Abstract
Expression of the tryptophanase (tna) operon of Escherichia coli is regulated by catabolite repression and by tryptophan-induced transcription antitermination. Tryptophan induction prevents Rho-dependent transcription termination in the leader region of the operon. Induction requires translation of a 24-residue leader peptide-coding region, tnaC, containing a single, crucial Trp codon. Studies with a lacZ reporter construct lacking the tnaC-tnaA spacer region suggest that, in the presence of excess tryptophan, the TnaC leader peptide acts in cis on the ribosome translating tnaC to inhibit its release. The stalled ribosome is thought to block Rho's access to the transcript. In this paper we examine the roles of the boxA sequence and the rut site in Rho-dependent termination. Deleting six nucleotides (CGC CCT) of boxA or introducing specific point mutations in boxA results in high-level constitutive expression. Some constitutive changes introduced in boxA do not change the TnaC peptide sequence. We confirm that deletion of the rut site results in constitutive expression. We also demonstrate that, in each constitutive construct, replacement of the tnaC start codon by a UAG stop codon reduces expression significantly, suggesting that constitutive expression requires translation of the tnaC coding sequence. Addition of bicyclomycin, an inhibitor of Rho, to these UAG constructs increases expression, demonstrating that reduced expression is due to Rho action. Combining a boxA point mutation with rut site deletion results in constitutive expression comparable to that of a maximally induced operon. These results support the hypothesis that in the presence of tryptophan the ribosome translating tnaC blocks Rho's access to the boxA and rut sites, thereby preventing transcription termination.
The enzyme tryptophanase catalyzes the degradation of l-tryptophan to indole, pyruvate, and ammonia (20, 39). Bacterial species that produce this enzyme can utilize tryptophan as a source of carbon, nitrogen, and energy (16). Tryptophanase can also catalyze the reverse reaction and synthesize l-tryptophan from indole and l-serine (or l-cysteine) or from pyruvate and ammonia (29, 47).
The tryptophanase (tna) operon from several bacterial species has been cloned and sequenced (7, 15, 17, 19, 24). In Escherichia coli, this operon contains two major structural genes, a promoter proximal gene, tnaA, encoding tryptophanase and a distal gene, tnaB, encoding a low-affinity tryptophan permease (7, 8). Preceding tnaA in the tna operon is a 319-nucleotide (nt) transcribed regulatory region that contains the coding region for 24-residue leader peptide TnaC. The 220-nt spacer region that separates tnaC from tnaA contains several transcription pause sites. Studies in vivo and in vitro have shown that these pause sites serve as regulated sites of Rho-dependent transcription termination (40, 41). In the presence of the inducer tryptophan, a transcription antitermination mechanism that increases transcription readthrough into the tnaA-tnaB structural gene region of the operon 10- to 100-fold is activated (41). Induction also requires the translation of tnaC. Thus, replacing the tnaC start codon by a stop codon or replacing the single, crucial Trp codon at position 12 by codons for other amino acids prevents induction (13; M. Eshoo and C. Yanofsky, unpublished results). In contrast, initiation of transcription of the tna operon is independent of tryptophan recognition and requires the binding of the cyclic AMP catabolite activator protein (CAP) (3, 4) at a CAP site located just upstream of the tna promoter (41).
Additional evidence supporting the essential role of the Rho factor in regulating tna operon expression comes from an analysis of Rho mutants (41), examination of Rho-inhibiting drugs (48), and the isolation of cis-acting mutants that express the tna operon constitutively (41). Mutations in rho that reduce Rho factor activity increase basal expression of the tna operon significantly (41, 48). Similarly, the drug bicyclomycin, an inhibitor of Rho action, increases expression of the tna operon (48, 52). cis-acting constitutive mutants, altered near the distal end of tnaC, also increase basal-level expression (41). The associated mutations are in a 9-nt sequence (CGC CCT TGA) that is homologous to the boxA sequences of the bacteriophage λ early region (9) and of rRNA operons (23). The boxA of these operons is necessary for antitermination or prevention of Rho-dependent termination and does not appear to be required for Rho-dependent termination. Several host factors called Nus factors are also involved in antitermination at sites of Rho-dependent termination in the above operons. In particular, in vitro studies with the rRNA operon have shown that a heterodimeric complex of NusB and NusE binds to boxA (26, 30). Mutations in boxA that prevent rRNA antitermination also interfere with the ability of boxA to bind to NusB and NusE (30). In addition, mutations altering either NusB or NusE factor prevent binding of the complex to boxA and are associated with reduced antitermination (37). Two other Nus factors, NusA and NusG, are believed to play an indirect role in antitermination by interacting with the transcribing polymerase (25, 30). Unlike findings with the rRNA operon, in vitro studies with the tna operon have shown that Rho-dependent transcription termination in the tna leader region is enhanced by the NusA factor (40).
Immediately following the boxA sequence and tnaC stop codon in the tna operon transcript, there is a sequence of approximately 22 nt that is rich in cytidylate residues. Comparable sequences, called Rho utilization or rut sites (1), are necessary for Rho binding and action in the phage λ early region (32). Interestingly, in phage λ the boxA sequence is embedded in part of the rut site (rutA) of the tR1 terminator (5). Deletion of the rut site in the spacer region of the tna operon results in semiconstitutive expression of the operon, suggesting that the rut site is required for efficient Rho-dependent termination (13).
The exact mechanism by which tryptophan induces antitermination in the tna operon is not known. Studies with a lacZ reporter construct lacking the spacer region between tnaC and tnaA suggest that, in the presence of inducer, the nascent TnaC peptide acts in cis on the ribosome translating tnaC to inhibit its release at the tnaC stop codon (21). The stalling of the translating ribosome at this stop codon presumably interferes with Rho binding or action. It has also been shown that a deletion in the tna operon leader region of Proteus vulgaris, one that places the start codon for TnaA near the tnaC stop codon, allows inducer inhibition of ribosome initiation at this start codon (18). The stalled ribosome apparently blocks translation initiation at tnaA.
The role of the tnaC stop codon in tna operon regulation in E. coli has been examined by replacing the natural stop codon, UGA, by UAG or UAA. These changes reduced both basal and induced expression of the tna operon (22), consistent with other evidence indicating that, in E. coli, peptide release factor 1 (RF1; recognizing UAG and UAA stop codons) terminates translation more efficiently than RF2 (recognizing UGA and UAA) (6, 44). It was shown that a mutation that alters RF1 increased basal-level expression of the tna operon in strains with UAG or UAA as the tnaC stop codon, but not in strains with UGA as the stop codon (22). Additionally, inactivation of the structural gene for RF3 increased basal-level expression of the tna operon at least threefold (21, 49), consistent with the role of RF3 in enhancing translation termination at UGA, UAG, and UAA stop codons (14, 27). These results support the hypothesis that, in the presence of tryptophan, the nascent TnaC peptide inhibits ribosome release at the tnaC stop codon, thereby preventing Rho-dependent termination.
