Abstract
Malaria is one of the most important infectious diseases worldwide. The causative of the most severe forms of malaria, Plasmodium falciparum, has developed resistances against all the available antimalarial drugs. In the present study, the phytochemical investigation of the green seaweed Halimeda macroloba has afforded two new compounds 1–2, along with 4 known ones 3–6. The structures of the compounds had been confirmed using 1& 2D-NMR and HRESIMS analyses. Extensive machine-learning-supported virtual-screening suggested cytochrome-C enzyme as a potential target for compound 2. Docking, absolute-binding-free-energy (ΔGbinding) and molecular-dynamics-simulation (MDS) of compound 2 revealed the strong binding interaction of this compound with cytochrome-C. In vitro testing for crude extract and isolated compounds revealed the potential in vitro inhibitory activity of both extract and compound 2 against P. falciparum. The crude extract was able to inhibit the parasite growth with an IC50 value of 1.8 ± 0.35 µg/mL. Compound 2 also showed good inhibitory activity with an IC50 value of 3.2 ± 0.23 µg/mL. Meanwhile, compound 6 showed moderate inhibitory activity with an IC50 value of 19.3 ± 0.51 µg/mL. Accordingly, the scaffold of compound 2 can be considered as a good lead compound for the future development of new antimalarial agents.
Keywords: malaria, Plasmodium falciparum, Halimeda macrolaba, cytochrome C, docking, molecular dynamics
1. Introduction
Infectious diseases impose a significant burden on global public health and economic stability [1]. Malaria is a potentially life-threatening parasitic disease caused by Plasmodium protozoa and accounting for approximately 229 million cases and 409,000 fatalities in 2019 [2]. Currently, two antimalarial drugs are used to control infection: artemisinin, obtained from Artemisia annua L., and quinine, obtained from Cinchona sp. [2]. The emergence of resistance on the part of mosquitoes to these antimalarial drugs, the weak development of new antimalarial drugs, the logistical problems of these drugs in poor malaria-endemic countries, and the lack of efficient and safe vaccines might increase the complications of malaria in the future [3]. Accordingly, malaria will continue to present challenges to the global goal of its control and eradication, which makes searching for effective, safe, and affordable alternatives an urgent necessity.
Antimalarial drug discovery from marine sources is currently targeted due to reports of such sources yielding effective and safe antimalarial phytochemicals owing to variations in structure and biological activity [4]. The imidazole alkaloid, paenidigyamycin A isolated from Ghanaian Paenibacillus polymyxa strain De2sh, inhibited P. falciparum 3D7 with IC50 = 36 nmol/L [5]. The bromo-pyrrole alkaloids isolated from Tedania braziliensis, a marine sponge, are made up of new pseudo-ceratidine and its derivatives and exhibited potent activity against sensitive (3D7 strain) and resistant (K1 strain) P. falciparum strains (0.96–1.24, 5.11–6.49, and 3–6 μmol/L) [6]. Two strains of P. falciparum (drug-resistant K1 and drug-sensitive FCR3) were inhibited also by bromo-tyrosine alkaloid, ceratinadin E (1.03 and 0.77 μg/mL respectively), and psammaplysin F (3.77 and 2.45 μg/mL respectively), produced by Okinawan marine sponge (Pseudoceratina sp.) [7]. From an Australian bryozoan, Orthoscuticella ventricose; two new orthoscuticellines, A and B; three new β-carboline alkaloids, orthoscuticellines C−E; and six other known compounds were isolated. These compounds showed moderate activity against the same parasite (12–21 μmol/L) [8]. A new linear diterpene, bifurcatriol, was isolated from an Irish brown alga, Bifurcaria bifurcate, and inhibited drug-resistant P. falciparum K1 at low concentration (0.65 µg/mL) [9]. Three potent antimalarial sesquiterpenoids, smenotronic acid, ilimaquinone, and pelorol, isolated from the sponge Hyrtios erectus, showed promising in vitro activity against chloroquine-resistant P. falciparum strain Dd2 at IC50 3.51, 2.11, and 0.8 µmol/L, respectively [10]. In in vitro antimalarial activity against P. falciparum 3D7, endoperoxide, sinuketal (isolated from soft corals Sinularia sp.), showed mild activity (IC50 = 80 µmol/L) [11]. Hoshinoamides A and B (isolated from cyanobacterium, Caldora penicillata) had moderate inhibitory activity against P. falciparum at an IC50 value of 0.52 and 1.0 μmol/L, respectively [12]. a new cyclic peptide, kakeromamide B, along with ulongamide A and lyngbyabellin A, isolated from the marine cyanobacterium Moorea producens collected from reef slopes off the shores of Tuvuca Island in Fiji, exhibited antiparasitic activity against P. falciparum blood-stages (EC50 = 8.9, 0.99 and 1.5 µmol/L, respectively) [13]. A macrolide with a 40-membered ring, palstimolide A, isolated from Central Pacific Ocean Cyanobacterium, exhibited interesting antiparasitic activities against the blood-stage of the P. falciparum Dd2 (IC50 = 223 nmol/L) [14]. Likewise, another new 24-membered polyhydroxy macrolide, bastimolide B, isolated along with a known bastimolide A from a cyanobacterium, Okeania hirsute, exhibited strong anti-plasmodial activity against chloroquine-sensitive P. falciparum strain HB3 (IC50 = 5.7 ± 0.7 μmol/L) [15]. Cholesterol and a new antimalarial mono-hydroxy acetylated sterol derivative, halymeniaol, were isolated from the marine alga Halymenia floresii. The former exhibited inhibitory activity against the chloroquine-resistant P. falciparum 3D7 strain (IC50 = 3.0 μmol/L) [16]. Sterols, kaimanol and saringosterol, with significant antimalarial activity against P. falciparum 3D7 strains (IC50 = 359 and 0.250 nmol/L respectively) were isolated from an Indonesian marine sponge, Xestospongia sp. n-hexane extract [17].
Marine plants, though rich in secondary metabolites with variant bioactivities, do not have a remarkable history of use in traditional medicine [18]. However, recent improvements in marine biology and engineering have aided in the investigation and scientific exploration of marine algae to identify and isolate novel compounds [18]. Marine seaweeds live in an environment with uneven circumstances of temperature, light, salinity, nutrients, and contaminants [19]. Therefore, they adapt to changing environmental conditions via production of a wide range of metabolites not found in organisms from terrestrial environments [19]. Hence, compounds extracted from marine seaweeds are an important source for the production of new medicines.
Halimeda macroloba is a widespread green seaweed in wide marine habitats which is always associated with coral reefs and therefore contains high amounts of calcium carbonate, causing it to be classified as a calcified or calcareous algae [20]. Halimeda attains a diversity of secondary metabolites which are still not fully explored, with few reported compounds which proved activity against human, fish, and shrimp pathogenic bacteria [21]. Halimeda macroloba generally grows in complex environmental conditions (relatively high salinity of seawater, high heavy metal content, and much susceptibility to surrounding organisms) [21]. This green seaweed proved antioxidant, antibacterial, and cytotoxic efficacies but has never been investigated against malaria, while other related species such as H. gracilis were [22]. In silico techniques including machine learning and artificial intelligence have significantly advanced during the last 15 years, to the point where they have become an integrated tool in the discovery and development of new therapeutics. These techniques have also facilitated the investigation of natural crude extracts to find out the potential bioactive chemical entity’s complex matrices [3,23].
