Abstract
A transposition mutant of Staphylococcus aureus was selected from the parent strain MT23142, a derivative of strain 8325. The site of transposition was near the 5′ terminus of the gene arlS. ArlS exhibits strong similarities with histidine protein kinases. Sequence analysis suggested that arlS forms an operon with upstream gene arlR. The predicted product of arlR is a member of the OmpR-PhoB family of response regulators. The arlS mutant formed a biofilm on a polystyrene surface unlike the parent strain and the complemented mutant. Biofilm formation was associated with increased primary adherence to polystyrene, whereas cellular adhesion was only slightly decreased. In addition, the arlS mutant exhibited increased autolysis and altered peptidoglycan hydrolase activity compared to the parental strain and to the complemented mutant. As it has been shown for coagulase-negative staphylococci that some autolysins are able to bind polymer surfaces, these data suggest that the two-component regulatory system ArlS-ArlR may control attachment to polymer surfaces by affecting secreted peptidoglycan hydrolase activity. Finally, the arlS mutant showed a dramatic decrease of extracellular proteolytic activity, including serine protease activity, in comparison to the wild-type strain and the complemented mutant, and cells grown in the presence of phenylmethylsulfonyl fluoride (a serine protease inhibitor) showed an increased autolysin activity. Since the locus arlR-arlS strikingly modifies extracellular proteolytic activity, this locus might also be involved in the virulence of S. aureus.
Staphylococcus aureus is a major pathogen in human infectious diseases. It appears to differ considerably from the coagulase-negative staphylococci (CoNS), such as Staphylococcus epidermidis and Staphylococcus saprophyticus, with respect to its pathogenicity in that it elaborates a large number of extracellular virulence factors and other proteins, while the CoNS, which are usually more indolent pathogens, generally produce few if any of these factors. CoNS cause infections associated with implanted foreign bodies such as intravascular catheters. Previous reports of adherence by staphylococci have concentrated primarily on the CoNS (8, 24, 33). However, an essential step in the establishment of any staphylococcal infection is attachment to the host tissues themselves or to foreign bodies within host tissues.
Environmental stimuli have been shown to play an important role in expression of virulence genes during pathogenesis. Because S. aureus has the ability to invade different tissues, it responds to environmental stimuli in order to survive and express virulence genes during the infection. To survive, the cells must monitor external conditions and adjust their structure, physiology, and behavior accordingly. Thus, bacteria have developed sophisticated signaling systems for eliciting adaptive responses to their environment. The molecular mechanisms responsible for stimulus-response coupling often involve two types of components encoded by an operon, a transmembrane sensor (histidine protein kinase) and its associated response regulator (50). In general, the transmembrane protein binds a specific ligand, the signal, and binding triggers autophosphorylation at a conserved histidine residue. The phosphorylated sensor then transfers the phosphate to an aspartic residue in the response regulator (39). The response regulator can in turn enhance or repress transcription of target genes, as occurs with PhoB in the phosphate regulon (28) and OmpR in the porin regulon (27, 55) of Escherichia coli. A two-component regulatory system, AgrC-AgrA, involved in the production of virulence factors was previously described for S. aureus (21, 37, 43). This locus has also been shown to affect autolysin expression (16). Peptidoglycan hydrolases, enzymes that hydrolyze either the glycan or the peptide moieties of peptidoglycan, participate in a number of important biological processes occurring during cell growth and division, including cell wall synthesis, daughter cell separation, and peptidoglycan turnover and recycling (1). As these hydrolases are also involved in lysis of bacteria, some have been called autolysins. Thus, the same two-component regulatory system can be involved in pathogenesis as well as in cellular physiology.
In order to find loci involved in the regulation of the multidrug efflux pump NorA of S. aureus, we used a library of Tn917LTV1 insertions in the chromosome of strain MT23142 with selection for higher and lower levels of resistance to tetraphenylphosphonium bromide (TPP), a substrate of NorA, than are found in MT23142. Overproduction of norA leads to resistance to drugs such as quinolones, ethidium bromide, and TPP (14, 35). MT23142 carries the flqB mutation. flqB, a cis-acting mutant of norA, is localized downstream of the initiation site of norA and overexpresses norA (35). One transposition mutant, BF15, obtained from the library showed a slight increase of resistance to TPP and modifies norA expression (14).
In addition to effects on norA expression, the mutant unexpectedly showed alterations in autolysis. In order to characterize this mutant, we analyzed the sequence of chromosomal DNA flanking the insertional site of Tn917LTV1. We found a new two-component regulatory system, ArlS-ArlR. Insertion of Tn917 in the arlS gene resulted in changes in adhesive properties, the rate of autolysis, and extracellular proteolytic activity. All these properties were complemented by the cloned arlR and arlS genes, indicating that the disruption of arlS was responsible for the mutant phenotype.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The S. aureus strains used in this study are listed in Table 1. Staphylococci were cultivated in Trypticase soy broth (TSB) at 37°C unless otherwise stated. E. coli cells were grown in Luria-Bertani medium. Cloning of S. aureus chromosomal DNA fragments was performed in E. coli DH5α (Gibco-BRL) using plasmid pGB2 (11).
TABLE 1.
S. aureus strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| ISP794 | 8325 pig-131 | 49 |
| MT23142 | 8325 pig-131 flqB | 35 |
| BF15 | 8325 pig-131 flqB arlS::Tn917LTV1 | 14 |
| BF16 | 8325 pig-131 arlS::Tn917LTV1 | This study |
| BF17 | 8325 pig-131 arlS::Tn917LTV1 complemented | This study |
| SH108 | 8325-4 atl::lacZ | 13 |
| BF18 | 8325-4 atl::lacZ arlS::Tn917LTV1 | This study |
Transposon mutagenesis.
Plasmid pLTV1 (6), which contains a derivative of Tn917, was introduced into S. aureus MT23142 at the permissive temperature (30°C). For generation of a library of transposon insertion mutants, a single colony was grown overnight at 30°C in TSB containing 1 μg of erythromycin per ml and 25 μg of lincomycin per ml (42). Five hundred milliliters of TSB was inoculated with 1 ml of the overnight culture, and the cells were grown to stationary phase at 42°C. The cells were centrifuged, resuspended in 5 ml of TSB, and frozen in aliquots at −70°C. The library was screened by plating on Trypticase soy agar containing 13 μg of TPP per ml. One mutant, resistant to TPP, was isolated and named BF15. Southern blot analysis of EcoRI-, HindIII-, MscI-, or HpaI-digested chromosomal DNA from BF15, using as a probe the cat gene of Tn917 (42), showed that BF15 contained a single transposon insertion (data not shown).
DNA manipulations.
DNA manipulations, Southern hybridization, and transformation of CaCl2-competent E. coli cells were performed using standard procedures (46). Plasmid DNA isolation was performed using the Qiagen Midiprep kit. S. aureus was transformed with plasmid DNA by electroporation (15). Chromosomal DNA from S. aureus was prepared as described previously (49). Transformation with high-molecular-weight chromosomal DNA was performed as previously described using phage Φ55 (49).
DNA sequencing and sequence analysis.
The ABI fluorescent system and Taq dye terminators (Qiagen) were employed for the sequencing reactions. DNA sequence analysis was performed using Genetics Computer Group (GCG) software. The predicted amino acid sequences of ArlS and ArlR proteins were compared with the sequences of the EMBL/GenBank/DDBJ data library using the TFASTA program (GCG).
Analyses of the Tn917 insertion site in BF15.
DNA flanking the Tn917 insertional site in the chromosome of BF15 was cloned using the strategy developed by Camilli et al. (6). Chromosomal DNA from BF15 was digested with EcoRI, and the flanking region of the transposon together with a partial sequence of Tn917 was self-ligated, creating plasmid pECO1. The nucleotide sequence of the chromosomal DNA in pECO1 was determined using a primer hybridizing within the transposon (42) and other synthetic oligonucleotide primers.