Although mutations in the boxA-like sequence (Fig. 1) result in constitutive expression (at least a threefold increase in basal expression) of the tna operon, it was not known whether tnaC translation influences the ability of boxA mutations to reduce transcription termination. It also was not known whether changes in the nucleotide sequence of boxA or the corresponding amino acid sequence or both are responsible for constitutive expression. In the present study we use combinations of point mutations and deletions to answer these questions and to further define the roles of boxA and the rut site (Fig. 1) in mediating Rho-dependent transcription termination. We show that some nucleotide changes that alter the boxA sequence but do not change the sequence of the TnaC peptide also result in constitutive expression of the tna operon. We also show that the constitutive expression exhibited by constructs with boxA or rut mutations is dependent on translation of the tnaC coding region. Combining a boxA mutation with a rut site deletion resulted in elevated tna operon expression comparable to that of the induced operon. These results are consistent with the participation of both the boxA nucleotide sequence and the rut site in Rho-dependent transcription termination in the intact tna operon and reveal the importance of tnaC translation to the regulatory mechanism controlling tna operon expression.
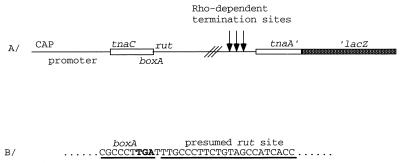
FIG. 1.
(A) Schematic diagram of the tnaA′-′lacZ fusion used in this study. This construct contains the tnaC leader region, the 220-nt spacer region, and the first 20 codons of tnaA joined to the ninth codon of lacZ. The mutagenized fragments in this study did not include the lacZ coding region. (B) Nucleotide sequence of boxA and the rut site (underlined). The TGA stop codon (boldface) is part of the boxA region. CAP, catabolite activator protein binding site.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The E. coli strains and the plasmids used in this study are listed in Table 1. Strains VK1300 to VK3100 are all single lysogens carrying λRS45 (21, 22, 38) with various inserts. To prepare these strains, the respective fusion constructs from pRS552 (Table 1) were independently crossed into phage λRS45 (38) and the recombinant phage genome was inserted into the chromosome of CY15076 (Table 1). Plasmids were introduced into various strains by transformation (34), with selection for the appropriate antibiotic resistance marker.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genetic characteristics | Reference or source |
|---|---|---|
| Strains | ||
| CY15076 | W3110 tnaA2 ΔlacU169 | 22 |
| PDG1114 | SVS1100 (λtnap tnaC265 tnaA′-′lacZ) | 13 |
| PDG1171 | SVS1100 (λtnap tnaC275 tnaA′-′lacZ) | 13 |
| PDG1172 | SVS1100 (λtnap tnaC-ACU24-tnaA′-′lacZ) | This study |
| PDG1173 | SVS1100 (λtnap tnaC-UCU24-tnaA′-′lacZ) | This study |
| PDG1181 | SVS1100 (λtnap tnaC-CGG12-AGC23-tnaA′-′lacZ) | This study |
| SVS1100 | W3110 bglR551 Δ(lac-argF)U169 | 41 |
| SVS1144 | SVS1100 (λtnap tnaA′-′lacZ) | 41 |
| VK800 | CY15076 (λtnap tnaC-UAG-tnaA′-′lacZ) | 22 |
| VK1300 | CY15076 (λtnap tnaC-UAG1-CGG12-AGC23-tnaA′-′lacZ) | This study |
| VK1400 | CY15076 (λtnap tnaC-Δ[CGC23CCU24]-tnaA′-′lacZ) | This study |
| VK1500 | CY15076 (λtnap tnaC-UAG1-Δ[CGC23CCU24]-tnaA′-′lacZ) | This study |
| VK1900 | CY15076 (λtnap tnaC-UAG1-Δ[bp 101–123]-tnaA′-′lacZ) | This study |
| VK2000 | CY15076 (λtnap tnaC-AGC23-Δ[bp 101–123]-tnaA′-′lacZ) | This study |
| VK2100 | CY15076 (λtnap tnaC-UAG1-AGC23-Δ[bp 101–123]-tnaA′-′lacZ) | This study |
| VK2200 | CY15076 (λtnap tnaC-UGA11-AGC23-tnaA′-′lacZ) | This study |
| VK2300 | CY15076 (λtnap tnaC-UGA18-AGC23-tnaA′-′lacZ) | This study |
| VK2301 | CY15076 (λtnap tnaC-UGA18-tnaA′-′lacZ) | This study |
| VK2400 | CY15076 (λtnap tnaC-CGA23-tnaA′-′lacZ) | This study |
| VK2500 | CY15076 (λtnap tnaC-CGU23-tnaA′-′lacZ) | This study |
| VK2600 | CY15076 (λtnap tnaC-CCA24-tnaA′-′lacZ) | This study |
| VK2700 | CY15076 (λtnap tnaC-CCG24-tnaA′-′lacZ) | This study |
| VK2800 | CY15076 (λtnap tnaC-UGA23-tnaA′-′lacZ | This study |
| VK2900 | CY15076 (λtnap tnaC-UGA24-tnaA′-′lacZ) | This study |
| VK3000 | CY15076 (λtnap tnaC-AGA23-tnaA′-′lacZ) | This study |
| VK3100 | CY15076 (λtnap tnaC-AGG23-tnaA′-′lacZ) | This study |
| Plasmids | ||
| pRS552 | pBR322 derivative, lac-based vector | 38 |
| pBE621 | trpT su9 (UGA suppressor) | 31 |
Media and enzyme assay.
Vogel and Bonner minimal medium (45) was used throughout. For β-galactosidase (β-Gal) assays (28), cultures were generally grown with shaking at 37°C in minimal medium plus 0.2% glycerol and 0.05% acid-hydrolyzed casein, with or without l-tryptophan (100 μg/ml). When appropriate, media were supplemented with 30 μg of kanamycin per ml, 15 μg of tetracycline per ml, or 20 μg of bicyclomycin per ml (a noninhibitory concentration). β-Gal assays were performed as described by Miller (28); β-Gal activity is reported in Miller units (28).
Site-directed mutagenesis.
