Abstract
Aptamers’ vast conformation ensemble consisting of interconverting substates severely impairs their performance and applications in biomedicine. Therefore, developing new chemistries stabilizing aptamer conformation and exploring the conformation–performance relationship are highly desired. Herein, we developed an 8-methoxypsoralen-based photochemically covalent lock to stabilize aptamer conformation via crosslinking the inter-stranded thymine nucleotides at TpA sites. Systematical studies and molecular dynamics simulations were performed to explore the conformation–performance relationship of aptamers, revealing that conformation-stabilized aptamers displayed better ability to bind targets, adapt to physiological environment, resist macrophage uptake, prolong circulation half-life, accumulate in and penetrate into tumor than their counterparts. As expected, conformation-stabilized aptamers efficiently improved the therapeutic efficacy of aptamer-drug conjugation on tumor-bearing mice. Collectively, our study has developed a general, simple and economic strategy to stabilize aptamer conformation and shed light on the conformation–performance relationship of aptamers, laying a basis for promoting their basic researches and applications in biomedicine.
Graphical Abstract
Graphical Abstract.
Locking aptamer conformation strengthens aptamer performance for biomedicine: an 8-methoxypsoralen-based photochemically covalent lock has been developed to stabilize aptamer conformation via crosslinking the inter-stranded thymine nucleotides at TpA sites. Locked aptamers present better binding affinity and in vivo performance than the unlocked aptamer.
INTRODUCTION
Single-stranded nucleic acid (ssNA) aptamers, generated from nucleic acid libraries by an in vitro directed evolved technique termed as SELEX (1–3), recognize a variety of targets by folding into specific secondary/tertiary structure (4,5). As chemical antibodies, aptamer have been extensively applied in biomedicine based on its easily chemical synthesis, accurate modification, excellent specificity, elegant programmability and good biocompatibility (6–10). However, each phosphodiester bond in ssNA contains five freely rotating single bonds. In addition, hydrogen bond in base pairing is dynamic. Thus, aptamer is inherently dynamic and has a vast conformation ensemble containing interconverting substates. Such flexible conformation usually causes binding affinity attenuation, off-target interaction, as well as susceptibility to exonuclease in biological fluids (11), thereby severely affecting the performance of aptamer in biomedicine.
The conformation of aptamers determines their performance and applications. Therefore, various strategies have been developed to facilitate aptamer folding into proper conformation and improve their performance (12–15). Among these strategies, cyclization of ssNA based on enzymatical or click chemical ligation is the main method facilitating the proper conformation formation of aptamers. Our group and other groups have demonstrated that the cyclized aptamers have no free ends of ssNA and reduce the possibility of misfolding, thereby facilitating their thermal stability, conformation stability and binding ability, which promotes aptamer applications (16–20). However, knowledge about the effect of the conformation stability of aptamer on their performance, especially in vivo performance, remains little. Moreover, current enzymatical or click chemical ligation is either time-consuming or expensive. Therefore, new chemistries that stabilize aptamer conformation and systematic studies that thoroughly elucidate the conformation–performance relationship of aptamer are highly desired and important for basic researches and applications of aptamer.
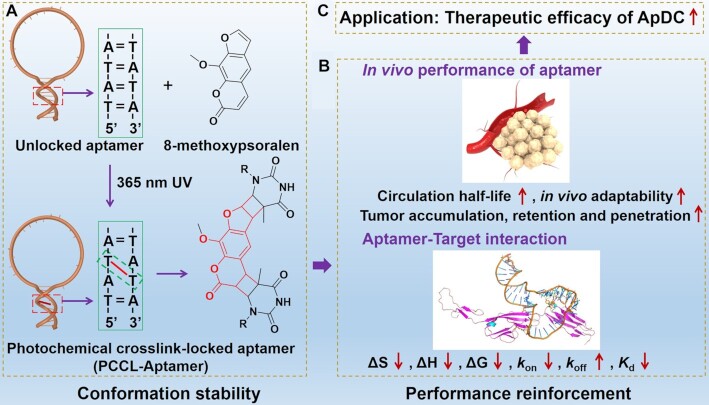
Purines and pyrimidines in nucleic acids can react with electrophilic molecules, for example alkylating agents, and form stable DNA crosslink, which prevents DNA duplex separation (21). Therefore, such molecules demonstrate their potential as molecular tools in modulating nucleic acid conformation. Latching on this idea, we herein reported an 8-methoxypsoralen-based photochemically covalent lock to stabilize aptamer conformation via covalently crosslinking the inter-stranded thymine nucleotides at 5'-TpA sites in the condition of ultraviolet radiation (Figure 1A) (22–24). To further elucidate the conformation–performance relationship of aptamer, the thermodynamic parameters, the binding affinity, the adaptability to physiological environment to resist the interference of exonuclease, flow shear stress, non-targeted proteins and macrophage uptake, the bloodstream circulation half-life, and the ability to accumulate in and penetrate into tumor of the photochemical crosslink-locked aptamer (PCCL-aptamer) were systematically studied and compared to those of the unlocked counterparts. Furthermore, the therapeutic efficacy of PCCL-aptamer-combretastatin A-4 (CA4) conjugation (PCCL-Ap-CA4) on tumor-bearing mice was also systematically investigated and compared to the unlocked aptamer-drug conjugation (ApDC). Our results clearly revealed that the photochemically covalent lock is general, simple and highly efficient strategy to stabilize aptamer conformation and strengthen its performance for biomedicine (Figure 1B, C).
Figure 1.
Illustration of 8-methoxypsoralen-based photochemical covalent lock to stabilize aptamer conformation via crosslinking the inter-stranded thymine nucleotides at TpA sites in aptamer (A). The conformation stability of aptamer can strengthen its performance (B) and applications in aptamer-based targeted therapy (C).
MATERIALS AND METHODS
Synthesis of photochemical crosslink-locked aptamer (PCCL-aptamer)
One OD DNA (Supplementary Table S1) was dissolved in 180 μl DPBS containing 5 mM MgCl2, and mixed with 20-fold equivalent 8-methoxypsoralen dissolved in 20 μl DMSO. The mixture was placed under ZF-8 UV Crosslinker (Shanghai Jiapeng Technology Co., Ltd.), and exposed to 365 nm UV-A illumination at 3.9 mW/cm2 for 15 min on ice. The DNA sample was then precipitated by adding 20 μl NaCl (3 M) and 500 μl cold ethanol, followed by standing at −20°C overnight. After centrifugation for 30 min (4°C, 12 000 rpm) and removal of the supernatant, the white pellet was dissolved in 0.1 M TEAA, and further purified on a C-18 column by high-performance liquid chromatography (Agilent 1260 Infinity) equipped with a UV detector and a fluorescence detector. The gradient elution procedure for PCCL-aptamer purification was optimized as follows: 5% acetonitrile (ACN) and 95% 0.1 M TEAA at 1.0 ml/min over 4 min, and then 5–55% ACN at 1.0 ml/min over 35 min, followed by 95% ACN at 1.0 ml/min over 5 min. The elution fractions with different retention time were collected and dried in vacuum for analysis. The PCCL-aptamers were evidenced by ESI-MS, which was performed Sangon Biotech (Shanghai, China).
Flow cytometry analysis of the binding ability of PCCL-aptamers
Briefly, adherent cell lines (HCT116 and HEK293) were seeded in 100-mm culture dish at appropriate density and allowed to grow to ∼90% confluence after 24 h incubation. After three washes with DPBS, the adherent cells were incubated with 2 mL 0.2% EDTA in DPBS for 3–5 min. After removing EDTA solution, cells were carefully washed twice with 2 ml DPBS and collected (Noting: after treating with 0.2% EDTA, cells are prone to detach from culture dish). After counting, 2 × 105 cells were incubated with FAM-labeled DNA at different concentrations in binding buffer for 30 min. Finally, cells were washed twice with washing buffer and resuspended in 500 μl washing buffer for flow cytometric analysis.
