Abstract
RepA, the replication initiator protein of the IncB plasmid pMU720, acts preferentially in cis. The cis activity of RepA is thought to be mediated by CIS, a 166-bp region of DNA separating the coding region of repA from the origin of replication (ori) of pMU720. To investigate the trans activity of RepA, the repA gene, without its cognate ori, was cloned on a multicopy plasmid, pSU39. The ori on which RepA acts was cloned on pAM34, a plasmid whose replicon is inactive without induction by isopropyl-β-d-thiogalactopyranoside (IPTG). Thus, in the absence of IPTG, replication of the pAM34 derivatives was dependent on activation of the cloned ori by RepA produced in trans from the pSU39 derivatives. The effect of CIS, when present either on the RepA-producing or the ori plasmid or both, on the efficiency of replication of the ori plasmid in vivo, was determined. The presence of CIS, in its native position and orientation, on the RepA-producing plasmid reduced the efficiency of replication of the ori plasmid. This inhibitory activity of CIS was sequence specific and involved interaction with the C-terminal 20 to 37 amino acids of RepA. By contrast, CIS had no effect when present on the ori plasmid. Initiation of replication from the ori in trans was independent of transcription into CIS.
Miniplasmid pMU720, a derivative of the large, low-copy-number, conjugative plasmid pMU707, is a member of incompatibility group B (8). Replication of pMU720 requires the synthesis of the RepA protein, which is rate limiting for replication. Analyses of pMU720 and the closely related IncI1 plasmid ColIb-P9 have shown that expression of repA is inhibited by a small antisense RNA, RNAI, which binds to its complementary sequence in the leader region of the repA mRNA (29, 38). Binding of RNAI to its target in repA mRNA prevents formation of a pseudoknot that activates translation of the repA mRNA (1–5, 39, 40, 44–46).
Initiation of replication of pMU720 and ColIb-P9 requires the presence in cis of two DNA sequences, the origin of replication (ori) and CIS; the latter separates the RepA coding sequence from ori (23, 30, 41). Members of the I complex (IncB, IncI1, IncIγ, IncK, and IncZ) as well as IncL/M and IncFII plasmids share this physical arrangement of the three genes required for replication, but the sequences of repA, CIS, and ori are not conserved in all of these plasmids. Thus, RepA, CIS, and ori of pMU720 show extensive homology to the corresponding sequences of ColIb-P9, R64-11, R144-3 (IncI1), R621a (IncIγ), and R387 (IncK) but no significant homology to the sequences of pIE545 (IncZ), R1, and R100 (IncFII) (6, 14, 18, 19, 26, 31, 33). The RepA proteins of pMU720 and the IncL/M plasmid pMU407.1 share ∼40% amino acid identity, the ori sequences of the two plasmids contain conserved motifs, but their CIS regions show no significant similarity to each other (6, 30).
CIS is composed of two domains. The repA-proximal domain (5′CIS) has strong transcription termination activity (23, 30) and is thought to be involved in the loading of RepA onto ori (30). The repA-distal domain (3′CIS) is a spacer whose function is to place sequences within ori at an appropriate distance and on the correct face of the DNA helix vis à vis the repA-proximal domain of CIS (30). The RepA protein of pMU720 does not appear to recognize a specific sequence(s) in CIS, as the CIS of pMU720 can be replaced by the CIS of both pMU407.1 and pIE545 (30). By contrast, RepA shows sequence specificity for ori, in that it activates replication at its cognate ori and the ori of pMU407.1, which shares conserved sequence motifs with the ori of pMU720 but not the ori of pIE545 (30).
The Rep protein of the I complex and IncFII plasmids acts in cis (12, 18, 21, 23–25); i.e., it preferentially activates the ori of the DNA molecule from which its mRNA was transcribed. Studies on the IncFII plasmid R1 have shown that this property of Rep is dependent on the presence of CIS, in its native position and orientation (18). In this study, we used a two-plasmid system, in which the ori is provided on one plasmid and repA is carried on another plasmid, to investigate the trans activity of RepA of pMU720 in vivo and the role played by CIS in this activity. We find that (i) RepA activity in trans is less efficient than activity in cis, (ii) the presence of CIS, in its native orientation, immediately upstream of ori has no effect on the efficiency of replication of the ori plasmid, (iii) the presence of CIS, in its native orientation, immediately downstream of the repA coding sequence of the RepA-producing plasmid reduces the efficiency of replication of the ori plasmid, (iv) this inhibitory activity of CIS appears to be sequence specific and involves interaction with the C-terminal amino acids of RepA, and (v) transcription, from an upstream promoter, into CIS of the ori plasmid is not required for initiation of replication in vivo.
MATERIALS AND METHODS
Bacterial strains, plasmids and phages.
The following strains of Escherichia coli K-12 were used in this study. E. coli JM101 [Δ(lac-proAB) supE thi F′ (traD36 proA+B+ lacIqZΔM15)] (20) was used for cloning and propagating M13 derivatives. XL1 Blue MRF′ [Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)] (Stratagene) was used to grow M13 derivatives which had undergone mutagenesis as described by Vandeyar et al. (42). JP3438 (thr-1 leuB6 thi-1 lacY1 gal-351 supE44 tonA21 hsdR4 rpoB364 recA56) was used for propagating pMU720 derivatives and for all copy number determinations.
Bacteriophage vectors used to clone fragments for DNA sequencing and mutagenesis were M13tg130, M13tg131 (15), and M13tg130BS, a derivative of M13tg130 which contains unique recognition sequences for restriction endonucleases SacII and BglII in its multiple cloning site. The plasmids used are described in Table 1.
TABLE 1.
Plasmids used in this study
| Plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| pMU720 | Miniplasmid derived from pMU707; Gal IncB | 8 |
| pMU604 | Miniplasmid derived from pMU407.1; Gal IncL/M | 11 |
| pMU605 | Miniplasmid derived from R64-11; Gal IncI1 | 11 |
| pMU2200 | Miniplasmid derived from pIE545; Gal IncZ | 31 |
| pMU2209 | Miniplasmid derived from R387; Gal IncK | 31 |
| pMU1598 | pAM34 carrying nt 1–2170 of pMU720 with a T643C copy up mutation; Ap Cm lacIq IncB | 30 |
| pMU1599 | pAM34 carrying nt 1–2170 of pMU720; Ap Cm lacIq IncB | 30 |
| pMU1583 | pSU39 carrying nt 1–1759 of pMU720 with T642C and T640C mutations; Km IncB; RepA plasmid | This study |
| pMU1584 | pSU39 carrying nt 1–1843 of pMU720 with T642C and T640C mutations; Km IncB; RepA plasmid | This study |
| pMU1585 | pSU39 carrying nt 1–1916 of pMU720 with T642C and T640C mutations; Km IncB; RepA plasmid | This study |
| pMU1586 | pAM34 carrying nt 1916–2170 of pMU720; Ap Cm lacIq; ori plasmid | This study |
| pMU1587 | pAM34 carrying nt 1840–2170 of pMU720; Ap Cm lacIq; ori plasmid | This study |
| pMU1588 | pAM34 carrying nt 1759–2170 of pMU720; Ap Cm lacIq; ori plasmid | This study |
| pMU1589 | pAM34 carrying nt 1825–2160 of pMU604; Ap Cm lacIq; ori plasmid | This study |
| pAM34 | pMB1 derivative in which the preprimer RNA is expressed from lacZpo; lacIq Ap Sp | 13 |
| pSU39 | p15A replicon; Km | 7 |
Abbreviations: Ap, ampicillin resistance; Cm, chloramphenicol resistance; Km, kanamycin resistance; Sp, spectinomycin resistance; nt, nucleotides. Mutations introduced into RepA and ori plasmids are described in Results.
