Abstract
Introduction Pharmacogenetic testing is proposed to minimize adverse effects when considered in combination with pharmacological knowledge of the drug. As yet, limited studies in clinical settings have investigated the predictive value of pharmacokinetic (pk) gene variation on therapeutic drug levels as a probable mechanism of adverse effects, nor considered the combined effect of pk gene variation and drug level on antidepressant treatment response.
Methods Two depression cohorts were investigated for the relationship between pk gene variation and antidepressant serum concentrations of amitriptyline, venlafaxine, mirtazapine and quetiapine, as well as treatment response. For the analysis, 519 patients (49% females; 46.6±14.1 years) were included.
Results Serum concentration of amitriptyline was associated with CYP2D6 (higher concentrations in poor metabolizers compared to normal metabolizers), of venlafaxine with CYP2C19 (higher concentrations in intermediate metabolizers compared to rapid/ultrarapid metabolizers) and CYP2D6 (lower metabolite-to-parent ratio in poor compared to intermediate and normal metabolizers, and intermediate compared to normal and ultrarapid metabolizers). Pk gene variation did not affect treatment response.
Discussion The present data support previous recommendations to reduce starting doses of amitriptyline and to guide dose-adjustments via therapeutic drug monitoring in CYP2D6 poor metabolizers. In addition, we propose including CYP2C19 in routine testing in venlafaxine-treated patients to improve therapy by raising awareness of the risk of low serum concentrations in CYP2C19 rapid/ultrarapid metabolizers. In summary, pk gene variation can predict serum concentrations, and thus the combination of pharmacogenetic testing and therapeutic drug monitoring is a useful tool in a personalized therapy approach for depression.
Key words: pharmacogenetics, depressive disorder, serum concentrations, pharmacokinetics
Introduction
Pharmacogenetics is the study of the response to drug therapy depending on a patientʼs genetic background 1 . In the context of precision medicine, pharmacogenetic (PGx) testing has gained significance as, in general, it can improve the effectiveness of drug prescriptions and minimize adverse effects 1 2 by considering a patientʼs genetic profile for drug selection or dose adjustment 3 4 5 . It is, therefore, increasingly suggested to include PGx in the selection of psychiatric medications 6 7 8 . Consequently, there is growing interest in PGx testing among clinicians and patients suffering from psychiatric disorders 3 9 . In general, about 1/3 of the patients with major depressive disorders do not respond to initial treatment and only a fraction achieves remission after the second trial of pharmacotherapy 10 11 12 ; therefore, patients aim to identify the most suitable medication according to their genetic variation by PGx testing 9 . A meta-analysis analyzing five randomized, controlled trials (RCT) with a total of 1737 patients proposed that pharmacogenetic-guided therapy using commercially available test kits, including pharmacokinetic (pk) genes, and also pharmacodynamic genes may improve clinical remission in patients suffering from major depressive disorders 5 . However, it is not clear if results emerging from RCTs with commercial kits are relevant for clinical settings; for example, due to restrictions in inclusion criteria and therapy protocols 3 13 and even more so, which genes are relevant. Applying PGx in clinical routine has begun only recently as there are several limitations that complicate the understanding of how and when using PGx in a clinical setting 6 7 14 . Limitations include the lack of a clear relationship between serum concentration and efficacy for some drugs, polypharmacy, high prevalence of comorbidities, overlapping phenotypes, the lack of guidelines on how to use pharmacogenetic information and the polygenic architecture of the response on antidepressants 1 3 6 7 14 15 . The Clinical Pharmacogenetics Implementation Consortium (CPIC) and the Dutch Pharmacogenetics Working Group have published peer-reviewed, evidence-based pharmacogenetic guidelines for CYP2D6 and CYP2C19 genotypes and dosing, in particular for tricyclic antidepressants and selective serotonin reuptake inhibitors 16 17 18 19 . The clinical utility for other pk enzymes has not been established yet 15 . Compared to pk genes, there is a lack of knowledge about pharmacodynamic genes and their impact on pharmacotherapy and consequently, there are no recommendations given yet 15 .
As the effects of pk genes are assumed to be mediated by therapeutic drug levels measured as serum concentrations, their effects should be larger with regard to serum concentrations than to treatment response. In contrast, serum concentrations that are modulated by genetics or somatic comorbidities should show stronger correlations with treatment response. However, studies investigating the relationship between pk genetics and serum concentrations in clinical settings, as well as studies investigating the combined effect of pk genetics and serum concentrations on treatment response, are lacking.
In consequence, in the present study, we investigated the influence of several genes (transporters, such as ATP-binding cassette transporters ( ABCG2 , ABCB1 , and ABCC2 ), cytochrome P450 (CYP) enzymes ( CYP3A5 , CYP3A4 , CYP2C19 , CYP2C9 , CYP1A2 , CYP2B6 , CYP2D6 ), uridine 5’-diphospho-glucuronosyltransferase (UGTs) ( UGT1A1 , UGT2B15 , and UGT2B7 ) and catechol-O-methyltransferase ( COMT ) affecting the pk of psychiatric drugs on serum concentrations of antidepressants and treatment response. Analyses were primarily performed based on diplotypes/phenotypes as the diplotype and/or the corresponding phenotype are proposed for prescribing. However, since the phenotype definition is a dynamic assignment that depends on the analyzed variants, we included post-hoc genotype and haplotype analyses. In the post-hoc analyses, we also include analyses investigating the combined effect of pk genes and serum concentrations on treatment response. The study was performed in clinical cohorts to evaluate the relevance of the observations for clinical settings.
