Abstract
The effects of rapid acute depletion of components of RNA polymerase II (Pol II) general transcription factors (GTFs) that are thought to be critical for formation of preinitiation complexes (PICs) and initiation in vitro were quantified in HAP1 cells using precision nuclear run-on sequencing (PRO-Seq). The average dependencies for each factor across >70 000 promoters varied widely even though levels of depletions were similar. Some of the effects could be attributed to the presence or absence of core promoter elements such as the upstream TBP-specificity motif or downstream G-rich sequences, but some dependencies anti-correlated with such sequences. While depletion of TBP had a large effect on most Pol III promoters only a small fraction of Pol II promoters were similarly affected. TFIIB depletion had the largest general effect on Pol II and also correlated with apparent termination defects downstream of genes. Our results demonstrate that promoter activity is combinatorially influenced by recruitment of TFIID and sequence-specific transcription factors. They also suggest that interaction of the preinitiation complex (PIC) with nucleosomes can affect activity and that recruitment of TFIID containing TBP only plays a positive role at a subset of promoters.
INTRODUCTION
The synthesis of pre-mRNA in eukaryotic cells is widely believed to begin with the assembly of the Pol II PIC over promoter sequences (1). The pathway for PIC assembly in vitro on linear DNA templates bearing TATA elements is well established for the minimum set of required GTFs (1–6). Template recognition begins with TBP binding to an upstream TATA element stabilized by TFIIA and TFIIB (7,8). Pol II and TFIIF then join the complex, which is completed by loading TFIIE and finally TFIIH. In the absence of negative superhelical tension in the template, the XPB subunit of TFIIH is needed to convert the closed PIC to open complex in an ATP-dependent manner (9). However, the precise role of XPB in metazoans is puzzling because inhibition of XPB by triptolide blocks initiation in vitro and in cells but elimination of XPB does not seem to have a major impact on Pol II transcription (10).
While this assembly pathway with the minimal GTF set is clear, it has long been appreciated that the large majority of Pol II promoters lack canonical TATA elements (11–14). PIC assembly in those cases has been at least partially attributed to promoter interactions of TFIID, the complex of TBP with 13 additional TBP-associated factors (TAFs). In metazoans, TBP generally participates in Pol II transcription through TFIID and not as free TBP (15). In vitro studies showed that TFIID protects TATA-containing promoter DNA from ∼40 bp upstream of the transcription start site (TSS) to 35 bp downstream of the TSS. Consistent with the potential importance of the downstream contacts in promoter recognition, consensus elements from roughly +25 to +35 have been described in human (16) and other metazoan Pol II promoters (17). Recent studies have shown that a distinctive sequence signature spanning the TFIID footprint is a common feature of human Pol II promoters (18). Nuclease protection experiments also show that the human Pol II PIC in vivo protects the sequences TFIID protects in vitro, regardless of the presence of a TATA element (19). Comprehensive cryo-EM analyses demonstrate that complete Pol II PICs assembled with TFIID on both TATA and TATA-less promoters all have the dimensions reported from earlier in vitro and in vivostudies (3,4,6,20–22).
This clear picture of the single structure of the Pol II PIC is, however, not in complete agreement with other work. TBP is apparently present at all Pol II promoters in vivo and in all reported PIC structures (3), but an earlier study showed that Pol II promoter recognition in vitro need not require TBP, even for a TATA promoter (23). Those results relied on the ability to prepare subcomplexes of TFIID that function in transcription but lack TBP as well as some of the TAFs. Another study showed that transcription of a TATA-less promoter in Drosophila requires TAFs 1 and 4 (24). TBP and a subset of TAFs can also be found in the SAGA complex that associates with promoters in yeast, but SAGA is now regarded as a co-activator that is not involved in promoter recognition itself (25,26).
The +1 nucleosome is located downstream of active human Pol II promoters. A recent study in human cells reported an average upstream boundary for that nucleosome of +42 relative to the TSS (18). The proximity of the +1 nucleosome to the downstream edge of TFIID in the PIC suggests the possibility that the nucleosome could be involved in directing PIC assembly. Consistent with this, it was recently shown by nuclease protection that a significant subset of human PICs are directly abutted to the +1 nucleosome (19). Earlier reports indicated that interaction of TAFs with modified histones in the +1 nucleosome can facilitate PIC assembly (27,28). All of these observations raise the possibility that PIC assembly at some promoters, particularly those most dependent on downstream TFIID-DNA contacts, may in part depend on +1 nucleosome interactions. After initiation, Pol II advances to the proximal paused state, controlled in part by the +1 nucleosome and the action of NELF and DSIF (29). A fraction of the paused complexes are released into productive elongation through phosphorylation of DSIF and NELF by P-TEFb (30). P-TEFb activity is controlled by regulated association with the 7SK snRNP (31). Any condition that leads to an inhibition of elongation leads to release of P-TEFb from the snRNP in an apparent compensatory mechanism (31).
Many unanswered questions remain on the relative roles of various GTFs and promoter sequence elements in directing the assembly of the human Pol II PIC and subsequent initiation. However, due to their indispensability for cell survival, the functions of the GTFs have not been well characterized in cells. To circumvent this limitation, we have successfully employed the proteolysis targeting chimeras (PROTAC) system to achieve rapid depletion of TBP, TAF1, TFIIB and XPB in human cells following the addition of the heterobifunctional degrader dTAGV-1 (32). Following selective degradation of each factor, we performed PRO-Seq to quantify nascent transcripts generated by all three active RNA polymerases. We determined the sequence composition surrounding promoters as a function of dependency on each factor for transcription. This allowed us to assess the functional relationships among promoter occupancy of TBP, the PIC and adjacent nucleosomes. Furthermore, we also analyzed the interplay between GTF dependency and the presence near promoters of recognition sites for selected sequence-specific transcription factors (TFs).
MATERIALS AND METHODS
Genome editing with CRISPR/Cas9
HAP1 cells were grown to 80% confluence in T-25 flasks at 37°C and 5% CO2 in IMDM (Gibco 12440053) supplemented with 10% FBS (Gibco 26140079). gRNAs were made by mixing 200 μM tracrRNA (IDT 1072532) and 200 μM of gene-specific crRNA (Supplementary Data File) with homology to the C-terminus of human TBP, TAF1, TAF4, TFIIB or XPB in nuclease-free duplex buffer (IDT 11010301) at 95°C for 5 min. RNP complexes were formed by combining gRNA (180 pmol) and HiFi Cas9 (60 pmol; IDT 1081060) at 37°C for 10 min. Donor templates were made by converting double-stranded-gBlock gene fragment (IDT) containing the FKBP12F36V sequence flanked by gene-specific homology arms to ssDNA (Takara 632666). A total of 200 000 cells (counted with Countess II FL ThermoFisher A27274) were electroporated in a Lonza 4D-Nucleofector using the EH-100 program with the RNP and ssDNA donor template (>5 μg). Cells were plated in a 24-well plate (Corning 3524) containing equilibrated IMDM and 30 μM HDR enhancer (IDT 1081072) for 24 h at 37°C and 5% CO2 after which the media was replaced with fresh IMDM only. Cells were allowed to recover for 5 days before they were trypsinized, counted and diluted in a 50 ml conical tube with IMDM to 1 cell/ml. Cells were plated in a 96-well plate (Corning 3988) at 37°C and 5% CO2 for 7 days. DNA from colonies formed by individual clones was isolated with QuickExtract (Lucigen QE09050) and genome editing was assessed by PCR (Supplementary Data File) and western blot. Clones were expanded and frozen at –80°C.
PRO-Seq
HAP1 cells were grown to 80% confluence in T-150 flasks at 37°C and 5% CO2 in IMDM (Gibco 12440053) supplemented with 10% FBS (Gibco 26140079). The Sf21 moth cell line was used for spike-in controls. These cells were incubated at 27°C in Sf-900 III SFM (Gibco 12658019). Nuclei isolations and PRO-Seq were performed as previously described (33,34). Briefly, dTAGV-1 (a gift from Nathanael S. Gray) was dissolved in DMSO and dTAGV-1 or DMSO only were added to HAP1 cells for 2 h prior to harvesting (the final concentration for DMSO was 0.1% and final concentration of dTAGV-1 was 400 nM). Two biological replicates were used for each treatment. However, one of the duplicates for TAF4 was lost during preparation. Just before completion of treatments of HAP1 cells, we prepared spike-in control moth cells by washing the cells with ice-cold PBS and lysing them with fresh lysis buffer (20 mM HEPES pH 7.6, 1% IGEPAL CA-630, 1 mM EDTA, 1 mM spermine, 1 mM spermidine, 1 mM DTT, 0.004 U/μl SUPERase-In [Ambion AM2696], 320 mM sucrose, 0.1% isopropanol-saturated PMSF and cOmplete EDTA-free protease inhibitor cocktail [Roche 11873580001]). At the two-hour mark of the HAP1 treatments, media was discarded, cells were washed with ice-cold PBS, and equal amounts of spike-in moth cells were introduced with lysis buffer into the HAP1 cells flasks. The amount of spike-in cells used was estimated to account for ∼1% of total HAP1 cells. Cells were quickly scrapped off the flasks before being layered on top of a sucrose cushion (20 mM HEPES pH 7.6, 1 M sucrose, 1 mM spermine, 1 mM spermidine, 0.1 mM EDTA, 1 mM DTT, 0.004 U/μl SUPERase-In, 0.1% isopropanol-saturated PMSF and cOmplete EDTA-free protease inhibitor cocktail). Cells were spun at 22 500 × g for 5 min at 4°C. Pelleted nuclei were resuspended in 60 μl storage buffer (20 mM HEPES pH 7.6, 5 mM MgCl2, 5 mM DTT and 25% glycerol) and stored at –80°C.