For flow cytometric analysis of suspension cells, 2 × 105 suspension cell (CCRF-CEM, K562) were incubated with fluorescent labeled aptamers at different concentrations in 200 μl of binding buffer at 4°C or 37°C, respectively. The cells were then washed twice with washing buffer and resuspended in 500 μl washing buffer for flow cytometric analysis. To identify the stability of aptamer-CCRF-CEM cells complexes, the complexes were further treated with SSB at different concentrations (0.5–2.5 μg/ml) for 30 min. After two washes with washing buffer, the cells were resuspended in 500 μl washing buffer for flow cytometric analysis.
For anti-interference assay, 2 × 105 CCRF-CEM cells were incubated with the mixture of 20 nM FAM-labeled aptamers (Sgc8, Sgc8m, PCCL-Sgc8m) and SSB at different concentrations. To ensure the anti-interference performance of PCCL-Sgc8m, aptamers were preincubated with the SSB at different concentrations (1.1–3.1 μg/ml) for 30 min at room temperature. After incubation, then cells were washed twice with washing buffer and resuspended in 500 μl washing buffer for flow cytometric analysis.
For macrophage uptake analysis, RAW264.7 cells (3 × 105) were seeded in 6-well cell plates and allowed to grow for 24 h. For macrophage uptake analysis, the cells were treated with 1 μg/ml lipopolysaccharide (LPS) for another 8 hours, and the differentiated RAW264.7 cells were first incubated with 200 nM Cy5-labeled aptamers for preassigned time (0.5, 1, 2 and 4 h) in DPBS supplemented with 1 mM Mg2+ at 37°C. To detach cells and remove the non-internalized aptamers on the cellular surface, these RAW 264.7 cells were then incubated with 0.25% trypsin for 5 min followed by thrice washing with DPBS. Finally, the cells were resuspended in 500 μl DPBS for flow cytometric analysis.
Flow cytometric analysis was performed with a BD FACSVerse™ system, and the data were analyzed with FlowJo_V10.
Ultracentrifugation spin-down assay
Aptamer deformability analysis was characterized via an ultracentrifugation spin-down assay. 300 uL FAM-labeled aptamer solution (100 nM) in DPBS containing 1 mM or 5 mM Mg2+ was loaded into the top holding unit of the 0.2-μm or 10-nm PES ultracentrifuge tubes. 50 × g of spin speed was set for ultracentrifuge tube with 0.2-μm pores for 3 min and the 800 × g of spin speed was set for ultracentrifuge tube with 10-nm pores for 3 min. After centrifugation, the samples in the filtrate were transferred in DPBS to make 300 μl. After the samples in the retentate were collected, the samples in the filter were extracted by adding DPBS to the top holding unit, followed by an ultracentrifugation. The samples in the retentate and in the filter were mixed and the volume was adjusted to 300 μl. The amount of DNA in the filter samples and the unfiltered samples were then determined by measuring the fluorescence intensity with a Fluoromax-4 spectrofluorometer.
Quantitative analysis of aptamer-cell interaction under flow shear stress condition
Quantitative analysis the interaction of aptamer and CCRF-CEM cells under shear stress condition was performed using a cone-plate viscometer, which consists of a stationary plate placed beneath a rotating cone (1° angle) maintained at 37°C (RotoVisco 1, Haake, Newington, NH). 2 × 105 CCRF-CEM cells and 50 nM aptamers were placed on the plate in DPBS supplemented with 1 mM or 5 mM MgCl2 buffer. Then cell suspensions were exposed to a predetermined shear stress for 30 min. After incubation, cells were collected and washed twice by DPBS for flow cytometric analysis. The data were analyzed with FlowJo_V10.
Surface plasmon resonance (SPR) assay
All SPR studies were performed on a Biacore 8K instrument (GE Healthcare Life Sciences). The Biacore 8K design incorporates eight channels, each consisting of a sensing lane paired with a reference lane for the subtraction of the nonspecifically bound analyte. PTK7 Ig3-4 was immobilized on a CM5 chip using the amine coupling method by the speed of flowing 0.2 mg/ml PTK7 Ig3-4, the flow rate was 10 μl/min. Aptamers (20, 40, 80, 160 or 320 nM) were allowed to pass over the surfaces at 15 μl/min for 180 s, followed by 180 s of dissociation in running buffer. The relative response at the end of dissociation was recorded in the sensogram and was used for binding evaluation. DPBS was used as a running buffer. Different concentration of aptamers produced different signal changes. All sensorgrams were displayed after the reference was subtracted with solvent correction procedures implemented. Data were fit to both steady state and a 1:1 kinetic model using Biacore evaluation software.
Microscale thermophoresis assay
Microscale thermophoresis assay was carried out to measure the binding constant between aptamer and PTK7 Ig3-4 at different concentration in DPBS supplemented with 1 mM MgCl2 or 5 Mm MgCl2. A serial of PTK7 Ig3-4 protein solutions from 79 μM to 2.4 nM were prepared by 2-fold serial dilution. Then, 10 μl each sample was mixed with 10 μl DNA solution (50 nM) for analysis. During the analysis, about 10 μl of each sample was filled into capillaries (NanoTemper Technologies, Lot: 892514), which were then detected in Monolith NT.115 instrument (NanoTemper Technologies, Germany) at different temperature. Measurements were conducted at 20% laser power. Data were analyzed with MO. Affinity Analysis.
Investigating the circulation half-life and histological distribution of various aptamers in healthy BALB/c nude mice
Healthy four- to six-week-old BALB/c nude mice were purchased from Hunan SJA Laboratory Animal Co. Ltd. (Changsha), and used under protocols approved by the Institution Animal Care and Use Committee of Hunan University (SYXK 2018-0006). The mice were injected with Cy5-labeled Sgc8, Sgc8m and PCCL-Sgc8m (equivalent Cy5, 4 nmol) via tail vein (n = 3) in the immobilizer, respectively. Then, 10 μl blood samples were collected from the tails of mice at 5, 10, 20, 30, 45, 60, 120 and 240 min post-injection, and stored in a sealed container supplemented with EDTA (2 mg/ml). The fluorescence signals of these blood samples were recorded with a Fluoromax-4 spectrofluorometer. After 240 min, mice were euthanized, and their organs were harvested for ex vivo imaging by an IVIS Lumina II in vivo imaging system (Caliper LifeSicence, USA). Cy5 fluorescent images and statistics of all organs were analyzed with IVIS Lumina II in vivo imaging system.
RESULTS AND DISCUSSION
Preparation of the photochemically covalent locked aptamer (PCCL-aptamer)
To prepare PCCL-aptamers, aptamer Sgc8 recognizing protein tyrosine kinase 7 (PTK7) (25), HG1-9 recognizing transferrin receptor (26), and TCO1 recognizing a cellular membrane protein (27) were chosen as candidates, and their nucleotides at 5’- or 3’-terminals were replaced with four ApT dinucleotides, providing potential sites for hosting 8-methoxypsoralen (Supplementary Table S1). After exposure to 365-nm UV (12 W, 15 min), the photochemical crosslink-locked Sgc8 (PCCL-Sgc8m), which was chosen for systematic analysis as a representative, could be generated with about a yield of 60% (Supplementary Figure S1). Our strategy was also successfully applied to covalently crosslink G-rich aptamer SL1, which binds to c-Met by folding into G-quadruplex (28), and aptamer Sgc8m with chemical modifications in backbone phosphodiester unit (e.g. phosphorothioate (PS), Sgc8m-4PS), in sugar unit (e.g. 2’-O-methylation, Sgc8m-4mA), or in nucleotide unit (e.g. fluorouracil, Sgc8m-5FU), demonstrating its generality for various aptamers (Supplementary Table S1). Of note, although multiple potential sites were introduced in these aptamers for crosslinking, however, only one 8-methoxypsoralen intercalated and crosslinked thymine nucleotides in DNA duplex, as determined by electrospray ionization mass spectrometry (ESI-MS) analysis (Supplementary Figure S2-S8). We deduced that the crosslink between two inter-stranded thymine nucleotides distorted local DNA at the crosslinked site, and thereby other sites could not hold 8-methoxypsoralen. Because some amino acid, such as methionine, tyrosine, histidine and tryptophan, were susceptible to 8-methoxypsoralen under UV irradiation (29,30), it should be careful when our strategy was used to photochemically crosslink aptamers containing these amino acid residues.