Media, enzymes, and chemicals.
The minimal medium used was half-strength buffer 56 (22) supplemented with 0.2% glucose, thiamine (10 μg/ml), and necessary growth factors. Luria-Bertani (LB [35]) agar containing basic fuchsin (3 μg/ml) was used as indicator medium for E. coli cells producing chloramphenicol acetyltransferase (CAT). Cells containing high levels of CAT produce dark red colonies on this medium, whereas those containing little or no CAT produce light colonies (32). Enzymes and chemicals of a suitable grade were purchased commercially and not purified further. [35S]dATPαS (>1,000 Ci/mmol) for use in sequencing was obtained from Amersham Corporation. LB-Km-Ap was LB agar supplemented with kanamycin (20 μg/ml) and ampicillin (50 μg/ml). Chloramphenicol was used at 10 μg/ml, isopropylthiogalactoside (IPTG) was used at 1 mM, and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) was used at 25 μg/ml.
Recombinant DNA techniques.
Plasmid and bacteriophage DNA were isolated and manipulated as described by Sambrook et al. (35). DNA was sequenced using a model 373 DNA sequencer and ABI Big Dye Terminator kits (Perkin-Elmer Corporation) or by the method of Sanger et al. (36), modified in that T7 DNA polymerase was used instead of the Klenow fragment and terminated chains were uniformly labeled with [35S]dATPαS. Oligonucleotide-directed in vitro mutagenesis reactions were performed on single-stranded M13 templates by the method of Vandeyar et al. (42). Oligonucleotides were purchased from Bresatec Ltd. or Gibco BRL. DNA sequencing was used to screen for and confirm the presence of mutations.
Construction of repA chimeras.
The coding sequence of the repA gene of the IncL/M plasmid pMU604 (repAL) was amplified to contain a unique BglII site at the 5′ end (at codon 10) and a HindIII site at the 3′ end, just downstream of the stop codon. The amplified fragment was inserted into the BglII/HindIII site of M13tg130BS, and its sequence was determined to ensure that it was free of errors. An error-free clone was used as the template for oligonucleotide-directed mutagenesis, which created unique SphI and SalI sites, at positions corresponding to where these sites occur in the coding sequence of the repA gene of the IncB plasmid pMU720. Creation of these two sites did not change the amino acid sequence of RepAL. RepAB-RepAL chimeras were constructed by exchanging corresponding fragments of the two repA genes. Thus, RepABLL-producing plasmid was made by joining the BglII/SphI fragment of pMU720 to the SphI/HindIII fragment of pMU604, whereas RepABLB plasmid was made by joining the BglI/SphI fragment of pMU720, the SphI/SalI fragment of pMU604 and the SalI/HindIII fragment of pMU720. RepAK/RepAB chimeras were generated by replacing the BamHI/SphI fragment of pMU720 by the BamHI/SphI fragment of the IncK plasmid pMU2209. This resulted in replacement of the IncB rnaI gene, the promoter-proximal sequence of repA (and thus the target of RNAI), and the sequence coding for the first 99 amino acids of RepA, by the corresponding sequences of the IncK plasmid.
Construction of RepA plasmids.
RepA plasmids were derived from pSU39 by insertion of the repA gene of pMU720, with or without the CIS sequence, into the multiple cloning site. These plasmids do not carry the ori of pMU720, so that the RepA protein they produce is freely available to drive the replication of the ori plasmid present in trans. Expression of repA in the RepA plasmids is derepressed due to the presence of promoter down mutations in the rnaI gene. Mutations were introduced by replacing the pMU720 sequences by those being tested.
Construction of ori plasmids.
The ori plasmids were derivatives of pMU1599 (30) in which the IncB replicon has been inactivated by deletion or mutation of repA. Those plasmids that contain repA have had their BamHI/SphI fragment, which carries the rnaI gene and the target for RNAI, replaced by corresponding fragment from the IncK replicon. Since RNAI of the IncK plasmid (RNAIK) does not recognize the target of RNAIB, this exchange ensured that expression of repA on the ori plasmid was regulated at the wild-type level, but expression of repA from the RepA plasmid present in trans remained derepressed. The ori plasmids contain the replicon from pAM34 (13), a modified pMB1 replicon in which the essential preprimer RNA is transcribed from the lacZ promoter operator. Since these plasmids contain the lacIq gene, replication of their pAM34 replicon requires the presence of a lac inducer, such as IPTG. Thus, in the absence of IPTG, replication of the ori plasmids is dependent on the RepA provided in trans by the RepA plasmids. The presence of a CAT reporter gene (derived from pACYC184) allows estimations of the copy numbers of ori plasmids to be made.
Introduction of ori and RepA plasmids into E. coli cells.
ori and RepA plasmids were cotransformed into E. coli K-12 strain JP3438 by the method of Chung et al. (10). Cells were plated onto medium containing half-strength buffer 56 (22), 0.2% glucose, 0.2% Casamino Acids, thiamine (10 μg/ml), ampicillin, chloramphenicol, and kanamycin, with and without IPTG, and incubated for 72 h at 37°C. Plates were checked after 48 and 72 h of incubation, and the number and size of colonies produced in the presence and absence of IPTG were compared. Single colonies from plates without IPTG were used for copy number estimations.
Measurement of CAT activity.
CAT activity of mid-log-phase cultures, grown in minimal medium containing 0.4% glucose, thiamine, leucine, threonine, kanamycin, ampicillin, and chloramphenicol, was assayed as described by Shaw (37). Cells were disrupted by sonication in a Braun Labsonic 2000 Sonicator and cellular debris removed by centrifugation before use in assays. Each assay was performed at least six times. CAT activity was expressed as units per milligram of protein.
Protein assay.
The concentration of protein in cleared cell lysates was determined by the method of Bradford (9), using bovine serum albumin as a standard.