Isolated nuclei (20 μl) were incubated with pre-heated nuclear run-on buffer (20 mM HEPES pH 7.6, 1.5% Sarkosyl, 5 mM MgCl2, 5 mM DTT, 100 mM KCl, 0.6 U/μl SUPERase-In and 0.06 mM of all four biotin-11-NTPs [Biotin-11-UTP, Jena NU-821-BIOX, Biotin-11-CTP Jena, NU-831-BIOX, Biotin-11-ATP, Jena NU-957-BIOX-L, Biotin-11-GTP, Jena NU-971-BIOX-L]). Nucleotide incorporation was allowed to proceed for 10 min at 37°C and RNA was isolated with Trizol LS (Ambion) following the manufacturer′s recommendations. The pellet was resuspended in 20 μl RNase-free water, incubated at 65°C for 2 min and immediately placed on ice. RNA was hydrolyzed for 20 min by adding 5 μl of ice-cold 1N NaOH and stopped with 25 μl 1M Tris pH 6.8. Biotinylated-RNA was incubated with M-280 streptavidin Dynabeads (Invitrogen 11206D) and washed at room temperature for 15 min three times with high salt buffer (50 Mm Tris pH 7.8, 2 M NaCl, 0.5% Triton X-100 and 1 mM EDTA) and twice with low salt buffer (20 mM Tris pH 7.8, 150 mM NaCl, 0.1% Triton X-100 and 1 mM EDTA). RNA was separated from Dynabeads with Trizol LS, precipitated and resuspended in 8 μl of 12.5 μM VRA3-4N adapter mix. The adapter was ligated by adding 12 μl of 3× Rnl1 mix (M0204) for 4 h at 37°C. Biotinylated-RNA was incubated with M-280 streptavidin Dynabeads, washed and isolated as described above. RNA deccaping and end repair reactions were performed by adding 10 μl of 2× RppH mix (NEB M0356) for 1 h at 37°C followed by the addition of 80 μl of 4x T4 PNK mix (NEB M0201) for 1 h at 37°C. RNA was isolated with Trizol LS, precipitated and resuspended in 8 μl of 12.5 μM VRA5-4N adapter mix. The adapter was ligated, and RNA was incubated with high-salt and low-salt buffer followed by Trizol isolation as described above. RNA was reverse transcribed with SuperScript IV (Thermofisher 18090010) (5 μM RP1 primer, 1 mM dNTP, 2× SSIV buffer, 10 mM DTT, 2 U/μl SUPERase-In and 20 U/μl SSIV enzym(E) at 45°C for 15 min, 50°C for 40 min, 55°C for 10 min, and 70°C for 15 min. Amplification of libraries was performed with 1 μM index primer (RPI-1 to RPI-19, we skipped RPI-17), 1 μM RP1 and 1× KAPA HiFi ready mix (Roche KK2600). We purified the amplified libraries with MinElute PCR purification kit (Qiagen 28004) and size selected for 135–600 bp using BluePippin 2% agarose gel cassette (Sage Science BDF2010). Samples were sequenced at the Iowa Institute of Human Genomics on an Illumina NovaSeq 6000 using 50 bp paired-end reads.
Western blotting
Protein fractions from drug or DMSO treated HAP1 cells were extracted as previously described (35). Briefly, cells were incubated in buffer A (10 mM HEPES, 10 mM KCl, 10 mM MgCl2, 1 mM EDTA, and 0.5% IGEPAL CA-630) on ice for 10 min. Cells were then spun down at 200 × g for 5 min at 4°C. The supernatant was collected (free factors) and mixed with loading buffer (20% Ficoll, 10% SDS, 50 mM Tris pH 7.6, 3% bromophenol blue, and 50 mM DTT). The pellet was resuspended in buffer B (10 mM HEPES, 450 mM KCl, 10 mM MgCl2, 1 mM EDTA, and 0.5% IGEPAL CA-630) on ice for 10 min. Cells were then spun down at 200 × g for 5 min at 4°C. The supernatant was collected (chromatin bound) and mixed with loading buffer. The pellet was resuspended in loading buffer (pellet). Also prepared were whole cell and nuclei (isolated as described above) lysates. Homogenized samples were heated at 95°C for 5 min, fractioned on SDS-PAGE, transferred onto 0.45 mM nitrocellulose membrane (GE Healthcare Life Sciences), and incubated overnight with anti-TAF1 (sc-735; 1:1000; Santa Cruz), anti-TFIIB (sc-56793; 1:1000; Santa Cruz), anti-TAF4 (sc-136093; 1:1000; Santa Cruz), anti-Cdk9 (sc-8338; 1:1000; Santa Cruz), anti-TBP (ab51841; 1:2000; Abcam), anti-FKBP12 (sc-133067; 1:1000; Santa Cruz), and anti-XPB (8746; 1:3000; Cell Signal). Proteins were visualized with Pierce ECL substrate (ThermoFisher Scientific 32106) in a UVP ChemStudio (Analytik Jena).
Processing of PRO-Seq data
PRO-Seq datasets were processed using the python pipeline RNAfastqtoBigWig (https://github.com/P-TEFb/RNAfastqtoBigWig) as previously described (36) to automate the work up of paired-end FASTQ files to the final generation of bigWig tracks. Briefly, fastq files were trimmed of Illumina 4N UMI RNA adapters using TrimGalore v0.6.0 (https://github.com/FelixKrueger/TrimGalore) with length parameter of 26 resulting in reads of 18–600 bp. Reads were aligned using bowtie v1.2.3 (37) to a concatenated genome containing the Spodoptera frugiperda (WGS number JQCY02) and human (UCSC assembly hg38) genomes. Next, reads with identical unique molecular identifiers (UMIs) were collapsed and the biotinylated NTP from the 3′ end was removed using the dedup program (https://github.com/P-TEFb/dedup). The output files were converted into bedGraphs with bedtools genomecov v2.27.1 (38). The read count for each sample was corrected by taking into account the library size and total spike-in reads as previously described (33) before generating bigWig tracks with the bedGraphToBigWig program.
Selection of TSRs
A number of bioinformatics tools used in this study are part of a group of programs called PolTools that can be found here https://github.com/GeoffSCollins/PolTools. The tsrPicker (https://geoffscollins.github.io/PolTools/tsrPicker.html) was used to identify 11 bp transcription start regions (TSR) from a combined dataset that contained all DMSO treated PRO-Seq datasets (Rep1 and Rep2 TBP-FKBP12F36V, Rep1 and Rep2 TAF1-FKBP12F36V, Rep1 and Rep2 TFIIB-FKBP12F36V, and Rep1 and Rep2 XPB-FKBP12F36V). We also generated a TAF4-FKBP12F36V tagged cell line that was treated with DMSO from which PRO-seq libraries were made and was included in the tsrPicker analysis for a total of nine PRO-Seq DMSO datasets. A modified blocklist from GenecodeV27 (Supplementary Data File – hg38 blocklist sheet) was used to remove Pol I and Pol III transcription units. Next, only individual genomic positions that contained a minimum number of reads of ≥100 were kept. From this filtered dataset, the program was allowed to find the base in the genome with the most 5′ end reads creating a strand-specific 11 bp TSR (±5 bp) with the MaxTSS in the center. This process was repeated allowing for 5 bp overlap (two TSRs cannot share the same MaxTSS) until no bases containing the minimum number of reads remain. The resulting TSR list was used to quantify the number of 5′ end reads within each 11 bp TSR for each of the individual datasets for DMSO and dTAGV-1. Only TSRs with ≥ 10 reads in each DMSO dataset were used and final counts were generated by adding DMSO or dTAGV-1 Rep1 and Rep2. Finally, Pol II specific TSRs were obtained by only considering those with a TFIIB dependency (DMSO/ dTAGV-1) ≥2. This gave rise to an All TSR list of 72 095. To find truQuant TSRs, we first applied the truQuant program (https://geoffscollins.github.io/PolTools/truQuant.html) to the all DMSO dataset to identify the most highly utilized TSS for each gene that is expressed (39). TSRs with a MaxTSS with the same genomic coordinate as the most utilized TSS identified by truQuant were denoted as truQuant TSRs (n = 10 273). Gene body annotations and quantifications as well as 150 bp pause regions were carried out by applying the truQuant program to the all DMSO dataset. The ggplot2 package in R was used to make point density correlations between All TSRs, truQuant TSRs, and gene body counts across replicates. tRNA TSRs were defined as an 11 bp region immediately upstream of the GENECODE V38 annotated mature 5′ end. tRNA TSRs with ≥10 reads in each DMSO dataset were considered active and used for further analysis.