Photochemically covalent lock efficiently stabilizes aptamer conformation and enhances its binding ability
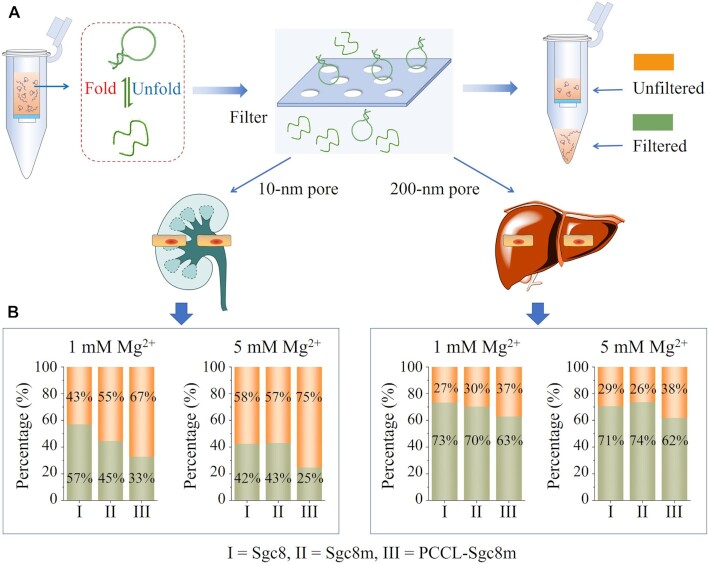
To characterize whether photochemically covalent lock could stabilize aptamer conformation, an ultracentrifugation spin-down assay was performed to investigate the deformation of the PCCL-Sgc8m and unlocked aptamers by analyzing their ability to pass through the 10-nm pores, which approach the size of glomerular filtration (31), and 200-nm pores, which fall in the range of the discontinuous endothelial pores in liver and spleen (Figure 2A) (32). After centrifugation, the fluorescence signal from the filtrate of the FAM-labeled PCCL-Sgc8m was lower than that of the FAM-labeled unlocked aptamers (Sgc8 and Sgc8m) (Figure 2B). Based on the previous report that a decreased flexibility can hamper a polymer to pass through a pore (33), our results displayed that PCCL-Sgc8m had lower deformability and were harder to pass the filters than the unlocked Sgc8m and Sgc8. Furthermore, melting temperature analysis revealed that the PCCL-aptamers had higher melting temperature than their unlocked counterparts (Supplementary Figure S9–S15). Among these PCCL-aptamers, PCCL-Sgc8m featured hairpin conformation and PCCL-SL1m featured G-quadruplex conformation were chosen as representatives for structure analysis with circular dichroism (CD) spectroscopy. CD analysis revealed that the photochemical crosslink had mariginal effect on their conformation, suggesting that the two aptamers maintained their conformation after crosslink (Supplementary Figure S16). Based on these results, it can be deduced that 8-methoxypsoralen-based photochemically covalent lock can be used to stabilize the conformation of various aptamer systems.
Figure 2.
Photochemically covalent lock efficiently stabilized aptamer conformation. (A) Schematics of conformation stability analysis of PCCL-Sgc8m, Sgc8m and Sgc8 in Dulbecco's phosphate-buffered saline (DPBS) containing 1 mM Mg2+ or 5 mM Mg2+ via ultracentrifugation spin-down assay to investigate their ability to pass through pores with 200-nm diameter, which falls in the range of the discontinuous endothelial pores in liver and spleen, or 10-nm diameter, which approaches the size of glomerular filtration. (B) The percentage of PCCL-Sgc8m, Sgc8m and Sgc8 in the unfiltered samples and the filtered samples after ultracentrifugation spin-down assay.
To elucidate the effect of PCCL on the binding affinity of aptamers, the thermodynamic parameters of the interaction between the three aptamers and PTK7 Ig3-4 spanning residues 219–410 of PTK7 were investigated by microscale thermophoresis analysis. PTK7 Ig3-4 was produced by baculovirus insect expression system, and purified by size-exclusion chromatography and anion-exchange chromatography (Supplementary Figure S17). As shown in Supplementary Table S2 and Supplementary Figure S18, the interaction of PCCL-Sgc8m and PTK Ig3-4 had more negative ΔS, ΔH and ΔG than the interaction of unlocked Sgc8m-PTK7 Ig3-4. The results reveal that the crosslink between inter-stranded thymine nucleotides induced by 8-methoxypsoralen allows aptamer to fold into proper secondary structure and decreases the probability to form alternative structures, which facilitates aptamer binding. The binding constants were further investigated by surface plasmon resonance (SPR). As shown in Supplementary Table S3 and Supplementary Figure S19, PCCL-Sgc8m displayed ∼3.6-fold enhancement in the dissociation constant (Kd) compared to aptamer Sgc8m. The photochemically covalent lock increased not only the association rate constant (kon), but also the dissociation rate constant (koff). It was deduced that the locked aptamer with less flexibility easily fell off from targets due to lack of conformational transition states.
To further explore the binding patterns at the molecular level of aptamer with PTK7 Ig3-4 in the locked state and the unlocked state, molecular dynamics simulations (MDS) were performed (34). As shown in Figure 3 and Supplementary Table S4, the binding interface profile of PCCL-Sgc8m and PTK7 Ig3-4 was different from that of Sgc8m and the target. The former involved in more hydrogen bonds than the latter. In addition, the binding energy of PCCL-Sgc8m and PTK7 Ig3-4 was ∼1.4-fold lower than that of Sgc8m and the target (Table 1). These results meant that the conformation stability of aptamer induced by the photochemically covalent lock could enhance the binding strength of aptamer to amino acid residues of PTK7 Ig3-4, and thereby enhance binding affinity of aptamer.
Figure 3.