The megaprimer PCR method (36) was used throughout to introduce mutations in the tna operon. First, the LACZ-RT primer, 5′-GCG ATT AAG TTG GGT AAC GCC AGG-3′, or the VK1 primer, 5′-CGG AAT TCA GCT TCT GTA TTG GTA AG-3′, was used along with the respective mutagenic primer to amplify a PCR product containing the chosen mutation. This product was then purified using a PCR purification kit (Qiagen Inc., Chatsworth, Calif.) and combined in the second PCR with the VK1 or LACZ-RT primer to synthesize the final PCR product. The final product was flanked with an EcoRI site at its 5′ end and a BamHI site at its 3′ end; it contained the mutant tnaC leader sequence followed by all or part of the tna spacer region and the coding sequence for the first 20 amino acids of TnaA (Fig. 1). This product was cloned into the pCRII vector (Invitrogen Co., San Diego, Calif.), and the sequence was confirmed (35). The resulting insert was cleaved with EcoRI and BamHI, purified using the GENECLEAN II kit (BIO 101 Inc., La Jolla, Calif.), and subcloned into the EcoRI- and BamHI-cleaved pRS552 vector (38).
RESULTS
Point mutations and a deletion in the boxA-like region of tnaC result in semiconstitutive expression of the tna operon.
There is a 9-nt sequence (CGC CCT TGA) at the end of the tnaC leader region (Fig. 1 and Table 2) which has strong homology to those of boxA of phage λ (9) and rRNA operons (23). The first 6 nt of this sequence, CGC CCT, encode Arg and Pro residues at positions 23 and 24, respectively, of the TnaC leader peptide. These 6 nt were deleted in construct VK1400 to examine the effects of removal of the presumed boxA sequence on Rho-dependent transcription termination. Deleting these 6 nt resulted in an eightfold increase in tna operon expression under noninducing conditions (Table 2). The addition of tryptophan did not increase the expression of this deletion construct. As reported previously, replacing the first nucleotide of the CGC CCT sequence, C, by A (Arg-to-Ser change in TnaC) (VK1600) resulted in a fourfold increase in the basal level of expression of the operon; this change allowed a twofold increase in induction (Table 2). Replacing the TnaC Trp residue of this construct by an Arg residue (PDG1181) resulted in higher basal-level expression than that of VK1600 but eliminated induction (Table 2). These results confirm that alterations of the nucleotides in the boxA-like region of tnaC can reduce Rho-dependent transcription termination in the tna operon; they also confirm that Trp12 is essential for induction (13).
TABLE 2.
Alteration of the boxA-like sequence in tnaC results in constitutive expression of the tna operon; constitutive expression depends on tnaC translationa
| Strain | tnaC codons | β-Gal activity (Miller units)
|
Ratiob | |
|---|---|---|---|---|
| −Trp | +Trp | |||
| boxA | ||||
| SVS1144 (wild type) | AUG1....UGG12....CGC23CCU24UGA.. | 390 ± 17 | 11,800 ± 1,390 | 30 |
| SVS1144 + bicyclomycin | 4,940 ± 290 | 18,080 ± 2,080 | ||
| VK1400 | Δ(CGC23CCU24) | 3,320 ± 344 | 3,770 ± 640 | |
| VK1400 + bicyclomycin | 4,580 ± 790 | 5,090 ± 1,275 | ||
| VK1600 | AGC23 | 1,870 ± 230 | 4,190 ± 430 | 2 |
| VK1600 + bicyclomycin | 2,670 ± 325 | 5,005 ± 1,205 | ||
| PDG1114 | CGG12 | 145 ± 9 | 130 ± 4 | |
| PDG1181 | CGG12...AGC23 | 3,210 ± 140 | 3,005 ± 285 | |
| PDG1171 | UAG1 | 13 ± 2 | 19 ± 3 | |
| PDG1171 + bicyclomycin | 2,275 ± 270 | 1,990 ± 220 | ||
| VK1500 | UAG1 Δ(CGC23CCU24) | 55 ± 8 | 51 ± 10 | |
| VK1700 | UAG1AGC23 | 5 ± 0.4 | 6 ± 0.8 | |
| VK1700 + bicyclomycin | 1,230 ± 60 | 1,300 ± 105 | ||
| VK1300 | UAG1CGG12AGC23 | 11 ± 1 | 10 ± 1 | |
| VK2200 | UGA11AGC23 | 150 ± 17 | 120 ± 10 | |
| VK2300 | UGA18AGC23 | 180 ± 11 | 190 ± 14 | |
Cultures were grown at 37°C in minimal medium (45) plus 0.2% glycerol and 0.05% acid-hydrolyzed casein, with (+) or without (−) l-Trp (100 μg/ml). When indicated, 20 μg of bicyclomycin/ml was added to the culture medium. At least five cultures of each type were assayed in each case. Nucleotide changes are underlined.
Ratio of activities for cultures grown with Trp and without Trp.
To determine whether translation of the tnaC coding region is required for the semiconstitutive expression observed in constructs with a boxA point mutation or deletion, the initiation codon AUG in these constructs was replaced by the stop codon UAG, giving derivatives VK1500 and VK1700. These changes resulted in a 60- to 370-fold decrease in basal-level expression (Table 2) and also eliminated induction. Thus constitutive expression by these boxA mutants requires tnaC translation. The addition of bicyclomycin, an inhibitor of Rho protein function (52), to VK1700, the strain in which the tnaC AUG is replaced by UAG and in which there is an altered boxA sequence, increased basal expression to a level approaching that of the construct with the start codon AUG (VK1600) (Table 2). These results suggest that, in the absence of translation, the tnaC coding sequence serves as a Rho-binding site and that in this context the boxA sequence is no longer required for termination. A UGA stop codon was introduced at position 11 or 18 of tnaC of the boxA mutant constructs (VK2200 and VK2300) to determine whether translation of a portion of tnaC is sufficient to allow constitutive tna operon expression. These changes increased basal-level expression only slightly, implying that translation of the first 18 codons of tnaC is insufficient to permit the semiconstitutive expression typical of the construct with the boxA mutation alone. Replacing the tnaC start codon by UAG in the construct in which Trp12 was replaced by Arg and in which there was a mutated boxA (VK1300) also eliminated the elevated basal-level expression typical of a construct with only the boxA change.
Moving the tnaC stop codon eliminates induction.