Methods
Patients
Wuerzburg Sample
Three hundred fifty-five patients (mean age 45.8±16.4 years; 52% female) admitted to the Department of Psychiatry, Psychosomatics and Psychotherapy of the University Hospital of Wuerzburg, due to a depressive episode (single major depressive episode, recurrent depression, or bipolar depression), were included in the study within the first five days after admission. Diagnosis of depressive episodes was ascertained by experienced psychiatrists according to the Diagnostic and Statistical Manual of Mental Disorders (DSM)-IV criteria. The Hamilton Depression Rating Scale (HAMD)-21 was used to assess symptom severity and effectiveness of the therapy. Only patients older than 18 years were included if they showed at least a moderate depressive episode (HAMD≥14) and were of Caucasian origin. Patients suffering from severe neurological or general medical conditions were excluded from the observational study. All patients were treated according to the doctorʼs choice, independent of study participation (naturalistic setting). Depressive symptom severity was graded weekly, and serum concentrations of the psychiatric drugs were determined in weeks 3, 5, and 7 of study participation and used to adjust doses. Adverse drug effects (grading “none”–“mild”–“medium”–“severe” and symptoms) were reported in therapeutic drug monitoring request forms.
Munich Sample
The Munich sample was collected within the Munich Antidepressant Response Signature (MARS) project 20 . Patients for the present study were selected from 1,105 Caucasian participants (mean age 47.6±14.1 years; 53% female) with available serum samples and a diagnosis of unipolar depression (single major depressive episode, recurrent depression). Patients were included in the naturalistic study within the first five days of admission as inpatients due to a depressive episode in the Max Planck Institute of Psychiatry (MPI-P, Munich, Germany) or in one of the participating clinical sites in Southern Bavaria, Germany. Trained psychiatrists diagnosed depressive episodes in accordance with the diagnostic criteria of DSM-IV. Patients with depressive syndromes secondary to any medical or neurological condition, patients with acute manic, hypomanic, or mixed affective symptoms, patients with a lifetime diagnosis of alcohol dependence, illicit drug abuse, or patients suffering from severe medical conditions were excluded from the study. HAMD-21 was used to grade depressive symptoms weekly until treatment week 6 and afterwards bi-weekly until discharge from the hospital. Serum concentrations of antidepressant drugs were used to adjust doses; therefore, TDM was performed according to the doctorʼs choice and not per protocol. Symptoms of adverse drug effects were reported within the weekly rating when a drug was changed due to adverse drug effects.
A more detailed demographic overview is given in Table 1 and Supplement 1.
Table 1 Demographic data of the patients included in the study. More detailed demographic data for amitriptyline, venlafaxine, mirtazapine, and quetiapine samples, respectively, are provided in Supplement 1.
| Combined sample | Wuerzburg sample | Munich sample | Patients included in the analyses | |||||
|---|---|---|---|---|---|---|---|---|
| N | Mean±SD (range) | N | Mean±SD (range) | N | Mean±SD (range) | N | Mean±SD (range) | |
| Included patients | 1 460 | 355 | 1 105 | 519 | ||||
| Age [years] | 1 435 | 47.19±14.68 (18–87) | 331 | 45.75±16.35 (18–85) | 1 104 | 47.63±14.13 (18–87) | 519 | 46.62±14.12 (18–80) |
| Male/female | 671/763 | 147/183 | 524/580 | 256/263 | ||||
| Nonsmoker/smoker | 808/460 | 208/120 | 600/340 | 305/177 | ||||
| Nonsmoker m/f | 377/431 | 87/121 | 290/310 | 144/161 | ||||
| Smoker m/f | 220/240 | 60/60 | 160/180 | 95/82 | ||||
| Daily number of cigarettes | 460 | 16.27±9.48 (0.5–60) | 120 | 15.01±9.33 (1–55) | 340 | 16.71±9.51 (0.5–60) | 177 | 16.55±9.30 (1–55) |
| Hamd baseline | 1 353 | 25.86±6.25 (14–46) | 329 | 26.07±7.27 (14–46) | 1 066 | 25.80±5.89 (14–45) | 481 | 25.82±6.77 (14–46) |
| Hamd out | 1 077 | 12.90±7.15 (0–38) | 293 | 10.20±6.48 (0–38) | 784 | 13.91±7.13 (0–37) | 509 | 12.82±7.16 (0–38) |
| Duration of disorder [years] | 1 379 | 11.91±12.27 (0–62) | 325 | 12.80±12.37 (0–60) | 1 054 | 11.63±12.24 (0–62) | 500 | 12.64±11.67 (0–51) |
| Resp/Nonresp (out) | 453/604 | 208/85 | 245/519 | 232/266 | ||||
| Rem/Nonrem (out) | 282/796 | 119/174 | 163/622 | 135/374 | ||||
N, number of patients; SD, standard deviation; m, male; f, female; HAMD, Hamilton Depression Rating Scale-21; Out, outcome time point; Resp, Response; Rem, Remission.
Written informed consent was obtained from each participant. Both studies were approved by the local ethics committees of the Universities of Wuerzburg (104/12, amended 128/15) and Munich (318/00, 21/03/2001) and carried out in accordance with the ethical principles of the Helsinki Declaration.
Treatment Response and Remission
Percentage reduction in the HAMD-21 score was used for analyses on clinical improvement. For post-hoc analyses on genotypes and haplotypes, treatment response was defined as ≥ 50% reduction in depressive symptoms from baseline, evaluated by HAMD-21 21 . Remission was defined as a HAMD score ≤ 7 22 . Week 7, the time point of discharge from the study (Wuerzburg sample) and week 6 (Munich sample), respectively, were chosen as outcome time points as an effective and consistent therapy can be expected. Accordingly, serum concentrations determined in week 7 (Wuerzburg) or week 6 (Munich) were used for the analyses.
Genotyping
Genotyping of pharmacokinetically relevant gene variants (SNPs and star allele coverage; supplement 2) was performed on a MassArray Analyzer 4 system (Agena Bioscience GmbH, Hamburg, Germany) based on Agena’s PGx 74 v1.0 Assay, VeriDose CYP2D6 CNV Assay and an additional self-designed panel using SpectroCHIP ® -384 Arrays and the iPLEX ® Pro chemistry following the instructions supplied by the manufacturer. Primer sequences of the self-designed assay are available on request.
Phenotypes were determined according to the CPIC specifications into poor metabolizers (PM), intermediate metabolizers (IM), normal metabolizers (NM), rapid metabolizers (RM), and ultrarapid metabolizers (UM) 23 . If no diplotype-phenotype table was available, diplotypes were used for analyses.