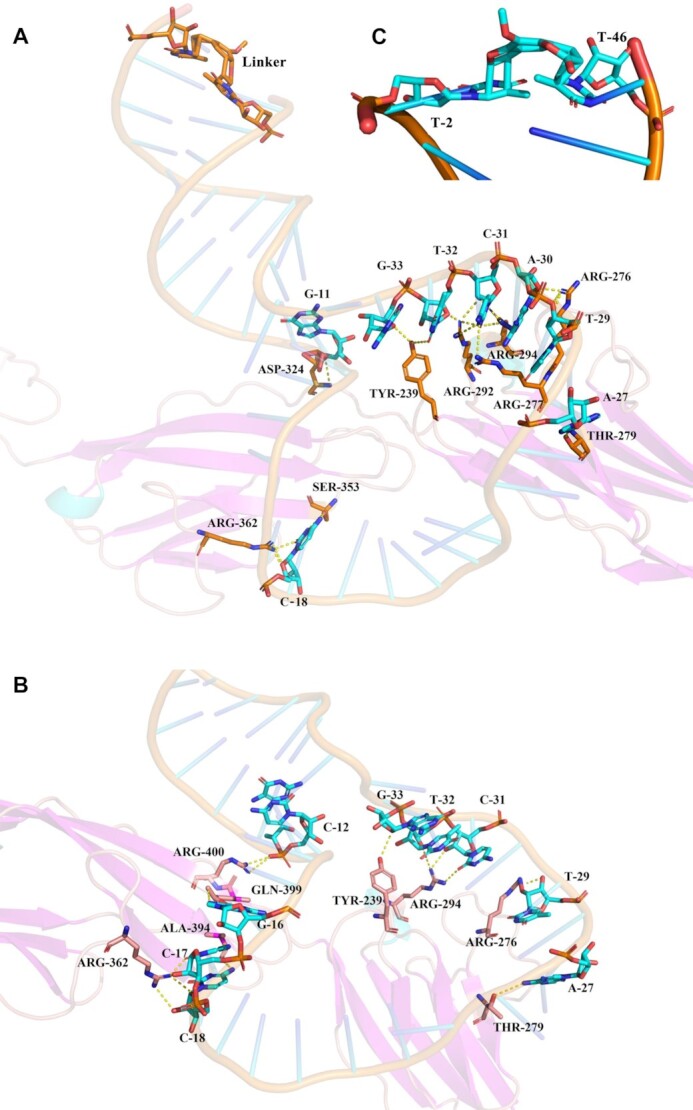
The binding interace of PCCL-Sgc8m-PTK7 Ig3-4 (A) and Sgc8m-PTK7 Ig3-4 (B) according to molecular dynamics simulations. (C) The structure of linker in (A). The crosslink site was set between T-2 and T-46 of aptamer.
Table 1.
The calculated binding energy of Sgc8m- or PCCL-Sgc8m-PTK7 Ig3-4 obtained from molecular simulation
| Ligand | Van der Waal (kKJ/mol) | Electro-static (kJ/mol) | Polar solvation (kJ/mol) | Non-polar solvation (kJ/mol) | Binding energy (kJ/mol) |
|---|---|---|---|---|---|
| Sgc8m | −57.11 ± 0.83 | −8.51 ± 0.46 | 7.15 ± 0.27 | −11.34 ± 0.49 | −69.81 ± 1.04 |
| PCCL-Sgc8m | −84.31 ± 1.07 | −12.58 ± 0.62 | 6.99 ± 0.57 | −9.14 ± 0.36 | −99.24 ± 0.36 |
Cell binding assays displayed that PCCL-Sgc8m had higher binding affinity compared to unlocked aptamers (Sgc8m and Sgc8) in Dulbecco's phosphate-buffered saline (DPBS) supplemented with 1 mM Mg2+ (Supplementary Figure S20A). However, of note, compared to Sgc8, PCCL-Sgc8m had strong binding on CCRF-CEM cells, but slightly weak binding on PTK7 Ig3-4. The results might be caused by the deduction that the matrix complexity of cell surface might interfere with the binding of the unlocked Sgc8. PCCL-Sgc8m remained recognition specificity, and bound to PTK7-positive CCRF-CEM cells and HCT116 cells, but not PTK7-negative K562 cells and HEK293 cells (Supplementary Figure S21–S25). The results suggest that photochemically covalent lock doesn’t affect the specificity of aptamer. The binding ability of PCCL-TCO1m (Supplementary Figure S3) on CCRF-CEM cells and PCCL-HG1m (Supplementary Figure S4) on K562 cells were also investigated by flow cytometric analysis and compared to their unlocked counterparts. As showed in Supplementary Figure S20B,C, the PCCL-aptamers had better binding affinity on cells than unlocked aptamers. Based on these results, it could be deduced that photochemically covalent lock could be used as a general strategy to stabilize aptamer conformation and increase its binding affinity.
PCCL-aptamer has better adaptability to physiological environment than its unlocked counterparts to resist the interferences of exonuclease, flow shear stress and non-targeted proteins
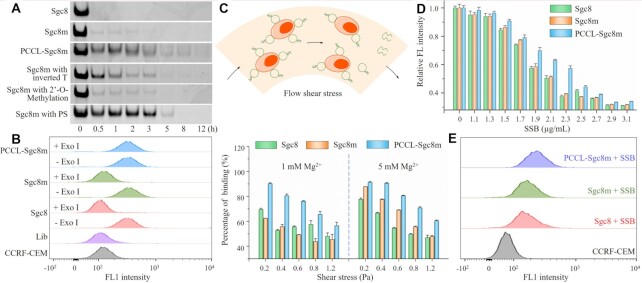
To recognize the intravascular or extravascular targets, aptamers should have good adaptability to physiological environment, and resist various interferences from exonuclease, flow shear stress and non-targeted proteins during travel in the body. Therefore, the stability of PCCL-Sgc8m, PCCL-TCO1m and PCCL-HG1m in exonuclease I or fetal bovine serum (FBS) was first analyzed by polyacrylamide gel electrophoresis (PAGE), and compared to their unlocked counterparts (Supplementary Table S1). As shown in Figure 4A and Supplementary Figure S26A, S27, these PCCL-aptamers displayed higher stability than these unlocked aptamers, which rapidly degraded in 0.5 U/μl exonuclease I (Exo I) or 50% FBS. PAGE analysis also revealed that the stability of PCCL-Sgc8m was comparable to or even better than Sgc8m modified with inverted thymine nucleotides, 2’-O-methylation or PS (Supplementary Figure S26A). After treatment with Exo I for 1 h or 50% FBS for 24 h, the binding ability of PCCL-Sgc8m had little decrease, however, the binding ability of Sgc8m and Sgc8 substantially decreased (Figure 4B and Supplementary Figure S26B, C). After treatment with 8-methoxypsoralen, Sgc8m1 and Sgc8m2, in which four ApT were replaced with 8A and 8T (Supplementary Table S1), could not resist the degradation of 0.5 U/μl Exo I, meaning that the crosslink induced by 8-methoxypsoralen was site selective (Supplementary Figure S28). The results suggest that the crosslink of the thymine nucleotides at TpA sites in DNA duplex induced by photochemically covalent lock cyclizes aptamers and enhances their stability in biological fluids.
Figure 4.
PCCL-aptamer had better adaptability to physiological environment than its unlocked counterparts. (A) PAGE analysis of the stability of PCCL-Sgc8m in 0.5 U/μl Exo I compared with the unlocked aptamers with or without chemical modifications. (B) Flow cytometric analysis of the binding of FAM-labeled PCCL-Sgc8m, Sgc8m and Sgc8 to CCRF-CEM cells after treatment with 0.5 U/μl Exo I for 1 h. FAM-labeled library (Lib) was used as negative control. The concentrations of DNA for the flow cytometric assay were 10 nM. (C) Evaluation of the interaction of FAM-labeled PCCL-Sgc8m, Sgc8m and Sgc8 with CCRF-CEM cells under well-defined shear conditions. (D) Flow cytometric analysis of the binding ability of FAM-labeled PCCL-Sgc8m, Sgc8m and Sgc8 with CCRF-CEM cells in the presence of SSB at different concentrations. In (C) and (D), the DNA concentration was 20 nM, and the data are presented as mean standard deviation (n = 3). (E) Flow cytometric analysis of the binding of FAM-labeled PCCL-Sgc8m, Sgc8m and Sgc8 with CCRF-CEM cells in the presence of 2.3 μg/ml SSB.
Next, we utilized a cone-plate viscometer to mimic a shear-flow environment and to study aptamer-cell interactions under well-defined shear conditions. When the shear stress was increased from 0.2 to 1.2 Pa, which is the physiological flow shear-stress range in vascular microcirculation and/or interstitial space (35), whether in DPBS supplemented by 1 mM Mg2+ or in DPBS supplemented by 5 mM Mg2+, the binding ability of the two unlocked aptamers (Sgc8 and Sgc8m) to CCRF-CEM cells decreased rapidly. However, the binding ability of PCCL-Sgc8m to cells in such flow shear conditions decreased slowly (Figure 4C). In addition, PCCL-TCO1m and PCCL-HG1m also displayed better adaptability to flow shear-stress than their unlocked parental aptamers (Supplementary Figure S29). These results demonstrate that PCCL-aptamer can resist the flow shear stress and bind well to targets based on the conformation stability.