It was shown in Table 2 that introducing stop codons at tnaC coding positions 11 and 18 eliminated the modest induction observed in VK1600, the construct with the boxA point mutation. UAG stop codons were also introduced at positions 23, 24, and 25 to examine their effect on basal-level expression and induction (constructs VK2800, VK2900, and VK800, respectively). These codon changes also alter the natural boxA sequence in tnaC. It can be seen in Table 3 that introducing UAG stop codons at positions 23, 24, and 25 altered basal-level expression. In addition, introducing stop codons at positions 23 and 24 eliminated induction. To determine if this lack of induction was due to a shortened TnaC peptide, a tryptophan-inserting UAG suppressor was introduced into strains VK2800 and VK2900 to restore translation through codon 24. Suppression of UAG23 was observed to restore a low level of induction in comparison to what was found for controls SVS1144 and VK800, whereas suppression of UAG24 did not (Table 3). These results suggest that amino acids at positions 23 and 24 of TnaC play a role in setting the basal and induced expression of the tna operon. In a previous report, suppression of the UAG stop codon in strain VK800 was shown to increase the basal expression of tna but not to affect induced expression (22). These findings do not distinguish whether a correct tnaC RNA sequence or TnaC peptide sequence or both are necessary for induction.
TABLE 3.
Introducing a UAG stop codon at position 23 or 24 prevents inductiona
| Strain | tnaC codons | β-Gal activity (Miller units)
|
Ratiob | |
|---|---|---|---|---|
| −Trp | +Trp | |||
| SVS1144 (wild type) | AUG..UGG12..CGC23CCU24UGA... | 570 ± 15 | 18,920 ± 365 | 33 |
| VK800 | UAG | 140 ± 18 | 2,100 ± 335 | 15 |
| VK2900 | UAG24 | 240 ± 25 | 280 ± 30 | |
| VK2900/pMY228 | su UAGc | 145 ± 14 | 160 ± 19 | |
| VK2800 | UAG23 | 930 ± 36 | 850 ± 60 | |
| VK2800/pMY228 | su UAGc | 540 ± 100 | 2,350 ± 279 | 4 |
Cultures were grown as for Table 2. More than five cultures of each type were assayed in each case. The nucleotide changes introduced are underlined.
Ratio of activities for cultures grown with Trp and without Trp.
su UAG, a tryptophan-inserting UAG suppressor.
Examination of other changes in the boxA-like region of tnaC, some of which do not change the TnaC amino acid sequence.
The finding that some mutational changes in the boxA region result in constitutive expression of the operon could indicate that the boxA nucleotide sequence must be properly recognized by some factor to obtain efficient Rho-dependent termination. To distinguish between effects of mutations at the DNA or RNA level and those at the protein level, changes that did not change the amino acid specified were introduced at tnaC codon 23. Replacing the CGC Arg codon by another Arg codon, either CGG, AGA, or AGG, resulted in a three- to sixfold increase in basal-level expression in comparison to that of the wild-type control (Table 4). These three changes also allow induction, but the basal and induced levels of expression differ from those of the parental control strain, SVS1144. Two additional changes to other Arg codons, CGA and CGT, reduced the basal level and induced levels two- to threefold. Replacing the CCT Pro codon by two other Pro codons, CCA and CCG, had no significant effect on the basal level but reduced induction about threefold. On the other hand, replacing the CCT Pro codon by ACT (Thr) or TCT (Ser) reduced basal expression by at least 2-fold and allowed 10-fold induction (Table 4). These findings establish that changes in the amino acid sequence of the TnaC peptide are not solely responsible for the altered operon expression observed in the various tnaC mutants; changes in the nucleotide sequence of the boxA region can also affect both basal-level expression and induced expression.
TABLE 4.
Some alterations in the nucleotide sequence of boxA that do not alter the TnaC amino acid sequence result in constitutive expression of the tna operona
| Strain | tnaC codons | β-Gal activity (Miller units)
|
Ratiob | |
|---|---|---|---|---|
| −Trp | +Trp | |||
| SVS1144 (wild type) | AUG.......CGC23CCU24UGA... | 520 ± 25 | 14,830 ± 1,495 | 29 |
| VK2400 | CGA23 | 360 ± 24 | 5,000 ± 995 | 14 |
| VK2500 | CGU23 | 250 ± 19 | 4,260 ± 720 | 17 |
| PDG1178 | CGG23 | 1,730 ± 85 | 17,860 ± 557 | 10 |
| VK3000 | AGA23 | 3,335 ± 260 | 10,080 ± 1,533 | 3 |
| VK3100 | AGG23 | 1,560 ± 231 | 8,520 ± 879 | 5 |
| VK2600 | CCA24 | 710 ± 66 | 5,670 ± 979 | 8 |
| VK2700 | CCG24 | 570 ± 112 | 5,970 ± 1,035 | 10 |
| PDG1172 | ACU24(Thr) | 150 ± 19 | 1,320 ± 245 | 9 |
| PDG1173 | UCU24(Ser) | 265 ± 25 | 1,250 ± 214 | 5 |
Cultures were grown as for Table 2. More than five cultures of each type were assayed in each case. CGA, CGU, CGG, AGA, and AGG are all Arg codons. CCA and CCG are Pro codons. The nucleotide changes introduced are underlined.
Ratio of activities for cultures grown with Trp and without Trp.
Deletion of the presumed rut site combined with a point mutation in the boxA region of tnaC results in near-maximal expression of the tna operon.
It was shown previously, and is confirmed in Table 5, that deletion of 22 nt (nt 101 through 123) from the tnaC-tnaA spacer region (VK1800), a presumed rut site (Fig. 1), results in an approximately eightfold increase in basal-level expression of the tna operon (13). Tryptophan induction of this construct increased expression only slightly (Table 5). Combining the rut deletion with the boxA mutation at codon 23 in construct VK2000 increased basal-level expression an additional twofold; addition of an inducer had little effect. Combining the rut deletion with a mutation replacing the tnaC start codon by a stop codon (construct VK1900) reduced basal expression appreciably, and there was no response to an inducer. The addition of bicyclomycin to the strain with this construct elevated expression four- to fivefold, implicating Rho factor in mediating the low level of expression seen in the absence of translation of tnaC. Replacing the tnaC start codon by a stop codon and combining this change with both the boxA change and the rut deletion (construct VK2100) allowed moderate expression but no induction. Apparently when both the boxA sequence and the rut site are altered, the absence of translation of tnaC also reduces operon expression, but not to the extent that it does when either the unaltered boxA or rut site is present. Addition of bicyclomycin to VK2100 increased expression only slightly.
TABLE 5.