Therapeutic drug monitoring
Therapeutic drug monitoring (TDM) was applied in both cohorts, according to the AGNP-TDM expert group consensus guideline 24 . Blood was drawn at trough levels at steady-state. Daily calibrations and internal quality control samples, integrated into each analytical series ensured correct results of the analyses. The laboratories (Wuerzburg and Munich) were certified by a quality control program 25 ; therefore, the results determined in different laboratories were compatible, giving the rationale for the joint analysis of the data. Sum serum concentrations for venlafaxine+O-desmethylvenlafaxine and amitriptyline+nortriptyline were considered, as they represent the active moieties (AM) of the drugs, for which therapeutic reference ranges are evaluated and which are relevant for treatment response 24 . Dose-corrected serum concentrations were calculated (serum concentration/dose; CD) 24 for regression analyses investigating the association between genetic markers and serum concentrations of the drugs to overcome the effect of the dose. Moreover, the metabolite-to-parent ratios (MPR) nortriptyline/amitriptyline, and O-desmethylvenlafaxine/venlafaxine were calculated 24 .
Dimensional outliers (≥4 SD from the mean) from serum concentrations and CD, as well as MPR, were set as missing data.
Statistical analyses
Statistical analyses were conducted with PLINK v1.9 26 and R v3.1.3 27 . Haplotype blocks based on sample-specific linkage disequilibrium (LD) pattern from Haploview v4.1 28 29 were defined for all analysed SNPs according to gene-specific haplotype tables from the PharmVar homepage (https://www.pharmvar.org/gene; supplement 2). Haplotype coverage for CYP2D6 and CYP2C19 aligns with the recommended tier 1 alleles for CYP2D6 and CYP2C19 for 5 of 10 and 2 of 3, respectively 30 31 . From a total of 101 genotyped single markers located within 26 genes, we restricted our analyses to genes affecting pk (n=14), thus including 86 variants. Of those, only 32 heterozygous SNPs in 14 genes with a minor allele frequency of more than 0.01 remained for analyses. All 32 variants reached a genotyping call rate above 95%, and genotype distribution did not deviate from Hardy-Weinberg-Equilibrium (p>0.001).
Diplotype/phenotype-based analyses to investigate differences in serum concentrations and clinical improvement depending on the diplotype or phenotype, respectively, were performed by Kruskal-Wallis tests. Diplotype/phenotype-based analyses to investigate differences in serum concentrations and remission depending on the diplotype or phenotype, respectively, were performed either by chi-squared tests or Fisherʼs exact tests. To adjust for alpha-error accumulation, nominal p-values were Bonferroni-corrected for the total number of genes (7x) and the number of analysed drug concentrations (4x) or MPR (2x), respectively. The significance threshold was set to p≤0.001, or p=0.002. Box-Plot diagrams were prepared in R v3.1.3 27 .
Results
To increase statistical power, all analyses were restricted to the combined sample. The combined sample comprised 355 patients from Wuerzburg and 1,105 from Munich; thus, there were 1,460 (mean age 47.2±14.7 years; 52% female) patients. For power reasons, only patients with available serum concentrations for amitriptyline (N=109), venlafaxine (N=258), mirtazapine (N=171), and quetiapine (N=193) were included in the analyses. A detailed demographic overview is given in Table 1 and supplement 1; the administered psychiatric drugs are summarized in supplement 3.
Serum Concentrations
CD AM of amitriptyline were associated with CYP2D6 phenotypes (p=0.033). Pairwise comparisons with continuity correction revealed significantly (116%) higher CD AM of amitriptyline in PM compared to NM (p=0.01) ( Table 2 , Fig. 1 , Supplement 4).
Table 2 Significant results in diplotype/phenotype analyses. To adjust for alpha-error accumulation, nominal p-values were Bonferroni-corrected for the total number of genes (7x) and the number of analysed drug concentrations or MPR (6x), respectively, in each analysis. The significance threshold was set to p≤0.001.
| Serum concentration | ||||
|---|---|---|---|---|
| Gene | P (Bonferroni) | Pairwise | ||
| Mean±SD CD [(ng/ml)/(mg/day)] | Adjusted p-value | |||
| CYP2D6 | CD Amitriptyline (N=109) | |||
| 7.90*10 -4 (0.033) | ||||
| NM | PM | |||
| 0.92±0.52 | 1.99±1.02 | 0.010 | ||
| CYP2C19 | CD Venlafaxine (N=256) | |||
| 5.67*10 -5 (0.002) | ||||
| IM | RM | |||
| 2.02±0.81 | 1.42±0.65 | 3.0*10 -4 | ||
| IM | UM | |||
| 2.02±0.81 | 1.38±0.66 | 0.035 | ||
| CYP2D6 | MPR O-desmethylvenlafaxine/Venlafaxine (N=129) | |||
| 1.22*10 -11 (5.12*10 -10 ) | ||||
| NM | IM | |||
| 5.32±3.14 | 2.07±1.21 | 3.3*10 -8 | ||
| NM | PM | |||
| 5.32±3.14 | 0.43±0.42 | 2.7*10 -5 | ||
| IM | PM | |||
| 2.07±1.21 | 0.43±0.42 | 6.0*10 -4 | ||
| UM | IM | |||
| 9.2±3.72 | 2.07±1.21 | 0.028 | ||
SD, standard deviation; CD, dose-corrected serum concentrations; MPR, metabolite-to-parent ratio; NM, normal metabolizer, IM intermediate metabolizer; PM, poor metabolizer; RM, rapid metabolizer; UM, ultrarapid metabolizer.
Fig. 1.