Then, we used single-stranded binding protein (SSB) to mimic non-targeted proteins in physiological environment based on its ability to non-specifically and strongly bind with ssDNA (36), and to study whether photochemically covalent lock enhanced the resistance of aptamer to the interferences from non-targeted proteins. After incubation with the mixture of 20 nM FAM-labeled aptamers and SSB varied from 0 to 3.1 μg/ml, which were preincubated for 30 min, CCRF-CEM cells were analyzed with flow cytometry. As shown in Figure 4D, PCCL-Sgc8m demonstrated a better ability to resist such interference than Sgc8 and Sgc8m. For example, PCCL-Sgc8m could well bind to CCRF-CEM cells even in the presence of SSB at 2.3 μg/ml, and the fluorescence signal intensity on the cell surface only decreased ∼42%. However, the fluorescence signal intensity on the cell surface decreased ∼60% after CCRF-CEM cells were incubated with the unlocked aptamers in the same conditions (Figure 4D, E). As expected, PCCL-TCO1m and PCCL-HG1m could well bind on the cell surface in the presence of SSB (Supplementary Figure S30). We also used SSB to interrogate the stability of aptamer-targeted cell complexes. As shown in Supplementary Figure S31, PCCL-Sgc8m-cell complexes were more stable than unlocked Sgc8-cell complexes in the presence of SSB. These results suggest that photochemically covalent lock can enhance the ability to resist the interference of non-targeted proteins than their unlocked counterparts based on the fact that the photochemical crosslink stabilizes aptamer conformation and increase their binding affinity.
PCCL-aptamer resists macrophage uptake and has a prolonged circulation half-life in healthy mice
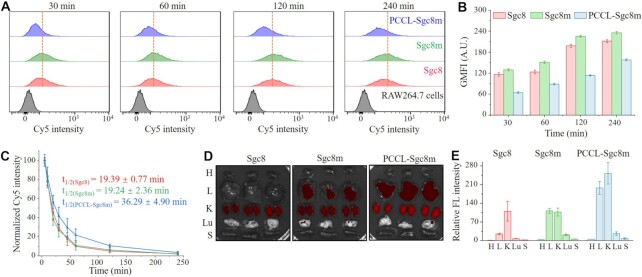
Given that a prolonged circulation half-life of aptamer can improve the chance to arrive the target sites, resulting in better targeting efficacy, we assessed macrophage uptake, in vivo circulation half-life and biodistribution patterns of PCCL-Sgc8m and the two unlocked aptamers. First, the macrophage uptake of aptamer was investigated by analyzing the endocytosis of cyanine 5 (Cy5)-labeled PCCL-Sgc8m and its unlocked counterparts in murine macrophage RAW 264.7 cells pre-stimulated with lipopolysaccharide (LPS). As shown in Figure 5A, B, PCCL-Sgc8m displayed less uptake in RAW 264.7 cells than the two unlocked aptamers. Supplemental Figure S32 also demonstrated that PCCL-TCO1m and PCCL-HG1m had decreased macrophage uptake compared to their unlocked counterparts. These data indicate that the stable conformation enable aptamer resist the uptake and the clearance by macrophages.
Figure 5.
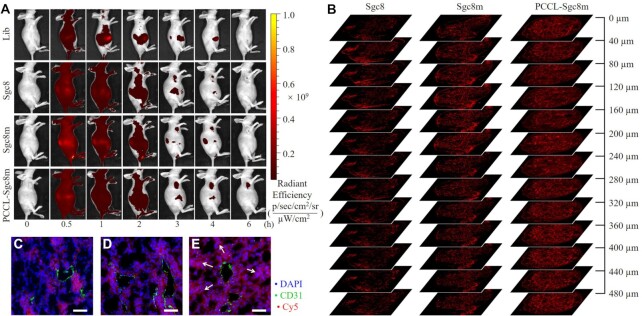
PCCL-aptamer resisted microphage uptake and had a prolonged circulation half-life than its unlocked counterparts. (A) Macrophage uptake analysis of Cy5-labeled PCCL-Sgc8m, Sgc8m and Sgc8 during a 4-h incubation period of with RAW264.7 cells pre-stimulated by LPS for 8 h. (B) Statistic analysis of geometric mean fluorescence intensity (GMFI) of RAW264.7 cells from three independent macrophage uptake analysis. (C) The pharmacokinetics analysis of Cy5-labeled PCCL-Sgc8m, Sgc8m and Sgc8 injected in normal mice at tail vein (n = 3). (D) Ex vivo imaging and quantification assay (E) of Cy5 fluorescence signal of major organs from the mice investigated in (C). H, heart; L, liver; K, kidney; Lu, lung; S, spleen.
The in vivo circulation half-life and the biodistribution patterns of the three aptamers were then investigated in healthy mice. The healthy mice were used to exclude the complicating effects induced by other diseases. After mice were received an injection of Cy5-labeled PCCL-Sgc8m, Sgc8 or Sgc8m at tail vein, the blood samples were collected from tail vein at multiple time points over a 4-h period and their fluorescence signals were measured. As shown in Figure 5C, PCCL-Sgc8m had a prolonged circulation half-life of ∼36.29 min, which was nearly 2-fold longer than that of the two unlocked aptamers. Such difference might be ascribed to the fact that the unlocked aptamer was more easily degraded than PCCL-aptamer in circulation system and released Cy5 labeled in the middle of aptamer into circulation system, thereby resulting in short circulation half-life. Then, major organs from these mice, which were euthanized after circulation half-life analysis, were imaged with in vivo imaging system (IVIS). The ex vivo fluorescent imaging demonstrated that PCCL-Sgc8m had a different distribution pattern in major organs with high accumulation in kidney and liver relative to the unlocked aptamers (Figure 5D, E). The reason for the different histological distribution profiles might be mainly derived from the prolonged circulation half-life and the low deformability of PCCL-Sgc8m, which did not only increase its accumulation of in liver, but also decreased its clearance in kidney.
PCCL-aptamer efficiently accumulates in and deeply penetrates into tumors
The accumulation of PCCL-Sgc8m and the unlocked aptamers in tumor was further investigated with IVIS at preassigned time points over a 6-h period after HCT116 tumor-bearing BALB/c nude mice were received an injection of Cy5-labeled aptamers at tail vein. Control mice were I.V. injected with Cy5-labeled library (Supplementary Table S1). As shown in Figure 6A, three aptamers could efficiently accumulate in tumor compared to the library. However, among the three aptamers, PCCL-Sgc8m had longest retention time in tumor (>6 h). Ex vivo imaging, which was performed at 6 h post-injection, also confirmed that PCCL-Sgc8m had more accumulation in tumor than two unlocked aptamers (Supplementary Figure S33). These results clearly reveal that PCCL-Sgc8m can more efficiently recognize, accumulate and reside in tumors than the unlocked aptamers.
Figure 6.
(A) In vivo whole-body fluorescence imaging of HCT116 tumor-bearing mice at different time points post-injection of Cy5-labeled Lib, Sgc8, Sgc8m and PCCL-Sgc8m via tail vein. The injection dose of Cy5-labeled DNA was 4 nmol. (B) The penetration depth of various aptamers in tumor was analyzed by imaging a series of tumor tissue sections from the HCT116 tumor-bearing mice receiving intravenous injection of Cy5-labeled Sgc8, Sgc8m and PCCL-Sgc8m at tail vein. Fluorescent imaging analysis of the extravascular diffusion of Sgc8 (C), Sgc8m (D) and PCCL-Sgc8m (E) in tumor at 6-h post injection. Cy5 channel, Sgc8, Sgc8m and PCCL-Sgc8m. FITC channel, CD31. DAPI channel, nucleus. White arrows denoted extravascular Cy5 diffusion. Scale bars: 50 μm.