Determination of plasmid copy number.
The copy number of ori plasmids was estimated by comparing CAT activity of cells carrying these plasmids to activity of cells carrying pMU1598. pMU1598 had been shown to have a copy number of ∼20 (30). The formula used was (CAT activity of ori plasmid)/CAT activity of pMU1598 × 20.
RESULTS
Does the presence of CIS affect the ability of ori to be activated by RepA provided in trans?
In pMU720, CIS is thought to facilitate the loading of RepA onto the ori (30). To determine whether CIS also performs this function when RepA is provided in trans, a two-plasmid system was developed. RepA was produced from a multicopy (∼20 copies per E. coli chromosomal equivalent) plasmid pSU39 (7) carrying an intact repA gene, so that both the transcription and the translation of repA mRNA were driven by its native sequences. Synthesis of RepA was modulated by the introduction of promoter down mutations into the gene for the antisense RNA, RNAI (43). The three mutations used changed the −10 box of the rnaI promoter from TATACT to TgTACT (RNAI.1), TgTgCT (RNAI.2), and TgTgCg (RNAI.3), causing progressive decrease in expression of rnaI and a corresponding increase in expression of repA (43). Expression of repA is fully derepressed in the RNAI.3 mutant (43, 45). Since RepA acts preferentially in cis, the RepA-producing plasmids were constructed so that they did not carry their cognate, active ori sequences, which might trap RepA protein and make it unavailable to activate replication of the ori plasmid in trans. The ori plasmids were derivatives of pAM34, which carries a fully repressible pMB1 replicon (13). This replicon is inactive in the absence of an inducer but can be switched on by the addition of IPTG. The ori sequences were inserted immediately downstream of a transcription terminator, to ensure that transcription from the β-lactamase gene, which is located upstream of the site of insertion, did not read through into ori. The ori plasmids carry a gene encoding a constitutively expressed CAT, which was used to monitor the plasmid copy number. All tests were carried out by cotransforming E. coli K-12 strain JP3438 with the ori and RepA plasmids and selecting for both plasmids, in the presence and absence of IPTG. CAT assays were carried out on transformants selected and grown in the absence of IPTG. RepA-ori plasmid combinations, which produced colonies only in the presence of IPTG, were scored as unable to activate replication of the ori plasmid in trans.
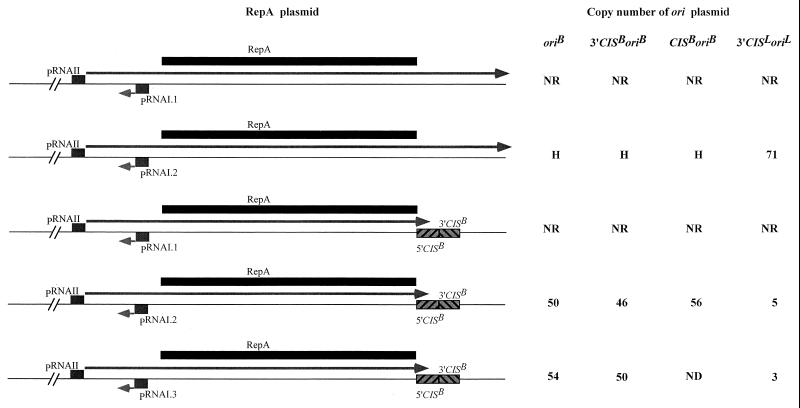
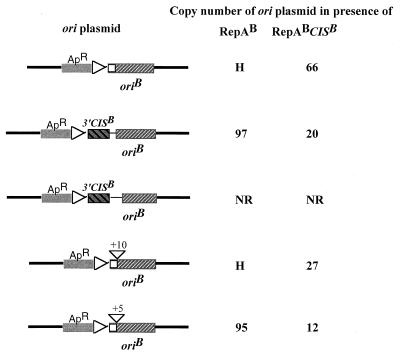
As shown in Fig. 1, RepA plasmids carrying the RNAI.1 mutation were unable to support the replication of the ori plasmids in trans. Introduction of the RNAI.2 and RNAI.3 mutation into the rnaI gene of the RepA plasmids resulted in an efficient activation of the ori present in trans. This suggests that the failure of the RNAI.1 mutants to activate replication of the ori plasmid was caused by their inability to produce enough RepA. The presence of 3′CIS or the intact CIS of the IncB replicon (CISB), in its native position and orientation on the oriB plasmid, had no effect on its efficiency of replication. However, the presence of CISB in its native position and orientation on the RepA plasmid resulted in a decrease in the copy number of the ori plasmid in trans. Unfortunately, it was impossible to measure the extent of this effect with oriB plasmids, because estimations of their copy number in the presence of the RepA plasmid that was not carrying CISB were not reproducible, ranging from ∼10 to ∼70 per chromosomal equivalent. Analysis on agarose gel of plasmid DNA extracted from cells cotransformed with the oriB plasmid and the RepA plasmid that was not carrying CISB revealed that the ratio of ori plasmid DNA to RepA plasmid DNA varied from colony to colony but did not appear to be less than 1. The cotransformants produced extremely small colonies, in both the presence and absence of IPTG, and exhibited segregational instability, as evidenced by the production of very small, dark red colonies showing extensive sectoring, on LB-Km-Ap plates containing basic fuchsin, an indicator for CAT. The sectored areas of these colonies were lighter colored and faster growing than the bulk of the colony. Upon restreaking on LB-Km-Ap-basic fuchsin plates, the cells from the red portion of the sectored colonies produced sectored colonies again, whereas cells from the light-colored sectors failed to grow. The latter cells did grow on LB-Km-basic fuchsin plates (i.e., in the absence of selection for the oriB plasmid), producing light-colored colonies. The simplest interpretation of these data is that the copy number of the oriB plasmid in the presence of RepA-producing plasmid that does not carry CIS is too high to be sustained, resulting in selection of faster-growing cells that have lost the oriB plasmid. In the IncB replicon, replacement of the ori by the corresponding sequence from the IncL/M replicon (oriL) reduced the copy number of the replicon (30), indicating that RepA interacts less efficiently with this heterologous ori than with its cognate ori. Therefore, we examined the copy number of the oriL plasmid in the presence of RepA-producing plasmid that does not carry CIS, to determine whether it was low enough to be sustainable. The copy number was found to be ∼70 chromosome equivalents/cell, and the estimations were reproducible; thus, the effect of CIS could be measured. The copy number of the oriL plasmid was reduced more than 10-fold when the RepA-producing plasmid carried CISB. These data are consistent with the notion that the segregational instability of the oriB plasmid in the presence of RepA plasmid lacking CISB is due to its frequency of replication being too high to be sustained.
FIG. 1.