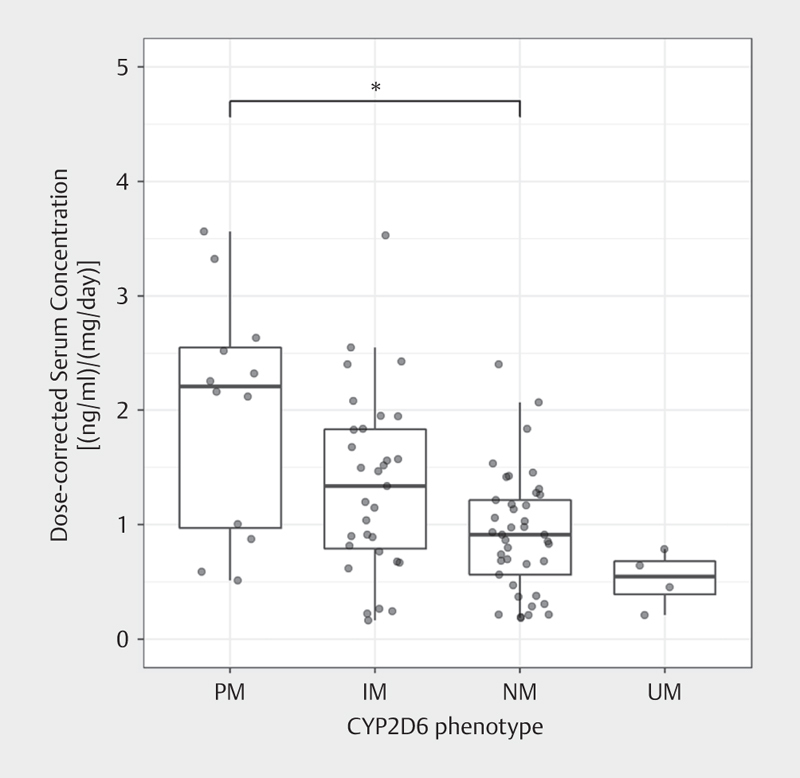
Dose-corrected serum concentrations of amitriptyline were associated with the CYP2D6 phenotype (Kruskal-Wallis test). Post-hoc analyses revealed significant differences in dose-corrected serum concentrations of amitriptyline between PM and NM.
CD AM of venlafaxine was associated with CYP2C19 phenotypes (p=0.002). CD AM were 42% higher in IM compared to RM (p=3.0*10 -4 ) and 46% higher in IM compared to UM (p=0.035) ( Table 2 , Fig. 2a , Supplement 4). The O-desmethylvenlafaxine/venlafaxine ratio was associated with CYP2D6 phenotypes (p=5.2*10 -10 ); MPR was higher in CYP2D6 UM compared to IM (344%, p=0.028), NM compared to PM (1137%, p=2.7*10 -5 ) and IM (157%, p=3.3*10 -8 ), and IM compared to PM (381%, p=6.0*10 -4 ) ( Table 2 , Fig. 2b , Supplement 4).
Fig. 2.
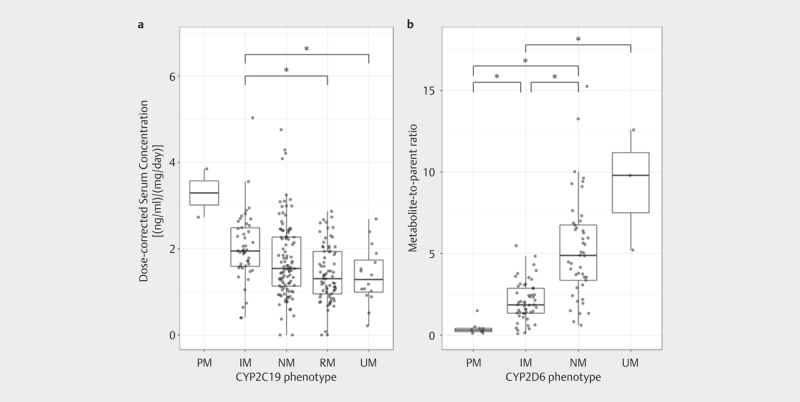
Dose-corrected serum concentrations of venlafaxine were associated with the CYP2C19 phenotypes (Kruskal-Wallis test) a , and the metabolite-to-parent ratio (MPR) of venlafaxine was associated with CYP2D6 phenotypes (Kruskal-Wallis test) b . Post-hoc analyses revealed significant differences in dose-corrected serum concentrations of venlafaxine between CYP2C19 IM and RM, and IM and UM; MPR was higher in CYP2D6 UM compared to IM, NM compared to PM and IM, and IM compared to PM.
CD of quetiapine and mirtazapine were not associated with the examined diplotypes/phenotypes (Supplement 4).
Post-hoc analyses on genotypes and haplotypes are summarized in Supplement 5.
Treatment Response and Remission
For amitriptyline, venlafaxine, quetiapine, and mirtazapine, none of the diplotypes/phenotypes showed a significant association with the response to drug therapy (supplement 4). However, CD of amitriptyline was nominally associated with ABCC2 , CD of venlafaxine with CYP2C9 , and CD of mirtazapine with CYP2D6 . In addition, in terms of remission, CD of amitriptyline was associated with ABCC2 (p=3.4*10 -4 ) (Supplement 4). Post-hoc analyses on genotypes and haplotypes are summarized in supplement 5.
Discussion
In this study, associations between pk gene variation and serum concentrations, as well as between pk gene variation and treatment response, were investigated in two depression cohorts in a clinical setting.