We were unable to clone one of two sides of the insertional site in BF15 by the method of Camilli et al. (6). As an alternative approach, chromosomal DNA from ISP794 was partially digested with Sau3A and ligated into plasmid pGB2 digested by BamHI. The plasmid containing the chromosomal DNA was selected by colony hybridization using a 1-kb probe specific for the insertional site in BF15. This probe was constructed by digesting pECO1 with SalI (site present in Tn917) and HpaI (site present in the flanking chromosomal DNA of BF15). A plasmid was isolated and named pBAM2-3. The nucleotide sequence of the chromosomal DNA in pBAM2-3 was determined using a primer from the chromosomal DNA sequence determined in pECO1 and other synthetic oligonucleotide primers.
Cloning of the wild-type arlR-arlS locus.
PCR amplification of a 2.4-kb product containing arlR and arlS was performed using Vent DNA polymerase (New England Biolabs) and chromosomal DNA of ISP794. Two primers containing the BamHI site (underlined), 5′-CTA TGG ATC CTA CAA TAG TGA AAA GTC-3′ (positions 29 to 55 according to Fig. 1) and 5′-GGG GGA TCC ACA GAA ATG ATA AAG AA-3′ (positions 2448 to 2473), were used. The conditions for PCR involved a 10-min step at 94°C followed by 30 cycles of 94°C for 45 s, 53°C for 45 s, and 72°C for 2 min 30 s and a 10-min final step at 72°C. The PCR product contained about 300 bp upstream and 100 bp downstream of the arlR-arlS locus.
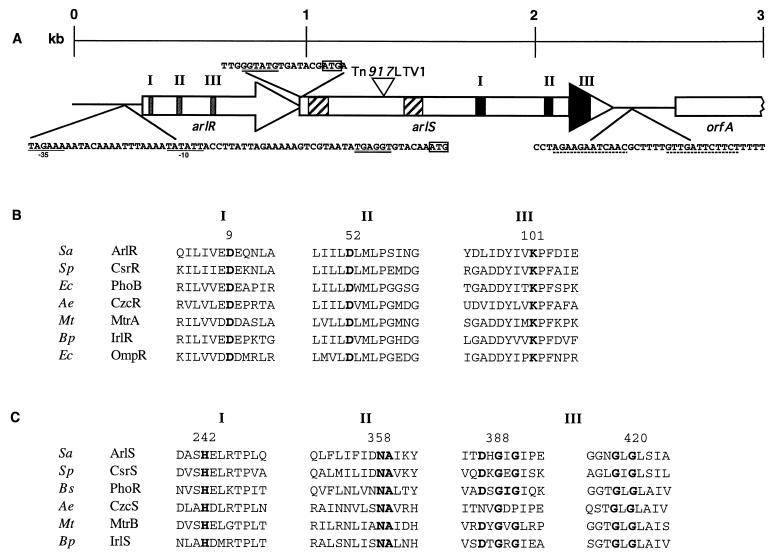
FIG. 1.
Schematic map of the arlR-arlS locus. (A) Diagram of the region of the BF15 chromosome including arlR, arlS, and flanking sequences. The site of the Tn917LTV1 insertion in strain BF15 is at position 1339. Arrows, direction of transcription for open reading frames arlR (positions 336 to 995) and arlS (positions 992 to 2347). In arlR, sequences encoding predicted receiver domains are indicated by shaded boxes (positions 363 to 639). In arlS, hatched boxes indicate two predicted hydrophobic membrane-spanning regions (positions 1028 to 1112 and 1442 to 1517) and the black areas represent predicted transmitter domains (positions 1718 to 2252). The −35 and −10 consensus sequences of the putative promoter are underlined, the ribosome-binding sites are double underlined, the start codons are boxed, and the transcription termination signal (positions 2505 to 2534) is indicated by dotted lines. (B) Amino acid sequence alignments of the putative ArlR with other response regulators presenting the strongest similarities with ArlR. Only regions of sequence similarity according to Stock et al. (50) are indicated. Boldface, highly conserved residues. Numbers refer to the amino acids of ArlR. Accession numbers and/or reference numbers for the protein sequences are as follows: CsrR, X98451, 26; PhoB, X04026, 29; CzcR, X98451, 56; MtrA, U01971, 57; IrlR, AF005358, 22; OmpR, 12. (C) Amino acid sequence alignments of the putative ArlS with other protein histidine kinases presenting the strongest similarities with ArlS. Only regions of sequence similarity according to Stock et al. (50) are indicated. Boldface, highly conserved residues. Numbers refer to the amino acids of ArlS. Accession numbers and reference numbers for the protein sequences are as follows: CsrS, AF082668, 26; PhoR, M23549, 47; CzcS, X98451, 56; MtrB, U14909, 57; IrlS, AF005358, 22. Abbreviations of organism names: Sa, S. aureus; Sp, Streptococcus pneumoniae; Ec, E. coli; Ae, Alcaligenes eutrophus; Mt, Mycobacterium tuberculosis; Bp, Burkholderia pseudomallei; Bs, B. subtilis.
PCR products were digested by BamHI and ligated into the BamHI site of pSK950 (36), which contains the attP site of staphylococcal phage L54a and which is capable of integrating specifically into the chromosomal attB site located just 3′ of the geh gene, which encodes staphylococcal lipase. Integration was facilitated by the presence of plasmid pYL112Δ19, which carries the L54a int gene encoding integrase. The resulting plasmid was first introduced at 30°C into a restriction-deficient derivative of S. aureus 8325-4 (RN4220) (25) carrying plasmid pYL112Δ19 and selected on tetracycline (3 μg/ml). The plasmid was integrated into the chromosome by two shifts at 42°C. Integration was verified by loss of lipase activity (48). High-molecular-weight DNA of RN4220 carrying the integrated plasmid was then transformed into BF16 using phage Φ55 by selection on 3 μg of tetracycline per ml to give strain BF17 (Table 1).
Biofilm formation on polystyrene.
The test for biofilm-forming capacity, described previously (10, 18), was performed. The cells were grown for 24 h at 37°C in 96-well polystyrene microtiter plates (Falcon). The cells were washed with phosphate-buffered saline (PBS), fixed with Bouin solution, and stained with safranin.
Primary adherence to a polystyrene surface.
Primary adherence to a polystyrene surface was determined as previously described (17), with some modifications. Overnight cultures were adjusted with PBS to an optical density at 578 nm (OD578) of 1.0. Five milliliters of each suspension was added to a polystyrene petri dish (Fisherbrand) and incubated for 30 min at 37°C. Petri dishes were washed at least five times with PBS. Cells were fixed with Bouin solution and Gram stained. Adherent bacterial cells were observed by oil immersion microscopy and counted (the mean of four different microscopic fields). All results shown are the means of at least two independent determinations.
Hemagglutination.
Hemagglutination assays were performed as previously described (45) using a suspension (1%) of human O erythrocytes (Sigma Chemical Co.) in PBS. Bovine serum albumin was added to a final concentration of 1% to avoid nonspecific hemagglutination. The bacterial suspension was adjusted to an OD600 of 1.0. The cells were centrifuged, washed once in PBS, and resuspended in the same volume of PBS. Twofold serial dilutions of cells were performed in a microtiter plate.
Triton X-100-induced autolysis assays.
Autolysis assays were performed as described by Mani et al. (30). Bacteria were grown in TSB containing 1 M NaCl to an OD580 of 0.6 to 0.8. The cells were pelleted by centrifugation and resuspended in the same volume containing 50 mM Tris-HCl (pH 7.5) and 0.1% Triton X-100. The cells were then incubated at 30°C with shaking, and the changes in A580 were measured. Results were normalized to OD580 at time zero (OD0), i.e., percent lysis at time t = [(OD0 − OD at time t)/OD0] × 100. All results shown are the means of at least two independent determinations.
Enzyme assays.