Consequently, we aimed in the present work to explore the phytochemical profile of the green seaweed Halimeda macroloba and test its possible anti-plasmodial potential via virtual screening and physics-based molecular simulation.
2. Results and Discussion
2.1. Phytochemical Investigation of Halimeda macroloba Seaweed
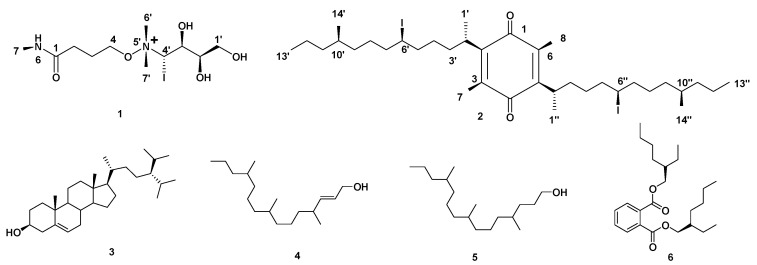
Based on the physicochemical and chromatographic properties, the spectral analyses from UV, 1H, and DEPT-Q NMR, as well as comparisons with the literature and some authentic samples, the crude ethanolic extract of Halimeda macroloba offered two new compounds, 4-O-(4‣-(dimethylamino)-4‣-iodobutan-5‣-yl-1‣,2‣,3‣-triol)-N-methylbutanamide 1, and 2,5-bis(6-iodo-10-methyltridecan-2-yl)-3,6-dimethylcyclohexa-2,5-diene-1,4-dione 2, along with four known compounds named as: 24-isopropyl cholesterol 3 [24], (E)-4,8,12-trimethylpentadec-2-en-1-ol 4 [25], 4,8,12-trimethylpentadecan-1-ol 5 [25], and bis(2-ethylhexyl) phthalate 6 [26] (Figure 1). Compound 3 was isolated here for the first time from the genus Halimeda (Figures S1–S14 and Figure 1).
Figure 1.
Structures of compounds isolated from Halimeda macroloba algae.
Compound 1 (Table 1, Figure 1, see Supplementary Figures S1–S4) was obtained as a yellow amorphous solid. The HRESIMS data for compound 1 showed an adduct pseudo-molecular ion peak at m/z 392.0809 [M + H]+, consistent with the molecular formula C11H25IN2O5 and suggesting one degree of unsaturation. The IR spectrum suggested the presence of one iodide atom at 540 cm−1. The 1H, DEPT-Q, HSQC, and HMBC NMR data (Table 1, Supplementary Figures S1–S4, Figure 1 and Figure 2), showed five characteristic resonances: two methylene groups at δH 2.38 (2H, br. s) δC 34.0, δH 2.11 (2H, br. s) δC 20.3, one oxy-methylene group at δH 3.45 (2H, m) δC 67.1, with one quaternary carbon at δC 180.8 and one downfield methyl group at δH 2.00 (3H, s) δC 24.2, suggesting the characteristic structure of the 4-hydroxy-N-methylbutanamide unit [27].
Table 1.
DEPT-Q (400 MHz) and 1H (100 MHz) NMR data of compounds 1 in CD3OD-d4; Carbon multiplicities were determined by the DEPT-Q experiments.
| δ C | δH (J in Hz) | |
|---|---|---|
| 1 | 180.8, qC | |
| 2 | 34.0, CH2 | 2.38, br. s |
| 3 | 20.3, CH2 | 2.11, br. s |
| 4 | 67.1, CH2 | 3.45, m |
| 5 | ||
| 6 | ||
| 7 | 24.2, CH3 | 2.00, s |
| 1′ | 64.3, CH2 | 3.76, 3.86, overlapped |
| 2′ | 72.1, CH | 3.80, overlapped |
| 3′ | 70.6, CH | 3.91, overlapped |
| 4′ | 54.2, CH | 3.37, overlapped |
| 5′ | ||
| 6′, 7′ | 53.7, 53.7, CH3 | 3.28, s |
qC, quaternary, CH, methine, CH2, methylene, CH3, methyl carbons.
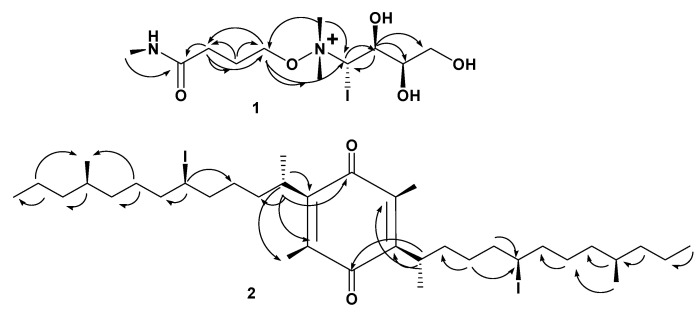
Figure 2.
Selected HMBC ( ) correlations of compounds 1, 2.
) correlations of compounds 1, 2.
Moreover, the 1H, DEPT-Q, HSQC, and HMBC NMR data (Table 1, Supplementary Figures S1–S4, Figure 1 and Figure 2) showed six characteristic resonances: one oxy-methylene group at δH 3.76, 3.86 (2H, overlapped) δC 64.3, two oxy-methine groups at δH 3.80 (1H, overlapped) δC 72.1, δH 3.91 (1H, overlapped) δC 70.6, one iodo-methine group at δH 3.37 (1H, overlapped) δC 54.2, and two methyl groups at δH 3.28 (6H, s) δC 53.7, 53.7, suggesting the characteristic structure of the 4-(dimethylamino)butane-1,2,3-triol [28]. Additionally, the HMBC experiment showed that the characteristic 4J HMBC correlation of proton H-4 δH 3.45 (2H, m) with C-6′, 7′ δC 53.7 confirmed the attachment of the 4-hydroxy-N-methylbutanamide unit of C-4 at N-5′ of the 4-(dimethylamino)-butane-1,2,3-triol unit (Figure 2). The NMR data for C-4′ at δH 3.37 (1H, overlapped) δC 54.2 suggested the attachment of iodide atom at C-4′. The relative stereochemistry of compound 1 was deduced using the J values and a Nuclear Overhauser Effect (NOE) experiment (Table 1). The NOEs observed between H-7, H-2′, H-3′, H-6′, H-7′ suggested that they are in the same plan, while the NOEs observed between H-4′ suggested they are on the other side of the compound skeleton. Accordingly, compound 1 was identified as 4-O-(4’-(dimethylamino)-4’-iodobutan-5’-yl-1’,2’,3’-triol)-N-methylbutanamide.