We report an association of the CYP2D6 phenotypes with CD AM of amitriptyline; therefore, CD AM of amitriptyline was higher in PM compared to NM. This supported previous recommendations that in CYP2D6 poor metabolizers, starting doses should be reduced, and dose adjustments should be guided via TDM 17 . In addition, our result on phenotypes also is supported by the results of post-hoc analyses on geno- and haplotypes. For the first time, we report an association of the CYP2D6 SNP rs1065852 with the CD AM of amitriptyline. Carriers of the A-allele showed higher serum concentrations of amitriptyline then the wild type (G). This is supported by the fact that rs1065852 is located in a conserved region of CYP2D6 and therefore, possibly important for protein function 32 . Modeling studies showed that the flexibility in the F-G loop, which may affect the entrance of a substrate into the protein, is lower in the mutant than the wild type, which may account for lower enzyme activity 32 . Consistent with this observation, in a previous genome-wide association study, rs1065852 was found to be associated with the metabolism of escitalopram 33 . Similarly, the minor (T) allele of rs3892097 was associated with higher amitriptyline serum concentrations compared to the major allele (C). SNP rs3892097 is the diagnostic variant for the *4 haplotype, resulting in a splicing defect and, therefore, a nonfunctional protein 34 35 36 . Variant rs1065852 is part of the CYP2D6 *10 haplotype with decreased function, as well as of the *4 haplotype with no function 34 37 . Consistent with this, CYP2D6 *4 was also associated with higher CD AM of amitriptyline in the present study. Besides CYP2C19, CYP2D6 is the major enzyme in the metabolism of amitriptyline and its active metabolite nortriptyline, responsible for the hydroxylation of amitriptyline and nortriptyline 17 38 . As only CYP2D6 seems to clinically affect total drug concentration, the present data support previous studies showing that compared to CYP2D6, CYP2C19 had less influence on total amitriptyline clearance 17 38 .
The main metabolizing enzymes for venlafaxine are CYP2D6 and CYP2C19 39 40 . CYP2D6 is mainly responsible for the formation of the active metabolite O-desmethylvenlafaxine; however, it was suggested that CYP2C19 might also contribute 39 40 . CYP2C19 and CYP3A4 catalyze the N-desmethylvenlafaxine formation; O-, and N-desmethylvenlafaxine are further metabolized via CYP2D6, CYP2C19, and CYP3A4 39 40 . CD AM of venlafaxine was lower in CYP2C19 RM and UM compared to IM. This phenotype also was supported by post-hoc geno-, and haplotype analyses. Thus, CD AM of venlafaxine was lower in carriers of CYP2C19 *17. The diagnostic SNP for this increased function haplotype 37 , rs12248560 40 , was also associated with lower CD AM . This variant is located in the promoter of the gene, and its minor allele is associated with increased CYP2C19 expression and activity 40 41 . Previously, lower serum concentrations of escitalopram were associated with the *17 haplotype 42 43 , but to the best of our knowledge, this is the first study reporting an association between CYP2C19 *17 and lower venlafaxine serum concentration. In accordance with our results, one study investigating the combined effect of CYP2D6/CYP2C19 on the pk of venlafaxine also reported an impact of CYP2C19 on venlafaxine serum concentration; however, contrary to the CPIC phenotype grouping 23 they merged the CYP2C19 *1/*17 group into the NM group 44 . Moreover, in contrast to our results, in combination with CYP2D6 phenotypes, CYP2C19 UM did not affect serum concentrations, but CYP2C19 PM did 44 . To investigate whether the association of CYP2C19 phenotypes with the active moiety is also true separately for the serum concentration of venlafaxine and its renally excreted and pharmacodynamically slightly different active metabolite O-desmethylvenlafaxine, we performed explorative analyses (Kruskal-Wallis tests as described in the methods section, however, with dose-corrected serum concentration of venlafaxine, and O-desmethylvenlafaxine as outcome parameter, respectively). Doing this, we found that CYP2C19 phenotypes were associated with a CD of venlafaxine (N=128, p=0.04; post-hoc tests: not significant) but not with a CD of O-desmethylvenlafaxine (N=130, p=0.26). As in the present, as well as in previous studies, CD AM of venlafaxine was not associated with CYP2D6 genotypes 45 46 ; our results extend the evidence that mainly CYP2C19 and not CYP2D6 affect total venlafaxine clearance. One study investigated the combined effect of CYP2D6/CYP2C19 phenotypes on CD AM of venlafaxine, reporting that both genes affect serum concentrations; however, they were not investigated separately 44 . CYP2D6, however, seems to influence the conversion from venlafaxine to O-desmethylvenlafaxine. Lower MPRs were present in CYP2D6 PM compared to IM and NM and in IM compared to NM and UM. In accordance, results of post-hoc analyses showed that minor alleles of rs1065852 and rs3892097, and in consequence, carriers of CYP2D6 *4 haplotype were associated with lower MPRs. Hence, present data suggest that CYP2D6 *4 shifts the metabolism towards a higher level of venlafaxine 45 46 . These results are in line with a previous study, reporting higher venlafaxine and lower O-desmethylvenlafaxine serum concentration in CYP2D6 PM compared to NM. In concordance with previous results, CYP2D6 did not affect the response to venlafaxine treatment 47 . Of note, an MPR below 0.3, indicative of CYP2D6 poor metabolizers 24 , was associated with more side effects 48 . In consequence, CYP2D6 may be relevant for the occurrence of side effects and less for treatment response to venlafaxine. However, in our analyses, MPR was not associated with adverse drug effects (Mann-Whitney-U test, p=0.18); possibly, as in our sample dosing was adjusted to the serum concentrations of the drugs. Thus, in the Wuerzburg sample, only mild (N=6) and medium (N=6) adverse drug effects were reported, and in the Munich sample, venlafaxine was discontinued only in one case due to adverse drug events.