To identify ectopic expression of 11 bp TSRs following TBP depletion, we again employed tsrPicker on TBP-DMSO-Rep1 and Rep2, and TBP-dTAGV-1-Rep1 and Rep2 PRO-Seq datasets. A concatenated list of TSRs was created from TSRs identified in each dataset (total TSRs = 138 644). Spike-in normalized 5′ ends were calculated for each of the TSRs and those showing a TBP-dependence ≤0.5 were deemed as appearing or increasing TSRs (total TSRs = 1496). TSRs nearest to an increasing TSR were annotated only if the distance between the MaxTSS of both TSRs was ≤200 bp in a strand independent manner.
Calculation of dependencies
Factor dependencies for TSRs were calculated by dividing the sum of 5′ ends of spike-in normalized reads present in a given TSR in DMSO Rep 1 and Rep 2 by the sum of 5′ ends of spike-in normalized reads present in the same TSR in dTAGV-1 Rep 1 and Rep 2. Dependencies for gene bodies were calculated by dividing the sum of 3′ ends of spike-in normalized reads present in a given gene body in DMSO Rep 1 and Rep 2 by the sum of 3′ ends of spike-in normalized reads present in the same gene body in dTAGV-1 Rep 1 and Rep 2.
Metaplots
Metaplots showing average 5′ or 3′ read densities across specific genomic regions for different dependency groups for each factor were calculated and plotted with MS Excel. Read through transcription averages were generated by using a custom python3 script: https://geoffscollins.github.io/PolTools/read_through_transcription.html. For analysis of 3′ reads around the CPS pause regions determined by truQuant in the regions were blocklisted and a 1000 bp running average was used to smooth the data. Also, distribution plots for A, C, G and T nucleotides were calculated for specific regions using a python3 script (https://geoffscollins.github.io/PolTools/base_distribution.html).
Motif discovery and web logos
Motifs were determined using MEME version 5.4.1 (40). An example command line used reads as follows: meme -brief 100 000 -dna -minw 6 -maxw 6 sequence.fasta. FIMO version 5.4.1 was used to scan chosen genomic intervals for the presence of motifs identified with MEME. An example command line used reads as follows: fimo –thresh 0.0001 MEMEmotif.txt sequence.fasta. For TBP-specificity motif analysis the strand was specified (fimo –thresh 0.0001 –norc MEMEmotif.txt sequence.fasta). CentriMo version 5.4.1 was used to determine the probability of finding motifs in particular locations in input sequences (41). An example command line used reads as follows: centrimo –verbosity 1 –local –score 5.0 –ethresh 10.0 sequence.fasta MEMEmotif.txt. The Inr web logo was constructed utilizing Web Logo 3 version 3.6.0 (42).
Feature analysis
Chromatin features from TBP DFF-ChIP data performed in HFF cells (19) where analyzed by extracting fragments of specific lengths whose center lay within genomic intervals described in Supplementary Figure S6D and counted using Bedtools v2.26 intersect program. The features analyzed were TBP/nucleosome with fragment lengths of 160–190 and fragment centers between +25 to +65, PIC/+1 nucleosome with fragment lengths of 210–250 and fragment centers between +50 to +90, TBP with fragment lengths of 30–60 and fragment centers between –50 to –10, and PIC with fragment lengths of 61–88 and fragment centers between –25 to –1. Pearson's correlation coefficient (r) were calculated across features using the final number of fragment centers for each feature.
FragMaps
fragMap.py (https://github.com/P-TEFb/fragMap) was used to make heatmaps displaying the average distribution and position for DFF-Seq fragments from Pol II Ser5P DFF-ChIP and TBP DFF-ChIP experiments performed in HFF cells (19). Fragments were analyzed across truQuant TSRs identified in HFF cells with PRO-Cap (43). Aspect ratio, black values, and color intensities were used as previously described (19).
Heatmaps
TSS heatmaps were generated by quantifying the spiked-in normalized 5′ reads over each base position within the indicated regions around the MaxTSS for each TSR. Python3 and R scripts were used to generate heatmaps in grayscale (https://geoffscollins.github.io/PolTools/region_heatmap.html) or color (log2 fold change)(https://geoffscollins.github.io/PolTools/region_fold_change_heatmap.html). For heatmaps in Figure 3 and Supplementary Figure S3, each row illustrates a TSR and each base is represented by 5 pixels. For heatmaps in Figure 4A, each row illustrates an average of 8 TSRs and each base is represented by 3 pixels. Heatmaps in Figure 4C were created using: https://geoffscollins.github.io/PolTools/gene_body_fold_change_heatmap.html. Fragment center heatmaps were generated using python3 and R scripts (https://github.com/P-TEFb/Heatmap) from previously published DFF-Seq data (19) around truQuant TSRs identified in this manuscript. For TBP DFF-ChIP heatmaps, each row illustrates an average of 10 TSRs and each base is represented by 3 pixels. Finally, for H3K4me3 DFF-ChIP heatmaps, each row illustrates an average of 10 TSRs and each base is represented by 1 pixel.
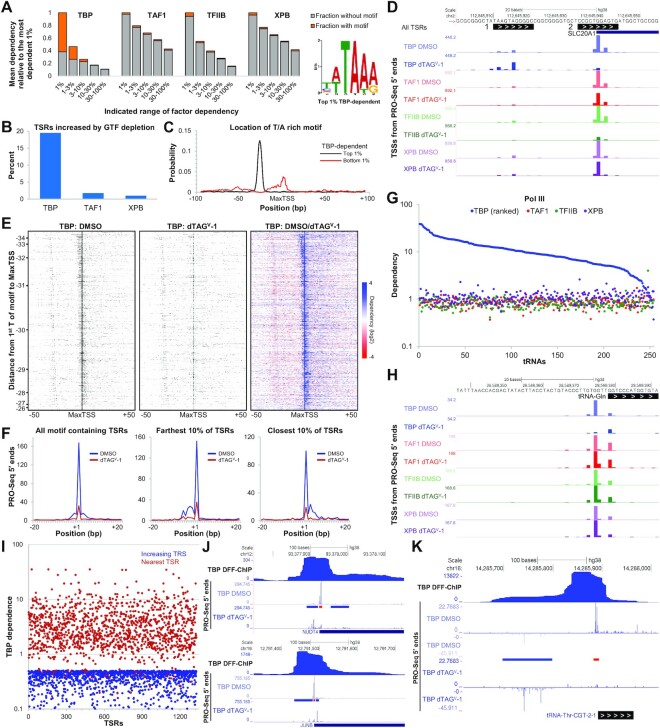
Figure 3.
TBP is only indispensable for driving transcription of TBP-specific motif containing TSRs. (A) The TBP-specific motif on the right was discovered using MEME in the –36 to –19 bp region upstream of the MaxTSS of the top 1% TBP-dependent All TSRs (450/720; P = 5.1e–329). All TSRs (n = 72 095) were divided into five dependency groups (1%, 1–3%, 3–10%, 10–30%, 30–100%, with 1% being the most dependent group) for each GTF. The mean dependency for each group was calculated and plotted relative to the 1% dependency group for each factor. The fraction of TSRs for each dependency group that contains the TBP-specific motif discovered using FIMO with P < 0.01 is depicted in orange. (B) Percentage of All TSRs (n = 72 095) with increased transcription following depletion of each factor. TFIIB is not included in the graph because non-dependent (DMSO/dTAGV-1 < 1) TSRs were determined to not be Pol II transcripts and were filtered out (see Materials and Methods). (C) The probability of occurrence of the TBP-specific motif determined with CentriMo is plotted as a function of distance from the MaxTSS of top 1% TBP-dependent All TSRs (black line, P = 1.0e–425; n = 720) or bottom 1% (red line, P = 2.4e–23; n = 720). (D) Genome browser view of the promoter region of SLC20A1. Two TSRs (1 and 2) with PRO-Seq 5′ ends tracks for each factor after DMSO or dTAGV-1 treatment are shown. (E) Heatmaps of PRO-Seq 5′ ends of the 1840 TSRs from All TSRs that contain the TBP-specific motif (P = 4.9e–179) and TBP-dependency ≥2.5. Each row represents an individual TSR. The TSRs are sorted by decreasing distance of the TBP-specific motif to the MaxTSS. The colored heatmap displays the change in each TSS upon depletion of TBP. (F) Line plots depicting the average number of PRO-Seq 5′ ends (TSSs) at each base position relative to the MaxTSS for the indicated fraction of TSRs plotted in (E). (G) Dependency of active tRNAs (n = 254) following depletion of GTFs. PRO-Seq 5′ ends of an 11 bp region immediately upstream of the GENECODE V38 annotated mature 5′ end were utilized. (H) Genome browser view of the promoter region of a tRNA-Gln showing PRO-Seq 5′ ends for each factor following treatment with DMSO or dTAGV-1. (I) Plot comparing the TBP dependence of the 1329 TSRs with a dependence ≤ 0.5 (increasing TSR) to the dependence of their closest neighboring TSR (nearby TSR). Genome Browser tracks as indicated for examples of TSRs impacted positively by reduction of a TBP-dependent Pol II promoter (J) or Pol III promoter (K). Regions where transcription is impacted positively (blue) and negatively (red) by depletion of TBP are indicated by bars between the PRO-Seq tracks.