Compound 2 (Table 2, Figure 1, see Supplementary Figures S5 and S6) was obtained as a white amorphous solid. The HRESIMS data for compound 2 showed an adduct pseudo-molecular ion peak at m/z 781.2919 [M + H]+, consistent with the molecular formula C36H63I2O2 and suggesting 5 degrees of unsaturation. The IR spectrum suggested the presence of an iodide atom at 540 cm−1. The 1H, DEPT-Q, HSQC, and HMBC NMR data (Table 2, Supplementary Figures S5 and S6, Figure 1 and Figure 2) showed eight characteristic resonances: six quaternary carbons at δC 187.8, 188.2, 145.0, 141.1, 141.0, and 140.7, and two methyl groups at δH 2.03, 2.04 (6H, s), and δC 12.5, suggesting the characteristic structure of the 3,6-dimethyl,1,4- benzoquinone unit [29]. The 1H, DEPT-Q, HSQC, and HMBC NMR data (Table 2, Supplementary Figures S5 and S6, Figure 1 and Figure 2) showed 28 characteristic resonances: six methyl groups at δH 1.27, 1.27 (6H, d, J = 4.2) δC 20.2, 20.3, δH 0.88, 0.87 (6H, d, J = 4.7) δC 12.8, 12.9, and δH 1.27, 1.26 (6H, t, J = 6.0, 6.1) δC 23.2, 23.2; sixteen methylene groups at δH 1.10–1.32 (4H, overlapped) δC 37.8, 38.0, δH 1.10–1.32 (4H, overlapped) δC 25.3, 25.3, δH 1.49–1.69 (4H, overlapped) δC 30.1, 30.2, δH 1.49–1.69 (4H, overlapped) δC 30.2, 30.2, δH 1.10–1.32 (4H, overlapped) δC 25.0, 25.0, δH 1.10–1.32 (4H, overlapped) δC 39.9, 39.9, δH 1.10–1.32 (4H, overlapped) δC 40.8, 40.8, δH 1.10–1.32 (4H, overlapped) δC 21.9, 22.0; and six methine groups at δH 2.5 (2H, overlapped) δC 28.5, 28.5, δH 3.66 (2H, m) δC 33.3, 33.4, δH 1.49–1.69 (2H, overlapped) δC 27.1, and 27.1, suggesting the characteristic structure of two units of 4-methyltridecane [16]. Additionally, the HMBC experiment showed the characteristic 2J HMBC correlation of proton H-2′, 2″ δH 2.50 (2H, overlapped) with C-2, 5 δC 145.0, 141.1, and confirmed the attachment of the two 4-methyltridecane units C-2′, C-2″ at C-2, C-5 of 3,6-dimethyl,1,4- benzoquinone unit (Figure 2). The NMR data for C-6′, 6″ at δH 3.66 (2H, overlapped) δC 33.3, 33.4 suggested the attachment of iodide atoms at C-6′, 6″. The NOEs observed between H-1′, H-1″ suggested that they are in the same plan, while the NOEs observed between H-6′, H-14′, H-6″, and H-14″ suggested they are on the other side of the compound skeleton. Accordingly, compound 2 was identified as 2,5-bis(6-iodo-10-methyltridecan-2-yl)-3,6-dimethylcyclohexa-2,5-diene-1,4-dione, which is similar to menzoquinone (isolated from Antarctic macroalgae) in structure, but different from the additional methyl and saturated 4-methyltridecane units at C-3 and 5, respectively [29].
Table 2.
DEPT-Q (400 MHz) and 1H (100 MHz) NMR data of compounds 2 in CDCl3-d; carbon multiplicities were determined by the DEPT-Q experiments.
| Position | δ C | δH (J in Hz) |
|---|---|---|
| 1 | 187.8, qC | |
| 2 | 145.0, qC | |
| 3 | 140.7, qC | |
| 4 | 188.2, qC | |
| 5 | 141.1, qC | |
| 6 | 141.0, qC | |
| 7,8 | 12.5, 12.5, CH3 | 2.03, 2.04, s |
| 1′, 1″ | 20.2, 20.3, CH3 | 1.27, d (4.2) |
| 2′, 2″ | 28.5, 28.5, CH | 2.50, overlapped |
| 3′, 3″ | 37.8, 38.0, CH2 | 1.10–1.32, overlapped |
| 4′, 4″ | 25.3, 25.3, CH2 | 1.10–1.32, overlapped |
| 5′, 5″ | 30.1, 30.2, CH2 | 1.49–1.69, overlapped |
| 6′, 6″ | 33.3, 33.4, CH | 3.66, m |
| 7′, 7″ | 30.2, 30.2, CH2 | 1.49–1.69, overlapped |
| 8′, 8″ | 25.0, 25.0, CH2 | 1.10–1.32, overlapped |
| 9′, 9″ | 39.9, 39.9, CH2 | 1.10–1.32, overlapped |
| 10′, 10″ | 27.1, 27.1, CH | 1.49–1.69, overlapped |
| 11′, 11″ | 40.8, 40.8, CH2 | 1.10–1.32, overlapped |
| 12′, 12′’ | 21.9, 22.0, CH2 | 1.10–1.32, overlapped |
| 13′, 13″ | 12.8, 12.9, CH3 | 0.88, 0.87, d (4.7) |
| 14′, 14″ | 23.2, 23.2, CH3 | 1.27, 1.26, t (6.0, 6.1) |
qC, quaternary, CH, methine, CH2, methylene, CH3, methyl carbons.
2.2. Predicting Possible Biological Activity
In order to predict the probable biological activity of the isolated compounds, we subjected their modeled structures to a machine learning-based virtual screening platform called Prediction of Activity Spectra of Substances (PASS; http://www.way2drug.com/passonline/products.php, accessed on 16 August 2022). This platform utilizes a pharmacophore-based screening algorithm to score the biological activities of query structure. Possible active score (Pa) > 0.5 indicates high probability of being active in the corresponding biological activity category [30]. After submitting all structures of the isolated compounds, compound 2 showed an interesting Pa score (0.892) for the cytochrome-C reductase inhibitor. This enzyme is involved in many essential biological processes in many eukaryotes, particularly in malaria [31,32]. Accordingly, this preliminary in silico screening directs our attention to test both the crude extract of H. macrolaba and the isolated compounds against P. falciparum in vitro. The results indicated that the extract inhibits the replication of P. falciparum in a dose-dependent manner with an IC50 value of 1.8 ± 0.35 µg/mL. Moreover, compound 2 also showed good inhibitory activity with an IC50 value of 3.2 ± 0.23 µg/mL, while compound 6 showed moderate inhibitory activity with an IC50 value of 19.3 ± 0.51 µg/mL (Table 3).
Table 3.
Antimalarial activity of H. macrolaba extract and its isolated compounds 1–6 expressed as IC50 values.
| Compound | IC50 (µg/mL) |
|---|---|
| 1 | >50 |
| 2 | 3.2 ± 0.23 |
| 3 | >50 |
| 4 | >50 |
| 5 | >50 |
| 6 | 19.3 ± 0.51 |
| Crude extract | 1.8 ± 0.35 |
| Chloroquine | 0.022 ± 0.018 |
2.3. Binding Mode Analysis and Absolute Binding Free Energy Calculation
According to the in vitro antimalarial inhibitory activity results, compound 2 is a potential antimalarial agent that probably targets and inhibits the parasite’s cytochrome-C reductase.