Effects of mutations in rnaI, which increase expression of repA, and the presence of CIS on the RepA plasmid, on the copy number of oriB and oriL plasmids. RNAI, antisense RNA regulating the expression of repA of pMU720 replicon; RNAII, repA mRNA; RNAI.1, RNAI.2, and RNAI.3 mutations that change the −10 box of the rnaI promoter from TATACT to TgTACT, TgTgCT, and TgTgCg, respectively. Copy number was determined by assaying the CAT activity of E. coli strain JP3438 carrying the ori plasmids, in the absence of IPTG. The values shown are the average of at least six independent determinations. ND, not done; NR, ori plasmid unable to replicate without the induction of the pAM34 replicon; H, copy number too high for accurate determination; →, RNA transcript.
Does 3′CIS play a role in reducing the trans activity of RepA?
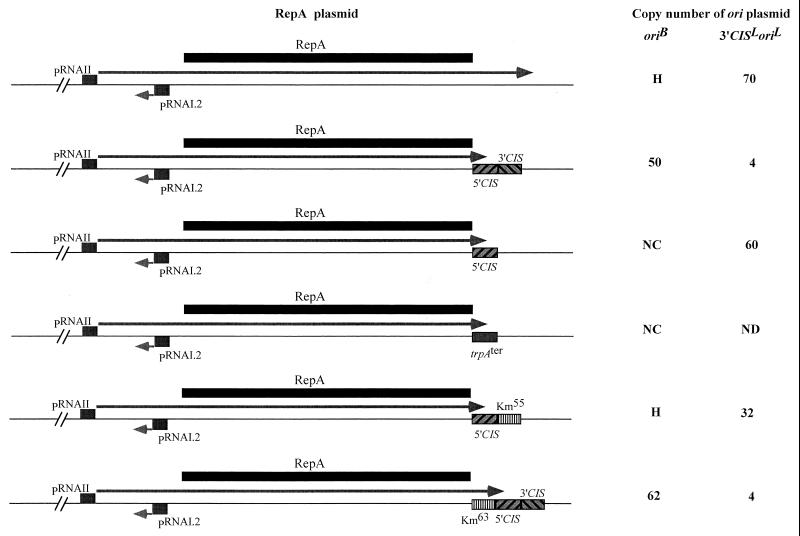
In the intact IncB replicon, where RepA activates the ori present in cis, the 3′CIS acts as a spacer for appropriately positioning 5′CIS and ori with respect to each other and can be replaced by an unrelated sequence of the same length without any loss in activity (30). Since this role should be superfluous when CIS is carried on a RepA-producing plasmid that has no cognate ori, we determined whether the presence of 3′CISB had any effect on the ability of RepA to activate ori in trans. Deletion of 3′CISB from the RepA-producing plasmid resulted in the loss of the inhibitory activity seen with the intact CISB (Fig. 2). Moreover, replacement of 3′CIS by the same spacer fragment (Km55) that was its fully functional substitute in cis (30) did not restore the inhibitory activity of CIS. Insertion of a 63-bp spacer (Km63) between repA coding sequence and the intact CIS had no effect on the trans activity of RepA (Fig. 2). By contrast, this insertion resulted in a three- to sixfold reduction in the copy number of the intact IncB replicon (30). Insertion of 5′CISB or the trpA transcription terminator, immediately downstream of the repA coding sequence of the RepA-producing plasmid, appeared to result in the runaway replication of the oriB plasmid, as no transformants could be detected even on the induction of the pAM34 replicon. Since both trpA terminator and 5′CISB produced the same effect, it is most likely due to prevention of read-through transcription from repA into the p15A replicon of the RepA plasmid. If, as seems likely, such read-through has a deleterious effect on the replication of the RepA plasmid, then obviation of this effect by efficient termination of repA transcripts would be expected to increase or stabilize the copy number of this plasmid and thus lead to an increase in the level of RepA available to the ori plasmid. Since the frequency of replication of the oriB plasmid in the presence of RepA plasmid that lacks CISB was already so high as to produce tiny, segregationally unstable colonies, even a small increase in this frequency might be expected to lead to runaway replication.
FIG. 2.
Effects of deletions, insertions, and substitutions in CIS of the RepA-producing plasmid on the copy number of oriB and oriL plasmids. Copy number was determined by assaying the CAT activity of E. coli strain JP3438 carrying the ori plasmids, in the absence of IPTG. The values shown are the average of at least six independent determinations. trpAter, 28-bp trpA transcription terminator; Km55 and Km63, 55- and 63-bp DNA fragments amplified from the coding sequence of aminoglycoside 3′-phosphotransferase; NC, no transformants detected, regardless of whether the pAM34 replicon was induced or not; ND, not done; H, copy number too high for accurate determination; →, RNA transcript.
Characterization of sequences involved in reducing the trans activity of RepA.
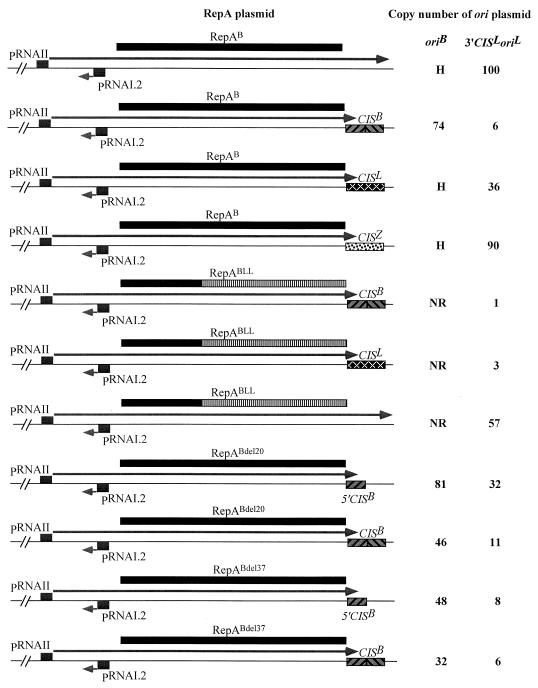
In the IncB replicon, CIS (CISB) could be replaced by the corresponding sequences from pMU407.1 (CISL) and pIE545 (CISZ), with only slight loss of function (30). However, replacement of CISB in the RepA-producing plasmid, by either CISL or CISZ, resulted in almost complete loss of inhibition of the trans activity of RepA (Fig. 3), suggesting that this effect is a result of sequence-specific interactions between RepA and CIS. To investigate this interaction further, the repA gene on the RepA-producing plasmid was replaced by a chimeric gene, in which the promoter and promoter-proximal portion of the coding sequence came from pMU720 and the remainder of the coding sequence was from pMU407.1. The resulting protein, RepABLL, is composed of the N-terminal 99 amino acids of RepAB and the C-terminal 247 amino acids of RepAL. RepABLL resembles RepAL in that it can activate oriL present in cis but fails to activate replication from oriB. The copy number of the replicon carrying the repABLL gene is sevenfold lower than that of the one with repAL-gene (data not shown). The activity of RepABLL in trans was reduced 57-fold by CISB and 19-fold by CISL (Fig. 3), suggesting that this protein, unlike RepAB, is able to recognize both the CIS sequences.