The penetration of Cy5-labeled PCCL-Sgc8m, Sgc8 and Sgc8m in tumor was also studied by analyzing the fluorescence signals of a series of tumor tissue sections, which were distributed in 480-micrometer range along z-axis. As shown in Figure 6B, strong red fluorescence signals were observed at every section depth for the sections from HCT116 tumor-bearing mice treated with Cy5-labeled PCCL-Sgc8m. However, as the depth increased along the z-axis for the sections from tumor-bearing mice treated with Cy5-labeled the two unlocked aptamers, the fluorescence signals of the sections decreased gradually (Figure 6B). These results revealed that PCCL-Sgc8m could penetrate deeply into tumor. We further analyzed the red fluorescence signals of Cy5-labeled aptamers around tumor blood vessels, which were labeled with green fluorescence signal. As shown in Figure 6C–E, the obvious red fluorescence signals could be observed far away from tumor blood vessels in tumor treated with Cy5-labeled PCCL-Sgc8m. However, in tumor treatment with Cy5-labeled Sgc8 or Sgc8m, the red fluorescence signals were only observed nearby tumor blood vessel. Taken together, these results demonstrated that PCCL-Sgc8m could penetrate more deeply into the tumor after extravasation from the tumor blood vessels than the two unlocked aptamers, although PCCL-Sgc8m had less deformability to pass through pores than the latter. The phenomenon could be explained by the results that PCCL-aptamer had stable conformation, prolonged circulation half-life, the ability to adapt to the physiological environment, thereby resulting in more accumulation and deep penetration in tumor. Therefore, PCCL-aptamer could be expected to efficiently deliver drug inside the tumor and achieve high drug concentration in tumor.
PCCL-aptamer improves the therapeutic efficacy of aptamer-drug conjugations in tumor-bearing mice
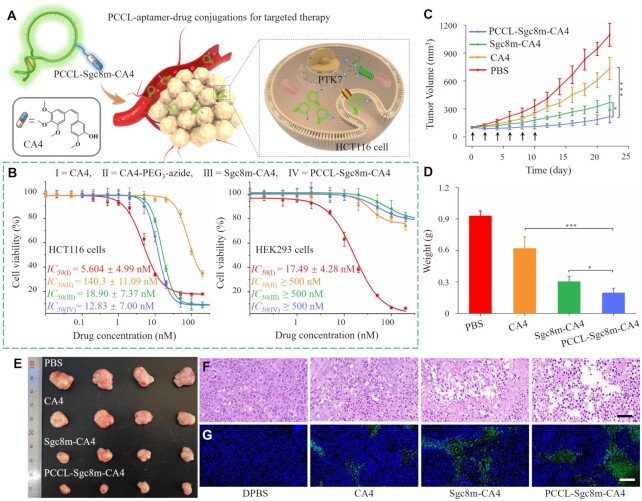
PCCL-aptamer possessed better binding affinity, adaptability to physiological environment, circulation half-life, and ability to accumulate in and penetrate into tumor of aptamers than unlocked aptamers based on conformation stability. Therefore, we final investigated whether PCCL-aptamer could improve the therapeutic efficacy of aptamer-drug conjugations. As shown in Supplementary Scheme S1, PCCL–aptamer–CA4 conjugation was synthesized by conjugating DBCO-modified Sgc8m with CA4-PEG3-azide via click chemistry, followed by photochemical crosslink with 8-methoxypsoralen. Of note, the order of click chemical conjugation and photochemical crosslink did not influence the synthesis of PCCL–Sgc8m–CA4. CA4-PEG3-azide was prepared according to our previous report (18). The resulting PCCL–aptamer–drug conjugations were purified by HPLC (Supplementary Figure S34), and evidenced by ESI-MS (Supplementary Figure S35). As expected, PCCL–Sgc8m–CA4 conjugation was more stable than Sgc8m–CA4 conjugation in the presence of Exo I (Supplementary Figure S36), and could recognize and enter HCT116 cells (Supplementary Figure S37).
To interrogate whether PCCL–aptamer could enhance the therapeutic efficacy of aptamer–drug conjugations (ApDCs) in vivo (Figure 7A), the cytotoxicity of PCCL–Sgc8m–CA4, Sgc8m–CA4, free CA4 and CA4–PEG3–azide on PTK7-positive HCT116 cells and PTK7-negative HEK293 cells was first evaluated. As shown Figure 7B, PCCL–Sgc8m–CA4 presented better cytotoxicity on HCT116 cells than Sgc8m–CA4, with an IC50 value of 12.83 ± 7.00 nM versus 18.90 ± 7.37 nM. Annexin V/propidium iodide apoptosis assay also confirmed that PCCL–Sgc8m–CA4 conjugation could efficiently induce cell apoptosis (Supplementary Figure S38). The results might from the fact that the stable PCCL–Sgc8m delivered more drug into cells than the unlocked Sgc8m. Free CA4 had strong and unselective cytotoxicity on both PTK7-positive HCT116 cells and PTK7-negative HEK293 cells, with IC50 values 5.60 ± 4.99 nM and 17.49 ± 4.28 nM, respectively, meaning that it has a narrow therapeutic window. Although, the two ApDCs had less cytotoxicity on HCT116 cells than free CA4, however, both PCCL–Sgc8m–CA4 and Sgc8m–CA4 had weak cytotoxicity on HEK293 cells with IC50 values of ≥500 nM, meaning that PCCL–Sgc8m, as well as Sgc8m could improve drug selectivity and decrease adverse effect.
Figure 7.
PCCL-aptamer improved the therapeutic efficacy of aptamer–CA4 conjugations on tumor-bearing mice. (A) Illustrating the application of the PCCL–aptamer–CA4 conjugation. (B) Cytotoxicity of free CA4, CA4–PEG3–azide, Sgc8m–CA4 and PCCL–Sgc8m–CA4 on HCT116 cells and HEK293 cells. (C) Tumor growth curves of HCT116 tumor-bearing xenografted mice after receiving 6 times of injection of PCCL–Sgc8m–CA4, Sgc8m–CA4, CA4 and DPBS via tail vein. Arrows denoted the administration. (D) Weight statistics of tumors from the euthanized mice after a 22-day treatment period. (E) Images of tumor from the euthanized mice after a 22-day treatment period. (F) H&E staining images and (G) TUNEL staining images of tumor sections from different mice groups. Scale bar for H&E staining: 20 μm, and scale bar for TUNEL staining: 100 μm. Blue, DAPI. Green, TUNEL. ***P < 0.001, *P < 0.05, ANOVA.
Next, the in vivo therapeutic efficacy of PCCL–Sgc8m–CA4 was evaluated after HCT116 tumor-bearing BALB/c mice were injected with DPBS, CA4 and two ApDCs at tail vein every 2 days, respectively. After six total injections, the mice were allowed to grow for another 12 days. As shown in Figure 7C–E, mice injected with DPBS displayed rapid growth in tumor volume (∼1095 mm3). Among mice treated with the CA4, Sgc8m–CA4 and PCCL–Sgc8m–CA4, PCCL–Sgc8m–CA4 showed the highest anticancer activity, resulting in a slow increase in tumor volume (∼216 mm3 in final tumor volume). In contrast, CA4 and Sgc8m–CA4 showed moderate anticancer activities (∼733 and ∼368 mm3 in final tumor volume, respectively). After treatment, the tumor tissue sections from each group were then analyzed with H&E staining and TUNEL staining. Figure 7F,G displayed that PCCL–Sgc8m–CA4 significantly caused tumor cell apoptosis. Furthermore, during the therapeutic period, there were not any decreases in weight of mice received injection of PCCL–Sgc8m–CA4 (Supplementary Figure S39). However, the weight of mice received injection of free CA4 decreased clearly, meaning adverse toxic effect. The H&E staining also confirmed that PCCL-Sgc8m-CA4 had little adverse effect on the main normal organs, such as heart, liver, spleen, lung and kidney (Supplementary Figure S40). The results suggest that PCCL-aptamer can enhance the therapeutic efficacy of ApDC in vivo, but has little toxicity on normal organs. The increased therapeutic efficacy from PCCL–Sgc8m–CA4 could be ascribed to the results that photochemically covalent lock stabilized aptamer conformation and strengthened its performance, making it more favorable in the application in biomedicine.