To study the binding mode of compound 2 with cytochrome-C reductase, the modeled structure of this compound was docked into the atovaquone-binding site of cytochrome bc1 reductase (PDB code: 4PD4; atovaquone is a quinone-based antimalarial drug) [33]. The resulting binding poses (10 poses) were almost identical with docking poses ranging from −10.83 to −9.94 kcal/mol (Table S1). Subsequently, these poses were subjected to molecular-dynamics simulation-based absolute binding free energy determination (ΔGbinding) to assess the pose with the highest affinity toward the enzyme’s binding site using the free energy perturbation method (FEP) [34]. The second binding pose, with a docking score of −10.52 kcal/mol, was the best in terms of affinity to the binding site, where it attained the lowest ΔGbinding value of −8.33 kcal/mol. Regarding the co-crystalized inhibitor atovaquone, its modeled structure was re-docked into the enzyme active site, and similarly to compound 2, ten poses for the docked structure were generated with docking scores ranging from −9.76 to −9.23 kcal/mol (Table S1). The ΔGbinding of these generated poses was also determined, where the first pose got the lowest value (−14.46 kcal/mol). The calculated RMSD of this pose with co-crystalized inhibitor atovaquone was 1.17 Å.
From the previous findings we can conclude that compound 2 has a comparable affinity with the co-crystalized ligand toward cytochrome-C reductase. Additionally, its lower affinity is comparable to the co-crystalized inhibitor, which might be attributed to its higher flexibility (i.e., higher number of rotatable bonds, 22 vs. 2 for atovaquone).
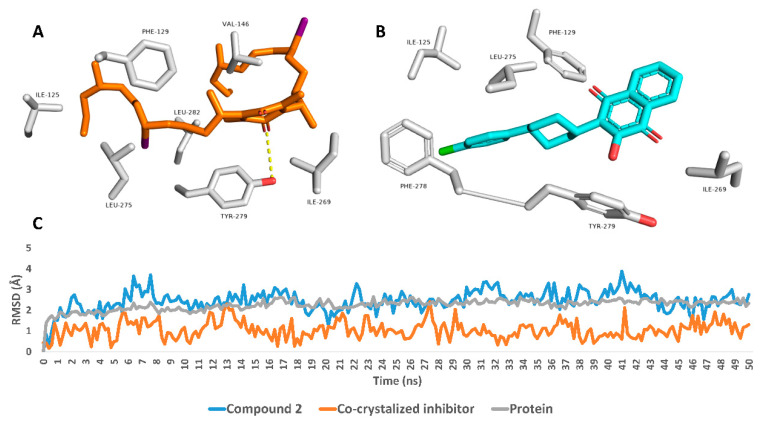
To study the dynamic binding mode of compound 2 and atovaquone inside the enzyme’s active site, their binding poses in terms of ΔGbinding were subjected to 50 ns long MDS. As shown in Figure 3, both structures achieved good binding stability over the course of simulation; however, atovaquone was more stable with an average RMSD of 1.34 Å (the average RMSD of compound 2 was 3.23 Å). The extracted top-populated binding poses of both structures revealed that both their binding and stability inside the binding pocket were achieved primarily via hydrophobic interactions. Atovaquone established hydrophobic interactions mainly with PHE-129, ILE-269, LEU-275, PHE-278, and TYR-279. Regarding compound 2, its structure was larger, more flexible, and richer in hydrophobic hydrocarbon moieties and attained two hydrophobic iodine atoms. Hence, it was able to establish more hydrophobic interactions inside the binding pocket, e.g., ILE-125, PHE-129, VAL-146, ILE-269, LEU-275, TYR-279, and LEU-282, moreover establishing a prominent single H-bond with TYR-279. Taken together, compound 2 can be considered a good scaffold for the future development of new antimalarial agents targeting cytochrome-C. However, this scaffold (i.e., compound 2) is not a drug-like molecule according to Lipinski’s and Veber’s rules of drug-likeness (Molecular weight > 500, LogP > 4.15, and the number of rotatable bonds > 10). The removal of one of the two long aliphatic sidechains can significantly improve the drugability of this scaffold. Accordingly, in our next stage of investigating this probable antimalarial molecule, its biological activity will be fully characterized and its essential structural features will be identified so that its structure can be modified synthetically to a drug-like molecule.
Figure 3.
Binding modes of compound 2 and atovaquone inside the cytochrome bc1’s binding site (PDB ID: 4PD4) (A and B, respectively). These poses were extracted as the top-populated poses from 50 ns MDS experiments. C: RMSDs of compound 2 and atovaquone inside the cytochrome bc1’s binding site over the 50ns long MDS runs.
To the best of our knowledge, this is the first study investigating the phytochemical environment of H. macrolaba with a focus on its anti-plasmodium efficacy. The phytochemical analyses led to the identification of six compounds, with the predominance of aliphatic hydrocarbon moieties as well as strong antimalarial activity (IC50 1.8 ± 0.354 µg/mL). Since bioactive extracts are categorized to be potent as antimalarials when their IC50 values are less than 10 µg/mL, H. macrolaba extract can be considered as a potential source of potent antimalarial agents [35]. Interestingly, a previous machine-learning study reported that the most persuasive physicochemical factor for antimalarial drugs to penetrate red blood cells is protein binding. The less a drug is bound to protein, the more it is freely available to penetrate the red blood cell. Drugs with aromatic hydrocarbons and/or aliphatic hydrocarbons may have a higher amount of freely available drug in the plasma to penetrate the red blood cells, facilitating their pharmacodynamic activities [36]. That might explain why compound 2 [2,5-bis (6-iodo-10-methyltridecan-2-yl)-3,6-dimethylcyclohexa-2,5-diene-1,4-dione], with its high number of aliphatic hydrocarbons, attained the highest binding score toward cytochrome c reductase. Moreover, this compound attains a quinone moiety that strengthens the antimalarial activity, which was supported by the reports of many quinone-derived compounds with potent antimalarial efficacies [37]. In a previous study, physcion and emodin (aromatic quinone derivatives) proved strong antimalarial activities with IC50 values of 0.9 and 1.9 µM, respectively [38]. Moreover, atovaquone is a well-known antimalarial quinone-based compound that is used in combination with proguanil (Malarone®) for the control of malaria infections worldwide [38]. Regarding compound 2, a new compound, this is the first report of a quinone-based aliphatic compound with potent antimalarial efficacy. Accordingly, and in addition to recent related published data, machine learning could be a powerful approach to be widely used in various scientific fields for finding valuable information from data. The aims of a machine-learning model progression can be employed to build a robust predictive model, especially as most antimalarial drugs are still orphan [36] and data about their safety are limited. More importantly, the target prediction software has become an integral part of the drug discovery platform, which reduces the time and effort required for screening huge libraries of chemical compounds to find out available drug candidates. Such in silico tools could be employed in drug discovery from natural sources, with the ability to prioritize possibly active metabolites in a natural extract; hence, isolation and identification efforts will be directed toward top-scoring candidates.
3. Materials and Methods
3.1. Seaweed Samples Collection and Identification
Specimens of the green seaweed Halimeda macroloba were collected during May 2021 from the littoral zone of shorelines in Savage City on the Red Sea coast, Egypt. The collected seaweed samples were washed well with seawater, then with tap water, and finally with distilled water to remove any impurities, sand particles, and salts on their surfaces. The samples were kept in sterile clean plastic bottles and transported chilled in an ice box to the laboratory. The specimens were kindly identified according to standard taxonomic keys, and a voucher specimen (2021-BuPD 82) was deposited at the Department of Pharmacognosy, Faculty of Pharmacy, Beni-Suef University, Egypt.