Heat-killed cells of S. aureus ISP794 were used as the standard substrate for lytic enzymes. Lytic activity was assayed by monitoring the rate of decrease in the turbidity of the cell suspension as previously described (52). Supernatants of cultures of different S. aureus strains were recovered at different growth times (OD600 = 0.5 and 0.9, and overnight). The supernatants were sterilized by filtration through a 0.45-μm-pore-size filter. Heated cells were suspended in 0.1 M phosphate buffer (pH 7.0; 10 mg [dry weight]/ml). One milliliter of cell suspension was mixed with 9 ml of culture supernatant. The mixture was incubated at 37°C with shaking, and the rate of change in turbidity was measured at 660 nm in a spectrophotometer. A reaction mixture lacking enzyme extract (TSB alone) was used as a control. The results were expressed as percent lysis as described for the autolysis assays.
Quantitative measurements of bacteriolytic activity were performed by mixing 8 ml of an appropriate dilution of overnight culture supernatant fluid with 500 μl of cell suspension. The decrease in OD660 was measured every 30 min for 3 h. Units of bacteriolytic activity are reported as the linear slope of a plot of OD660 versus time. Bacteriolytic specific activities were expressed as units per milligram of protein.
Proteolytic activities were determined using the insoluble proteolytic substrate azocoll (Calbiochem) as previously described (22, 44) with some modifications. The substrate (4 mg/ml) was suspended in 100 mM phosphate buffer (pH 7.0). Five hundred to 750 μl of an overnight culture supernatant was added to 500 μl of the substrate suspension. The mixtures were incubated for 2 h at 37°C with shaking. After incubation, the assay mixtures were centrifuged and the absorbances at 520 nm were determined. Proteases were distinguished through the use of different protease inhibitors (phenylmethylsulfonyl fluoride [PMSF], EDTA, and p-hydroxymercuribenzoate [PHMB]). These inhibitors were added for 45 min to the culture supernatant before adding the proteolytic substrate azocoll. One unit of protease activity was arbitrarily defined as the proteolysis which resulted in the release of 0.001 absorbance unit of dye for 2 h. Protease-specific activities were expressed as units per milligram of protein.
β-Galactosidase activity was determined as previously described (31). Protein concentrations were determined by the Bradford method (Bio-Rad). All results shown are the means of at least two independent determinations.
Protein isolation and SDS-PAGE.
Proteins were isolated as previously described (17, 33) by cultivating the staphylococcal strain on a sterile dialysis membrane (Millipore VS) overlaid on Trypticase soy agar. Five hundred microliters of an overnight culture (precultivated in TSB) adjusted in PBS buffer to an OD578 of 1.0 was placed on the membrane. After 20 h of growth, extracellular proteins were isolated by adding 100 μl of PBS to the membranes and scraping the growth into a microcentrifuge tube; the bacteria were separated from the extracellular proteins by centrifugation. The protein concentration was measured by the Bradford method (Bio-Rad). Fifty micrograms of proteins was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (11% separation gel, 4% stacking gel). Proteins were stained with Coomassie brilliant blue R250.
Identification of the arlR-arlS locus in other S. aureus strains.
Two primers chosen inside the arlR-arlS locus, 5′-AAT TTT ACG TCG TCA GCC ACA A-3′ (positions 677 to 699 according to Fig. 1) and 5′-GAT TTT TAG ATG TCA GAT CCG T-3′ (positions 1985 to 2006), were used to amplify the chromosomal DNA of six different clinical strains. Ten microliters of the PCR products was loaded on an agarose gel, and the presence of the product of 1.3 kb was determined.
Antimicrobial susceptibility testing.
MICs were determined by a serial twofold dilution method on Mueller-Hinton agar as previously described (15).
Nucleotide sequence accession number.
The entire nucleotide sequence of arlR and arlS was deposited in the GenBank/EMBL nucleotide sequence data library under accession no. AF165314.
RESULTS
Sequence analysis of the insertional site in BF15.
During our studies on the multidrug efflux pump NorA of S. aureus, we selected Tn917 insertion mutant BF15 (Table 1) using plasmid pLTV1 (14). We found that Tn917 was inserted within an open reading frame which we have named arlS for autolysis-related locus, sensor protein. Sequencing of the DNA upstream of arlS revealed the presence of a second open reading frame, which we named arlR for autolysis-related locus, regulator protein (Fig. 1A). These open reading frames were both preceded by consensus ribosome-binding sites and were oriented in the same direction (Fig. 1A). A putative promoter was found 36 bp upstream of arlR (Fig. 1A). The overlap between arlR and arlS and the absence of an obvious transcriptional terminator or promoter sequence upstream of arlS suggested that the two genes might be transcribed as a single message, but additional transcriptional analysis will be needed. A predicted stem-loop structure likely to act as a factor-independent transcription termination signal was observed 159 bp downstream of the arlS stop codon (Fig. 1A). The putative arlS gene product is predicted to initiate with an ATG start codon and to encode a 52.4-kDa protein (termed ArlS) containing 451 amino acids. The putative arlR gene product is predicted to initiate with an ATG start codon and to encode a 25.5-kDa protein containing 219 amino acids. The G+C content of this region was low (30%), which is consistent with that of the S. aureus genome.
The predicted amino acid sequence of ArlR shared strong similarities with those of response regulators (Fig. 1B). The response regulator family is defined by a conserved domain of about 120 amino acids that is generally located in the N terminus of each of these proteins (50). The residues that correspond to Asp-9, Asp-52, and Lys-101 in ArlR are invariant among response regulators (50) (Fig. 1B). These residues are located in the active site of the regulator. The response regulators may be placed into subfamilies based on their C-terminal domains (50). ArlR belongs to the PhoB-OmpR family (Fig. 1B). This family has a conserved C terminus that binds to specific DNA sequences upstream of the regulated promoters. In the intact protein, phosphorylation of the N-terminal domain serves to stimulate DNA-binding activity (50).
The predicted amino acid sequence of ArlS had strong similarities with sequences of all of the regions conserved in sensor proteins (Fig. 1C). The histidine protein kinase family is defined by a region of conserved sequence generally located near the C terminus (50). Within the C terminus, there are several residues that are strongly conserved in three different regions (Fig. 1C). The conserved histidine in region I (position 242 for ArlS) is by homology predicted to be the site of phosphorylation of ArlS (50). A second domain (II) is necessary for kinase activity (51). The last domain (III) contains a G box (53). In addition, ArlS contains two hydrophobic membrane-spanning domains (Fig. 1A), suggesting that this protein is membrane associated like many of the histidine kinases (50). These results strongly suggest that the protein ArlS is a sensor phosphorylating its regulator, ArlR. However, we have no direct evidence of such function, nor are the environmental factors that modulate expression of ArlS known. The arlR-arlS locus had the strongest similarity with locus csrS-csrR of group A streptococci: ArlS and CsrS showed 32% amino acid identity and 43% similarity for the entire protein, and ArlR and CsrR showed 54% identity and 63% similarity for the entire protein. The csrSR locus has been shown to be involved in the regulation of hyaluronic acid capsule synthesis in group A streptococci (26).
To determine if the arlR-arlS locus was also present in other clinical strains of S. aureus, this locus was amplified by PCR using chromosomal DNA from six epidemiologically unrelated clinical S. aureus strains from one hospital and a primer set with one primer within arlR and the other within arlS (see Materials and Methods). The expected PCR product of 1.3 kb was amplified in all six strains (data not shown), indicating that this locus, identified in laboratory strains of S. aureus, could be present in most or all members of the species.
Complementation.
To prove that the BF15 phenotype is due to transposon insertion, the erythromycin resistance marker from the Tn917 mutant BF15 was reintroduced into the wild-type parent strains ISP794 and MT23142 by using phage Φ55. Analyses of two erythromycin-resistant transformants showed properties similar to those of BF15 (data not shown), indicating that the BF15 phenotype was linked to the transposon. One transformant in the wild-type strain ISP794 carrying the arlS mutation (BF16) was used for this study (Table 1).