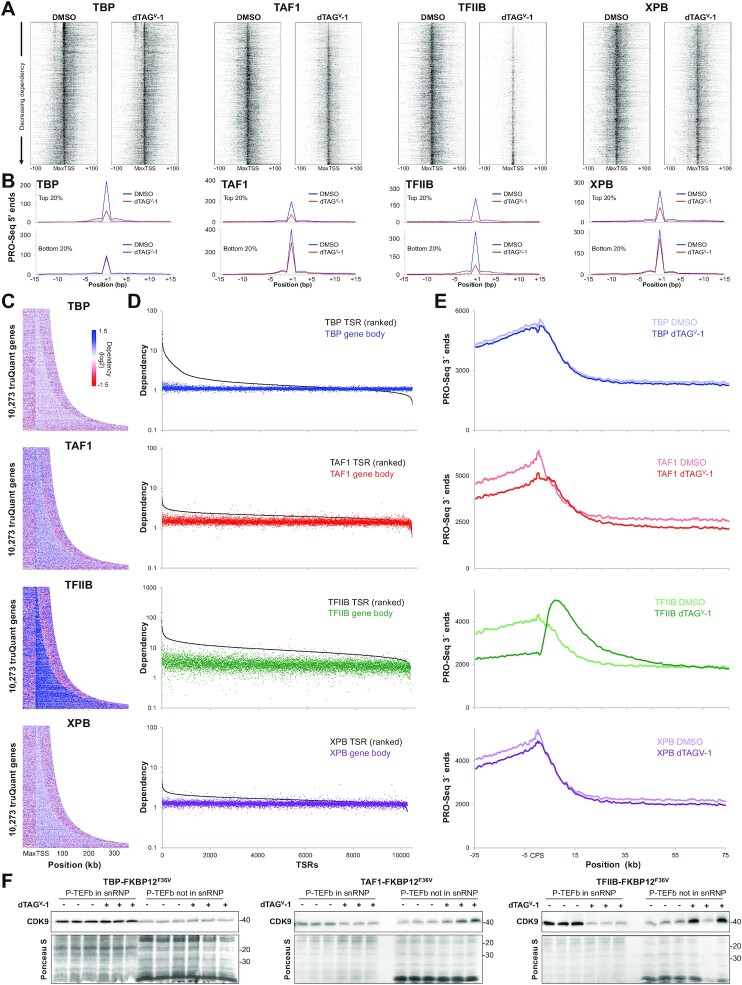
Figure 4.
Distinct effects of GTF depletion around the promoter, in the gene body and downstream of the CPS. (A) Heatmaps of PRO-Seq 5′ ends sorted by decreasing dependency centered at the MaxTSS of truQuant TSRs (n = 10 273) for each factor. (B) Plots depicting the average PRO-Seq 5′ ends at each base position in DMSO and dTAGV-1 treated cells for the top and bottom 20% dependent truQuant TSRs for each factor. (C) Dependency heatmaps of truQuant TSR gene bodies sorted by gene length and buffered with 50 kb upstream of the MaxTSS and downstream of the CPS. (D) Dependency plots of truQuant TSRs and their corresponding gene bodies. Genes are ordered left to right by their TSR dependency. (E) Average PRO-Seq 3′ ends across all truQuant genes at the indicated positions around the CPS after DMSO or dTAGV-1 treatment. (F) Western blot analysis of Cdk9 levels from cells treated with DMSO or dTAGV-1 extracted with buffer containing 150 mM KCl or 450 mM KCl to obtain free (P-TEFb in snRNP) or chromatin associated Cdk9 (P-TEFb not in snRNP), respectively. This experiment was performed in triplicate. The nitrocellulose membrane below the Cdk9 band was cut and stained with Ponceau S for a loading control.
RESULTS
Rapid depletion of TBP, TAF1, TFIIB and XPB
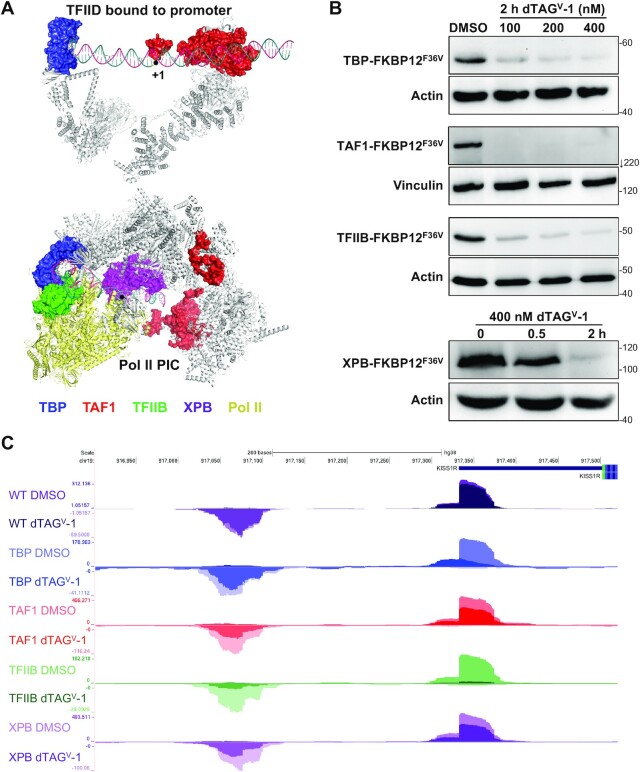
With the ultimate goal of examining the function of core PIC components in transcription initiation in cells at all active human promoters, four HAP1 cell lines were generated in which endogenous TBP, TAF1, TFIIB and XPB were individually fused with the 12 kDa FKBP12F36V protein at their C-termini. As illustrated by recent cryo-EM structures (3,6,21), the first three factors play key roles in the recognition of promoter elements during the formation of PICs while XBP is involved in the actual initiation event (Figure 1A). Near complete depletion of all tagged proteins was achieved within two hours of dTAGV-1 treatment (Figure 1B). Given that TAF1 and TBP are both components of TFIID (6,44), the effects of TAF1 depletion in TBP protein stability and vice versa was determined. After the short 2 h depletion, loss of TBP led to only an ∼30% decrease in TAF1 protein levels in whole cell lysates (Supplementary Figure S1A). Likewise, TAF1 depletion caused only about a 30% reduction in TBP protein levels (Supplementary Figure S1B). Depletion of either protein, however, did not significantly affect TAF4 (another component of TFIID) or TFIIB levels (Supplementary Figure S1A, B).
Figure 1.
GTFs targeted for rapid depletion. (A) Cryo-EM structures of TFIID bound to promoter DNA (6MZM) (6) and assembled PIC containing TFIID and TFIIH (7ENC) (3). Targeted proteins have been colored in the structures as follows: TBP (blue), TAF1 (red), TFIIB (green), and XPB (purple). (B) Depletion of TBP, TAF1 and TFIIB at different concentrations of dTAGV-1. Also shown is XPB depletion over time. Protein levels were determined by western blots of whole cell lysates utilizing GTF-specific antibodies, except the anti-FKBP12 antibody was used for XPB. Actin or vinculin was used as a loading control. (C) Genome browser view of tracks depicting sense and divergent PRO-Seq transcript pileups for wildtype (WT) HAP1 cells or the indicated FKBP12 tagged HAP1 cell lines at the KISS1R locus. Cells were treated for 2 h with DMSO as a control (light colors) or 400 nM dTAGV-1 to deplete the tagged factor (dark color) before PRO-Seq was performed.
To determine if depletion of human TBP or TAF1 had any effect on the association of TAF1, TAF4, TBP or TFIIB with chromatin, the amounts of free and chromatin bound factors were quantified. Cells with or without a 2 h dTAGV-1 treatment were lysed with a detergent containing buffer at 150 mM KCl and the nuclei were separated from the cytosol which contains non-chromatin bound factors (free). The nuclei were further extracted with 450 mM KCl (chromatin bound) to quantify the level of chromatin association. The extracted nuclei were pelleted and resuspended in SDS (pellet). Western blotting of the fractions revealed that in untreated cells TAF1 and TAF4 were mostly chromatin bound while TFIIB was mostly free with only low levels of all factors remaining in the high salt extracted nuclear pellet (Supplementary Figure S1C, D). dTAGV-1 treatment of TBP-tagged (Supplementary Figure S1C) or TAF1-tagged cell lines (Supplementary Figure S1D) led to near complete depletion of the targeted factor. Just as with whole cell lysates, there were no significant changes in TAF4 and TFIIB levels after depletion of TBP or TAF1 (Supplementary Figure S1C, D). Following TAF1 depletion, TBP was slightly reduced in the cytosol and nuclear fractions (Supplementary Figure S1D), while there were no appreciable changes in TAF1 protein levels detected after TBP depletion (Supplementary Figure S1C).