3.2. Chemicals and Reagents
The solvents employed in this work included n-hexane (n-hex., boiling point b.p. 60–80 °C), dichloromethane (DCM), ethyl acetate (EtOAC), ethanol, and methanol (MeOH), all purchased from El-Nasr Company for Pharmaceuticals and Chemicals (Cairo, Egypt). Deuterated solvents used for chromatographic and spectroscopic analyses were purchased from Sigma-Aldrich (Saint Louis, MO, USA), including methanol-d4 (CD3OD-d4), dimethyl sulfoxide-d6 (DMSO-d6), and chloroform-d (CDCL3-d). Column chromatography (CC) was performed using silica gel 60 (63–200 μm, E. Merck, Sigma-Aldrich), and silica gel GF254 for thin-layer chromatography (TLC) (El-Nasr Company for Pharmaceuticals and Chemicals, Cairo, Egypt) was employed for vacuum liquid chromatography (VLC). Thin-layer chromatography (TLC) was carried out using pre-coated silica gel 60 GF254 plates (E. Merck, Darmstadt, Germany; 20 cm × 20 cm, 0.25 mm in thickness). Spots were visualized by spraying with dragendorff reagent (DRR) (solution a: 0.85 g basic bismuth nitrate in 10 mL glacial acetic acid and 40 mL water under heating, solution b: 8 g potassium iodide in 30 mL water, then mix a + b (1:1), then 1 ml of the mixture was mixed with 2 mL glacial acetic acid and 10 mL water) and para-anisaldehyde (PAA) reagent (85:5:10:0.5 absolute EtOH:sulfuric acid:G.A.A.:para-anisaldehyde), followed by heating at 110 °C [39].
3.3. Spectral Analyses
Proton 1H and Distortionless Enhancement by Polarization Transfer-Q (DEPT-Q) 13C NMR spectra were recorded at 400 and 100 MHz, respectively. Tetramethylsilane (TMS) was used as an internal standard in CDCL3-d and CD3OD-d4, utilizing residual solvent peaks (δH = 7.26; and δC 77.2) and (δH = 3.34; 4.78; and δC 49.9) as references, respectively. Measurements were performed on a Bruker Advance III 400 MHz with BBFO Smart Probe and a Bruker 400 MHz EON Nitrogen-Free Magnet (Bruker AG, Billerica, MA, USA). Carbon multiplicities were determined using a DEPT-Q experiment. The ultraviolet radiation (UV) spectrum in methanol was obtained using a Shimadzu UV 2401PC spectrophotometer (Shimadzu Corporation-UV-2401PC/UV-2501PC, Kyoto, Japan). Infrared (IR) spectra were measured using a Jasco FTIR 300E infrared spectrophotometer. HRESIMS data were obtained using an Acquity Ultra Performance Liquid Chromatography system coupled with a Synapt G2 HDMS quadrupole time-of-flight hybrid mass spectrometer (Waters, Milford, MA, USA).
3.4. Extraction and Fractionation of Halimeda Macroloba
The Halimeda macroloba samples (0.25 kg) were collected and air-dried in the shade for one month, dried, then finely powdered using an OC-60B/60B grinding machine (60–120 mesh, Henan, Mainland China) [40,41]. The powder was extracted by maceration using 70% ethanol (5 L, 3×, seven days each) at room temperature and concentrated under vacuum at 45 °C using a rotary evaporator (Buchi Rotavapor R-300, Cole-Parmer, Vernon Hills, IL, USA) to afford 50 g crude extract [42,43]. The dry extract was suspended in 100 mL distilled water (H2O), and successively portioned with solvents of different polarities (n-Hex., DCM). The organic phase in each step was separately evaporated under reduced pressure to afford the corresponding fractions I (5.0 g) and II (25.0 g), respectively, while the remaining mother liquor was then concentrated down to give the aqueous fraction (20.0 g). All the resulting fractions were kept at 4 °C for biological and phytochemical investigations [44,45].
3.5. Isolation and Purification of Compounds
Fractions I and II (30 g) were gathered according to TLC, then subjected to VLC fractionation using silica gel GF254 (column 6 cm × 30 cm, 100 g). Elution was performed using DCM:EtoAc gradient mixtures in order of increasing polarities (0–20%, 1%, 250 mL each). The effluents from the column were collected in fractions (250 mL each), and each fraction was concentrated and monitored by TLC using the system DCM:EtoAc 9.5:0.5 and PAA reagent. Similar fractions were grouped and concentrated to provide five sub-fractions (I1–I5); each one was further purified on a Sephadex LH20 column (0.25–0.1 mm, 100 cm × 0.5 cm, 100 gm) and eluted with MeOH to afford compound 2 (20 mg), 3 (7 mg), 4 (10 mg), 5 (7 mg), and 6 (10 mg). Finally, the liquid fraction (5 g) was further purified on a Sephadex LH20 column (0.25–0.1 mm, 100 cm × 0.5 cm, 100 gm), where it was eluted with MeOH to afford compound 1 (2 g).
4-O-(4′-(dimethylamino)-4′-iodobutan-5′-yl-1′,2′,3′-triol)-N-methylbutanamide (1): yellow powder; [UV (MeOH) λmax (logε) 280 (5.5), 270 (6.0), 300 (4.5) nm; IR υmax (KBr) 3600, 3500, 3400, 3100, 1640, 1550, 1350, 1300, 1000, 540 cm−1; NMR data; see Table 1; HRESIMS m/z 392.0809 [M + H]+ (calc. for C11H25IN2O5, 392.0808).
2,5-bis(6-iodo-10-methyltridecan-2-yl)-3,6-dimethylcyclohexa-2,5-diene-1,4-dione (2): white powder; [UV (MeOH) λmax (logε) 282 (5.5), 273 (6.0), 306 (4.5) nm; IR υmax (KBr) 3100, 1720, 1670, 1465, 540 cm−1; NMR data; see Table 2; HRESIMS m/z 781.2919 [M + H]+ (calc. for C36H63I2O2, 781.2917).
3.6. Antimalarial Assay
The anti-plasmodial effect of the seaweed extract on P. falciparum erythrocytic replication in vitro was investigated using the Malstat assay [3]. The details of this method can be found in the supplementary file.
3.7. Biological Activity Prediction
Prediction of the probable biological activities of the isolated compounds was carried out using the (www.way2drug.com, accessed on 16 August 2022) [46]. The full details of this virtual screening method are provided in the supplementary file.
3.8. Molecular Docking, Δgbinding Calculation, and Molecular Dynamics Simulation
Docking, ΔGbinding calculation, and molecular dynamics simulation experiments were carried out as previously described [47]. The detailed methods can be found in the supplementary file.
4. Conclusions
Phytochemical investigation of the green seaweed Halimeda macroloba led to the isolation of two new compounds and four previously reported ones. The compounds’ structures were confirmed using 1D, 2D NMR, and HRESIMS analyses. The cytochrome-C enzyme, the critical target for the malaria parasite, was identified as a possible target for compound 2 after a machine learning-based virtual screening. The mode of interaction of compound 2′s scaffold inside the active site of cytochrome-C was putatively determined using comprehensive molecular docking and MDS experiments. In addition, compound 2′s affinity to the active site was calculated in terms of absolute binding free energy (ΔGbinding = −14.46 kcal/mol). In vitro against P. falciparum showed the potential inhibitory activity of compound 2 against the parasite. Accordingly, compound 2’s scaffold can be considered as a promising lead compound for future antimalarial drug development.