In order to confirm the properties of the two-component regulatory proteins, the chromosomal DNA region containing the arlR-arlS locus was amplified by PCR. The product was cloned into pSK950 and integrated into the chromosome of BF16. One representative transformant, S. aureus BF17, was characterized. As detailed in following sections, introduction of the arlR-arlS locus into the chromosome complemented all of the phenotypic properties of the arlS mutant, indicating that the disruption of arlS itself was responsible for the mutant phenotype.
Colony morphology.
S. aureus ISP794 and MT23142 formed smooth, shiny colonies on Trypticase soy agar whereas colonies of BF16 were smaller, dull, and sticky. The complemented arlS mutant BF17 had a morphology similar to that of the wild-type strains.
Biofilm formation on polystyrene.
In a previously described biofilm assay, primary attachment and accumulation of multilayered cell clusters, which together lead to biofilm formation, can be measured. After growth of the strains in microtiter plates, the appearance of the culture of BF16 was different from that of the wild type. In contrast to the wild type, which exhibited no affinity for the polystyrene surface, leading to a minimal adherent-cell density by safranin staining when the culture fluid was removed from the microtiter plates, BF16 formed an adhesive film of bacteria on the surface (Fig. 2). The complemented mutant BF17, like the parent strain, did not form a film on the polystyrene surface.
FIG. 2.
Quantitative assay of biofilm formation in polystyrene microtiter plates. Lane 1, wild-type strain ISP794; lane 2, mutant BF16; lane 3, complemented mutant BF17.
In order to determine whether the biofilm formation in the arlS mutant was due to a modification of primary adherence to polystyrene, the numbers of cells bound to polystyrene for the three strains were measured. The number of attached cells with BF16 (410 cells for four oil immersion microscopic fields) was fourfold higher than that with wild-type strain ISP794 (100 cells), demonstrating that BF16 had enhanced binding to polystyrene. The complemented mutant BF17 (130 cells) bound to polystyrene at a level comparable to that of the wild type. As the hemagglutination assay is a commonly used assay to demonstrate bacterial adherence, it was applied to the wild-type strain, the arlS mutant, and the complemented mutant, and no hemagglutination was observed with intact washed cells (data not shown).
Cell aggregation is indicative of the ability of strains to express intercellular adhesion and to accumulate in multilayered cell clusters. In cultures grown overnight in TSB, BF16, observed by phase-contrast microscopy, failed to produce cell clusters. Wild-type strain ISP794 and complemented mutant BF17 formed small cell clusters of five cells. Intercellular adhesion seems to be slightly impaired in mutant BF16. Together, these results indicate that insertion of Tn917 in arlS is responsible for the altered adhesive and aggregation properties of BF16.
Extracellular proteins.
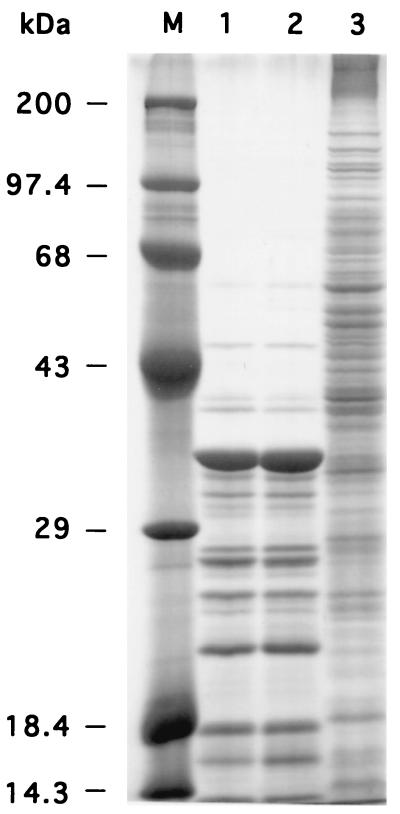
Using SDS-PAGE, we compared the patterns of secreted proteins (Fig. 3). In order to concentrate secreted proteins, cells were grown on a dialysis membrane. Extracellular proteins were isolated by adding PBS to the overnight culture. The pattern of secreted proteins for BF16 differed strikingly from that for the wild-type parent strain ISP794 and is similar to that for a cellular lysate (Fig. 3). One likely possibility to explain the pattern observed with the mutant cells is that these proteins originated from the cytoplasm rather than the supernatant due to increased lysis of the BF16 cells in stationary phase.
FIG. 3.
SDS-PAGE analysis of extracellular proteins from different S. aureus strains. Samples were loaded as follows: lane 1, parent strain ISP794; lane 2, strain MT23142; lane 3, mutant BF16. Lane M, protein standard.
Autolysis assays.
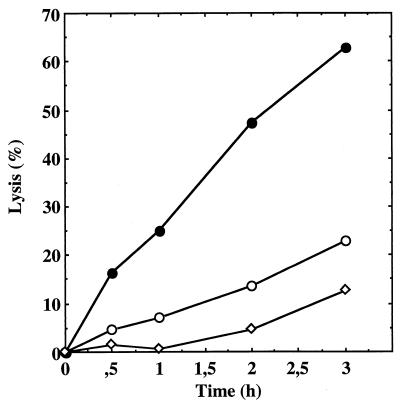
To study Triton X-100-induced autolysis, S. aureus strains were grown in TSB containing 1 M NaCl and resuspended in a medium containing 0.1% Triton X-100. For the wild-type strain, a small degree of lysis was observed (Fig. 4). Under these conditions, the BF16 mutant exhibited an increased Triton X-100-induced autolysis rate (63% autolysis in 3 h) compared to the parent strain ISP794 (23% autolysis in 3 h). The complemented mutant BF17 exhibited a slow autolysis rate (13% autolysis in 3 h) (Fig. 4).
FIG. 4.
Autolysis of whole cells of S. aureus ISP794 (○), BF16 (●), and complemented mutant BF17 (◊) by Triton X-100. The results are expressed as lysis percentages as described in Materials and Methods.
The major autolysin of S. aureus 8325-4 is the 137-kDa Atl protein (13, 52). Atl is a bifunctional protein that has glucosaminidase (GL) and amidase (AM) domains, which are separated by proteolytic processing to generate two extracellular lytic enzymes, a 51-kDa GL and a 62-kDa AM (38). When atl is disrupted in SH108 by introduction of the plasmid pSA107 (13), almost all autolysins disappear on the zymograph, implying that most of the autolysins of S. aureus are the products of a simple gene, and proform processing results in the multiple bands visualized on the zymograph (13). The arlS mutation was introduced into SH108 by transformation of high-molecular-weight DNA from BF15 with phage Φ55 and by selection on 5 μg of chloramphenicol per ml to give strain BF18 (Table 1). For both strain SH108 (atl) and strain BF18 (atl arlS), the autolysis rate was very low (4% in 3 h) (data not shown).
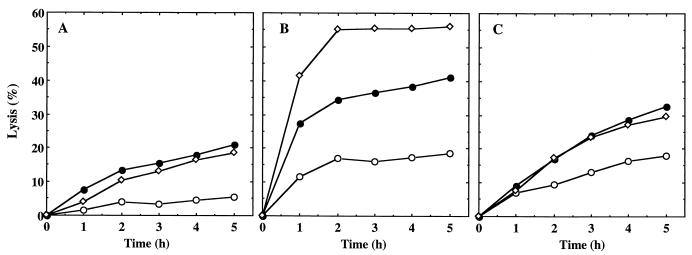
The bacteriolytic activity of the extracellular proteins contained in the culture supernatant was investigated using as lysis targets heat-killed cells of S. aureus ISP794 (Fig. 5). Supernatants were recovered at different phases of growth: OD600, 0.5 (early exponential phase); OD600, 0.9 (late exponential phase); and overnight (stationary phase). First, for the supernatant of the parent strain, minimal lysis was observed with cells in early exponential phase (5% lysis in 5 h) (Fig. 5A), and slight lysis was observed with the supernatant from the late exponential and stationary phases (20% lysis in 5 h). In contrast, the supernatant of BF16 produced more-extensive lysis of S. aureus cells (Fig. 5B). With the supernatant from early exponential phase, slight lysis was observed (18% lysis in 5 h). Lysis was more pronounced with the supernatant from late exponential phase (41% lysis in 5 h) and was higher still with the stationary-phase supernatant (56% lysis in 5 h). For the transformant BF17, the lysis was similar to that of the wild type (Fig. 5C). Since the mutant BF16 lysed extensively in stationary phase when cells were grown on a dialysis membrane (Fig. 3), we could not determine the relative amounts and diversity of murein hydrolases excreted by the different strains using zymography.