FIG. 3.
Effects of heterologous and homologous RepA-CIS combinations and C-terminal deletions in RepA on the copy number of oriB and oriL plasmids. Copy number was determined by assaying the CAT activity of E. coli strain JP3438 carrying the ori plasmids, in the absence of IPTG. The values shown are the average of at least six independent determinations. NR, ori plasmid unable to replicate without the induction of the pAM34 replicon; H, copy number too high to be determined accurately; →, RNA transcript.
To determine whether the sequence, or part of the sequence, recognizing CIS lies within the C-terminal portion of RepA, deletion derivatives missing the last 20 (RepABdel20) and the last 37 (RepABdel37) amino acids were tested. IncB replicons carrying these mutant repA genes are unable to replicate (I. W. Wilson, J. Praszkier, and A. J. Pittard, unpublished data) unless expression of repA is increased by the introduction of the RNAI.1 mutation. However, the efficiency of replication of these repA/rnaI mutants is severely affected, as evidenced by the 11-fold (repABdel20) and 59-fold (repABdel37) reduction in their copy number (data not shown). Comparison of the trans activity of RepABdel20 produced from the plasmid carrying 5′CISB with one produced from the plasmid carrying an intact CISB showed that the inhibitory effect of CISB (3-fold with 3′CISL oriL) was much lower than that seen with the intact RepAB (17-fold with 3′CISL oriL) (Fig. 3). Deletion of further 17 amino acids from this protein (RepABdel37) resulted in almost complete loss of the inhibitory effect of CISB (Fig. 3; compare RepABdel37 5′CISB with RepABdel37 CISB).
Does the presence of the repA gene on the ori plasmid increase its ability to respond to RepA produced in trans?
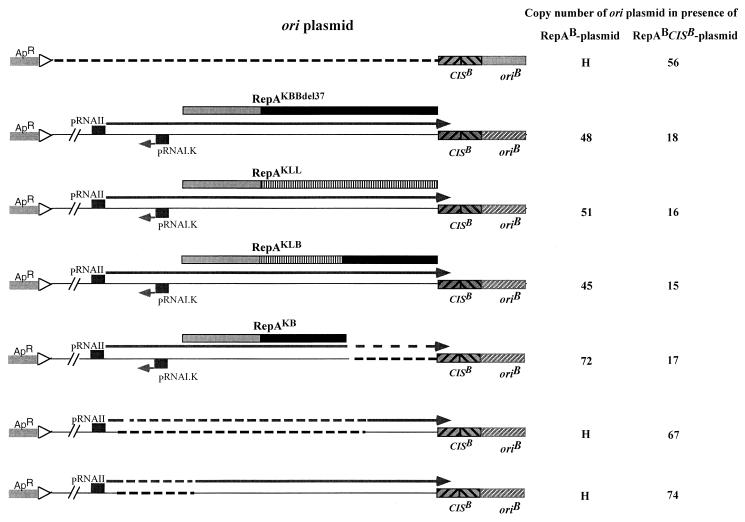
It has been suggested that the mRNA for the Rep protein of the IncFIC replicon of plasmid P307, which is closely related to the replicon of pMU720, is required not only for the synthesis of the initiator protein but also to prime initiation of DNA synthesis (16). It was postulated that the repA mRNA is processed at the junction of the RepA coding sequence and CIS-ori, producing a replication primer that is used by the polymerase for initiation of leading-strand synthesis (16). However, as there was no transcription directed towards the ori region of the plasmids used in the assays described above, synthesis of such a primer is not essential for activation of ori in trans. To determine whether transcription of repA mRNA, upstream of ori, enhances its ability to be activated by RepA produced in trans, IncB replicons encoding mutant RepA proteins unable to initiate replication from the ori present in cis were used as ori plasmids. To prevent RNAI (a trans-acting molecule) produced by these replicons from repressing the synthesis of RepA from the RepA-producing plasmid, the DNA fragment encoding their rnaI gene and the promoter-proximal part of their repA gene was replaced by the corresponding fragment from the IncK plasmid, pMU2209. RNAIK does not interact with the repA mRNA of the IncB plasmid pMU720 (31). Exchange of the DNA fragment carrying rnaI resulted also in replacement of the N-terminal 99 amino acids of RepAB by the corresponding sequence from RepAK. However, these two proteins are so similar that only two amino acids are changed by this exchange (30).
All three repA genes used in these experiments differed in their 3′ coding sequences. The first gene had the sequence from pMU720, but its termination codon was present 112 bp upstream of the wild-type position (repABdel37). The second gene had the sequence from pMU604 (repAKLL), and the third gene contained the wild-type sequence from pMU720 (repAKLB). Despite these differences, all three ori plasmids replicated with very similar efficiencies, resulting in copy numbers of 45 to 51 and 15 to 18 in the absence and presence, respectively, of CIS on the RepA-producing plasmid (Fig. 4). These copy numbers are significantly lower than those seen when the only pMU720 sequences present on the ori plasmid are CIS and ori and there is no transcription reading into CIS (Fig. 4, compare lines 2 to 4 with line 1). Furthermore, deletion of the coding sequence for the last 121 amino acids of RepA (RepAKB) from the repA gene of the ori plasmid had no significant effect on its copy number. However, deletion of a sequence encompassing the translation initiation region of repA increased the copy number of the ori plasmid to levels similar to those seen with ori alone, regardless of whether all or only part of the coding sequence of RepA was present on the ori plasmid (Fig. 4). These data suggest that transcriptional activity immediately upstream of CIS-ori has no effect on the efficiency of replication of the ori plasmid, but that synthesis of replication-defective RepA from the ori plasmid reduces the copy number of this plasmid. However, the possibility that removal of translational activity had a polar effect on the level of transcription reading into CIS, which contributed to the observed increase in the copy number of the ori plasmids, cannot be discounted.
FIG. 4.
Effects of deletions and substitutions in its repA coding sequence, and the presence of repA transcription and translation initiation signals, on the ability of the ori plasmid to be activated for replication by RepA provided in trans. Copy numbers were determined by assaying the CAT activity of E. coli strain JP3438 carrying the ori plasmids, in the absence of IPTG. The values shown are the average of at least six independent determinations. H, copy number too high to be determined accurately. ApR, β-lactamase gene; ▹, transcription terminator; →, RNA transcript; ––––, region deleted; RNAI.K, replacement of the sequence encoding the IncB RNAI and its target by the corresponding sequence from the IncK replicon.
