samples were stained with 2% neutral uranyl acetate for 5 min in the dark and viewed under a Tecnai Spirit transmission electron microscope (FEI) at 120 kV.
RNA Extraction and cDNA Synthesis.
Total RNA from five planthoppers, 50 salivary glands, one rice leaf, 400 μL of exosome fractions, or saliva from 50 planthoppers was extracted using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. One microgram of total RNA was reverse transcribed to cDNA with Moloney murine leukemia virus (M-MLV) reverse transcriptase and random primers (Promega).
Protein Expression and Antibody Preparation.
The N fragment (1 to 416 aa) and C fragment (417 to 982 aa) of exportin 6, RSV NP, and VPS37a were cloned and constructed into the vector pET28a to generate His-tagged or Flag-tagged recombinant proteins. The primers are listed in SI Appendix, Table 1. The recombinant plasmids were transformed into E. coli BL21(DE3) for expression. After 10 h of induction with 0.5 mmol/L isopropyl-β-D-thiogalactoside (IPTG) at 16 °C, the cells were pelleted by centrifugation and then sonicated for 30 min on ice. Then, the supernatant from the sonicated cells was used for Co-IP assays. The polypeptide from 937 to 961 aa of exportin 6 was synthesized and served as an antigen for the production of a rabbit polyclonal antibody by the Beijing Protein Innovation Company. The specificity of the antibody was tested for the recognition of the recombinantly expressed His-tagged C fragment of exportin 6 through Western blot analysis.
Co-IP and Mass Spectrometry.
Protein G beads (50 μL) were mixed with 5 μg of monoclonal anti-NP or IgG antibodies (Merck) and then incubated with 7 mL of saliva from viruliferous planthoppers overnight. The antibody–protein complex was dissociated from the beads with elution buffer (Thermo Fisher Scientific) and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). After the heavy chain and light chain of immunoglobulin were removed, the remaining gels were used for LC-MS/MS analysis on a Q-Exactive instrument (Thermo Fisher Scientific) at the Beijing Protein Innovation Company. Three biological replicates were prepared. The band between 55 kDa and 72 kDa was isolated for LC-MS/MS analysis.
The in vitro Co-IP assays to identify the interactions between the recombinantly expressed NP and fragments of exportin 6 or VPS37a and the in vivo Co-IP assays to identify the binding of RSV NP to exportin 6 in saliva and whole bodies of planthoppers were performed as previously described (2).
Absolute Real-Time qPCR.
The RNA levels of four genomic RNAs of RSV (RNA1, NC_003755; RNA2, NC_003754; RNA3, NC_003776; and RNA4, NC_003753) and NP in saliva exosomes, hemolymph exosomes, or saliva were determined using absolute qPCR on a LightCycler 480 II (Roche). The primers are shown in SI Appendix, Table 1. The standard curves were obtained as described previously (33). The linear regression equation, along with the coefficient of determination (R2), was used to evaluate the standard curves. The copy numbers were determined from the standard curves. Two different batches for each sample were prepared.
Real-Time qPCR.
qPCR was used to measure the RNA levels of RSV NP (GenBank accession no. DQ299151) and the transcript levels of CD63 (evm.model.Contig253.61 in L. striatellus genome dataset), syntenin (evm.model.Contig104.45), SMPD (evm.model.Contig371.22.2), VPS37a (evm.model.Contig226.21), Rab27a (evm.model.Contig31.98), VAMP7 (evm.model.Contig389.14), and exportin 6 (RZF34382.1). The transcript levels of planthopper translation elongation factor 2 (EF2) (evm.model.Contig0.299) and rice tubulin (XP_015649724.2) were quantified to normalize the cDNA templates of planthoppers and rice. The primers for each gene are listed in SI Appendix, Table 1. The relative RNA or transcript level of each gene to EF2 or tubulin was reported as the mean ± SEM. Differences were statistically evaluated using Student’s t test to compare the two means and one-way ANOVA followed by Tukey’s test for multiple comparisons in SPSS 19.0.
Double-Stranded RNA Synthesis and Injection.
The dsRNAs for CD63 (398 bp), syntenin (457 bp), SMPD (666 bp), VPS37a (515 bp), Rab27a (271 bp), VAMP7 (340 bp), and exportin 6 (785 bp) were synthesized using the T7 RiboMAX Express RNAi System (Promega) following the manufacturer’s instructions. The corresponding PCR primers are shown in SI Appendix, Table 1. A total of 23 nL of 6 μg/μL dsRNA for each gene or a mixture of dsRNAs for CD63, syntenin, and SMPD was injected into viruliferous third-instar nymphs using a Nanoliter 2000 microinjector (World Precision Instruments). The control group was injected with dsRNA for GFP. Four days after injection, the insects were fed one healthy rice seedling for 3 d or 1 d (for injection of dsRNA for exportin 6). The whole bodies and salivary glands of the insects and plants were collected for RNA extraction and qPCR. A total of 7 to 9 biological replicates of the insect samples and 9 to 15 biological replicates of the rice were prepared. The saliva of 15 insects secreted in PBS buffer was collected for 1 d at 7 dpi and used for absolute qPCR. A total of 7 to 11 biological replicates were prepared.