CONCLUSIONS
In this study, we have developed an 8-methoxypsoralen-based photochemical lock strategy to stabilize aptamer conformation by crosslinking two inter-stranded thymine nucleotides at TpA sites. Such crosslinking could decrease the deformability and stabilize aptamer conformation, and thereby strengthen the binding ability, the adaptability to physiological environment to resist various interferences (i.e. exonuclease, shear stress, non-targeted proteins and macrophage uptake), the circulation half-life, and the ability to accumulate in and penetrate into tumor of aptamers. Based on the gained performances, PCCL-aptamer-based ApDC has demonstrated better in vivo antitumor effect than unlocked aptamer-based ApDC. In addition, our strategy for stabilizing conformation of aptamer is simple, economic and efficient, and does not need special chemical modification, providing a general strategy to stabilize aptamer conformation. Furthermore, the in-depth exploration of the conformation–performance relationship of aptamers could be expected to facilitate elucidating the structure–function of aptamers, and promote the applications of aptamers in biomedicine.
DATA AVAILABILITY
All relevant data are included in the paper and/or its supplementary information files.
Supplementary Material
Contributor Information
Fang Zhou, Molecular Science and Biomedicine Laboratory (MBL), State Key Laboratory of Chemo/Biosensing and Chemometrics, College of Chemistry and Chemical Engineering, Aptamer Engineering Center of Hunan Province, Hunan University, Changsha, Hunan 410082, China; The Cancer Hospital of the University of Chinese Academy of Sciences (Zhejiang Cancer Hospital), Institute of Basic Medicine and Cancer (IBMC), Chinese Academy of Sciences Hangzhou, Zhejiang 310022, China.
Peng Wang, Molecular Science and Biomedicine Laboratory (MBL), State Key Laboratory of Chemo/Biosensing and Chemometrics, College of Chemistry and Chemical Engineering, Aptamer Engineering Center of Hunan Province, Hunan University, Changsha, Hunan 410082, China; The Cancer Hospital of the University of Chinese Academy of Sciences (Zhejiang Cancer Hospital), Institute of Basic Medicine and Cancer (IBMC), Chinese Academy of Sciences Hangzhou, Zhejiang 310022, China.
Jianghuai Chen, Molecular Science and Biomedicine Laboratory (MBL), State Key Laboratory of Chemo/Biosensing and Chemometrics, College of Chemistry and Chemical Engineering, Aptamer Engineering Center of Hunan Province, Hunan University, Changsha, Hunan 410082, China.
Zhijia Zhu, Molecular Science and Biomedicine Laboratory (MBL), State Key Laboratory of Chemo/Biosensing and Chemometrics, College of Chemistry and Chemical Engineering, Aptamer Engineering Center of Hunan Province, Hunan University, Changsha, Hunan 410082, China.
Youshan Li, Molecular Science and Biomedicine Laboratory (MBL), State Key Laboratory of Chemo/Biosensing and Chemometrics, College of Chemistry and Chemical Engineering, Aptamer Engineering Center of Hunan Province, Hunan University, Changsha, Hunan 410082, China.
Sujuan Wang, Molecular Science and Biomedicine Laboratory (MBL), State Key Laboratory of Chemo/Biosensing and Chemometrics, College of Chemistry and Chemical Engineering, Aptamer Engineering Center of Hunan Province, Hunan University, Changsha, Hunan 410082, China.
Shanchao Wu, Molecular Science and Biomedicine Laboratory (MBL), State Key Laboratory of Chemo/Biosensing and Chemometrics, College of Chemistry and Chemical Engineering, Aptamer Engineering Center of Hunan Province, Hunan University, Changsha, Hunan 410082, China.
Yingyu Sima, Molecular Science and Biomedicine Laboratory (MBL), State Key Laboratory of Chemo/Biosensing and Chemometrics, College of Chemistry and Chemical Engineering, Aptamer Engineering Center of Hunan Province, Hunan University, Changsha, Hunan 410082, China.
Ting Fu, Molecular Science and Biomedicine Laboratory (MBL), State Key Laboratory of Chemo/Biosensing and Chemometrics, College of Chemistry and Chemical Engineering, Aptamer Engineering Center of Hunan Province, Hunan University, Changsha, Hunan 410082, China; The Cancer Hospital of the University of Chinese Academy of Sciences (Zhejiang Cancer Hospital), Institute of Basic Medicine and Cancer (IBMC), Chinese Academy of Sciences Hangzhou, Zhejiang 310022, China.
Weihong Tan, Molecular Science and Biomedicine Laboratory (MBL), State Key Laboratory of Chemo/Biosensing and Chemometrics, College of Chemistry and Chemical Engineering, Aptamer Engineering Center of Hunan Province, Hunan University, Changsha, Hunan 410082, China; The Cancer Hospital of the University of Chinese Academy of Sciences (Zhejiang Cancer Hospital), Institute of Basic Medicine and Cancer (IBMC), Chinese Academy of Sciences Hangzhou, Zhejiang 310022, China; Institute of Molecular Medicine (IMM), Renji Hospital, Shanghai Jiao Tong University School of Medicine, and College of Chemistry and Chemical Engineering, Shanghai Jiao Tong University Shanghai 200240, China.
Zilong Zhao, Molecular Science and Biomedicine Laboratory (MBL), State Key Laboratory of Chemo/Biosensing and Chemometrics, College of Chemistry and Chemical Engineering, Aptamer Engineering Center of Hunan Province, Hunan University, Changsha, Hunan 410082, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [21890744, 21974040, 22177028]; China National Postdoctoral Program for Innovative Talents [BX20220309]; State Key Laboratory of Chemo/Biosensing and Chemometrics and Hunan University. Funding for open access charge: National Natural Science Foundation of China [21974040].
Conflict of interest statement. None declared.