The finding that synthesis of replication-defective RepA from the ori plasmid reduces its copy number raised the possibility that these proteins are able to bind to ori and block initiation of replication by the replication-proficient RepA produced in trans. Since these proteins are produced in cis, they should be loaded onto ori more efficiently than the protein produced from the RepA plasmid. However, comparison of an ori plasmid carrying repAKBBdel37 to one carrying repAKBB showed that their copy numbers were not significantly different from each other (data not shown). The latter plasmid is able to replicate in the absence of RepA in trans, with a copy number of 7, which is comparable to that of the wild-type pMU720 replicon. This copy number increased ∼10-fold in presence of a RepA-producing plasmid and ∼4-fold in presence of one in which CISB is present immediately downstream of the repA coding sequence. A plasmid which was unable to replicate in the absence of a RepA in trans because its 3′CISB was deleted reached copy number not significantly different from those of the ori plasmids carrying repAKBBdel37 and repAKBB, in the presence of RepA-producing plasmids (data not shown). Deletion of the 3′CIS is thought to reduce the efficiency of loading of RepA onto ori present in cis (30). Thus, ori plasmids that produce RepA replicate with a lower copy number than ones that do not, irrespective of whether the protein is replication proficient or not. Moreover, all of these plasmids replicate with similar copy numbers, suggesting that the same mechanism is operating in all of them to lower their frequency of initiation of replication.
Effects of mutations involving the DnaA box on the copy number of the ori plasmid.
The ori of pMU720 contains the sequence 5′ TTATCCACA 3′, which is a consensus sequence for a binding site of the DnaA protein (DnaA box). Although this sequence is not essential for replication of pMU720, its deletion lowers the copy number of the wild-type plasmid 3-fold and the copy number of the RNAI.1 mutant 1.6-fold (30). Deletion of the DnaA box from the ori plasmid reduced its copy number 3.3-fold (Fig. 5). As found with ori present in cis, deletion of additional 10 bp inactivated the ori. Insertion of 10 bp immediately downstream of the DnaA box reduced the copy number of the ori plasmid 2.4-fold, whereas insertion of 5 bp reduced it 5.5-fold (Fig. 5). These data show that the DnaA box is required for efficient activation of ori in trans and that its effectiveness is largely lost when it is moved by as little as one turn of the DNA helix. Moving the box by half a helical turn had a greater effect than moving it by a full turn, indicating that it must be positioned on the correct face of the helix relative to other sequences within ori.
FIG. 5.
Effects of mutations in and immediately downstream of the DnaA box on the ability of the ori plasmid to be activated for replication by RepA provided in trans. Copy numbers were determined by assaying the CAT activity of E. coli strain JP3438 carrying the ori plasmids, in the absence of IPTG. The values shown are the average of at least six independent determinations. NR, ori plasmid unable to replicate without the induction of the pAM34 replicon; H, copy number too high to be determined accurately; the thin line, region deleted; □, DnaA box; ▿, 5- or 10-bp insertion; ▹, transcription terminator.
DISCUSSION
RepA protein of IncB and IncI1 plasmids is cis acting, preferentially activating the origin of replication on the DNA molecule from which its mRNA was transcribed (23–25). We have constructed a two-plasmid system for studying the activity of RepA, when it is present in trans to the ori on which it acts. Using this system, we show that in the absence of a cognate ori in cis, RepA does activate an ori present in trans. However, this activation requires a much higher level of repA expression than activation of ori in cis, confirming that some aspect of the interaction between RepA and the ori in trans is inefficient. The level of repA expression necessary to initiate replication from an ori in trans is unsustainable when the ori is present in cis, as IncB replicons carrying the RNAI.2 mutation cannot be recovered, presumably due to their runaway replication (data not shown).
The presence of CIS, in its native position and orientation, on the RepA-producing plasmid reduced the copy number of the ori plasmid. This effect required the presence of 3′CIS and involved sequence-specific interaction between CIS and the C terminus of RepA. Thus, it is unlike the interaction between RepA and CIS that is proposed to be the first step in the loading of this protein onto the ori in cis (30). These data suggest that CIS has an additional role, which involves the trapping and/or inactivation of the initiator protein. The current model for the control of frequency of replication initiation of IncB and other I-complex plasmids involves translational regulation of the synthesis of RepA. For this system to work satisfactorily, it is necessary for RepA to be synthesized de novo and to be modified during its first round of initiation, so that it is not available for initiation of subsequent rounds of DNA synthesis. The observed interaction between RepA and CIS might ensure that the initiator protein does not participate in more than one round of replication. The finding that this interaction is sequence specific solves the apparent anomaly posed by the observation that the 3′CIS acts as a nonspecific spacer for loading of RepA in cis (30) yet its sequence is very highly conserved in the I-complex plasmids (14, 31). Studies on the replication initiator protein of the IncFII plasmid NR1 also showed that the presence in cis of the CIS-ori region results in an apparent “titration” of the protein (12). Because this effect was lost when the sequence encoding 3′CIS ori was deleted, it was suggested that the “titration” of Rep was the consequence of its binding to ori. Our data show that, at least in the case of RepA of the IncB plasmid and the chimera composed of N-terminal sequence of IncB and C-terminal sequence of IncL/M proteins, the presence of CIS alone is sufficient to reduce the amount of RepA available for activation of ori in trans.
The only sequence required in trans to the RepA-producing plasmid was ori, supporting the notion that in the intact replicon CIS is involved in loading of RepA onto the ori, but not in the initiation of replication. The efficiency of initiation of replication was not influenced by the presence or absence of transcription into the region upstream of ori, indicating that such transcripts are not involved in priming of DNA synthesis. Initiation of replication of IncFII plasmids was also shown to be independent of transcription into the ori region (12, 27).
Synthesis of RepA protein in cis to ori reduced the copy number of the ori plasmid, regardless of whether the protein was competent for replication or not. The finding that all constructs tested, including one missing the 3′CIS and thus predicted to be impaired in the loading of RepA onto ori in cis, showed similar reduced levels of activation by RepA provided in trans indicates that the mechanism responsible for this phenomenon is unlikely to involve competition between cis- and trans-produced RepA for binding sites in ori. This effect was seen even with proteins carrying C-terminal deletions, showing that its cause is different to the interaction that reduced the availability of protein from the RepA-producing plasmid. It has been suggested that loading of RepA in cis might involve bending of DNA to bring ori and the RepA positioned on 5′CIS close together (30). If that is the case, then it is likely that in this closed-configuration ori is less easily accessible to RepA produced in trans.