Mechanical Injection of Saliva Exosomes in Rice Seedlings.
Saliva exosomes were microinjected into the midribs of healthy 4-wk-old rice leaves through a glass needle using a Nanoliter 2000 microinjector (World Precision Instruments). Each leaf was microinjected five times with 23 nL each time at an interval of 1 cm. The leaves were collected at 3, 6, and 9 dpi for RSV NP measurement with qPCR. One leaf per replicate and 13 to 20 biological replicates were prepared. Disease symptoms were observed after 4 wk of inoculation. RSV crude extracts from viruliferous planthoppers were prepared as described previously (19) and used as positive control.
Phylogenetic Analysis.
The homologous exportin proteins from other insect species, Danio rerio, mice, and humans were downloaded from the National Center for Biotechnology Information. A neighbor-joining phylogenetic tree was constructed using the pairwise deletion and p-distance model in Mega X software. Bootstrap analysis with 1,000 replicates was performed to evaluate the internal support for the tree topology.
Survival Curves and Feeding Behavior of Planthoppers.
After the injection of dsRNAs for exportin 6, the third-instar viruliferous nymphs were raised on rice seedlings. The number of living insects was recorded daily, and the survival rate was calculated within 12 d in one group with 14 to 20 insects. The control group was injected with dsRNA for GFP. Sixteen biological replicates were prepared. Survival curves were assessed by the Kaplan–Meier method, and differences were evaluated according to the Mantel–Cox log-rank statistic test. The feeding behavior of exportin 6 knockdown planthoppers and the control group was analyzed using electrical penetration graph (EPG) technology. The waveforms of 25 individuals were recorded continuously for 8 h using a Giga-8 EPG amplifier (Wageningen University, Wageningen, The Netherlands) as previously described (20). Time ratios of waveforms were reported as the means with SE. Differences between the two groups were statistically evaluated using Student’s t test.
Supplementary Material
Acknowledgments
We thank the Center for Biological Imaging, Institute of Biophysics, Chinese Academy of Sciences for the electron microscopy and we are grateful to Li Wang for her help in making EM samples and taking EM images. This work was supported by grants from the National Natural Science Foundation of China (Nos. 32090012 and 32102207).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2207848119/-/DCSupplemental.
Data, Materials, and Software Availability
All study data are included in the article and/or supporting information.
References
- 1.Hogenhout S. A., Ammar D., Whitfield A. E., Redinbaugh M. G., Insect vector interactions with persistently transmitted viruses. Annu. Rev. Phytopathol. 46, 327–359 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Ma Y., et al. , Membrane association of importin α facilitates viral entry into salivary gland cells of vector insects. Proc. Natl. Acad. Sci. U.S.A. 118, e2103393118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vora A., et al. , Arthropod EVs mediate dengue virus transmission through interaction with a tetraspanin domain containing glycoprotein Tsp29Fb. Proc. Natl. Acad. Sci. U.S.A. 115, E6604–E6613 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou W., et al. , Exosomes serve as novel modes of tick-borne flavivirus transmission from arthropod to human cells and facilitates dissemination of viral RNA and proteins to the vertebrate neuronal cells. PLoS Pathog. 14, e1006764 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Q., et al. , Exosomes mediate horizontal transmission of viral pathogens from insect vectors to plant phloem. eLife 10, e64603 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pegtel D. M., Gould S. J., Exosomes. Annu. Rev. Biochem. 88, 487–514 (2019). [DOI] [PubMed] [Google Scholar]
- 7.van Niel G., D’Angelo G., Raposo G., Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 19, 213–228 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Morrow I. C., et al. , Flotillin-1/reggie-2 traffics to surface raft domains via a novel golgi-independent pathway. Identification of a novel membrane targeting domain and a role for palmitoylation. J. Biol. Chem. 277, 48834–48841 (2002). [DOI] [PubMed] [Google Scholar]
- 9.de Jong O. G., et al. , Cellular stress conditions are reflected in the protein and RNA content of endothelial cell-derived exosomes. J. Extracell. Vesicles 1, 18396 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meckes D. G. Jr., et al. , Modulation of B-cell exosome proteins by gamma herpesvirus infection. Proc. Natl. Acad. Sci. U.S.A. 110, E2925–E2933 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paggetti J., et al. , Exosomes released by chronic lymphocytic leukemia cells induce the transition of stromal cells into cancer-associated fibroblasts. Blood 126, 1106–1117 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kowal J., et al. , Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. U.S.A. 113, E968–E977 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toriyama S., Rice stripe virus: Prototype of a new group of viruses that replicate in plants and insects. Microbiol. Sci. 3, 347–351 (1986). [PubMed] [Google Scholar]