FIG. 5.
Lytic activity of S. aureus culture supernatants at different growth phases: OD600, 0.5 (○); OD600, 0.9 (●); overnight (◊). The results are expressed as lysis percentages as described in Materials and Methods. (A) Parent strain ISP794. (B) Mutant BF16. (C) Complemented mutant BF17.
The supernatants of late-exponential- and stationary-phase cultures produced extensive lysis of S. aureus cells (Fig. 5B), indicating that secreted autolysins were increased. However, we also verified that the susceptibility of the cell wall to autolysin and peptidoglycan hydrolase binding to the cell wall were not modified. To determine the susceptibility of cell walls to autolysins, heat-killed cells from early-exponential-phase (OD600, 0.5) and overnight cultures of the atl mutant strain SH108 and the atl arlS double mutant BF18 were incubated with an overnight supernatant of wild-type strain ISP794. Cells from SH108 and BF18 exhibited similar rates of autolysis (data not shown), indicating that the mutation in arlS does not alter the composition of the cell wall leading to a hypersusceptibility to autolysins. In order to investigate the relative capacities of cell walls from wild-type and mutant cells to bind autolysins, heat-killed cells from early-exponential-phase (OD600, 0.5) and overnight cultures of the atl mutant SH108 and the atl arlS double mutant BF18 were incubated with an overnight supernatant of the strain BF16 at 0°C for 60 min. The cells were removed by centrifugation and filtration, and the unbound autolytic activity was determined by using heat-killed cells of ISP794. The results indicate that autolysins bound readily to cell walls from both strains and that there was no detectable difference in binding due to the arlS mutation (data not shown).
It was important to establish whether the increased autolysin activity associated with the cells was due to an overproduction of autolysins, an increase of translocation enzyme, or a decrease of proteases inactivating autolysins. To determine the effect of the arlS mutation on atl transcription, we used strain SH108, which contains a transcriptional fusion between atl and lacZ (13). β-Galactosidase activities of wild-type strain SH108 and mutant BF18 (arlS) (Table 1) were measured at different phases of growth: OD600, 0.5 (early exponential phase); OD600, 0.9 (late exponential phase); and overnight (stationary phase). No differences between the wild type and the mutant were observed (data not shown), indicating that expression of atl is not modified in the arlS mutant.
To rule out the possibility that the phenotype of the mutant was due to an increase in enzyme translocation from the cytoplasm to the cell wall, the intracellular concentration of lytic enzymes was determined. Cells at an OD600 of 0.5 were collected, washed, homogenized with a glass beater, and centrifuged. The supernatant was used for lytic activity measurement using heat-killed cells. The intralytic activity of mutant BF16 was similar to that of wild-type strain ISP794 (data not shown), suggesting that the translocation of the autolysins was not involved in the phenotype observed in the mutant.
Extracellular proteolytic activity.
As proteases are known to modify autolysis and cell wall turnover (1, 22, 34, 58), supernatants from overnight cultures were assayed for proteolytic activity using the substrate azocoll. Total extracellular proteolytic activity of the mutant strain BF16 was undetectable by this method, whereas those of wild-type strain ISP794 and complemented mutant BF17 were at least 30-fold higher (Table 2). By using different protease inhibitors, we were able to quantify the different types of proteases present in the culture supernatant. Serine proteases are inhibited by hydroxyl-reactive organofluorides such as PMSF, whereas metalloproteases requiring divalent metal cations are inhibited by chelators such as EDTA (40). Both serine proteases and metalloproteases are present in the wild-type strain and the complemented mutant, whereas no detectable activity was observed in the arlS mutant (Table 2). As thiol proteases contain a sulfhydryl group in the active site, they are only active in the presence of reducing agents such as dithiothreitol (DTT) and are inhibited by mercurials and heavy metals (2, 40). Thus, thiol protease activities were determined in the presence of DTT (Table 2). The activity in the presence of PMSF was similar to that in its absence, and inhibitors of thiol proteases (PHMB and iodoacetamide) inhibited the proteolytic activities of the wild-type strain and the complemented mutant, indicating that only thiol proteases were measured in the presence of DTT. The thiol protease activity of mutant strain BF16 was 70-fold lower than those of the wild type and the complemented mutant (Table 2).
TABLE 2.
Extracellular protease activity determined in the culture supernatant
| Protease | Additives | Activitya (U/mg of protein) of strain:
|
||
|---|---|---|---|---|
| ISP794 | BF16 | BF17 | ||
| Total protease | 440 | ≤15 | 580 | |
| Serine protease | 380 | ≤15 | 470 | |
| Metalloprotease | 110 | ≤15 | 96 | |
| Thiol protease | DTT, EDTA | 11,100 | 160 | 13,300 |
| DTT, EDTA, PMSF (1 mM) | 10,700 | 100 | 12,900 | |
| DTT, EDTA, PHMB (3 mM) | 230 | 100 | 110 | |
| DTT, EDTA, iodoacetamide (1 mM) | 130 | 90 | NDb | |
Units of serine protease and metalloprotease were determined by subtraction of units obtained with PMSF (1 mM) and EDTA (1 mM), respectively, from the total units of proteolytic activity. Thiol protease activities were measured in the presence of DTT (1 mM) and EDTA (1 mM).
ND, not determined.
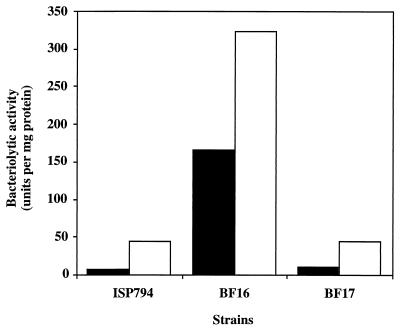
In order to see the effect of PMSF on autolysis, the three strains were grown overnight in the absence and in the presence of PMSF. The culture supernatants were then tested for their bacteriolytic activities (Fig. 6). Bacteriolytic activities of wild-type strain ISP794 and the complemented mutant BF17 were increased 6- and 4.5-fold, respectively, in the presence of PMSF, suggesting that inhibition of serine proteases increases autolysins in the culture supernatant. In contrast, bacteriolytic activity of the mutant strain in the presence of PMSF was less than twofold higher than that in the absence of PMSF.
FIG. 6.
Effect of PMSF on the bacteriolytic activity of the culture supernatant. Cells were grown for 18 h in the absence (■) or in the presence (□) of PMSF. The culture supernatants were filtered and tested for bacteriolytic activities. All results shown are the means of at least two independent determinations.
Growth in the presence or absence of NaCl and KCl.
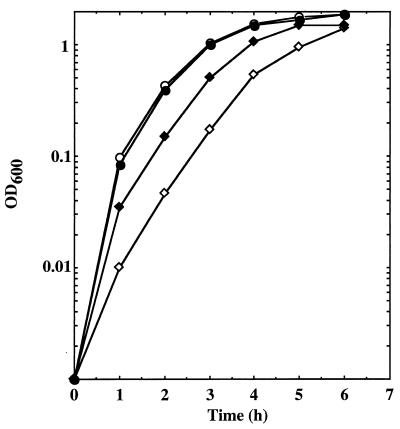
As NaCl has been shown to induce autolysis, we determined the effect of NaCl and KCl on the growth of mutant and wild-type strains. Overnight cultures of the parent (ISP794) and mutant (BF16) strains were diluted 1/100 in TSB with or without NaCl (50 mM) and grown with shaking at 37°C. Values of OD600 were measured every hour (Fig. 7). The parent strain grew normally in the presence as well as in the absence of NaCl. For BF16, growth was strikingly slower without supplemented NaCl and was partially restored in the presence of NaCl (Fig. 7). BF16 growth in the presence of KCl (50 mM) was similar to that in the presence of NaCl (50 mM) (data not shown). Thus, growth of BF16 is most impaired in the absence of supplemented NaCl or KCl.