Quantitative analysis of actively transcribed promoters genome-wide
PRO-Seq (34,45) was performed in each of the four cell lines after treatment with DMSO as a control or 400 nM dTAGV-1 for two hours prior to the isolation of nuclei. We employed our previously described method using a lysis buffer with EDTA to rapidly halt transcription as cells are lysed leading to accurate retention and positioning of engaged transcription complexes (33). Because paired end sequencing was performed the location of the paused Pol II near promoters can be obtained from 3′ end reads and the location of the TSS from 5′ end reads. Immunoblot analyses of the isolated nuclei demonstrated the dramatic loss of the tagged proteins (Supplementary Figure S1E), similar to those observed with whole cell lysates (Figure 1B). A constant, small amount of Sf21 moth nuclei was spiked-in to each flask of lysed HAP1 cells, enabling us to reliably quantify absolute global changes in transcription. To quantitatively ascertain the changes in levels of transcription for all promoters in the genome following depletion of the GTFs, a list of transcription start regions (TSRs) was compiled using PRO-Seq data and a bioinformatics tool, tsrPicker. This method identifies 11 bp TSRs with the MaxTSS positioned in the center (see MATERIALS AND METHODS for more details) which is likely generated by a single PIC (18). The data queried was a combination of the 5′ reads which arise from TSSs in promoter regions from highly correlated (Supplementary Figure S2C) DMSO control PRO-Seq datasets (∼280 million total reads – Supplementary Data File). Inclusion in the list required at least 100 5′ total reads in the combined DMSO datasets and at least 10 reads in each DMSO dataset, resulting in 72 095 total TSRs (henceforth referred to as All TSRs). A sub-list of TSRs was also created that contains the most highly utilized promoter for each expressed gene by employing the truQuant analysis tool (39). Of the 72 095 TSRs, 10 273 were determined to contain the MaxTSS for each transcribed gene (henceforth referred to as truQuant TSRs).
Two biological replicates were performed for each tagged cell line in the presence or absence of the drug. Using All TSRs, truQuant TSRs and truQuant gene bodies, correlation of the library size and spike-in normalized replicates showed strong linear correlations as determined by Pearson's correlation coefficient (Supplementary Figure S2A). Because of the spike-in normalization the correlation plots for PRO-Seq signals should have slopes close to 1 since absolute levels of transcription are determined. To analyze how effective the spike-in normalization was we calculated the slopes for Rep1 versus Rep2 truQuant TSR data and found that the average slope for the all four datasets was 0.96 ± 0.17 (DMSO) and 0.86 ± 0.28 (VHL). This indicates that the spike-ins did an excellent job determining the absolute transcription levels. Additionally, a control PRO-Seq experiment in non-tagged HAP1 cells was performed to corroborate that any effects following drug treatments were specifically due to depletion of the GTFs and not secondary off-target drug effects. The replicates correlated very well and importantly correlation of the control and dTAGV-1 treated wildtype cells was also high indicating that dTAGV-1 has no significant effects on transcription by itself (Supplementary Figure S2B). Although we observed no growth defects in the tagged lines, to determine if tagging the GTFs had a direct effect on transcription we calculated Pearson's correlation coefficients for both replicas of wildtype HAP1 cells and all replicas of DMSO treated tagged GTF lines (Supplementary Figure S2C). The strong correlations indicated that GTF tagging did not have a significant effect on transcription in the undepleted state. Genome browser tracks depicting pileups of forward and reverse nascent transcripts around the KISS1R promoter provide examples of the effects of GTF depletion (Figure 1C). The promoter-proximal paused transcripts in the sense and divergent direction are reduced to different extents upon loss of each of the GTFs. Addition of dTAG to wild type cells had a negligible effect on nascent transcript levels. However, depletion of all factors had a significant effect in transcript levels with TBP and TFIIB having greater effects than TAF1 and XPB.
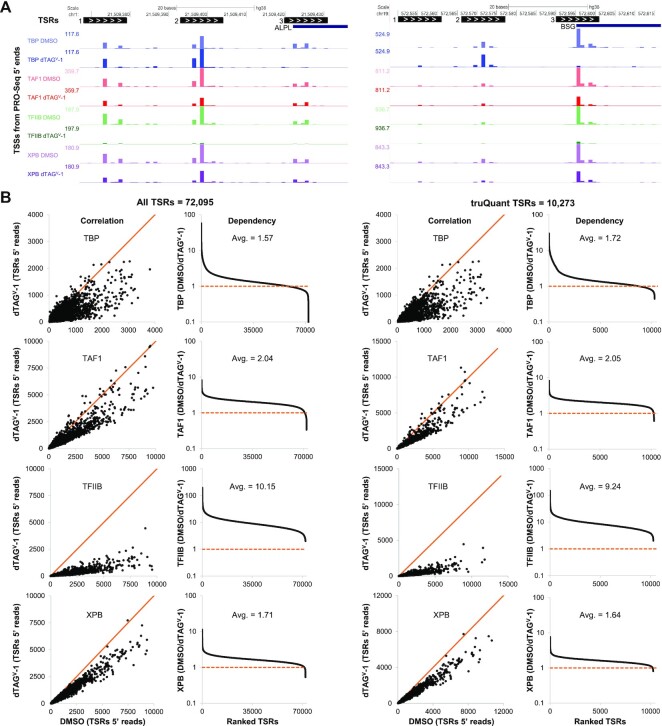
Figure 2A depicts genome browser views of 5′ reads in the promoter regions of two genes, ALPL and BSG, and three TSRs identified for each of them. The reproducibility of 5′ end quantification can be seen from the consistency of the relative reads from each individual TSS across the DMSO datasets. Transcription from all three TSRs in both genes are reduced after TAF1, XPB, and TFIIB depletion, with the latter resulting in the most dramatic decrease (Figure 2A). For both genes, only TSR #3 is TBP-dependent while TSR #2 shows an increase in initiation following TBP depletion. This phenomenon will be further analyzed and discussed in later sections. Initial quantification of the effects of GTF depletion on either All TSRs or truQuant TSRs was carried out by determining the sum of the spike-in normalized reads for each TSR from control (DMSO) and GTF-depleted (dTAGV-1) cells. A correlation of the two values for each TSR was plotted as well as a calculated dependency. Depletion of each of the four factors had a negative impact on almost all promoters, as is clear from the correlation plots where the large majority of data points are displaced from the orange line representing no change. Depletion of TFIIB had by far the greatest effect (Figure 2B). A dependency was calculated by dividing the sum of 5′ reads for each TSR (DMSO/ dTAGV-1) and plotted after sorting from most to least dependent. The average dependency on TFIIB was between 9.2 (truQuant) and 10.2 (All), and average dependencies on TAF1 and XPB were between 1.6 and 2.4 for both sets of TSRs (Figure 2B). TBP had a more disparate effect with 14% (All) or 16% (truQuant) of promoters with a dependency of 2 or greater while the remainder had only negligible changes or dependencies less than 1, meaning initiation increased upon depletion (Figure 2B).
Figure 2.
Global Pol II transcription is reduced after GTF depletion. (A) tsrPicker algorithm (see Materials and Methods) was implemented on nine combined DMSO PRO-Seq datasets to identify 11 bp TSRs (black boxes) across the genome. White arrows inside TSRs indicate the direction of transcription. TSRs identified in two genes, ALPL and BSG, each with three TSRs, are shown as examples. Also depicted are PRO-Seq 5′ ends tracks (TSSs) for each factor after DMSO or dTAGV-1 treatment. (B) Effects of GTF depletion on All TSRs (n = 72 095) or truQuant TSRs (n = 10 273). For all factors the sum of reads for the two replicas were used. Correlation plots compare the sum of the spike-in and sequence depth normalized PRO-Seq 5′ ends for each TSR in control (DMSO) and GTF-depleted (dTAGV-1) cells. Also shown are plots sorted by high to low dependency (DMSO/ dTAGV-1) of each TSR following depletion of each factor. Avg. refers to the average dependency. Solid orange lines depict a slope of 1.
TBP is only required for transcription of a small fraction of TSRs
To determine the sequence motif most associated with TBP dependency, the –36 to –19 bp region upstream of the MaxTSS of the top 1% dependent All TSRs was examined with MEME (40). A six base pair motif was found to be present in >60% (450/720, P = 5.1e–329) of these TSRs (Figure 3A). Because it is significantly different from the so called TATA element, we call it the TBP-specific motif. For the members of the All TSRs set with the top 3% of dependency, 49% (1065/2160, P = 1.8e–312) had the TBP-specific motif upstream. As the occurrence of the motif decreased, so did the dependency on TBP (Figure 3A). As expected, the functions of the rest of the GTFs are primarily independent of the presence of the TBP-specific motif (Figure 3A). Additionally, among the TBP dependent promoters, strong dependency is mainly restricted to the top 1% whereas with the other GTFs broad transcriptional effects lead to closer mean dependencies across groups (Figure 3A). Almost identical results were obtained with truQuant TSRs where the same TBP-specific motif and similar enrichment was found upstream of the MaxTSS of TBP-dependent truQuant TSRs (Supplementary Figure S3A).