Combining a deletion of the presumed rut site with a point mutation in the boxA-like region results in near-maximum expression of the tna operon; tnaC translation is required for constitutive expressiona
| Strain | tnaC codon and spacer region nt | β-Gal activity (Miller units)
|
Ratiob | |
|---|---|---|---|---|
| −Trp | +Trp | |||
| (nt 101–123) | ||||
| SVS1144 (wild type) | AUG1...CGC23CCU24UGAU(UU..CC...CC...C..CCA)... | 410 ± 32 | 12,890 ± 1,268 | 31 |
| VK1800 | Δ(nt 101–123) | 3,450 ± 312 | 4,420 ± 698 | |
| VK1800 + bicyclomycin | 3,120 ± 630 | 3,880 ± 330 | ||
| VK2000 | AGC23 Δ(nt 101–123) | 8,970 ± 766 | 9,640 ± 1,027 | |
| VK1900 | UAG1 Δ(nt 101–123) | 480 ± 57 | 480 ± 72 | |
| VK1900 + bicyclomycin | 2,360 ± 65 | 2,320 ± 44 | ||
| VK2100 | UAG1...AGC23 Δ(nt 101–123) | 2,365 ± 154 | 2,320 ± 243 | |
| VK2100 + bicyclomycin | 3,075 ± 200 | 3,100 ± 560 | ||
Cultures were grown as for Table 2. More than five cultures of each type were assayed in each case. The nucleotide changes introduced are underlined.
Ratio of activities for cultures grown with Trp and without Trp.
Expression of the tna operon does not correlate with changes in the stability of the tnaC secondary structure.
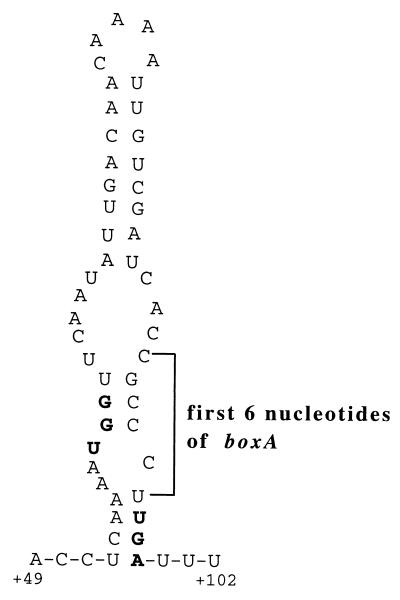
Nucleotides at positions 52 through 99 of the tna leader transcript are predicted to fold and form a relatively stable hairpin structure (ΔG = −9.6 kcal/mol) (40) (Fig. 2 and Table 6). Some of the mutational changes we have examined could exert their effect by altering the stability of this hairpin structure. To explore this possibility, the stabilities of leader mRNA secondary structures for the mRNA segment comprising nt 52 to 99 were predicted using the Zuker MFOLD program (43, 50). In Table 6, it can be seen that many of the mutations that result in noninducibility decrease the stability of this tnaC mRNA secondary structure. However, decreased stability also is predicted for transcripts of some mutants that constitutively express the tna operon (Table 6). In addition, some changes in the latter group of mutants resulted in a significant increase in the stability of the mRNA secondary structure (Table 6). In the class of mutations that have little or no effect on tna expression, there is no significant change in the predicted stability of the leader mRNA structure (Table 6). Considering all of the predicted changes in this leader mRNA structure, it seems unlikely that changes in its stability are primarily responsible for the phenotypes of the various mutants.
FIG. 2.
Predicted RNA secondary structure present at the end of the tnaC transcript. The RNA secondary structure was predicted using the MFOLD program of Zuker et al. (50, 51). The free energy of formation of the wild-type structure is −9.6 kcal/mol. Trp codon UGG and the stop codon UGA are in boldface. The UGA stop codon is part of the 9-nt sequence of boxA (CGC CCU UGA) in the tna operon of E. coli.
TABLE 6.
Changes in the stability of the tnaC secondary structure do not correlate with expression of the tna operon
| Mutational change | tna operon expression | Change in tnaC secondary structure stabilitya (kcal/mol) |
|---|---|---|
| UUC12 (W→F) | Noninducible | +2.9 |
| UGA12 (W→stop) | Noninducible | +4.2 |
| UAG12 (W→stop) | Noninducible | +3.6 |
| AUG12 (W→M) | Noninducible | +3.6 |
| UGC13 (F→C) | Noninducible | +3.1 |
| UUU15 (I→F) | Noninducible | +0.6 |
| AAU15 (I→N) | Noninducible | +2.8 |
| Δ(CGC23CCU24) | Constitutive | +1.7 |
| AGC23 (R→S) | Constitutive | −0.8 |
| CGG23 (R→R) | Constitutive | +0.8 |
| AGA23 (R→R) | Constitutive | −1.3 |
| AGG23 (R→R) | Constitutive | −0.8 |
| UGG13 (F→W) | Constitutive | −6 |
| AUA15 (I→I) | Constitutive | +2.6 |
| AUC15 (I→I) | Constitutive | −2.1 |
| AUG15 (I→M) | Constitutive | +1.7 |
| CAC16 (D→H) | Constitutive | +5.5 |
| UUC16 (D→F) | Constitutive | +5.7 |
| CGA23 (R→R) | Regulated | −1.3 |
| CGU23 (R→R) | Regulated | −1.6 |
| CCA24 (P→P) | Regulated | +0.1 |
| CCG24 (P→P) | Regulated | +0.1 |
| UUU13 (F→F) | Regulated | 0 |
| GAU16 (D→D) | Regulated | +1.9 |
| AAC14 (N→N) | Regulated | 0 |
| ACU24 (P→T) | Regulated | +0.3 |
| UCU24 (P→S) | Regulated | +0.3 |
DISCUSSION
In the present report, we extend our analysis of the role of the boxA-like sequence and the rut site in mediating transcription termination in the tna operon leader region. We show that deleting 6 nt of the boxA sequence or substituting a Ser for Arg codon 23 results in elevated constitutive (at least threefold increase in the basal-level) expression of the tna operon (Table 2). In further analyses with these constitutive boxA mutant constructs, we observed that eliminating translation of all or part of the tnaC coding sequence reversed the elevated expression of the tna operon associated with the boxA change and reduced expression well below that of the wild-type parental construct (Table 2). Addition of bicyclomycin, an inhibitor of Rho activity (48, 52), increased expression from these constructs, confirming that the reduced basal expression observed in the absence of translation is due to Rho action (Table 2). Interestingly, bicyclomycin addition had only a slight stimulatory effect on the expression of boxA deletion or missense mutant constructs which exhibit moderately high basal-level expression, whereas it had a significant effect on the wild-type construct and on constructs with very low basal levels of expression (Tables 2 and 5). In Table 3, introduction of a stop codon at position 23 or 24 of tnaC drastically changed the boxA sequence. These changes resulted in alterations in the basal level and loss of tryptophan induction. In addition, suppression of UAG23 slightly increased (fourfold) induced expression whereas suppression of UAG24 had no effect on basal or induced expression. These results suggest that the nature of the amino acids at positions 23 and 24 of tnaC and perhaps the nucleotide sequence in boxA contribute to the basal level of tna expression observed in wild-type cultures.