FIG. 7.
Growth curve of the parent strain ISP794 and mutant BF16 in the presence or absence of NaCl. Results are for ISP794 with (●) or without (○) NaCl (50 mM) and for BF16 with (⧫) or without (◊) NaCl (50 mM). OD600 was measured every hour.
Susceptibility to heavy metals and cell wall-active antimicrobials.
Because the arlR-arlS locus, particularly the response regulator ArlR, has similarity with other regulatory systems involved in heavy-metal physiology (Fig. 1B) (irl [23], czc [56], pco [3], cop [32], cut [54]), MICs of Cu2+, Cd2+, Zn2+, Co2+, and Ni2+ were tested. No differences between mutant BF16 and wild-type strain ISP794 were observed (data not shown), suggesting that the arlR-arlS locus is not involved in heavy-metal physiology.
For BF16, the MICs of ampicillin, oxacillin, methicillin, cycloserine, and vancomycin, antibiotics that inhibit the synthesis of peptidoglycan, did not differ from those for the wild-type parent strains ISP794 and MT23142 (data not shown).
DISCUSSION
We have identified a new locus, arlR-arlS, encoding a two-component regulatory system. Disruption of arlS by insertion of Tn917 resulted in increased adherence to a polymer such as polystyrene. CoNS colonize polymer surfaces by formation of multilayered cell clusters, which are embedded in an extracellular matrix (10). The colonized bacteria together with the extracellular matrix, which is largely composed of cell wall teichoic acids, are referred to as the biofilm (20). Biofilm formation takes place in two phases: rapid attachment of the bacteria to the surface, followed by a more prolonged accumulation phase that involves cell proliferation and intercellular adhesion (18). The formation of a biofilm on polystyrene by the arlS mutant strain BF16 was due to a strong primary attachment, whereas the intercellular adhesion, as tested by determining cell aggregation, was only slightly impaired. This property is due to the disruption of arlS, since in the presence of the integrated arlR-arlS locus the arlS mutant BF16 loses its ability to bind polystyrene.
We also found that the arlS mutant had an increased rate of Triton X-100-induced autolysis compared to that of the parental strain ISP794. The supernatants of late-exponential- and stationary-phase cultures produced extensive lysis of S. aureus cells (Fig. 5B), indicating that secreted autolysins were increased. It has been previously demonstrated that autolysins can mediate primary attachment to polystyrene surfaces (18, 19). Homologs of Atl, AtlE in S. epidermidis and Aas in S. saprophyticus, have the ability to bind polystyrene, fibronectin, and vitronectin, a protein of the intercellular matrix (18, 19). In S. saprophyticus, Aas is able to bind sheep erythrocytes (19). As expected from previous results for S. aureus (45), we found no hemagglutination when we tested washed cells of either parent or mutant. This suggests that autolysins of S. aureus do not likely agglutinate erythrocytes. AtlE binds vitronectin but not fibronectin, unlike Aas (19), indicating that these autolysins have different binding properties. Mutants of S. aureus and S. epidermidis that lack the extracellular AM activity of Atl and AtlE, respectively, have also been shown to form cell clusters in liquid culture (7, 18, 52). Because our arlS mutant shows an opposite phenotype, the presence of extracellular peptidoglycan hydrolases and a slight decrease of cell cluster formation, we postulate that an alteration in arlS mediates increased primary attachment to polystyrene and decreased cell clusters by increased expression of autolysins. It might be paradoxical that a strain, such as our mutant, presenting decreased cell aggregation also forms a biofilm on polystyrene surfaces. Since autolysin production is enhanced in this mutant, decreased cell aggregation in this case might reflect the role of peptidoglycan hydrolases in cell division rather than in intercellular adhesion (58).
The increased autolysis rate could be due to increased autolysin activity associated with the cells (by overproduction of autolysins, increase of the translocation enzyme, or decrease of proteases inactivating autolysins) or to an increased susceptibility of the cell wall to autolysin. All these possibilities were tested, and the only observed difference between the wild-type strain and its arlS mutant was in the extracellular proteolytic activity. In mutant strain BF16 all proteases, including serine, metalloproteases, and thiol proteases, were strikingly decreased in comparison to the wild-type strain and the complemented mutant (Table 2). It has been shown that secreted autolysins are susceptible to inactivation by serine proteases (1, 22, 58). In Staphylococcus haemolyticus, addition of subtilisin (a serine protease of Bacillus subtilis) to exponentially growing cells causes a loss of cell wall lytic activity (58). As the mutant strain BF16 does not produce detectable serine protease activity, we can postulate that this deficiency of protease increases the levels of secreted autolysins and is responsible for the increased autolysis of the mutant cells. Furthermore, when the S. aureus cells were grown overnight in the presence of PMSF (a serine protease inhibitor), the bacteriolytic activities of the culture supernatant from the wild-type strain and the complemented mutant were significantly increased (6- to 4.5-fold), whereas that of the mutant was only slightly increased (less than 2-fold) (Fig. 6). This suggests that, in the absence of serine protease, the level of secreted autolysin increases.
We observed that growth of the arlS mutant is salt dependent. Both 50 mM NaCl and 50 mM KCl increased the growth rate of the mutant but not of the parent strain. NaCl at 300 mM did not further increase growth rates (data not shown). It has been proposed that Na+s but not K+s induce autolysis. This activation is probably due in some cases to the displacement of autolysins bound to teichoic acid in the cell wall (9, 30). As NaCl induces autolysis, mutant BF16 should grow slower in the presence of NaCl. However, the opposite was true since BF16 grew faster with NaCl (Fig. 7). The B. subtilis lyt-15 mutant has a low rate of wall turnover but a normal rate of autolysis in low-salt medium in comparison to its parent strain. The presence of 200 mM NaCl or KCl in the culture medium normalizes the cell wall turnover of this mutant (9). Thus, in mutant BF16, we can speculate that the cell wall turnover, in addition to autolysis, is also impaired and that the presence of Na+ restores a normal rate of cell wall turnover.
Because of their potential to destroy the cell wall, peptidoglycan hydrolases must be tightly controlled. In S. aureus, two two-component regulatory systems have been found to control autolysins. First, mutations in the virulence regulatory genes agr and sar affect autolysis. These loci modulate peptidoglycan hydrolase expression and/or activity (16). When the agrC-agrA locus is disrupted, low-molecular-weight cell wall-associated autolysins (32 kDa) are produced at a lower level than those of the parental strain, whereas high-molecular-weight autolysins (>75 kDa) are overproduced. It is not yet known how AgrC-AgrA acts on autolysin activities. Second, locus lytS-lytR was shown to modify autolysis in S. aureus (4). Similarly the lytS mutant was characterized by the diminished activity of several peptidoglycan hydrolases and substantial increases in the activities of several high-molecular-weight extracellular proteins (4). It has been suggested that this locus modifies the expression of 25-kDa autolysin LrgB (5).
The V8 serine protease of S. aureus is known to be involved in virulence because of its ability to cleave and inactivate immunoglobulin G antibodies in vitro. This and other proteases have been proposed to function in blocking the action of antibodies by cleaving and inactivating them (41). Another role for proteases may involve protection against antimicrobial peptides such as the neutrophil defensins and the platelet microbiocidal proteins. These peptides seem to play important roles in host defense. Because these antimicrobial peptides are subject to proteolytic inactivation, the production of extracellular proteases by the staphylococci may represent a bacterial defense system (41). Since the arlR-arlS locus modifies extracellular proteolytic activity, it is tempting to speculate that this locus might be involved in the virulence of S. aureus. It is not yet known whether ArlR acts directly on protease gene transcription or through interactions with other genes. The mechanism of action of the two-component regulatory system on proteases and other virulence factors will be further studied.