The TBP binding motif is also a site of initiation
Interestingly, about 20% of All TSRs became more active after TBP depletion (Figure 3B). To better understand why this is the case, local TBP motif enrichment analysis (CentriMo) (41) of the bottom 1% TBP-dependent (most increased) of All TSRs was performed (Figure 3C). In contrast to the top 1% TBP-dependent of All TSRs, which contain the TBP-specific motif upstream of the MaxTSS, the bottom 1% TBP-dependent of All TSRs showed an enrichment of the TBP-specific motif overlapping the TSR itself (Figure 3C) indicating that the TBP binding motif is frequently used as an initiation site. Figure 3D and Supplementary Figure S3B show examples of both a top 1% and a bottom 1% TBP-dependent All TSR present in the same promoter region of the gene SLC20A1 and SHISA2, respectively. The main promoter for each gene (TSR #2) is TBP-dependent given the significant reduction in 5′ reads after TBP depletion (Figure 3D, Supplementary Figure S3B). The TBP-specific motif located upstream of TSR #2, called by our analysis as TSR #1, is presumably the TBP binding site and also an initiation site as evidenced by the presence of mapped 5′ reads in each of our PRO-Seq data sets (Figure 3D, Supplementary Figure S3B). TBP depletion led to an increase in initiation in TSR #1 not observed after depletion of the other GTFs (Figure 3D, Supplementary Figure S3B). Indeed, after depletion of the rest of the GTFs, both TSRs show a decrease in transcription relative to each other with TFIIB depletion causing the most dramatic decrease (Figure 3D, Supplementary Figure S3B). The distribution of the position of the TBP-specific motif upstream of TSSs varies within the –35 to –25 bp window (46). This is easily visualized in heatmaps depicting 5′ reads in the 100 bp surrounding the MaxTSS of the fraction of All TSRs that had TBP dependencies >2.5 sorted by decreasing distance from the TBP-specific motif to the MaxTSS (Figure 3E). Initiation in the region corresponding to the TBP binding motif is seen to increase after TBP depletion. Taken together, we believe that for PICs that drive transcription in a TBP dependent manner, TBP binds to the TBP-specific motif occluding it from being used as an initiation site. Depleting TBP enables formation of a PIC over this region that drives initiation in a TBP independent manner.
This analysis additionally demonstrates that the further away the TBP binding motif is from the MaxTSS, the more initiation occurs upstream of the MaxTSS compared to downstream (top of the heatmap, Figure 3E, F). On the other hand, for TSRs where the motif is closest to the MaxTSS, initiation occurs more frequently downstream of the MaxTSS compared to upstream (bottom of the heatmap, Figure 3E, F). The same results were observed when analyzing truQuant TSRs (Supplementary Figure S3C). This region where initiation occurs surrounding the MaxTSS, that is most affected following TBP depletion, is about ±5 bp in size (Figure 3E, F, Supplementary Figure S3C). This provides further evidence that the PIC supports transcription of a ±5 bp region around a primary TSS (18).
Distinct TBP requirements for Pol I, II, and III transcription
Next, given that TBP is also a major component of the multiprotein complexes that mediate transcription from Pol I and Pol III promoters (47), we analyzed the effects of TBP depletion on nascent RNAs originating from these promoters. For Pol I, we specifically examined the 45S preribosomal rDNA loci whereas for Pol III we looked at tRNAs and the small nuclear RNA (snRNA) genes U6, 7SK and 7SL. Also included in the analysis are the Pol II transcribed snRNA genes U1–U5, U11 and U12. For Pol I, we observed only a modest reduction (avg. dependency of 1.5) in transcription initiating from the main TSS of the 45S preribosomal rDNA loci (Supplementary Figure S3D). As expected, depletion of TAF1, TFIIB, or XPB did not lead to any major changes in rRNA transcription (Supplementary Figure S3D). For Pol III promoters, two distinct effects were observed after TBP depletion. tRNAs were highly sensitive to loss of TBP (avg. dependency of 11.5), with some tRNAs showing almost complete abrogation of transcription from their main TSS (Figure 3G, H). In contrast, the U6, 7SK and 7SL were not affected to any significant degree (avg. dependency of 1.1) (Supplementary Figure S3E). As controls, we also quantified PRO-Seq 5′ reads of these Pol III genes following depletion of TAF1, TFIIB and XPB. As expected, there were no significant changes (Figure 3G, Supplementary Figure S3E). All analyzed Pol II transcribed snRNA genes were dependent on TBP (average dependency of 2.8), TFIIB (average dependency of 8.4), and XPB (average dependency of 2.2) (Supplementary Figure S3E). However, loss of TAF1 had no major effect in transcript levels (average dependency of 1.2 and 0.7, respectively). This may be explained by the observation that the TFIID complex that forms at these genes lacks TAF1 (48).
To explore further how depletion of TBP had a positive influence on existing TSRs and to determine if any new TSRs appeared, we discovered TSRs from each of the four TBP datasets (both replicas with and without dTAGV-1). Of the 138 645 TSRs in the analysis, 1496 had a TBP dependence of ≤0.5, meaning they increased 2-fold upon depletion, and 1329 of these were found within 200 bp of a TBP-dependent promoter. The TBP-dependency of positively affected TSRs and the nearby TSR were compared by plotting both dependencies for all 1329 TSR pairs (Figure 3I). The average TBP-dependence of the nearby TSRs was 4.0 which is much higher than the average of 1.6 for all reported TSRs in Figure 2. We found that some of the nearby TBP-dependent promoters were Pol II driven and some were Pol III driven. Examples of each are shown in Figure 3J and K. Using recently reported DFF-ChIP data that detects TBP PICs and associated nucleosomes (19) it appears that the positively affected TSRs lie under the region protected by TBP containing complexes (Figure 3J and K). For Pol II the TBP PIC covers ∼65–85 bp and the downstream nucleosome adds about 150 bp. For Pol III over tRNA genes the TBP PICs over Pol III transcribed tRNA promoters are similar except that associated nucleosomes are found upstream (49). The DFF-ChIP was carried out in a different cell type, but its use here is justified in a later section. These results support the idea that depletion of TBP leads to loss of a fairly stable complex which increases access to otherwise buried promoters. We did not find any examples of truQuant TSRs driving genes that might be upregulated due to TBP depletion, strengthening our hypothesis that 2 h of depletion does not cause significant secondary effects.
To investigate the effects of GTF depletion on the distribution of TSSs in the 200 bp region surrounding the MaxTSS of truQuant TSRs, heatmaps of 5′ ends were generated and sorted by decreasing factor dependency (Figure 4A). Black values were set to the same value for the DMSO or dTAGV-1 data for each factor. This setting facilitates visualizing weaker TSSs but the MaxTSSs were saturated. In addition, for each factor the total number of 5′ ends were plotted for a 30 bp region around the MaxTSS for the top and bottom 20% of truQuant TSRs (Figure 4B). The most TBP dependent genes, which typically contain the TBP-specific motif, are also some of the most highly transcribed (top of the TBP DMSO heatmap; Figure 4A, B). TBP depletion mainly impacts the MaxTSS of these genes and the initiation occurring in the immediate upstream and downstream regions. About ∼30 bp upstream of the MaxTSS, the use of the TBP-specific motif as initiation site can be seen (Figure 4A). Initiation in this region is not TBP-dependent but rather experiences increased initiation after TBP-depletion as demonstrated earlier (Figure 3E, 4A). Depletion of the rest of the GTFs results in similar broad effects in which initiation at the MaxTSS and surrounding areas are reduced, with TFIIB having the most acute effect (Figure 4A). The TSS heatmaps and narrow TSS plots demonstrate that unlike TBP TAF1, TFIIB, and XPB have more impact on weaker promoters (Figure 4A, B).
Effects of factor depletion on transcription in and downstream of gene bodies
We were also interested in understanding the effects of GTF depletion on Pol II across genes bodies and downstream of the major transcript cleavage and polyadenylation sites (CPS). Dependency heatmaps of PRO-Seq 3′ ends with genes sorted by increasing gene length, indicate only a modest reduction in signal over genes bodies after depletion of TBP, TAF1 and XPB (Figure 4C). However, depletion of TFIIB had a stronger negative effect. We then compared the dependencies of each truQuant TSR to dependencies calculated for their gene bodies which was simply the ratio of the sum of 3′ end reads (DMSO/dTAGV-1) over the entire gene body (see Materials and Methods for details). The data was plotted after sorting from high to low TSR dependence (left to right) so that the TSR and gene body dependencies could be compared for each gene. The results revealed a less severe average reduction of reads in gene bodies than promoters after depletion of each factor (Figure 4D). Interestingly, TFIIB depletion resulted in a substantial increase in transcripts downstream of the CPS compared to control. This difference is evident beginning roughly 5 kb downstream of the CPS, reaching its maximum height at ∼10 kb and slowly declining over ∼50 kb (Figure 4E). This phenotype was observed regardless of gene length (Figure 4C) and is illustrated in several examples (Supplementary Figure S4A). Only TAF1 had a similar although more modest effect (Figure 4E).