Strengths and limitations
The present observational study in two independent cohorts provides real-life data from naturalistic settings and the results, thus, are relevant for clinical settings. We did our analyses based on diplotypes/phenotypes as the diplotype and/or the corresponding phenotype are proposed for prescribing. However, since the phenotype definition is a dynamic assignment that can change over time and depending on the analyzed variants, the phenotype may be variable, we also included post-hoc genotype and haplotype analyses (supplement 5). However, this design also had some limitations. The inclusion criteria in both samples were not exactly the same. The selected sample from the Munich cohort only included unipolar depressed patients, whereas the Wuerzburg sample also comprised cases with a bipolar diagnosis. Including also bipolar patients possibly affected the analyses regarding treatment response. The sample only comprised Caucasians; therefore, our results are valid only in the Caucasian population. We did not control for any inhibiting or inducing medications; thus, possibly inhibiting or inducing drugs resulted in phenotypically different characteristics than genetically determined, which possibly affected our results. In the amitriptyline, venlafaxine, quetiapine, and mirtazapine sample, 8, 17, 37, and 19 patients received CYP2D6 inhibiting drugs (bupropion, doxepin, duloxetine, fluoxetine, melperone, paroxetine, perazine), no patient received a CYP2C19 inhibiting drug, and 1, 2, 1, and 3 patients received carbamazepine (inductive for CYP2B6, CYP2C9, CYP3A4, UGT, p-glycoprotein) 49 . In addition, we performed an explorative post-hoc analysis excluding patients with interacting drugs for CYP2D6 to consider phenoconversion effects (Supplement 6). The results did not differ from the results of our main analyses. Analyses were not controlled for smoking status, which may have affected mirtazapine results as smoking affects mirtazapine metabolism due to the involvement of CYP1A2 50 51 . For the Wuerzburg sample, no severe adverse effect was reported, as dosing was adjusted to serum concentrations of the drugs. For the MARS sample, adverse drug effects were reported only in case of a change in drug therapy. In the amitriptyline, venlafaxine, quetiapine, and mirtazapine sample only in 1, none, 1, and 3 patients, respectively, the drug was changed due to adverse drug effects; however, dosing was adjusted to serum concentrations of the drugs as well. Due to the limited data, further analyses on adverse drug effects and genetic data were not conducted. While physicians were not aware of post-hoc determined PGx data but were aware of online-determined TDM results, dosing thus was adjusted to the serum concentrations of the drugs. However, this may have biased results of the response analyses in a direction against correlations between phenotypes and treatment response. In the Munich sample, a bias due to the selection of patients for TDM, for example, non-response, cannot be excluded, while in the Wuerzburg sample, TDM was determined by protocol (weeks 3, 5, and 7). This may have biased results against correlations between phenotypes and serum concentrations on one side and treatment response on the other. All these limitations thus may have contributed to reducing our power to detect correlations and thus finally argue for the robustness of those observed.
Conclusion
The present study supports previous recommendations that in CYP2D6 poor metabolizers, starting doses should be reduced, and dose adjustments should be guided via TDM 17 . Therapy in venlafaxine-treated patients was affected by CYP2C19 . CD AM was lower in CYP2C19 RM and UM compared to IM. While CYP2C19 testing is not recommended for venlafaxine yet 52 , we propose including CYP2C19 in routine testing for venlafaxine-treated patients to improve therapy outcomes by raising awareness of the risk of low serum concentrations and potentially non-response in CYP2C19 rapid/ultrarapid metabolizers. Thus, our findings primarily support the role of pk gene variation for serum concentrations and because of their strong relation with adverse effects, also for adverse effects.
We are aware that the present results are based on restricted sample size. Since, however, such real-life samples are rare, we believe that our results are valuable with respect to providing initial guidance for the application of PGx in combination with TDM in routine clinical practice. They may thus contribute to further meta-analysis and the development of precision medicine in psychiatry.
Ethical approval
All procedures performed in the analysis involving human participants were in accordance with the ethical standards of the institutional research committees and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Author contributions
Project administration: J. Deckert, E. Binder, A. Menke
Data collection: C. Wurst, S. Stonawski, N. Rost, A. Menke, S. Lucae, T. Brückl
Analysis and interpretation of the data: M. Scherf-Clavel, H. Weber
Writing – original draft preparation: M. Scherf-Clavel, H. Weber
Writing – review and editing: M. Scherf-Clavel, H. Weber, C. Wurst, S. Stonawski, L. Hommers, S. Unterecker, C. Wolf, K. Domschke, N. Rost, T. Brückl, S. Lucae, M. Uhr, E. Binder, A. Menke, J. Deckert
All authors have approved of the contents of this manuscript and provided consent for publication.
Funding Statement
Funding N. Rost is supported by the International Max Planck Research School of Translational Psychiatry (IMPRS-TP) and received funding from the Bavarian Ministry of Economic Affairs, Regional Development and Energy (BayMED, PBN_MED-1711-0003).
Conflicts of Interest J. Deckert is the co-recipient of a grant from the Bavarian Ministry of Economic Affairs, Regional Development and Energy (BayMED, MED-1604-0010) and an investigator in a European grant (Horizon 2020 SME programme of the European Union ref 696802) to P1Vital. A. Menke has given educational lectures for the health insurance company AOK, Servier, Recordati, Neuraxpharm, and Medice. K. Domschke is a member of the Janssen Pharmaceuticals, Inc. Steering Committee Neurosciences. M. Scherf-Clavel, H. Weber, C. Wolf, S. Stonawski, C. Wurst, L. Hommers, S. Unterecker, N. Rost, T. Brückl, S. Lucae, M. Uhr and E. Binder have no conflicts of interest.