All I-complex, IncFII, and FIC plasmids examined to date encode a DnaA box at the 5′ end of ori (14, 26, 31, 34). Although this box is not part of minimal ori, its deletion or inactivation reduces the efficiency of replication of IncB and IncFII plasmids (28, 30). Deletion of the DnaA box reduced the copy number of the ori plasmid, showing that the efficiency of activation of ori in trans is also dependent on this sequence. Analysis of binding between RepA and ori of the IncB plasmid in vitro showed that the protein binds immediately downstream of the DnaA box (T. Betteridge, J. Praszkier, J. Yang, and A. J. Pittard, unpublished data). Therefore, the finding that moving the DnaA box by one turn of the helix reduced the copy number of the ori plasmid, and that moving the box by half a helical turn reduced it even more, suggests cooperative interaction between DnaA and RepA. Binding in vitro of DnaA protein to the DnaA box located at the 5′ end of the ori of the IncFII plasmid R1 required the presence of RepA, which is indicative of cooperative interaction between the two proteins (17).
The use of a repressible replicon as delivery vector for ori allowed us to distinguish between RepA-ori combinations that resulted in lack of replication of the ori plasmid and those that led to too high a level of replication. Thus, dependence of cotransformation of E. coli cells by ori and RepA plasmids on induction of the repressible replicon was a clear indication of failure of replication by the ori plasmid. On the other hand, the inability to detect cotransformants in both the presence and absence of the inducer was taken as evidence of high level of uncontrolled replication by the ori plasmid. This view was supported by the finding that RepA-ori combinations in which the ori plasmids replicated with high copy number formed small colonies, both in the presence and in the absence of the inducer. Replacement of the oriB by oriL, which has been shown to reduce the copy number of the IncB replicon (30), led to an increase in the size of the colonies and a reduction in the copy number of the ori plasmid. Similarly, although cotransformation of E. coli cells by oriB and RepA-producing plasmids carrying 5′CIS failed, replacement of oriB by oriL or deletion of the C-terminal amino acids of RepA led to high efficiency of transformation. Analysis of these transformants showed that the ori plasmid was replicating with high efficiency, reaching a copy number of ∼50 to 60 per chromosomal equivalent (Fig. 2 and 3). In the intact IncB replicon, upward fluctuations in copy number result in increased gene dosage of rnaI. The consequent increase in the concentration of the antisense RNA, RNAI, leads to inhibition of synthesis of RepA, and thus a decrease in frequency of initiation of replication. This feedback loop is missing in our two-plasmid system, because expression of repA from the RepA-producing plasmid is not sensitive to fluctuations in the copy number of the ori plasmid. Consequently, efficient activation of ori in trans may lead to runaway replication of the ori plasmid.
ACKNOWLEDGMENTS
This work was supported by a grant from the National Health and Medical Research Council.
We thank Iain Wilson for providing del20 and del37 mutants of repA, and we thank Thu Betteridge for excellent technical assistance.
REFERENCES
- 1.Asano K, Kato A, Moriwaki H, Hama C, Shiba K, Mizobuchi K. Positive and negative regulations of plasmid ColIb-P9 repZ gene expression at the translational level. J Biol Chem. 1991;266:3774–3781. [PubMed] [Google Scholar]
- 2.Asano K, Mizobuchi K. Copy number control of IncIα plasmid ColIb-P9 by competition between pseudoknot formation and antisense RNA binding at a specific RNA site. EMBO J. 1998;17:5201–5213. doi: 10.1093/emboj/17.17.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asano K, Mizobuchi K. An RNA pseudoknot as the molecular switch for translation of the repZ gene encoding the replication initiator of IncIα plasmid ColIb-P9. J Biol Chem. 1998;19:11815–11825. doi: 10.1074/jbc.273.19.11815. [DOI] [PubMed] [Google Scholar]
- 4.Asano K, Moriwaki H, Mizobuchi K. An induced mRNA secondary structure enhances repZ translation in plasmid ColIb-P9. J Biol Chem. 1991;266:24549–24556. [PubMed] [Google Scholar]
- 5.Asano K, Niimi T, Yokoyama S, Mizobuchi K. Structural basis for binding of the plasmid ColIb-P9 antisense Inc RNA to its target RNA with the 5′-rUUGGCG-3′ motif in the loop sequence. J Biol Chem. 1998;19:11826–11838. doi: 10.1074/jbc.273.19.11826. [DOI] [PubMed] [Google Scholar]
- 6.Athanasopoulos V, Praszkier J, Pittard A J. The replication of an IncL/M plasmid is subject to antisense control. J Bacteriol. 1994;177:4730–4741. doi: 10.1128/jb.177.16.4730-4741.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartolomé B, Jubete Y, Martínez E, de la Cruz F. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene. 1991;102:75–78. doi: 10.1016/0378-1119(91)90541-i. [DOI] [PubMed] [Google Scholar]
- 8.Bird P I, Pittard J. Demonstration of a third incompatibility function on plasmids already incompatible with group P and group I plasmids. Plasmid. 1983;9:191–200. doi: 10.1016/0147-619x(83)90020-3. [DOI] [PubMed] [Google Scholar]
- 9.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 10.Chung C T, Niemela S, Miller R H. One step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci USA. 1989;86:2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davey R B D, Bird P I, Nikoletti S M, Praszkier J, Pittard J. The use of mini-Gal plasmids for the rapid incompatibility grouping of conjugative R plasmids. Plasmid. 1984;11:234–242. doi: 10.1016/0147-619x(84)90029-5. [DOI] [PubMed] [Google Scholar]
- 12.Dong X, Womble D D, Rownd R H. In-vivo studies on the cis-acting replication initiator protein of IncFII plasmid NR1. J Mol Biol. 1988;202:495–509. doi: 10.1016/0022-2836(88)90281-1. [DOI] [PubMed] [Google Scholar]
- 13.Gil D, Bouché J-P. ColE1-type vectors with fully repressible replication. Gene. 1991;105:17–22. doi: 10.1016/0378-1119(91)90508-9. [DOI] [PubMed] [Google Scholar]
- 14.Hama C, Takizawa T, Moriwaki H, Urasaki Y, Mizobuchi K. Organization of the replication control region of plasmid ColIb-P9. J Bacteriol. 1990;172:1983–1991. doi: 10.1128/jb.172.4.1983-1991.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kieny M P, Lathe R, Lecocq J P. New versatile cloning and sequencing vectors based on bacteriophage M13. Gene. 1983;26:91–99. doi: 10.1016/0378-1119(83)90039-2. [DOI] [PubMed] [Google Scholar]