- 14.Xiong R., Wu J., Zhou Y., Zhou X., Identification of a movement protein of the tenuivirus rice stripe virus. J. Virol. 82, 12304–12311 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi M., Toriyama S., Hamamatsu C., Ishihama A., Nucleotide sequence and possible ambisense coding strategy of rice stripe virus RNA segment 2. J. Gen. Virol. 74, 769–773 (1993). [DOI] [PubMed] [Google Scholar]
- 16.Toriyama S., Takahashi M., Sano Y., Shimizu T., Ishihama A., Nucleotide sequence of RNA 1, the largest genomic segment of rice stripe virus, the prototype of the tenuiviruses. J. Gen. Virol. 75, 3569–3579 (1994). [DOI] [PubMed] [Google Scholar]
- 17.Whitfield A. E., Falk B. W., Rotenberg D., Insect vector-mediated transmission of plant viruses. Virology 479-480, 278–289 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Li J., Zhao W., Wang W., Zhang L., Cui F., Evaluation of Rice stripe virus transmission efficiency by quantification of viral load in the saliva of insect vector. Pest Manag. Sci. 75, 1979–1985 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Chen X., et al. , A plant virus ensures viral stability in the hemolymph of vector insects through suppressing prophenoloxidase activation. MBio 11, e01453–e01420 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang W., et al. , Flotillin 2 facilitates the infection of a plant virus in the gut of insect vector. J. Virol. 96, e0214021 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Niel G., et al. , The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis. Dev. Cell 21, 708–721 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baietti M. F., et al. , Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 14, 677–685 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Trajkovic K., et al. , Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 319, 1244–1247 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Stefani F., et al. , UBAP1 is a component of an endosome-specific ESCRT-I complex that is essential for MVB sorting. Curr. Biol. 21, 1245–1250 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Ostrowski M., et al. , Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 12, 19–30, 1–13 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Fader C. M., Sánchez D. G., Mestre M. B., Colombo M. I., TI-VAMP/VAMP7 and VAMP3/cellubrevin: Two v-SNARE proteins involved in specific steps of the autophagy/multivesicular body pathways. Biochim. Biophys. Acta 1793, 1901–1916 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Fraser K. B., et al. , LRRK2 secretion in exosomes is regulated by 14-3-3. Hum. Mol. Genet. 22, 4988–5000 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu J., et al. , Genome sequence of the small brown planthopper, Laodelphax striatellus. Gigascience 6, 1–12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cautain B., Hill R., de Pedro N., Link W., Components and regulation of nuclear transport processes. FEBS J. 282, 445–462 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Izdebska M., Gagat M., Grzanka D., Halas M., Grzanka A., The role of exportin 6 in cytoskeletal-mediated cell death and cell adhesion in human non-small-cell lung carcinoma cells following doxorubicin treatment. Folia Histochem. Cytobiol. 52, 195–205 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Onuma A., Fujioka Y. A., Fujii W., Sugiura K., Naito K., Expression and function of exportin 6 in full-grown and growing porcine oocytes. J. Reprod. Dev. 65, 407–412 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thran J., Poeck B., Strauss R., Serum response factor-mediated gene regulation in a Drosophila visual working memory. Curr. Biol. 23, 1756–1763 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Zhao W., Wang Q., Xu Z., Liu R., Cui F., Distinct replication and gene expression strategies of the Rice Stripe virus in vector insects and host plants. J. Gen. Virol. 100, 877–888 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Kakutani T., Hayano Y., Hayashi T., Minobe Y., Ambisense segment 3 of rice stripe virus: The first instance of a virus containing two ambisense segments. J. Gen. Virol. 72, 465–468 (1991). [DOI] [PubMed] [Google Scholar]
- 35.Xiong R., Wu J., Zhou Y., Zhou X., Characterization and subcellular localization of an RNA silencing suppressor encoded by Rice stripe tenuivirus. Virology 387, 29–40 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Zhu Y., Hayakawa T., Toriyama S., Complete nucleotide sequence of RNA 4 of rice stripe virus isolate T, and comparison with another isolate and with maize stripe virus. J. Gen. Virol. 73, 1309–1312 (1992). [DOI] [PubMed] [Google Scholar]
- 37.Mao Q., et al. , Filamentous structures induced by a phytoreovirus mediate viral release from salivary glands in its insect vector. J. Virol. 91, e00265-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramírez B. C., Haenni A. L., Molecular biology of tenuiviruses, a remarkable group of plant viruses. J. Gen. Virol. 75, 467–475 (1994). [DOI] [PubMed] [Google Scholar]
- 39.Takahashi Y., Tomiyama M., Hibino H., Omura T., Conserved primary structures in core capsid proteins and reassembly of core particles and outer capsids between rice gall dwarf and rice dwarf phytoreoviruses. J. Gen. Virol. 75, 269–275 (1994). [DOI] [PubMed] [Google Scholar]
- 40.Pulkkinen L. I. A., Butcher S. J., Anastasina M., Tick-borne encephalitis virus: A structural view. Viruses 10, 350 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao W., Yang P., Kang L., Cui F., Different pathogenicities of Rice stripe virus from the insect vector and from viruliferous plants. New Phytol. 210, 196–207 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.