The muted decrease in gene body transcripts compared to short paused transcripts could be due in part to an increase in paused Pol II that is released into productive elongation. The positive transcription elongation factor b (P-TEFb), composed of cyclin T1 or T2 and the cyclin dependent kinase Cdk9, allows Pol II to transition into the transcription elongation phase (50). There are two main P-TEFb states within the cell: sequestered in an inactive form in the 7SK snRNP or released so that it can functionally associate with chromatin. The 7SK snRNP is found in the cytosol after a low salt detergent lysis of cells and released P-TEFb is found associated with the resulting nuclei (51). Western blots show an accumulation of Cdk9 associated with chromatin after TFIIB depletion (Figure 4F). TAF1 depletion also led to accumulation of Cdk9 associated with chromatin but to a lesser extent than TFIIB depletion. Collectively, our data illustrates that gene bodies do not experience the same level of loss of PRO-Seq signal as do regions of promoter-proximal pausing following GTF depletion, presumably due to compensation by an increase in P-TEFb activity. The major effect downstream of the CPS seen after depletion of TFIIB and to a lesser extent TAF1 is likely due to a kinetic delay in Pol II termination, also resulting from increased P-TEFb activity. This point will be explored further in the Discussion.
Because release into productive elongation leads to a reduction of paused Pol II and P-TEFb activity can change, especially after TFIIB depletion, we wondered if there was a relationship between the level of productive elongation from each promoter and its factor dependency. The number of 3′ ends/1000 bp across each gene body was determined and plotted after sorting the dependency of the 5′ ends in the 150 bp promoter region (Supplementary Figure S4B). The most TFIIB-dependent promoter regions had significantly lower productive elongation before the depletion. There was a similar relationship for TAF1. TBP was different with many of the most dependent promoters having high levels of productive elongation. These effects can be rationalized given that TFIIB and TAF1 depletion caused a release of active P-TEFb from the 7SK snRNP, but TBP did not (Figure 4F). This increase in active P-TEFb could have a greater effect on genes that initially had low levels of productive elongation and this would lead to an apparent slightly greater dependence for those promoters. However, even for TFIIB this effect is somewhat minor compared to the large dependencies across all genes.
Relationship of GTF-dependence and promoter sequence composition
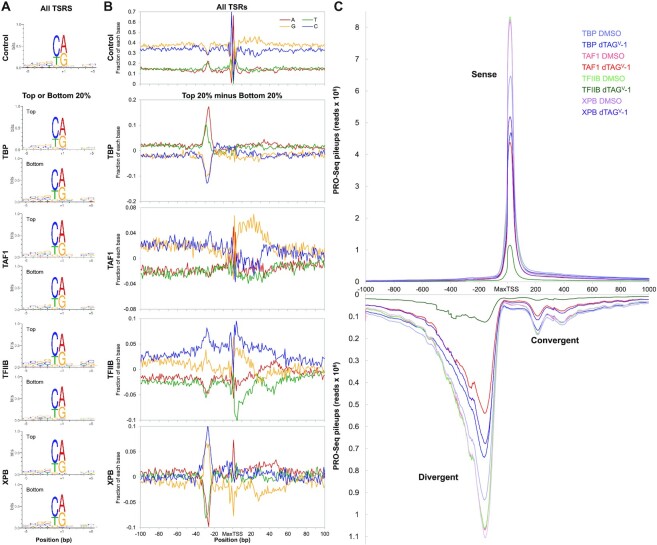
To investigate any sequence preferences for GTFs, we divided All TSRs into quintiles based on dependency for each factor. We then compared the sequence composition of the top 20% of dependent TSRs, presumably reflecting the most favorable sequences for that factor, and the bottom 20% of dependent TSRs, reflecting the least favorable sequences. A preference for the Inr was found regardless of dependency for all factors (Figure 5A) which is in accordance with previous results showing the Inr to be generally present in most TSSs (18). The top 20% of TAF1, TFIIB, and XPB-dependent TSRs had a slightly higher presence for the Inr compared to the bottom 20%. Again TBP was the outlier with the most dependent TSRs having the weaker Inr.
Figure 5.
Sequence preference for GTFs. (A) Sequence logo analysis performed on ±5 bp region from the MaxTSS of the top and bottom 20% dependent TSRs for each factor (n = 14 419 of All TSRs). (B) Base distribution plot for A, C, G and T nucleotides ±100 bp around the MaxTSS of each TSR in the All TSRs dataset (n = 72 095). Also shown are base distribution plots resulting from the subtraction of the bottom 20% from the top 20% dependent TSRs for each GTF. (C) Sense and divergent PRO-Seq transcript pileups for ±1000 bp regions flanking the MaxTSS of truQuant TSRs (n = 10 273).
Promoters have positional sequence biases that can be seen from a base distribution plot ±100 bp around the MaxTSSs in the All TSR dataset (Figure 5B, all TSRs). This broad view highlights a number of features that include peaks of T and A between –30 and –25, a very distinguishable Inr at the MaxTSS, and an enrichment of a stretch of G bases and a reduction in C downstream of the MaxTSS around +6 to +32 (Figure 5B). Our results faithfully recapitulate the base distribution that was observed when analyzing over 170 000 TSSs in the human genome using PRO-Cap (18). To determine if there are any sequence determinates for factor dependency, the average base distributions for the top quintile was subtracted from the bottom quintile for each factor. Each factor had a distinctive base distribution (Figure 5B). As expected (11), the main sequence element found for TBP was the A/T rich region between –30 and –25. The difference base distribution for TAF1 shows high GC content overall and an enrichment of G’s stretching +6 to +32 downstream of the MaxTSS. This region harbors the G-rich downstream promoter element (DPE) previously identified in Drosophila and shown to be enriched in highly transcribed mammalian randomized promoter libraries (16,17). The TFIIB difference base distribution shows high GC content overall in this region. TFIIB and XPB difference base distribution plots exhibit a number of similarities (Figure 5B). In contrast to TBP, both TFIIB and XPB disfavor A and T bases between –30 and –25 (Figure 5B). Very similar results were obtained when the top and bottom dependency quintiles were compared for the truQuant TSRs (Supplementary Figure S5).
To determine the consequences of GTF depletion on divergent and convergent transcription, we quantified the number of sense and anti-sense pileup reads in a 2000 bp region surrounding truQuant TSRs. Both divergent and convergent transcription behave in a similar manner to sense transcription following GTF depletions (Figure 5C). Note that the scale for divergent and convergent reads is about 10% of that for sense. The divergent peak is wider than the sense peak due to differences in the distance between the two promoters or sets of promoters at individual genes. Convergent transcripts are interestingly found downstream of the +1 and +2 nucleosome for the sense promoter similar to what was found in an earlier study (52).
To examine co-dependencies for the depleted GTFs, a comparison of the overlap of the top 5% dependent truQuant TSRs (n = 514) for each factor pair was performed (Supplementary Figure S6A). The red line indicates the expected overlap if results were random (5% overlap). TFIIB had the highest co-dependencies with the other GTFs and its co-dependency with XPB was striking. TBP and XPB had the lowest co-dependency. These results suggest that XPB might be most effective when there is no strong recruitment of TFIID by TBP.
Relationship between GTF dependency and the nearby sequence motifs
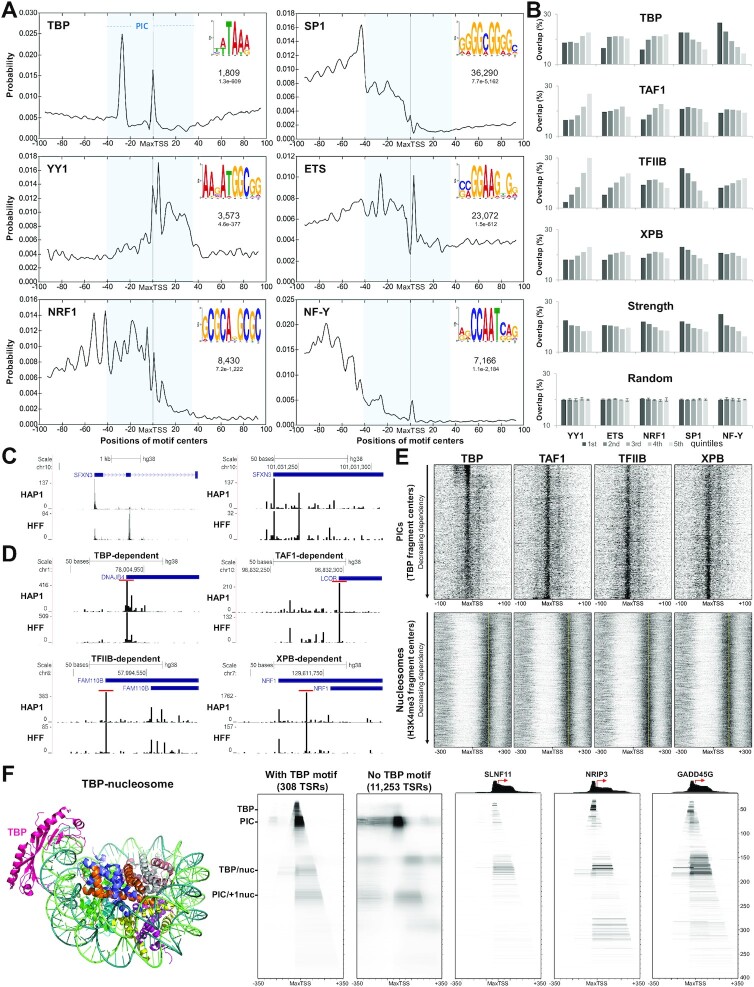
To determine if there were any sequence motifs that correlated with factor dependencies, extensive MEME analyses were performed on different regions around TSRs with high factor dependency. The only binding motif that correlated with the regions identified in Figure 5B was the TBP-specific motif upstream of the most TBP-dependent TSRs. Therefore, MEME was used to discover the most prevalent motifs across the 200 bp regions surrounding the MaxTSSs of All TSRs and the distribution of the motifs was determined with CentriMo (41) (Figure 6A). The TBP binding motif was found in the expected upstream position, but in addition was found over the MaxTSS (Figure 6A). The other motifs discovered by MEME were for the specific transcription factors YY1, ETS, NRF1, SP1 and NF-Y (Figure 6A). SP1 and ETS were much more prevalent than the other sites. SP1, ETS, NRF1 and NF-Y were found primarily upstream of the MaxTSS and YY1 was found over the Inr and downstream.