- 16.Maas R, Wang C. Role of the RepA1 protein in RepFIC plasmid replication. J Bacteriol. 1997;179:2163–2168. doi: 10.1128/jb.179.7.2163-2168.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masai H, Arai K. RepA and DnaA proteins are required for initiation of R1 plasmid replication in vitro and interact with the oriR sequence. Proc Natl Acad Sci USA. 1987;84:4781–4785. doi: 10.1073/pnas.84.14.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masai H, Arai K-I. RepA protein and oriR-dependent initiation of R1 plasmid replication: identification of a rho-dependent transcription terminator for cis-action of repA protein. Nucleic Acids Res. 1988;16:6493–6514. doi: 10.1093/nar/16.14.6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masai H, Laziro Y, Arai K-I. Definition of oriR, the minimum DNA segment essential for initiation of R1 plasmid replication in vitro. Proc Natl Acad Sci USA. 1983;80:6814–6818. doi: 10.1073/pnas.80.22.6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- 21.Miki T, Easton A M, Rownd R H. Cloning of replication, incompatibility, and stability functions of R plasmid NR1. J Bacteriol. 1980;141:87–99. doi: 10.1128/jb.141.1.87-99.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monod J, Cohen-Bazire G, Cohen M. Sur la biosynthèse de la β-galactosidase (lactase) chez Escherichia coli. La specificité de l'induction. Biochim Biophys Acta. 1951;7:585–599. doi: 10.1016/0006-3002(51)90072-8. [DOI] [PubMed] [Google Scholar]
- 23.Mori A, Ito K, Mizobuchi K, Nakamura Y. A transcription terminator signal necessary for plasmid ColIb-P9 replication. Mol Microbiol. 1995;17:291–301. doi: 10.1111/j.1365-2958.1995.mmi_17020291.x. [DOI] [PubMed] [Google Scholar]
- 24.Murthy S. Studies on the cis action of the RepA protein of a group B plasmid of the enteric bacteria. B.Sc. (Honors) thesis. Melbourne, Victoria, Australia: The University of Melbourne; 1997. [Google Scholar]
- 25.Nikoletti S M. Molecular studies on incompatibility properties of I complex plasmids. Ph.D. thesis. Melbourne, Victoria, Australia: The University of Melbourne; 1986. [Google Scholar]
- 26.Ohtsubo H, Ryder T B, Maeda Y, Armstrong K, Ohtsubo E. DNA replication of the resistance plasmid R100 and its control. Adv Biophys. 1986;21:115–133. doi: 10.1016/0065-227x(86)90018-3. [DOI] [PubMed] [Google Scholar]
- 27.Ortega S, Lanka E, Diaz R. The involvement of host replication proteins and of specific origin sequences in the in vitro replication of miniplasmid R1 DNA. Nucleic Acids Res. 1986;14:4865–4879. doi: 10.1093/nar/14.12.4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ortega-Jiménez S, Giraldo-Suárez R, Fernández-Tresguerres M E, Berzal-Herranz A, Díaz-Orejas R. DnaA dependent replication of plasmid R1 occurs in the presence of point mutations that disrupt the dnaA box of oriR. Nucleic Acids Res. 1992;20:2547–2552. doi: 10.1093/nar/20.10.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Praszkier J, Bird P, Nikoletti S, Pittard A J. Role of countertranscript RNA in the copy number control system of an IncB miniplasmid. J Bacteriol. 1989;171:5056–5064. doi: 10.1128/jb.171.9.5056-5064.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Praszkier J, Pittard A J. Role of CIS in replication of an IncB plasmid. J Bacteriol. 1999;181:2765–2722. doi: 10.1128/jb.181.9.2765-2772.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Praszkier J, Wei T, Siemering K, Pittard J. Comparative analysis of the replication regions of the IncB, IncK, and IncZ plasmids. J Bacteriol. 1991;173:2393–2397. doi: 10.1128/jb.173.7.2393-2397.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Proctor G N, Rownd R H. Rosanilins: indicator dyes for chloramphenicol-resistant enterobacteria containing chloramphenicol acetyltransferase. J Bacteriol. 1982;150:1375–1382. doi: 10.1128/jb.150.3.1375-1382.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ryder T B, Davison D B, Rosen J I, Ohtsubo E, Ohtsubo H. Analysis of plasmid genome evolution based on nucleotide-sequence comparison of two related plasmids of Escherichia coli. Gene. 1982;17:299–310. doi: 10.1016/0378-1119(82)90146-9. [DOI] [PubMed] [Google Scholar]
- 34.Saadi S, Maas W K, Hill D F, Bergquist P L. Nucleotide sequence analysis of RepFIC, a basic replicon present in IncFI plasmids P307 and F, and its relation to the RepA replicon of IncFII plasmids. J Bacteriol. 1987;169:1836–1846. doi: 10.1128/jb.169.5.1836-1846.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 36.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaw V W. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 1975;43:737–755. doi: 10.1016/0076-6879(75)43141-x. [DOI] [PubMed] [Google Scholar]
- 38.Shiba K, Mizobuchi K. Posttranscriptional control of plasmid ColIb-P9 repZ gene expression by a small RNA. J Bacteriol. 1990;172:1992–1997. doi: 10.1128/jb.172.4.1992-1997.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siemering K R, Praszkier J, Pittard A J. Interaction between the antisense and target RNAs involved in the regulation of IncB plasmid replication. J Bacteriol. 1993;175:2895–2906. doi: 10.1128/jb.175.10.2895-2906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siemering K R, Praszkier J, Pittard A J. Mechanism of binding of the antisense and target RNAs involved in the regulation of the IncB plasmid replication. J Bacteriol. 1994;176:2677–2688. doi: 10.1128/jb.176.9.2677-2688.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanaka K, Sakai H, Honda Y, Nakamura T, Higashi A, Komano T. Structural and functional features of cis-acting sequences in the basic replicon of plasmid ColIb-P9. Nucleic Acids Res. 1992;20:2705–2710. doi: 10.1093/nar/20.11.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vandeyar M A, Weiner M P, Hutton C J, Batt C A. A simple and rapid method for the selection of oligodeoxynucleotide-directed mutants. Gene. 1988;65:129–133. doi: 10.1016/0378-1119(88)90425-8. [DOI] [PubMed] [Google Scholar]
- 43.Wilson I W. Molecular analysis of the replication control system of IncB plasmids. Ph.D. Melbourne, Victoria, Australia: The University of Melbourne; 1995. [Google Scholar]
- 44.Wilson I W, Praszkier J, Pittard A J. Mutations affecting pseudoknot control of the replication of B group plasmids. J Bacteriol. 1993;175:6476–6483. doi: 10.1128/jb.175.20.6476-6483.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilson I W, Praszkier J, Pittard A J. Molecular analysis of RNAI control of repB translation in IncB plasmids. J Bacteriol. 1994;176:6497–6508. doi: 10.1128/jb.176.21.6497-6508.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson I W, Siemering K R, Praszkier J, Pittard A J. Importance of structural differences between complementary RNA molecules to control of replication of an IncB plasmid. J Bacteriol. 1997;179:742–753. doi: 10.1128/jb.179.3.742-753.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]