We examined the specific role of the boxA RNA sequence by introducing mutational changes in Arg codon 23, some of which did not alter the amino acid specified at this position. Three such mutations, which introduced the Arg codons CGG, AGA, and AGG at codon position 23 of tnaC, resulted in elevated basal-level expression of the tna operon (Table 4). This result establishes that the RNA sequence itself, specifically the sequence of the boxA-like region of tnaC, plays a role in determining the extent of Rho-mediated termination in the tna operon leader region. The boxA sequence at the end of the tnaC leader region does not behave like a typical boxA sequence. Indeed, the boxA sequence in the tna operon overlaps the tnaC stop codon, UGA. Furthermore, the boxA sequences of phage λ (9, 10) and rRNA operons (2, 23) are required for transcription antitermination, not termination.
A sequence resembling a Rho utilization (rut) site is located immediately downstream of the boxA sequence (41). We confirm the report that deleting this rut site elevates expression of the tna operon in the absence of inducer (Table 5) (13). Interestingly, the rut site responsible for Rho-dependent transcription termination at the λtR1 terminator contains boxA in one of its two discrete rutA sites (5). We analyzed the relative contribution of boxA and the rut site in Rho-dependent termination with a construct containing the rut deletion combined with a substitution of Ser codon AGC for Arg codon CGC in boxA. The combined mutations resulted in near-maximal expression of the tna operon with or without tryptophan; thus their effects are additive (Table 5). These findings suggest that, in the absence of tryptophan, both boxA and the rut site contribute to efficient Rho-dependent transcription termination in the tna operon. Rho may bind directly to boxA and the rut site, or the binding of Rho at the rut site may be enhanced by interactions with a cellular factor(s) bound at boxA. Interaction of boxA with NusA, NusB, or NusE factor alone appears to have been ruled out. Indeed, previous studies with a nusA1 (41) mutant strain have shown no defect in tna operon regulation. In addition, nusA1, nusB100, and nusE100 mutant strains (33, 46) were examined for tna operon expression by measuring tryptophanase (7) levels in cultures grown with or without inducing levels of tryptophan; enzyme levels in these mutants were indistinguishable from that of the wild-type control (data not shown). NusA, NusB, and NusE could bind to boxA as a complex; in this case, any particular mutation in any one of these factors might have a negligible effect on tryptophanase regulation. In any event, since protein factors do bind at lambda's boxA, it is likely that they could influence Rho's activity through interaction. This possibility is under continuing examination.
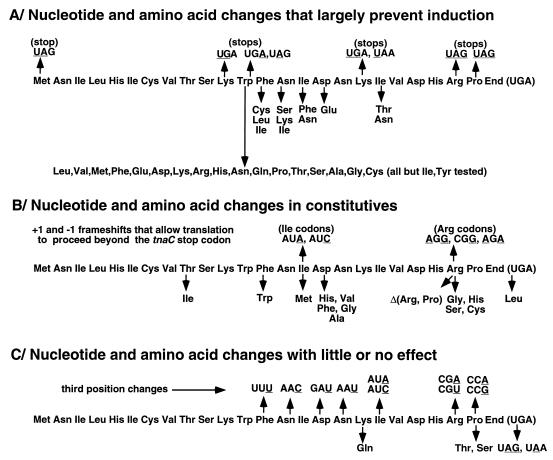
A summary of the nucleotide and amino acid changes that have been introduced to date in tnaC and its peptide product is presented in Fig. 3. In the first group of mutations (Fig. 3A), substitution of a stop codon for a sense codon at codon position 1, 11, 12, 18, 23, or 24 prevents tryptophan induction, consistent with the importance of synthesis of the TnaC peptide in relieving Rho-dependent transcription termination (Fig. 3A). Tryptophan inducibility also is lost when some tnaC codons conserved between E. coli, P. vulgaris, and Enterobacter aerogenes (12) are replaced by codons specifying different amino acids (Fig. 3A). Most importantly, replacing Trp12 by a codon specifying a different amino acid eliminates induction. However, when a stop codon at position 12 is suppressed by a tRNA that inserts tryptophan, induction is restored (12, 13).
FIG. 3.
Nucleotide and amino acid changes in tnaC and the TnaC peptide that do or do not affect tna operon expression. Nucleotide changes that do not lead to amino acid changes are underlined and are above the TnaC leader peptide sequence. The results come from this study, from references 11 to 13, 41, and 42, and from unpublished data of M. Eshoo and C. Yanofsky.
The second group of mutations results in partial constitutive (at least a threefold increase in basal-level) expression of the operon; many of these mutations remain inducible in the presence of tryptophan (Fig. 3B). Some of these changes alter the TnaC amino acid sequence, while others, such as some of the Arg codon replacements described in this report, do not. This class also includes frameshift mutations and a mutation (replacement of UAA with UGA) that allows translation to proceed beyond the natural tnaC stop codon. Interestingly, some mutations that change residues in the conserved amino acid sequence near Trp residue 12 also result in constitutive expression (12). These findings establish that the nucleotide sequence of the tnaC coding region must play a role in establishing the low Rho-dependent basal-level expression of the operon. Nucleotide changes that alter amino acid coding specificity could act either at the nucleotide sequence level or by altering the amino acid sequence of the TnaC peptide.
The third group of mutations (Fig. 3C) introduces changes in tnaC or its product that have little or no effect on basal or induced expression of the tna operon (Fig. 3C). Some of these changes (e.g., in the Arg23 or Pro24 codon) appear to increase Rho-dependent termination, but tryptophan-mediated induction is retained (Table 4). Other changes (e.g., replacement of UAG or UAA with UGA) are known to increase the efficiency of ribosome release at the tnaC stop codon.
An analysis of the mutational changes made in this and other studies (12, 13, 21, 41, 42) shows a lack of correlation between the stability of the tnaC mRNA secondary structure and expression of the tna operon. These observations suggest that changes in the stability of the tnaC leader mRNA cannot alone account for the phenotypes of the respective tna mutants.