REFERENCES
- 1. Tuerk C., Gold L.. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990; 249:505–510. [DOI] [PubMed] [Google Scholar]
- 2. Ellington A.D., Szostak J.W.. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990; 346:818–822. [DOI] [PubMed] [Google Scholar]
- 3. Shangguan D., Li Y., Tang Z., Cao Z.C., Chen H.W., Mallikaratchy P., Sefah K., Yang C.J., Tan W.. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:11838–11843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hermann T., Patel D.J.. Adaptive recognition by nucleic acid aptamers. Science. 2000; 287:820–825. [DOI] [PubMed] [Google Scholar]
- 5. Boehr D.D., Nussinov R., Wright P.E.. The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 2009; 5:789–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu C., Zhao J., Tian F., Cai L., Zhang W., Feng Q., Chang J., Wan F., Yang Y., Dai B.et al.. Low-cost thermophoretic profiling of extracellular-vesicle surface proteins for the early detection and classification of cancers. Nat. Biomed. Eng. 2019; 3:183–193. [DOI] [PubMed] [Google Scholar]
- 7. Yu H., Alkhamis O., Canoura J., Liu Y., Xiao Y.. Advances and challenges in small-molecule DNA aptamer isolation, characterization, and sensor development. Angew. Chem. Int. Ed. 2021; 60:16800–16823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pusuluri A., Krishnan V., Lensch V., Sarode A., Bunyan E., Vogus D.R., Menegatti S., Soh H.T., Mitragotri S.. Treating tumors at low drug doses using an aptamer-peptide synergistic drug conjugate. Angew. Chem. Int. Ed. 2019; 58:1437–1441. [DOI] [PubMed] [Google Scholar]
- 9. Gray B.P., Kelly L., Ahrens D.P., Barry A.P., Kratschmer C., Levy M., Sullenger B.A.. Tunable cytotoxic aptamer-drug conjugates for the treatment of prostate cancer. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:4761–4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mayer G. The chemical biology of aptamers. Angew. Chem. Int. Ed. 2009; 48:2672–2689. [DOI] [PubMed] [Google Scholar]
- 11. Eaton B.E., Gold L., Zichi D.A.. Let's get specific: the relationship between specificity and affinity. Chem. Biol. 1995; 2:633–638. [DOI] [PubMed] [Google Scholar]
- 12. Schmidt K.S., Borkowski S., Kurreck J., Stephens A.W., Bald R., Hecht M., Friebe M., Dinkelborg L., Erdmann V.A.. Application of locked nucleic acids to improve aptamer in vivo stability and targeting function. Nucleic Acids Res. 2004; 32:5757–5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhao L., Qi X., Yan X., Huang Y., Liang X., Zhang L., Wang S., Tan W.. Engineering aptamer with enhanced affinity by triple helix-based terminal fixation. J. Am. Chem. Soc. 2019; 141:17493–17497. [DOI] [PubMed] [Google Scholar]
- 14. Giusto Di, A. D., King G.C.. Construction, stability, and activity of multivalent circular anticoagulant aptamers. J. Biol. Chem. 2004; 279:46483–46489. [DOI] [PubMed] [Google Scholar]
- 15. Liu M., Yin Q., Chang Y., Zhang Q., Brennan J.D., Li Y.. In vitro selection of circular DNA aptamers for biosensing applications. Angew. Chem. Int. Ed. 2019; 58:8013–8017. [DOI] [PubMed] [Google Scholar]
- 16. Kuai H., Zhao Z., Mo L., Liu H., Hu X., Fu T., Zhang X., Tan W.. Circular bivalent aptamers enable in vivo stability and recognition. J. Am. Chem. Soc. 2017; 139:9128–9131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jiang Y., Pan X., Chang J., Niu W., Hou W., Kuai H., Zhao Z., Liu J., Wang M., Tan W.. Supramolecularly engineered circular bivalent aptamer for enhanced functional protein delivery. J. Am. Chem. Soc. 2018; 140:6780–6784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhou F., Wang P., Peng Y., Zhang P., Huang Q., Sun W., He N., Fu T., Zhao Z., Fang X.et al.. Molecular engineering-based aptamer-drug conjugates with accurate tunability of drug ratios for drug combination targeted cancer therapy. Angew. Chem. Int. Ed. 2019; 58:11661–11665. [DOI] [PubMed] [Google Scholar]
- 19. Ji D., Lyu K., Zhao H., Kwok C.K.. Circular L-RNA aptamer promotes target recognition and controls gene activity. Nucleic Acids Res. 2021; 49:7280–7291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Litke J.L., Jaffrey S.R.. Highly efficient expression of circular RNA aptamers in cells using autocatalytic transcripts. Nat. Biotechnol. 2019; 37:667–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Noll D.M., Mason T.M., Miller P.S.. Formation and repair of interstrand cross-links in DNA. Chem. Rev. 2006; 106:277–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Derheimer F.A., Hicks J.K., Paulsen M.T., Canman C.E., Ljungman M.. Psoralen-induced DNA interstrand cross-links block transcription and induce p53 in an ataxia-telangiectasia and rad3-related-dependent manner. Mol. Pharmacol. 2009; 75:599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cimino G.D., Gamper H.B., Isaacs S.T., Hearst J.E.. Psoralens as photoactive probes of nucleic acid structure and function: organic chemistry, photochemistry, and biochemistry. Ann. Rev. Biochem. 1985; 54:1151–1193. [DOI] [PubMed] [Google Scholar]
- 24. Rajendran A., Endo M., Katsuda Y., Hidaka K., Sugiyama H.. Photo-cross-linking-assisted thermal stability of DNA origami structures and its application for higher-temperature self-assembly. J. Am. Chem. Soc. 2011; 133:14488–14491. [DOI] [PubMed] [Google Scholar]
- 25. Shangguan D., Cao Z., Meng L., Mallikaratchy P., Sefah K., Wang H., Li Y., Tan W.. Cell-specific aptamer probes for membrane protein elucidation in cancer cells. J. Proteome Res. 2008; 7:2133–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang N., Bing T., Shen L., Feng L., Liu X., Shangguan D.. A DNA aptameric ligand of human transferrin receptor generated by Cell-SELEX. Int. J. Mol. Sci. 2021; 22:8923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tang Z., Shangguan D., Wang K., Shi H., Sefah K., Mallikratchy P., Chen H.W., Li Y., Tan W.. Selection of aptamers for molecular recognition and characterization of cancer cells. Anal. Chem. 2007; 79:4900–4907. [DOI] [PubMed] [Google Scholar]
- 28. Ueki R., Sando S.. A DNA aptamer to c-Met inhibits cancer cell migration. Chem. Commun. 2014; 50:13131–13134. [DOI] [PubMed] [Google Scholar]
- 29. Schmitt IrisM., Chimenti Sergio, Gasparro FrancisP.. Psoralen-protein photochemistry-a forgotten field. J. Photochem. Photobiol. B. 1995; 27:101–107. [DOI] [PubMed] [Google Scholar]
- 30. Veronese F.M., Schiavon O., Bevilacqua R., Bordin F., Rodighiero G.. Photoinactivation of enzymes by linear and angular furocoumarins. Photochem. Photobiol. 1982; 36:25–30. [DOI] [PubMed] [Google Scholar]
- 31. Choi C.H., Zuckerman J.E., Webster P., Davis M.E.. Targeting kidney mesangium by nanoparticles of defined size. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:6656–6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kong S.M., Costa D.F., Jagielska A., Van Vliet K.J., Hammond P.T.. Stiffness of targeted layer-by-layer nanoparticles impacts elimination half-life, tumor accumulation, and tumor penetration. Proc. Natl. Acad. Sci. U.S.A. 2021; 118:e2104826118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fox M.E., Szoka F.C., Fréchet J.M.. Soluble polymer carriers for the treatment of cancer: the importance of molecular architecture. Acc. Chem. Res. 2009; 42:1141–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huang M., Li T., Xu Y., Wei X., Song J., Lin B., Zhu Z., Song Y., Yang C.. Activation of aptamers with gain of function by small-molecule-clipping of intramolecular motifs. Angew. Chem. Int. Ed. 2021; 60:6021–6028. [DOI] [PubMed] [Google Scholar]
- 35. Shi P., Wang X., Davis B., Coyne J., Dong C., Reynolds J., Wang Y.. In situ synthesis of an aptamer-based polyvalent antibody mimic on the cell surface for enhanced interactions between immune and cancer cells. Angew. Chem. Int. Ed. 2020; 59:11892–11897. [DOI] [PubMed] [Google Scholar]
- 36. Harami G.M., Kovacs Z.J., Pancsa R., Palinkas J., Barath V., Tarnok K., Malnasi-Csizmadia A., Kovacs M.. Phase separation by ssDNA binding protein controlled via protein-protein and protein–DNA interactions. Proc. Natl. Acad. Sci. U.S.A. 2020; 117:26206–26217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are included in the paper and/or its supplementary information files.