In conclusion, these data indicate that a new two-component regulatory system, ArlS-ArlR, may control the rate of autolysis as well as the attachment to a polymer by affecting the secreted peptidoglycan hydrolase activity.
ACKNOWLEDGMENTS
We thank Georges Rapoport, André Klier, and Michel Débarbouillé for helpful discussions and critical reading and Tarek Msadek and Steven J. Projan for critical reading of this manuscript. We also thank Andrew Camilli for the gift of the plasmid pLTV1, Gordon L. Archer for plasmid pSK950, and Simon J. Foster for providing S. aureus strain SH108.
This work was supported by U.S. Public Health Service grant AI23988 (to D.C.H.) from the National Institutes of Health and by research funds from the Institut Pasteur and Centre National de la Recherche Scientifique.
REFERENCES
- 1.Archibald A R, Hancock I C, Harwood C R. Cell wall structure, synthesis, and turnover. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria. Washington, D.C.: American Society for Microbiology; 1993. pp. 381–410. [Google Scholar]
- 2.Arvidson S, Holme T, Lindholm B. Studies on extracellular proteolytic enzymes from Staphylococcus aureus. I. Purification and characterization of one neutral and one alkaline protease. Biochim Biophys Acta. 1973;302:135–148. doi: 10.1016/0005-2744(73)90016-8. [DOI] [PubMed] [Google Scholar]
- 3.Brown N L, Barrett S R, Camakaris J, Lee B T O, Rouch D A. Molecular genetics and transport analysis of the copper-resistance determinant (pco) from Escherichia coli plasmid pRJ10004. Mol Microbiol. 1995;17:1153–1166. doi: 10.1111/j.1365-2958.1995.mmi_17061153.x. [DOI] [PubMed] [Google Scholar]
- 4.Brunskill E W, Bayles K W. Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J Bacteriol. 1996;178:611–618. doi: 10.1128/jb.178.3.611-618.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunskill E W, Bayles K W. Identification of LytSR-regulated genes from Staphylococcus aureus. J Bacteriol. 1996;178:5810–5812. doi: 10.1128/jb.178.19.5810-5812.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Camilli A, Portnoy D A, Youngman P. Insertional mutagenesis of Listeria monocytogenes with a novel Tn917 derivative that allows direct cloning of DNA flanking transposon insertions. J Bacteriol. 1990;172:3738–3744. doi: 10.1128/jb.172.7.3738-3744.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chatterjee A N, Wong W, Young F E, Gilpin R W. Isolation and characterization of a mutant of Staphylococcus aureus deficient in autolytic activity. J Bacteriol. 1976;125:961–967. doi: 10.1128/jb.125.3.961-967.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheesbrough J S, Finch R G, Burden R P. A prospective study of the mechanisms of infection associated with hemodialysis catheters. J Infect Dis. 1986;154:579–589. doi: 10.1093/infdis/154.4.579. [DOI] [PubMed] [Google Scholar]
- 9.Cheung H-Y, Freese E. Monovalent cations enable cell wall turnover of the turnover-deficient lyt-15 mutant of Bacillus subtilis. J Bacteriol. 1985;161:1222–1225. doi: 10.1128/jb.161.3.1222-1225.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christensen G D, Simpson W A, Younger J J, Baddour L M, Barrett F F, Melton D M, Beachey E H. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J Clin Microbiol. 1985;22:996–1006. doi: 10.1128/jcm.22.6.996-1006.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Churchward G, Belin D, Nagamine Y. A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene. 1984;31:165–171. doi: 10.1016/0378-1119(84)90207-5. [DOI] [PubMed] [Google Scholar]
- 12.Comeau D E, Ikenaka K, Tsung K, Inouye M. Primary characterization of the protein products of Escherichia coli OmpB locus: structure and regulation of synthesis of the OmpR and EnvZ proteins. J Bacteriol. 1985;164:578–584. doi: 10.1128/jb.164.2.578-584.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foster S J. Molecular characterization and functional analysis of the major autolysin of Staphylococcus aureus 8325/4. J Bacteriol. 1995;177:5723–5725. doi: 10.1128/jb.177.19.5723-5725.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fournier B, Aras R, Hooper D C. Expression of the multidrug resistance transporter NorA from Staphylococcus aureus is modified by a two-component regulatory system. J Bacteriol. 2000;182:664–671. doi: 10.1128/jb.182.3.664-671.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fournier B, Hooper D C. Mutations in topoisomerase IV and DNA gyrase of Staphylococcus aureus: novel pleiotropic effects on quinolone and coumarin activity. Antimicrob Agents Chemother. 1998;42:121–128. doi: 10.1128/aac.42.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujimoto D F, Bayles K W. Opposing roles of the Staphylococcus aureus virulence regulators, Agr and Sar, in Triton X-100- and penicillin-induced autolysis. J Bacteriol. 1998;180:3724–3726. doi: 10.1128/jb.180.14.3724-3726.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heilmann C, Gerke C, Perdreau-Remington F, Götz F. Characterization of Tn917 insertion mutants of Staphylococcus epidermidis affected in biofilm formation. Infect Immun. 1996;64:277–282. doi: 10.1128/iai.64.1.277-282.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heilmann C, Hussain M, Peters G, Götz F. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol Microbiol. 1997;24:1013–1024. doi: 10.1046/j.1365-2958.1997.4101774.x. [DOI] [PubMed] [Google Scholar]
- 19.Hell W, Meyer H-G W, Gatermann S G. Cloning of aas, a gene encoding a Staphylococcus saprophyticus surface protein with adhesive and autolytic properties. Mol Microbiol. 1998;29:871–881. doi: 10.1046/j.1365-2958.1998.00983.x. [DOI] [PubMed] [Google Scholar]
- 20.Hussain M, Wilcox M H, White P J. The slime of coagulase-negative staphylococci: biochemistry and relation to adherence. FEMS Microbiol Rev. 1993;104:191–208. doi: 10.1111/j.1574-6968.1993.tb05867.x. [DOI] [PubMed] [Google Scholar]
- 21.Janzon L, Ardvidson S. The role of the delta-lysin (hld) in the regulation of virulence genes by the accessory gene regulator (agr) in Staphylococcus aureus. EMBO J. 1990;9:1391–1399. doi: 10.1002/j.1460-2075.1990.tb08254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jolliffe L K, Doyle R J, Streips U N. Extracellular proteases modify cell wall turnover in Bacillus subtilis. J Bacteriol. 1980;141:1199–1208. doi: 10.1128/jb.141.3.1199-1208.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones A L, DeShazer D, Woods D E. Identification and characterization of a two-component regulatory system involved in invasion of eukaryotic cells and heavy-metal resistance in Burkholderia pseudomallei. Infect Immun. 1997;65:4972–4977. doi: 10.1128/iai.65.12.4972-4977.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karchmer A W, Archer G L, Dismukes W E. Staphylococcus epidermidis causing prosthetic valve endocarditis: microbiologic and clinical observations as guides to therapy. Ann Intern Med. 1983;98:447–455. doi: 10.7326/0003-4819-98-4-447. [DOI] [PubMed] [Google Scholar]
- 25.Kreiswirth B N, Lofdalh S, Betley M J, O'Reilly M, Schlievert P M, Bergdoll M S, Novick R P. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature (London) 1983;305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 26.Levin J C, Wessels M R. Identification of csrR/csrS, a genetic locus that regulates hyaluronic acid capsule synthesis in group A Streptococcus. Mol Microbiol. 1998;30:209–219. doi: 10.1046/j.1365-2958.1998.01057.x. [DOI] [PubMed] [Google Scholar]