In conclusion, it is apparent that both the TnaC peptide and the sequence of its encoding transcript play a role in Rho-dependent transcription termination and tryptophan-induced antitermination in the tna operon of E. coli. The studies described define some of the features of the boxA sequence and the rut site in Rho-dependent transcription termination and their relationship to translation of the leader peptide coding region.
ACKNOWLEDGMENTS
We thank the Fujisawa Pharmaceutical Co. Ltd., Osaka, Japan, for providing samples of bicyclomycin. We are indebted to Virginia Horn for assistance with E. coli strain construction. We are also grateful to Kurt Gish, Ajith Kamath, and Peter Margolis for their helpful comments.
These studies were supported by grant GM09738 to C.Y. from the United States Public Health Service. K.V.K. was a Postdoctoral Fellow.
REFERENCES
- 1.Bektesh S L, Richardson J P. A rho-recognition site on phage lambda cro gene mRNA. Nature. 1980;283:102–104. doi: 10.1038/283102a0. [DOI] [PubMed] [Google Scholar]
- 2.Berg K L, Squires C, Squires C L. Ribosomal RNA operon anti-termination. Function of leader and spacer region box B-box A sequences and their conservation in diverse microorganisms. J Mol Biol. 1989;209:345–358. doi: 10.1016/0022-2836(89)90002-8. [DOI] [PubMed] [Google Scholar]
- 3.Bilezikian J, Kaempfer R, Magasanik B. Mechanism of tryptophanase induction in Escherichia coli. J Mol Biol. 1967;27:495–506. doi: 10.1016/0022-2836(67)90054-x. [DOI] [PubMed] [Google Scholar]
- 4.Botsford J L, DeMoss R D. Catabolite repression of tryptophanase in Escherichia coli. J Bacteriol. 1971;105:303–312. doi: 10.1128/jb.105.1.303-312.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen C Y, Richardson J P. Sequence elements essential for rho-dependent transcription termination at lambda tR1. J Biol Chem. 1987;262:11292–11299. [PubMed] [Google Scholar]
- 6.Craigen W J, Caskey C T. Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature. 1986;322:273–275. doi: 10.1038/322273a0. [DOI] [PubMed] [Google Scholar]
- 7.Deeley M C, Yanofsky C. Nucleotide sequence of the structural gene for tryptophanase of Escherichia coli K-12. J Bacteriol. 1981;147:787–796. doi: 10.1128/jb.147.3.787-796.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edwards R M, Yudkin M D. Location of the gene for the low-affinity tryptophan-specific permease of Escherichia coli. Biochem J. 1982;204:617–619. doi: 10.1042/bj2040617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedman D I, Olson E R. Evidence that a nucleotide sequence, “boxA,” is involved in the action of the NusA protein. Cell. 1983;34:143–149. doi: 10.1016/0092-8674(83)90144-7. [DOI] [PubMed] [Google Scholar]
- 10.Friedman D I, Olson E R, Johnson L L, Alessi D, Craven M G. Transcription-dependent competition for a host factor: the function and optimal sequence of the phage λboxA transcription antitermination signal. Genes Dev. 1990;4:2210–2222. doi: 10.1101/gad.4.12a.2210. [DOI] [PubMed] [Google Scholar]
- 11.Gish K, Yanofsky C. Inhibition of expression of the tryptophanase operon in Escherichia coli by extrachromosomal copies of the tna leader region. J Bacteriol. 1993;175:3380–3387. doi: 10.1128/jb.175.11.3380-3387.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gish K, Yanofsky C. Evidence suggesting cis action by the tnaC leader peptide in regulating transcription attenuation in the tryptophanase operon of Escherichia coli. J Bacteriol. 1995;177:7245–7254. doi: 10.1128/jb.177.24.7245-7254.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gollnick P, Yanofsky C. tRNATRP translation of leader peptide codon 12 and other factors that regulate expression of the tryptophanase operon. J Bacteriol. 1990;172:3100–3107. doi: 10.1128/jb.172.6.3100-3107.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grentzmann G, Brechemier-Baey D, Heurgue V, Mora L, Buckingham R. Localization and characterization of the gene encoding release factor RF3 in Escherichia coli. Proc Natl Acad Sci USA. 1994;91:5848–5852. doi: 10.1073/pnas.91.13.5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirahara T, Suzuki S, Horinouchi S, Beppu T. Cloning, nucleotide sequences, and overexpression in Escherichia coli of tandem copies of a tryptophanase gene in an obligately symbiotic thermophile, Symbiobacterium thermopilum. Appl Environ Microbiol. 1992;58:2633–2642. doi: 10.1128/aem.58.8.2633-2642.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hopkins F G, Cole S W. A contribution to the chemistry of proteid. Part II. The constitution of tryptophanase, and the action of bacteria upon it. J Physiol (London) 1903;29:451–466. doi: 10.1113/jphysiol.1903.sp000968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamath A V, Yanofsky C. Characterization of the tryptophanase operon of Proteus vulgaris. J Biol Chem. 1992;267:19978–19985. [PubMed] [Google Scholar]
- 18.Kamath A V, Yanofsky C. Roles of the tnaC-tnaA spacer region and Rho factor in regulating expression of the tryptophanase operon of Proteus vulgaris. J Bacteriol. 1997;179:1780–1786. doi: 10.1128/jb.179.5.1780-1786.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawasaki K, Yokota A, Oita S, Kobayashi C, Yoshikawa S, Kawamoto S, Takao S, Tomita F. Cloning and characterization of a tryptophanase gene from Enterobacter aerogenes SM-18. J Gen Microbiol. 1993;139:3275–3281. doi: 10.1099/00221287-139-12-3275. [DOI] [PubMed] [Google Scholar]
- 20.Kazarinoff M N, Snell E E. Essential arginine residues in tryptophanase from Escherichia coli. J Biol Chem. 1977;252:7598–7602. [PubMed] [Google Scholar]
- 21.Konan K V, Yanofsky C. Regulation of the Escherichia coli tna operon: nascent leader peptide control at the tnaC stop codon. J Bacteriol. 1997;179:1774–1779. doi: 10.1128/jb.179.5.1774-1779.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Konan K V, Yanofsky C. Role of ribosome release in regulation of tna operon expression in Escherichia coli. J Bacteriol. 1999;181:1530–1536. doi: 10.1128/jb.181.5.1530-1536.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li S C, Squires C L, Squires C. Antitermination of E. coli rRNA transcription is caused by a control region segment containing lambda nut-like sequences. Cell. 1984;38:851–860. doi: 10.1016/0092-8674(84)90280-0. [DOI] [PubMed] [Google Scholar]