Acknowledgments
The authors would like to thank the Deanship of Scientific Research at Umm Al-Qura University for supporting this work by Grant Code: 22UQU4331312DSR05. The authors would like to thank Deraya University for the use of its laboratory space.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/molecules27175617/s1, Figure S1: 1H NMR spectrum of compound 1 measured in CD3OD-d4 at 400 MHz; Figure S2: DEPT-Q NMR spectrum of compound 1 measured in CD3OD-d4 at 100 MHz, Figure S3: HSQC spectrum of compound 1 measured in CD3OD-d4; Figure S4: HMBC spectrum of compound 1 measured in CD3OD-d4; Figure S5: 1H NMR spectrum of compound 2 measured in CDCL3-d at 400 MHz; Figure S6: DEPT-Q NMR spectrum of compound 2 measured in CDCL3-d at 100 MHz; Figure S7: 1H NMR spectrum of compound 3 measured in CDCL3-d at 400 MHz; Figure S8: DEPT-Q NMR spectrum of compound 3 measured in CDCL3-d at 100 MHz; Figure S9: 1H NMR spectrum of compound 4 measured in CDCL3-d at 400 MHz; Figure S10: DEPT-Q NMR spectrum of compound 4 measured in CDCL3-d at 100 MHz; Figure S11: 1H NMR spectrum of compound 5 measured in CDCL3-d at 400 MHz; Figure S12: DEPT-Q NMR spectrum of compound 5 measured in CDCL3-d at 100 MHz; Figure S13: 1H NMR spectrum of compound 5 measured in CDCL3-d at 400 MHz; Figure S14: DEPT-Q NMR spectrum of compound 5 measured in CDCL3-d at 100 MHz. References [48,49,50,51,52,53,54] are cited in the Supplementary Materials.
Author Contributions
Conceptualization: A.H.E., E.M.Z., U.R.A. and G.P.; methodology: A.H.E., A.M.S., E.M.Z., C.J.N., H.S., F.A. and A.A.; software: U.R.A. and A.M.S.; formal analysis: R.S. and A.A.; investigation: U.R.A., A.M.S. and H.S.; resources: G.P., R.S. and A.A.; data curation: A.H.E., A.M.S., E.M.Z., C.J.N., F.A. and A.A.; writing—original draft: A.H.E. and A.M.S.; writing—review and editing: all authors; supervision: U.R.A.; project administration: U.R.A., R.S. and A.A.; funding acquisition: U.R.A., R.S. and A.A. All the authors have read and agreed to the published version of the manuscript. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
The authors would like to thank the Deanship of Scientific Research at Umm Al-Qura University for supporting this study.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mohamad S.A., Zahran E.M., Fadeel M.R.A., Albohy A., Safwat M.A. New acaciin-loaded self-assembled nanofibers as mpro inhibitors against bcv as a surrogate model for SARS-CoV-2. Int. J. Nanomed. 2021;16:1789. doi: 10.2147/IJN.S298900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alhadrami H.A., Thissera B., Hassan M.H., Behery F.A., Ngwa C.J., Hassan H.M., Pradel G., Abdelmohsen U.R., Rateb M.E. Bio-Guided Isolation of Antimalarial Metabolites from the Coculture of Two Red Sea Sponge-Derived Actinokineospora and Rhodococcus spp. Mar. Drugs. 2021;19:109. doi: 10.3390/md19020109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ibrahim S.R., Mohamed G.A., Al Haidari R.A., El-Kholy A.A., Zayed M.F. Potential anti-malarial agents from endophytic fungi: A review. Mini-Rev. Med. Chem. 2018;18:1110–1132. doi: 10.2174/1389557518666180305163151. [DOI] [PubMed] [Google Scholar]
- 4.Abdelmohsen U.R., Balasubramanian S., Oelschlaeger T.A., Grkovic T., Pham N.B., Quinn R.J., Hentschel U. Potential of marine natural products against drug-resistant fungal, viral, and parasitic infections. Lancet Infect. Dis. 2017;17:e30–e41. doi: 10.1016/S1473-3099(16)30323-1. [DOI] [PubMed] [Google Scholar]
- 5.Osei E., Kwain S., Mawuli G.T., Anang A.K., Owusu K.B.-A., Camas M., Camas A.S., Ohashi M., Alexandru-Crivac C.-N., Deng H. Paenidigyamycin A, potent antiparasitic imidazole alkaloid from the Ghanaian Paenibacillus sp. DE2SH. Mar. Drugs. 2018;17:9. doi: 10.3390/md17010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parra L.L., Bertonha A.F., Severo I.R., Aguiar A.C., de Souza G.E., Oliva G., Guido R.V., Grazzia N., Costa T.R., Miguel D.C. Isolation, derivative synthesis, and structure–activity relationships of antiparasitic bromopyrrole alkaloids from the marine sponge Tedania brasiliensis. J. Nat. Prod. 2018;81:188–202. doi: 10.1021/acs.jnatprod.7b00876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kurimoto S.-i., Ohno T., Hokari R., Ishiyama A., Iwatsuki M., Ōmura S., Kobayashi J.i., Kubota T. Ceratinadins E and F, new bromotyrosine alkaloids from an Okinawan marine sponge Pseudoceratina sp. Mar. Drugs. 2018;16:463. doi: 10.3390/md16120463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kleks G., Duffy S., Lucantoni L., Avery V.M., Carroll A.R. Orthoscuticellines A–E, β-Carboline Alkaloids from the Bryozoan Orthoscuticella ventricosa Collected in Australia. J. Nat. Prod. 2020;83:422–428. doi: 10.1021/acs.jnatprod.9b00933. [DOI] [PubMed] [Google Scholar]
- 9.Smyrniotopoulos V., Merten C., Kaiser M., Tasdemir D. Bifurcatriol, a new antiprotozoal acyclic diterpene from the brown alga Bifurcaria bifurcata. Mar. Drugs. 2017;15:245. doi: 10.3390/md15080245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ju E., Latif A., Kong C.-S., Seo Y., Lee Y.-J., Dalal S.R., Cassera M.B., Kingston D.G. Antimalarial activity of the isolates from the marine sponge Hyrtios erectus against the chloroquine-resistant Dd2 strain of Plasmodium falciparum. Z. Nat. C. 2018;73:397–400. doi: 10.1515/znc-2018-0025. [DOI] [PubMed] [Google Scholar]
- 11.Qin G.-F., Tang X.-L., Sun Y.-T., Luo X.-C., Zhang J., Van Ofwegen L., Sung P.-J., Li P.-L., Li G.-Q. Terpenoids from the soft coral Sinularia sp. collected in Yongxing Island. Mar. Drugs. 2018;16:127. doi: 10.3390/md16040127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwasaki A., Tadenuma T., Sumimoto S., Shiota I., Matsubara T., Saito-Nakano Y., Nozaki T., Sato T., Suenaga K. Hoshinoamides A and B, acyclic lipopeptides from the marine cyanobacterium Caldora penicillata. J. Nat. Prod. 2018;81:2545–2552. doi: 10.1021/acs.jnatprod.8b00643. [DOI] [PubMed] [Google Scholar]