shared first authorship
shared last authorship
Supplementary Material
References
- 1.Malsagova K A, Butkova T V, Kopylov A T et al. Pharmacogenetic testing: A tool for personalized drug therapy optimization. Pharmaceutics. 2020;12:1240. doi: 10.3390/pharmaceutics12121240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Menke A. Precision pharmacotherapy: Psychiatry’s future direction in preventing, diagnosing, and treating mental disorders. Pharmgenomics Pers Med. 2018;11:211–222. doi: 10.2147/PGPM.S146110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vest B M, Wray L O, Brady L A et al. Primary care and mental health providers’ perceptions of implementation of pharmacogenetics testing for depression prescribing. BMC Psychiatry. 2020;20:518. doi: 10.1186/s12888-020-02919-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenblat J D, Lee Y, McIntyre R S. The effect of pharmacogenomic testing on response and remission rates in the acute treatment of major depressive disorder: A meta-analysis. J Affect Disord. 2018;241:484–491. doi: 10.1016/j.jad.2018.08.056. [DOI] [PubMed] [Google Scholar]
- 5.Bousman C A, Arandjelovic K, Mancuso S G et al. Pharmacogenetic tests and depressive symptom remission: A meta-analysis of randomized controlled trials. Pharmacogenomics. 2019;20:37–47. doi: 10.2217/pgs-2018-0142. [DOI] [PubMed] [Google Scholar]
- 6.Drozda K, Müller D J, Bishop J R. Pharmacogenomic testing for neuropsychiatric drugs: Current status of drug labeling, guidelines for using genetic information, and test options. Pharmacotherapy. 2014;34:166–184. doi: 10.1002/phar.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bousman C A, Menke A, Müller D J. Towards pharmacogenetic-based treatment in psychiatry. J Neural Transm (Vienna) 2019;126:1–3. doi: 10.1007/s00702-018-01968-9. [DOI] [PubMed] [Google Scholar]
- 8.International Society for Psychiatric Genetics (ISPG) Genetic testing statement: A statement from the international society of psychiatric genetics. 2019https://ispg.net/genetic-testing-statement/ cited: 22 December, 2020
- 9.Liko I, Lai E, Griffin R J et al. Patients’ perspectives on psychiatric pharmacogenetic testing. Pharmacopsychiatry. 2020;53:256–261. doi: 10.1055/a-1183-5029. [DOI] [PubMed] [Google Scholar]
- 10.Gartlehner G, Hansen R A, Morgan L C et al. Comparative benefits and harms of second-generation antidepressants for treating major depressive disorder: An updated meta-analysis. Ann Intern Med. 2011;155:772–785. doi: 10.7326/0003-4819-155-11-201112060-00009. [DOI] [PubMed] [Google Scholar]
- 11.Chen P. Singapore: Springer Singapore; 2019. Optimized Treatment Strategy for Depressive Disorder. In: Fang Y ed, Depressive Disorders: Mechanisms, Measurement and Management; pp. 201–217. [DOI] [PubMed] [Google Scholar]
- 12.Gaynes B N, Rush A J, Trivedi M H et al. The STAR*D study: Treating depression in the real world. Cleve Clin J Med. 2008;75:57–66. doi: 10.3949/ccjm.75.1.57. [DOI] [PubMed] [Google Scholar]
- 13.Wisniewski S R, Rush A J, Nierenberg A A et al. Can phase III trial results of antidepressant medications be generalized to clinical practice? A STAR*D report. Am J Psychiatry. 2009;166:599–607. doi: 10.1176/appi.ajp.2008.08071027. [DOI] [PubMed] [Google Scholar]
- 14.Eap C B. Personalized prescribing: A new medical model for clinical implementation of psychotropic drugs. Dialogues Clin Neurosci. 2016;18:313–322. doi: 10.31887/DCNS.2016.18.3/ceap. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bousman C A, Bengesser S A, Aitchison K J et al. Review and consensus on pharmacogenomic testing in psychiatry. Pharmacopsychiatry. 2021;54:5–17. doi: 10.1055/a-1288-1061. [DOI] [PubMed] [Google Scholar]
- 16.Hicks J K, Bishop J R, Sangkuhl K et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin Pharmacol Ther. 2015;98:127–134. doi: 10.1002/cpt.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hicks J K, Sangkuhl K, Swen J J et al. Clinical pharmacogenetics implementation consortium guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin Pharmacol Ther. 2017;102:37–44. doi: 10.1002/cpt.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Relling M V, Klein T E. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin Pharmacol Ther. 2011;89:464–467. doi: 10.1038/clpt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swen J J, Nijenhuis M, de Boer A et al. Pharmacogenetics: from bench to byte--an update of guidelines. Clin Pharmacol Ther. 2011;89:662–673. doi: 10.1038/clpt.2011.34. [DOI] [PubMed] [Google Scholar]
- 20.Hennings J M, Owashi T, Binder E B et al. Clinical characteristics and treatment outcome in a representative sample of depressed inpatients - findings from the Munich Antidepressant Response Signature (MARS) project. J Psychiatr Res. 2009;43:215–229. doi: 10.1016/j.jpsychires.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 21.Rush A J, Kraemer H C, Sackeim H A et al. Report by the ACNP Task Force on response and remission in major depressive disorder. Neuropsychopharmacology. 2006;31:1841–1853. doi: 10.1038/sj.npp.1301131. [DOI] [PubMed] [Google Scholar]
- 22.Zimmerman M, Chelminski I, Posternak M. A review of studies of the Hamilton depression rating scale in healthy controls: Implications for the definition of remission in treatment studies of depression. J Nerv Ment Dis. 2004;192:595–601. doi: 10.1097/01.nmd.0000138226.22761.39. [DOI] [PubMed] [Google Scholar]
- 23.CPIC - Clinical Pharmacogenetics Implementation Consortium. 2021. Cited: 15 March, 2022
- 24.Hiemke C, Bergemann N, Clement H W et al. Consensus guidelines for therapeutic drug monitoring in neuropsychopharmacology: Update 2017. Pharmacopsychiatry. 2018;51:9–62. doi: 10.1055/s-0043-116492. [DOI] [PubMed] [Google Scholar]
- 25.INSTAND Gesellschaft zur Förderung der Qualitätssicherung in medizinischen Laboratorien e. V. 2020https://www.instand-ev.de/ueber-instand-ev/instand-ev.htmlcited: February 10, 2020