- 24.Martin K, Morlin G, Smith A, Nordike A, Eisenstark A, Golomb M. The tryptophanase gene cluster of Haemophilus influenzae type b: evidence for horizontal gene transfer. J Bacteriol. 1998;180:107–118. doi: 10.1128/jb.180.1.107-118.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mason S W, Greenblatt J. Assembly of transcription elongation complexes containing the N protein of phage lambda and the Escherichia coli elongation factors NusA, NusB, NusG, and S10. Genes Dev. 1991;5:1504–1512. doi: 10.1101/gad.5.8.1504. [DOI] [PubMed] [Google Scholar]
- 26.Mason S W, Li J, Greenblatt J. Direct interaction between two Escherichia coli transcription antitermination factors, NusB and ribosomal protein S10. J Mol Biol. 1992;223:55–66. doi: 10.1016/0022-2836(92)90715-v. [DOI] [PubMed] [Google Scholar]
- 27.Mikuni O, Ito K, Moffat J, Matsumura K, McCaughan K, Nobukuni T, Tate W, Nakamura Y. Identification of the prfC gene, which encodes peptide-chain-release factor 3 of Escherichia coli. Proc Natl Acad Sci USA. 1994;91:5798–5802. doi: 10.1073/pnas.91.13.5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller J. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 29.Newton W A, Snell E E. Catalytic properties of tryptophanase, a multifunctional pyridoxal phosphate enzyme. Proc Natl Acad Sci USA. 1964;51:382–389. doi: 10.1073/pnas.51.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nodwell J R, Greenblatt J. Recognition of boxA antiterminator RNA by the E. coli antitermination factors NusB and ribosomal protein S10. Cell. 1993;72:261–268. doi: 10.1016/0092-8674(93)90665-d. [DOI] [PubMed] [Google Scholar]
- 31.Raftery L A, Egan J B, Cline S W, Yarus M. Defined set of cloned termination suppressors: in vivo activity of isogenic UAG, UAA, and UGA suppressor tRNAs. J Bacteriol. 1984;158:849–859. doi: 10.1128/jb.158.3.849-859.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richardson L V, Richardson J P. Rho-dependent termination of transcription is governed primarily by the upstream rho utilization (rut) sequences of a terminator. J Biol Chem. 1996;271:21597–21603. doi: 10.1074/jbc.271.35.21597. [DOI] [PubMed] [Google Scholar]
- 33.Robledo R, Atkinson B L, Gottesman M E. Escherichia coli mutations that block transcription termination by phage HK022 Nun protein. J Mol Biol. 1991;220:613–619. doi: 10.1016/0022-2836(91)90104-e. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarkar G, Sommer S S. The “megaprimer” method of site-directed mutagenesis. BioTechniques. 1990;8:404–407. [PubMed] [Google Scholar]
- 37.Sharrock R A, Gourse R L, Nomura M. Defective antitermination of rRNA transcription and derepression of rRNA and tRNA synthesis in the nusB5 mutant of Escherichia coli. Proc Natl Acad Sci USA. 1985;82:5275–5279. doi: 10.1073/pnas.82.16.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 39.Snell E E. Tryptophanase: structure, catalytic activities, and mechanism of action. Adv Enzymol. 1975;42:389–446. doi: 10.1002/9780470122877.ch6. [DOI] [PubMed] [Google Scholar]
- 40.Stewart V, Landick R, Yanofsky C. Rho-dependent transcription termination in the tryptophanase operon leader region of Escherichia coli K-12. J Bacteriol. 1986;166:217–223. doi: 10.1128/jb.166.1.217-223.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stewart V, Yanofsky C. Evidence for transcription antitermination control of tryptophanase operon expression in Escherichia coli K-12. J Bacteriol. 1985;164:731–740. doi: 10.1128/jb.164.2.731-740.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stewart V, Yanofsky C. Role of leader peptide synthesis in tryptophanase operon expression in Escherichia coli K-12. J Bacteriol. 1986;167:383–386. doi: 10.1128/jb.167.1.383-386.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stiegler P, Carbon P, Zuker M, Ebel J P, Ehresmann C. Structural organization of the 16S ribosomal RNA from E. coli. Topography and secondary structure. Nucleic Acids Res. 1981;9:2153–2172. doi: 10.1093/nar/9.9.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uno M, Ito K, Nakamura Y. Functional specificity of amino acid at position 246 in the tRNA mimicry domain of bacterial release factor 2. Biochimie. 1996;78:935–943. doi: 10.1016/s0300-9084(97)86715-6. [DOI] [PubMed] [Google Scholar]
- 45.Vogel H J, Bonner D M. Acetylornithase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 46.Ward D F, DeLong A, Gottesman M E. Escherichia coli nusB mutations that suppress nusA1 exhibit lambda N specificity. J Mol Biol. 1983;168:73–85. doi: 10.1016/s0022-2836(83)80323-4. [DOI] [PubMed] [Google Scholar]
- 47.Watanabe T, Snell E E. Reversibility of the tryptophanase reaction: synthesis of tryptophan from indole, pyruvate and ammonia. Proc Natl Acad Sci USA. 1972;69:1086–1090. doi: 10.1073/pnas.69.5.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yanofsky C, Horn V. Bicyclomycin sensitivity and resistance affect Rho factor-mediated transcription termination the tna operon of Escherichia coli. J Bacteriol. 1995;177:4451–4456. doi: 10.1128/jb.177.15.4451-4456.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yanofsky C, Horn V, Nakamura Y. Loss or overproduction of polypeptide release factor 3 influences expression of the tryptophanase operon of Escherichia coli. J Bacteriol. 1996;178:3755–3762. doi: 10.1128/jb.178.13.3755-3762.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zuker M. Computer prediction of RNA structure. Methods Enzymol. 1989;180:262–288. doi: 10.1016/0076-6879(89)80106-5. [DOI] [PubMed] [Google Scholar]
- 51.Zuker M, Jaeger J A, Turner D H. A comparison of optimal and suboptimal RNA secondary structures predicted by free energy minimization with structures determined by phylogenetic comparison. Nucleic Acids Res. 1991;19:2707–2714. doi: 10.1093/nar/19.10.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zwiefka A, Kohn H, Widger W R. Transcription termination factor rho: the site of bicyclomycin inhibition in Escherichia coli. Biochemistry. 1993;32:3564–3570. doi: 10.1021/bi00065a007. [DOI] [PubMed] [Google Scholar]