- 13.Sweeney-Jones A.M., Gagaring K., Antonova-Koch J., Zhou H., Mojib N., Soapi K., Skolnick J., McNamara C.W., Kubanek J. Antimalarial peptide and polyketide natural products from the Fijian marine cyanobacterium Moorea producens. Mar. Drugs. 2020;18:167. doi: 10.3390/md18030167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keller L., Siqueira-Neto J.L., Souza J.M., Eribez K., LaMonte G.M., Smith J.E., Gerwick W.H. Palstimolide A: A complex polyhydroxy macrolide with antiparasitic activity. Molecules. 2020;25:1604. doi: 10.3390/molecules25071604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shao C.-L., Mou X.-F., Cao F., Spadafora C., Glukhov E., Gerwick L., Wang C.-Y., Gerwick W.H. Bastimolide B, an antimalarial 24-membered marine macrolide possessing a tert-butyl group. J. Nat. Prod. 2018;81:211–215. doi: 10.1021/acs.jnatprod.7b00917. [DOI] [PubMed] [Google Scholar]
- 16.Meesala S., Gurung P., Karmodiya K., Subrayan P., Watve M.G. Isolation and structure elucidation of halymeniaol, a new antimalarial sterol derivative from the red alga Halymenia floresii. J. Asian Nat. Prod. Res. 2018;20:391–398. doi: 10.1080/10286020.2017.1342636. [DOI] [PubMed] [Google Scholar]
- 17.Murtihapsari M., Salam S., Kurnia D., Darwati D., Kadarusman K., Abdullah F.F., Herlina T., Husna M.H., Awang K., Shiono Y. A new antiplasmodial sterol from Indonesian marine sponge, Xestospongia sp. Nat. Prod. Res. 2021;35:937–944. doi: 10.1080/14786419.2019.1611815. [DOI] [PubMed] [Google Scholar]
- 18.Zahran E.M., Sayed A.M., Abdelwahab M.F., Albohy A., Abdulrazik B.S., Ibrahim A.M., Bringmann G., Abdelmohsen U.R. Identifying the specific-targeted marine cerebrosides against SARS-CoV-2: An integrated computational approach. RSC Adv. 2021;11:36042–36059. doi: 10.1039/D1RA07103C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zahran E.M., Albohy A., Khalil A., Ibrahim A.H., Ahmed H.A., El-Hossary E.M., Bringmann G., Abdelmohsen U.R. Bioactivity potential of marine natural products from Scleractinia-associated microbes and in silico anti-SARS-CoV-2 evaluation. Mar. Drugs. 2020;18:645. doi: 10.3390/md18120645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hillis-Colinvaux L. Advances in Marine Biology. Volume 17. Elsevier; Amsterdam, The Netherlands: 1980. Ecology and taxonomy of Halimeda: Primary producer of coral reefs; pp. 1–327. [DOI] [Google Scholar]
- 21.Latifah L.A., Soekamto N.H., Tahir A. Green Algae Halimeda macroloba in Spermonde Archipelago: Phytochemical and In Vitro Antibacterial Studies. Pharmacogn. J. 2020;12:1000–1004. doi: 10.5530/pj.2020.12.141. [DOI] [Google Scholar]
- 22.Suganya S., Ishwarya R., Jayakumar R., Govindarajan M., Alharbi N., Kadaikunnan S., Khaled J., Al-Anbr M., Vaseeharan B. New insecticides and antimicrobials derived from Sargassum wightii and Halimeda gracillis seaweeds: Toxicity against mosquito vectors and antibiofilm activity against microbial pathogens. S. Afr. J. Bot. 2019;125:466–480. doi: 10.1016/j.sajb.2019.08.006. [DOI] [Google Scholar]
- 23.Orfali R., Rateb M.E., Hassan H.M., Alonazi M., Gomaa M.R., Mahrous N., GabAllah M., Kandeil A., Perveen S., Abdelmohsen U.R. Sinapic acid suppresses SARS-CoV-2 replication by targeting its envelope protein. Antibiotics. 2021;10:420. doi: 10.3390/antibiotics10040420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dai J., Sorribas A., Yoshida W.Y., Kelly M., Williams P.G. Topsentinols, 24-isopropyl steroids from the marine sponge Topsentia sp. J. Nat. Prod. 2010;73:1597–1600. doi: 10.1021/np100374b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Handley J.T. Ph.D. Thesis. University of Tasmania; Tasmania, Australia: 2003. Studies of the Secondary Metabolite Content of Tasmanian Caulerpa Species. [Google Scholar]
- 26.Lotfy M.M., Hassan H.M., Hetta M.H., El-Gendy A.O., Mohammed R. Di-(2-ethylhexyl) Phthalate, a major bioactive metabolite with antimicrobial and cytotoxic activity isolated from River Nile derived fungus Aspergillus awamori. Beni-Suef Univ. J. Basic Appl. Sci. 2018;7:263–269. doi: 10.1016/j.bjbas.2018.02.002. [DOI] [Google Scholar]
- 27.Oliferenko P.V., Oliferenko A.A., Poda G.I., Osolodkin D.I., Pillai G.G., Bernier U.R., Tsikolia M., Agramonte N.M., Clark G.G., Linthicum K.J. Correction: Promising Aedes aegypti Repellent Chemotypes Identified through Integrated QSAR, Virtual Screening, Synthesis, and Bioassay. PLoS ONE. 2014;9:e64547. doi: 10.1371/annotation/1dc83f26-8045-4b63-8a86-24c7b71a7f79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith H.E., Warren M.E., Ingersoll A.W. Optically Active Amines. I. N-Isopropylidene Derivatives of Optically Active Open Chain Primary Amines and Their Rotatory Powers. J. Am. Chem. Soc. 1962;84:1513–1514. doi: 10.1021/ja00867a053. [DOI] [Google Scholar]
- 29.Ankisetty S., Nandiraju S., Win H., Park Y.C., Amsler C.D., McClintock J.B., Baker J.A., Diyabalanage T.K., Pasaribu A., Singh M.P. Chemical investigation of predator-deterred macroalgae from the Antarctic Peninsula. J. Nat. Prod. 2004;67:1295–1302. doi: 10.1021/np049965c. [DOI] [PubMed] [Google Scholar]
- 30.Filimonov D., Lagunin A., Gloriozova T., Rudik A., Druzhilovskii D., Pogodin P., Poroikov V. Prediction of the biological activity spectra of organic compounds using the PASS online web resource. Chem. Heterocycl. Compd. 2014;50:444–457. doi: 10.1007/s10593-014-1496-1. [DOI] [Google Scholar]
- 31.Krungkrai J. The multiple roles of the mitochondrion of the malarial parasite. Parasitology. 2004;129:511–524. doi: 10.1017/S0031182004005888. [DOI] [PubMed] [Google Scholar]
- 32.Charlwood D. Malaria Drug Trials–An Ethical Dilemma? Parasitol. Today. 1999;15:346. doi: 10.1016/S0169-4758(99)01492-1. [DOI] [PubMed] [Google Scholar]