- 27.Maeda S, Mizuno T. Evidence for multiple OmpR-binding sites in the upstream activation sequence of the ompC promoter in Escherichia coli: a single OmpR-binding site is capable of activating the promoter. J Bacteriol. 1990;172:501–503. doi: 10.1128/jb.172.1.501-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Makino K, Shinagawa H, Amemura M, Kinura S, Nakata A, Ishihama A. Regulation of the phosphate regulon of Escherichia coli. Activation of pstS transcription by PhoB protein in vitro. J Mol Biol. 1988;203:85–95. doi: 10.1016/0022-2836(88)90093-9. [DOI] [PubMed] [Google Scholar]
- 29.Makino K, Shinagawa H, Amemura M, Nakata A. Nucleotide sequence of the phoB gene, the positive regulatory gene for the phosphate regulon of Escherichia coli K12. J Mol Biol. 1986;190:37–44. doi: 10.1016/0022-2836(86)90073-2. [DOI] [PubMed] [Google Scholar]
- 30.Mani N, Tobin P, Jayaswal R K. Isolation and characterization of autolysis-defective mutants of Staphylococcus aureus created by Tn917-lacZ mutagenesis. J Bacteriol. 1993;175:1493–1499. doi: 10.1128/jb.175.5.1493-1499.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 32.Mills S D, Jasalavitch C A, Cooksey D A. A two-component regulatory system required for copper-inducible expression of the copper resistance operon of Pseudomonas syringae. J Bacteriol. 1993;175:1656–1664. doi: 10.1128/jb.175.6.1656-1664.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muller E, Hübner J, Gutierrez N, Takeda S, Goldmann D A, Pier G B. Isolation and characterization of transposon mutants of Staphylococcus epidermidis deficient in capsular polysaccharide/adhesin and slime. Infect Immun. 1993;61:551–558. doi: 10.1128/iai.61.2.551-558.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neumann V C, Health H E, Lebanc P A, Sloan G L. Extracellular proteolytic activation of bacteriolytic peptidoglycan hydrolases of Staphylococcus simulans biovar staphylolyticus. FEMS Microbiol Lett. 1993;110:205–212. doi: 10.1111/j.1574-6968.1993.tb06321.x. [DOI] [PubMed] [Google Scholar]
- 35.Ng E Y, Trucksis M, Hooper D C. Quinolone resistance mediated by norA: physiologic characterization and relationship to flqB, a quinolone resistance locus on the Staphylococcus aureus chromosome. Antimicrob Agents Chemother. 1994;38:1345–1355. doi: 10.1128/aac.38.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niemeyer D M, Pucci M J, Thanassi J A, Sharma V K, Archer G L. Role of mecA transcriptional regulation in the phenotypic expression of methicillin resistance in Staphylococcus aureus. J Bacteriol. 1996;178:5464–5471. doi: 10.1128/jb.178.18.5464-5471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Novick R P, Ross H F, Projan S J, Kornblum J, Kreiswirth B, Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993;12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oshida T, Sugai M, Komatsuzawa H, Hong Y-M, Suginaka H, Tomasz A. A Staphylococcus aureus autolysin that has an N-acetylmuramoyl-L-alanine amidase domain and an endo-β-N-acetylglucosaminidase domain: cloning, sequence, analysis, and characterization. Proc Natl Acad Sci USA. 1995;92:285–289. doi: 10.1073/pnas.92.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parkinson J S. Signal transduction schemes of bacteria. Cell. 1993;73:857–871. doi: 10.1016/0092-8674(93)90267-t. [DOI] [PubMed] [Google Scholar]
- 40.Pero J, Sloma A. Proteases. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria. Washington, D.C.: American Society for Microbiology; 1993. pp. 939–952. [Google Scholar]
- 41.Projan S J, Novick R P. The molecular basis of pathogenicity. In: Crossley K B, Archer G L, editors. The staphylococci in human disease. New York, N.Y: Churchill Livingstone; 1997. pp. 55–81. [Google Scholar]
- 42.Quirk P G, Dunkley E A, Lee P, Krulwich T A. Identification of a putative Bacillus subtilis rho gene. J Bacteriol. 1993;175:647–654. doi: 10.1128/jb.175.3.647-654.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Recsei P, Kreiswirth B, O'Reilly M, Schlievert P, Gruss A, Novick R P. Regulation of exoprotein gene expression in Staphylococcus aureus by agr. Mol Gen Genet. 1987;202:58–61. doi: 10.1007/BF00330517. [DOI] [PubMed] [Google Scholar]
- 44.Robinson J M, Keating M S, Sloan G L. The characteristics of extracellular protein secretion by Staphylococcus staphylolyticus. J Gen Microbiol. 1980;118:529–533. doi: 10.1099/00221287-118-2-529. [DOI] [PubMed] [Google Scholar]
- 45.Rupp M E, Archer G L. Hemagglutination and adherence to plastic by Staphylococcus epidermidis. Infect Immun. 1992;60:4322–4327. doi: 10.1128/iai.60.10.4322-4327.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 47.Seki T, Yoshikawa H, Takahashi H, Saito H. Nucleotide sequence of the Bacillus subtilis phoR gene. J Bacteriol. 1988;170:5935–5938. doi: 10.1128/jb.170.12.5935-5938.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smeltzer M S, Hart M E, Iandolo J J. Quantitative spectrophotometric assay for staphylococcal lipase. Appl Environ Microbiol. 1992;58:2815–2819. doi: 10.1128/aem.58.9.2815-2819.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stahl M L, Pattee P A. Confirmation of protoplast fusion-derived linkages in Staphyloccus aureus by transformation with protoplast DNA. J Bacteriol. 1983;154:406–412. doi: 10.1128/jb.154.1.406-412.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stock J B, Ninfa A J, Stock A M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stock J B, Surette M G, Levit M, Park P. Two-component signal transduction systems: structure-function relationships and mechanisms of catalysis. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C.: American Society for Microbiology; 1995. pp. 25–51. [Google Scholar]
- 52.Sugai M, Komatsuzawa H, Akiyama T, Hong Y-M, Oshida T, Miyake Y, Yamaguchi T, Suginaka H. Identification of endo-β-N-acetylglucosaminidase and N-acetylmuramyl-l-alanine amidase as cluster-dispersing enzymes in Staphylococcus aureus. J Bacteriol. 1995;177:1491–1496. doi: 10.1128/jb.177.6.1491-1496.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taylor S S, Knighton D R, Zheng J, Ten Eyck L F, Sowadski J M. Structural framework for the protein kinase family. Annu Rev Cell Biol. 1992;8:429–462. doi: 10.1146/annurev.cb.08.110192.002241. [DOI] [PubMed] [Google Scholar]
- 54.Tseng H C, Cheng C W. A cloned ompR-like gene of Streptomyces lividans 66 suppresses defective melC1, a putative copper-transfer gene. Mol Microbiol. 1991;5:1187–1196. doi: 10.1111/j.1365-2958.1991.tb01892.x. [DOI] [PubMed] [Google Scholar]
- 55.Tsung K, Brissette R E, Inouye M. Identification of the DNA-binding domain of the OmpR protein required for transcriptional activation of the ompF and ompC genes of Escherichia coli by in vivo DNA footprinting. J Biol Chem. 1989;264:10104–10109. [PubMed] [Google Scholar]
- 56.van der Lelie D, Schwuchow T, Schwidetzky U, Wuertz S, Baeyens W, Mergeay M, Nies D H. Two-component regulatory system involved in transcriptional control of heavy-metal homeostasis in Alcaligenes eutrophus. Mol Microbiol. 1997;23:493–503. doi: 10.1046/j.1365-2958.1997.d01-1866.x. [DOI] [PubMed] [Google Scholar]
- 57.Via L E, Curcic R, Mudd M H, Dhandayuthapani S, Ulmer R J, Deretic V. Elements of signal transduction in Mycobacterium tuberculosis: in vitro phosphorylation and in vivo expression of the response regulator MtrA. J Bacteriol. 1996;178:3314–3321. doi: 10.1128/jb.178.11.3314-3321.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yabu K, Nishiyama Y, Ochiai T. Protease-induced multicell formation in Staphylococcus haemolyticus. Microbiol Immunol. 1997;41:799–803. doi: 10.1111/j.1348-0421.1997.tb01930.x. [DOI] [PubMed] [Google Scholar]