- 26.Purcell S, Neale B, Todd-Brown K et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.RCoreTeam R: A language and environment for statistical computing., R Core Teamhttps://www.R-project.org/ R Foundation for Statistical Computing, Vienna, Austria 2014
- 28.Barrett J C, Fry B, Maller J et al. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 29.Broad Institute of MIT and Harvard Haploview. 2006https://www.broadinstitute.org/haploview/haploview cited: 10 January, 2020
- 30.Pratt V M, Cavallari L H, Del Tredici A L et al. Recommendations for clinical CYP2D6 genotyping allele selection: A joint consensus recommendation of the Association for Molecular Pathology, College of American Pathologists, Dutch Pharmacogenetics Working Group of the Royal Dutch Pharmacists Association, and the European Society for Pharmacogenomics and Personalized Therapy. J Mol Diagn. 2021;23:1047–1064. doi: 10.1016/j.jmoldx.2021.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pratt V M, Del Tredici A L, Hachad H et al. Recommendations for clinical CYP2C19 genotyping allele selection: A report of the Association for Molecular Pathology. J Mol Diagn. 2018;20:269–276. doi: 10.1016/j.jmoldx.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 32.Xin J, Yuan M, Peng Y et al. Analysis of the deleterious single-nucleotide polymorphisms associated with antidepressant efficacy in major depressive disorder. Front Psychiatry. 2020;11:151. doi: 10.3389/fpsyt.2020.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ji Y, Schaid D J, Desta Z et al. Citalopram and escitalopram plasma drug and metabolite concentrations: genome-wide associations. Br J Clin Pharmacol. 2014;78:373–383. doi: 10.1111/bcp.12348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Owen R P, Sangkuhl K, Klein T E et al. Cytochrome P450 2D6. Pharmacogenet Genomics. 2009;19:559–562. doi: 10.1097/FPC.0b013e32832e0e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gaedigk A, Ingelman-Sundberg M, Miller N A et al. The Pharmacogene Variation (PharmVar) Consortium: Incorporation of the Human Cytochrome P450 (CYP) Allele Nomenclature Database. Clin Pharmacol Ther. 2018;103:399–401. doi: 10.1002/cpt.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaedigk A, Sangkuhl K, Whirl-Carrillo M et al. The Evolution of PharmVar. Clin Pharmacol Ther. 2019;105:29–32. doi: 10.1002/cpt.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clinical Pharmacogenetics Implementation Consortium CPIC® Guideline for Tricyclic Antidepressants and CYP2D6 and CYP2C19. 2020https://cpicpgx.org/guidelines/guideline-for-tricyclic-antidepressants-and-cyp2d6-and-cyp2c19/ cited: 05 January, 2020
- 38.Jiang Z P, Shu Y, Chen X P et al. The role of CYP2C19 in amitriptyline N-demethylation in Chinese subjects. Eur J Clin Pharmacol. 2002;58:109–113. doi: 10.1007/s00228-002-0445-6. [DOI] [PubMed] [Google Scholar]
- 39.Sangkuhl K, Stingl J C, Turpeinen M et al. PharmGKB summary: Venlafaxine pathway. Pharmacogenet Genomics. 2014;24:62–72. doi: 10.1097/FPC.0000000000000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whirl-Carrillo M, McDonagh E M, Hebert J M et al. Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther. 2012;92:414–417. doi: 10.1038/clpt.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sim S C, Risinger C, Dahl M L et al. A common novel CYP2C19 gene variant causes ultrarapid drug metabolism relevant for the drug response to proton pump inhibitors and antidepressants. Clin Pharmacol Ther. 2006;79:103–113. doi: 10.1016/j.clpt.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 42.Rudberg I, Mohebi B, Hermann M et al. Impact of the ultrarapid CYP2C19*17 allele on serum concentration of escitalopram in psychiatric patients. Clin Pharmacol Ther. 2008;83:322–327. doi: 10.1038/sj.clpt.6100291. [DOI] [PubMed] [Google Scholar]
- 43.Huezo-Diaz P, Perroud N, Spencer E P et al. CYP2C19 genotype predicts steady state escitalopram concentration in GENDEP. J Psychopharmacol. 2012;26:398–407. doi: 10.1177/0269881111414451. [DOI] [PubMed] [Google Scholar]
- 44.Kringen M K, Bråten L S, Haslemo T et al. The influence of combined CYP2D6 and CYP2C19 genotypes on venlafaxine and O-desmethylvenlafaxine concentrations in a large patient cohort. J Clin Psychopharmacol. 2020;40:137–144. doi: 10.1097/JCP.0000000000001174. [DOI] [PubMed] [Google Scholar]
- 45.Veefkind A H, Haffmans P M, Hoencamp E. Venlafaxine serum levels and CYP2D6 genotype. Ther Drug Monit. 2000;22:202–208. doi: 10.1097/00007691-200004000-00011. [DOI] [PubMed] [Google Scholar]
- 46.Hermann M, Hendset M, Fosaas K et al. Serum concentrations of venlafaxine and its metabolites O-desmethylvenlafaxine and N-desmethylvenlafaxine in heterozygous carriers of the CYP2D6*3, *4 or *5 allele. Eur J Clin Pharmacol. 2008;64:483–487. doi: 10.1007/s00228-007-0453-7. [DOI] [PubMed] [Google Scholar]
- 47.Ng C, Sarris J, Singh A et al. Pharmacogenetic polymorphisms and response to escitalopram and venlafaxine over 8 weeks in major depression. Hum Psychopharmacol. 2013;28:516–522. doi: 10.1002/hup.2340. [DOI] [PubMed] [Google Scholar]
- 48.Shams M E, Arneth B, Hiemke C et al. CYP2D6 polymorphism and clinical effect of the antidepressant venlafaxine. J Clin Pharm Ther. 2006;31:493–502. doi: 10.1111/j.1365-2710.2006.00763.x. [DOI] [PubMed] [Google Scholar]
- 49.Springer-Verlag GmbH, Heidelberg. PSIAC. https://www.psiac.de/; cited: 18 March, 2019
- 50.Kroon L A. Drug interactions with smoking. Am J Health Syst Pharm. 2007;64:1917–1921. doi: 10.2146/ajhp060414. [DOI] [PubMed] [Google Scholar]
- 51.Scherf-Clavel M, Samanski L, Hommers L G et al. Analysis of smoking behavior on the pharmacokinetics of antidepressants and antipsychotics: Evidence for the role of alternative pathways apart from CYP1A2. Int Clin Psychopharmacol. 2019;34:93–100. doi: 10.1097/YIC.0000000000000250. [DOI] [PubMed] [Google Scholar]
- 52.Fabbri C, Serretti A. Genetics of treatment outcomes in major depressive disorder: Present and future. Clin Psychopharmacol Neurosci. 2020;18:1–9. doi: 10.9758/cpn.2020.18.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