- 33.Kaur K., Jain M., Reddy R.P., Jain R. Quinolines and structurally related heterocycles as antimalarials. Eur. J. Med. Chem. 2010;45:3245–3264. doi: 10.1016/j.ejmech.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 34.Ngo S.T., Nguyen H.M., Quan P.M., Truong V.K., Tung N.T., Vu V.V. Assessing potential inhibitors of SARS-CoV-2 main protease from available drugs using free energy perturbation simulations. RSC Adv. 2020;10:40284–40290. doi: 10.1039/D0RA07352K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Endang A.S., Alim I., Tjut S.D., Raden W.N. Antimalarial Activity of Microalgae Extracts Based on Inhibition of PfMQO, a Mitochondrial Plasmodium falciparum Enzyme. Pharmacogn. J. 2019;11:1477–1482. doi: 10.5530/pj.2019.11.228. [DOI] [Google Scholar]
- 36.Pornputtapong N., Suriyapakorn B., Satayamapakorn A., Larpadisorn K., Janviriyakul P., Khemawoot P. In silico analysis for factors affecting anti-malarial penetration into red blood cells. Malar. J. 2020;19:215. doi: 10.1186/s12936-020-03280-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patel O.P.S., Beteck R.M., Legoabe L.J. Antimalarial application of quinones: A recent update. Eur. J. Med. Chem. 2021;210:113084. doi: 10.1016/j.ejmech.2020.113084. [DOI] [PubMed] [Google Scholar]
- 38.Alhadrami H.A., Sayed A.M., El-Gendy A.O., Shamikh Y.I., Gaber Y., Bakeer W., Sheirf N.H., Attia E.Z., Shaban G.M., Khalifa B.A. A metabolomic approach to target antimalarial metabolites in the Artemisia annua fungal endophytes. Sci. Rep. 2021;11:2770. doi: 10.1038/s41598-021-82201-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stahl E. Thin-Layer Chromatography. Springer; Berlin, Germany: 1965. A Laboratory Handbook. [Google Scholar]
- 40.Alzarea S.I., Elmaidomy A.H., Saber H., Musa A., Al-Sanea M.M., Mostafa E.M., Hendawy O.M., Youssif K.A., Alanazi A.S., Alharbi M. Potential anticancer lipoxygenase inhibitors from the red sea-derived brown algae sargassum cinereum: An in-silico-supported In-Vitro Study. Antibiotics. 2021;10:416. doi: 10.3390/antibiotics10040416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elmaidomy A.H., Alhadrami H.A., Amin E., Aly H.F., Othman A.M., Rateb M.E., Hetta M.H., Abdelmohsen U.R., Hassan H.M. Anti-inflammatory and antioxidant activities of terpene-and polyphenol-rich Premna odorata leaves on alcohol-inflamed female wistar albino rat liver. Molecules. 2020;25:3116. doi: 10.3390/molecules25143116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elmaidomy A.H., Abdelmohsen U.R., Alsenani F., Aly H.F., Shams S.G.E., Younis E.A., Ahmed K.A., Sayed A.M., Owis A.I., Afifi N. The anti-Alzheimer potential of Tamarindus indica: An in vivo investigation supported by in vitro and in silico approaches. RSC Adv. 2022;12:11769–11785. doi: 10.1039/D2RA01340A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Al-Warhi T., Zahran E.M., Selim S., Al-Sanea M.M., Ghoneim M.M., Maher S.A., Mostafa Y.A., Alsenani F., Elrehany M.A., Almuhayawi M.S. Antioxidant and Wound Healing Potential of Vitis vinifera Seeds Supported by Phytochemical Characterization and Docking Studies. Antioxidants. 2022;11:881. doi: 10.3390/antiox11050881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elmaidomy A.H., Mohyeldin M.M., Ibrahim M.M., Hassan H.M., Amin E., Rateb M.E., Hetta M.H., El Sayed K.A. Acylated iridoids and rhamnopyranoses from premna odorata (lamiaceae) as novel mesenchymal–epithelial transition factor receptor inhibitors for the control of breast cancer. Phytother. Res. 2017;31:1546–1556. doi: 10.1002/ptr.5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alnusaire T.S., Sayed A.M., Elmaidomy A.H., Al-Sanea M.M., Albogami S., Albqmi M., Alowaiesh B.F., Mostafa E.M., Musa A., Youssif K.A. An In Vitro and In Silico Study of the Enhanced Antiproliferative and Pro-Oxidant Potential of Olea europaea L. cv. Arbosana Leaf Extract via Elastic Nanovesicles (Spanlastics) Antioxidants. 2021;10:1860. doi: 10.3390/antiox10121860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kc G.B., Bocci G., Verma S., Hassan M.M., Holmes J., Yang J.J., Sirimulla S., Oprea T.I. A machine learning platform to estimate anti-SARS-CoV-2 activities. Nat. Mach. Intell. 2021;3:527–535. doi: 10.1038/s42256-021-00335-w. [DOI] [Google Scholar]
- 47.Albohy A., Zahran E.M., Abdelmohsen U.R., Salem M.A., Al-Warhi T., Al-Sanea M.M., Abelyan N., Khalil H.E., Desoukey S.Y., Fouad M.A. Multitarget in silico studies of Ocimum menthiifolium, family Lamiaceae against SARS-CoV-2 supported by molecular dynamics simulation. J. Biomol. Struct. Dyn. 2022;40:4062–4072. doi: 10.1080/07391102.2020.1852964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lagunin A., Stepanchikova A., Filimonov D., Poroikov V. PASS: Prediction of activity spectra for biologically active substances. Bioinformatics. 2000;16:747–748. doi: 10.1093/bioinformatics/16.8.747. [DOI] [PubMed] [Google Scholar]
- 49.Seeliger D., de Groot B.L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput. Aided Mol. Des. 2010;24:417–422. doi: 10.1007/s10822-010-9352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bowers K.J., Chow D.E., Xu H., Dror R.O., Eastwood M.P., Gregersen B.A., Klepeis J.L., Kolossvary I., Moraes M.A., Sacerdoti F.D., et al. Scalable algorithms for molecular dynamics simulations on commodity clusters; Proceedings of the SC’06: Proceedings of the 2006 ACM/IEEE Conference on Supercomputing; Tampa, FL, USA. 11–17 November 2006; New York, NY, USA: IEEE; 2006. p. 43. [Google Scholar]
- 51.Release S. 3: Desmond Molecular Dynamics System, DE Shaw Research, New York, NY, 2017. Maestro-Desmond Interoperability Tools, Schrödinger; New York, NY, USA: 2017. [Google Scholar]
- 52.Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R.D., Kalé L., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim S., Oshima H., Zhang H., Kern N.R., Re S., Lee J., Rous B., Sugita Y., Jiang W., Im W. CHARMM-GUI free energy calculator for absolute and relative ligand solvation and binding free energy simulations. J. Chem. Theory Comput. 2020;16:7207–7218. doi: 10.1021/acs.jctc.0c00884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ngo S.T., Tam N.M., Quan P.M., Nguyen T.H. Benchmark of Popular Free Energy Approaches Revealing the Inhibitors Binding to SARS-CoV-2 Mpro. J. Chem. Inf. Model. 2021;61:2302–2312. doi: 10.1021/acs.jcim.1c00159. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.