Figure 6.
Relationship between GTF dependencies with chromatin structure and occurrence of transcription factor motifs. (A) Plots show the probability of finding the TF motif (top right) within ±100 bp from the MaxTSS of All TSRs (n = 72 095) as determined with CentriMo. The numbers below the TF motif indicates the number of sequences containing the motif and the E-value (statistical significance of the motif after adjusting for multiple tests), respectively. The blue rectangle depicts the size and position of a standard TFIID containing PIC. (B) All TSRs were divided into quintiles based on GTF dependency, strength or randomly. The plots indicate the total number of TSRs from each quintile that contained a particular TF motif as determined by FIMO (P < 0.0001). Also quantified are the overlap between TSRs containing a TF motif and randomly generated quintile subgrouping, performed in triplicate. Error bars represent standard deviation. 1st, 2nd, 3rd, 4th and 5th refer to most to least dependent for each factor, strongest to weakest in strength or random subgrouping. (C) Comparison of truQuant genes identified in HAP1 cells with PRO-Seq (top track) and in HFF cells with PRO-Cap (bottom track). Only 5′ ends (TSSs) are shown. The left panel shows an example of a promoter region where the position of the MaxTSS identified is different between HFF and HAP1. The right panel shows the distribution of TSSs for the MaxTSS identified in HAP1 cells for the same gene. (D) Comparison of promoter regions in HAP1 cells and HFFs that were highly dependent on TBP, TAF1, TFIIB or XPB. The TSR identified by truQuant in both cell types is shown (red bar). (E) Heatmaps of centers of 60–70 bp fragments (PICs) from TBP DFF-ChIP performed in HFF cells (19) sorted by decreasing factor dependency centered at MaxTSS of truQuant TSRs from HAP1 cells. Also shown are heatmaps of the centers of 140–160 bp fragments (nucleosomes) from H3K4me3 DFF-ChIP performed in HFF cells (19) sorted by decreasing factor dependency centered at the MaxTSS of truQuant TSRs of HAP1 cells. Yellow dashed lines show the position of the average center of all +1 nucleosomes (∼+125 bp). (F) TBP-nucleosome cryo-EM structure (7OHB) (22) (left panel). TBP-DFF-ChIP data represented as fragMaps depicting the average coverage of fragments of various lengths (y-axis) at each position ±350 bp surrounding a MaxTSS (x-axis) from experiment performed in HFF cells (19). Five fragMaps are shown (from left to right): HFF cells truQuant TSRs determined from PRO-Cap data (43) that contain a TBP motif within the –36 to –19 region upstream of the MaxTSS (n = 308), truQuant TSRs without the TBP motif (n = 11 253), and examples of genes that contain an upstream TBP motif (SLNF11, NRIP3 and GADD45G). TBP-DFF-ChIP tracks are shown above the gene fragMap examples. Red arrows indicate the direction of transcription from the MaxTSS. Nuc (nucleosome).
Next, All TSRs or truQuant TSRs were partitioned into dependency quintiles for each factor (most to least dependent), quintiles of TSR strength (strongest to weakest), or random quintiles, and the number of TSRs that contained the TF motifs were quantified (Figure 6B, All TSRs; Supplementary Figure S6B, truQuant TSRs). A random sort of TSRs was performed three times and the distribution of the five motifs across the 5 quintiles was 20% with a standard deviation of 0.3% for All TSRs and 0.9% for truQuant TSRs. This provides a baseline to interpret significance of the other sorts. Interestingly, even though YY1-motifs correlated with strength, they inversely correlated with dependencies of all factors especially TAF1 and TFIIB. On the other hand, the SP1-motifs positively correlated with TBP, TFIIB, and XPB dependencies. ETS motifs inversely correlated with TFIIB-dependency and to a lower degree with TAF1-dependencies. Curiously, while NF-Y-motifs correlated with TBP dependency, they inversely correlated with TFIIB dependency. Finally, all motifs tested correlated with TSR strength to varying degrees (Figure 6B). Evidently, factor dependencies are influenced by neighboring TF binding sites.
Effects of GTF depletion on transcription surrounding the major TSRs and the relationship to nearby chromatin
We recently developed a method to examine transcription complexes and their interactions with nearby chromatin that utilizes the DNA Fragmentation Factor (DFF) to digest native nuclei followed by immunoprecipitation and sequencing of DNA fragments (DFF-ChIP) (19). The method accurately positions PICs and H3K4me3 modified nucleosomes and sites of transcription factor binding (49,53). We wanted to compare the existing DFF-ChIP data (19) to GTF dependency, but because the DFF-ChIP data was generated in primary human foreskin fibroblasts (HFFs), we examined how similar transcription was in HFFs and HAP1 cells. Of the 10 273 truQuant genes that were identified in HAP1 cells, 9712 overlapped with those found in HFF cells. In a small number of cases the dominant promoter driving each gene was different between the two cell types, likely due to differences in the specific transcription factor milieu. However, the promoter used in HAP1 cells was also found in HFFs and likely used similar GTFs to achieve initiation. As an example, the HAP1 promoter for SFXN3 was not the main promoter in HFFs, but the fine detail of the spread of PRO-Seq 5′ ends (TSSs) across the region was highly reproducibly detected (Figure 6C). The distribution of TSSs around the main TSR for genes that were highly dependent on the four GTFs were very similar, as illustrated by four example genes (Figure 6D). Because of the high similarity in promoter usage between HAP1 and HFF cells we believe that comparing the two sets of data are justified. The high similarity between the two datasets also indicates that our use of HAP1 PRO-Seq 5′ ends for TSSs is appropriate because the HFF dataset was a PRO-Cap dataset that specifically enriches for actual start sites.
To examine the relationship of PIC occupancy to GTF dependency, heatmaps were generated from the positions of centers of 60–70 bp fragments from TBP DFF-ChIP-seq data collected from HFF cells (19). The heatmaps cover a 200 bp region flanking the MaxTSS of truQuant TSRs that are rank ordered by dependency for each factor determined in tagged GTF HAP1 cells (Figure 6E, top). Fragment centers around –7 result from PICs that are initiating around +1. The top 10% of TBP dependent TSRs have a relatively high concentration of PICs centered around –7 and in addition, PICs centered about 35 bp upstream of the MaxTSS that are initiating in the TBP-specific motif (Figure 6E). The amount of PIC fragment centers around –7 is higher for the most TBP-dependent TSRs, adding support to the finding that TBP dependent TRSs are some of the most highly transcribed TSRs (Figure 4A). The bottom 90% of TBP dependent TSRs contain almost no TBP-PIC initiating in the –30 to –25 region, correlating with the absence of a TBP-binding motif. These bottom 90%, but not the top 10% of TBP dependent TSRs, also have another PIC centered downstream of the TSS such that for these PICs TBP would be positioned over the Inr and initiation would be at about +25 as seen in Figure 4A. The other factors displayed different relationships with the position of PICs. The patterns seen in the TAF1-, TFIIB- and XPB-dependent sorts are somewhat reversed from those seen from the TBP dependency sort. The upstream PIC over the TBP-binding motif and the most prevalent PICs are concentrated at the bottom of these heatmaps instead of the top as seen for TBP (Figure 6E, top) in support of the TSS strengths seen in Figure 4A and B.
To analyze the relationship between the position and occupancy of the +1 nucleosome with GTF dependency, heatmaps of H3K4me3 DFF-ChIP from HFF cells were utilized in a similar manner. Centers of 140–160 bp fragments resulting from DFF digestion (19) were mapped to a 600 bp region centered on the MaxTSS of truQuant TSRs determined in HAP1 cells. A nucleosome-depleted region (NDR) is evident in these maps, in addition to a well positioned +1 nucleosome and a more diffuse –1 nucleosome flanking the MaxTSS (Figure 6E, bottom) as expected for active promoters (54). The top 10% of truQuant TBP-dependent TSRs have lower +1 nucleosome occupancy (Figure 6E, bottom) as found in recent studies (55,56). Interestingly, the exact position of the +1 nucleosome was dependent on the factor sort. The +1 nucleosome was closer to the MaxTSS for the least TBP-dependent promoters. This can be seen by comparing the nucleosome occupancy to the average +1 nucleosome position (yellow line) (Figure 6E). TAF1-dependency was the opposite with the +1 nucleosome closer to the MaxTSS for the most dependent promoters.
Evidence for a direct TBP/nucleosome interaction
The TBP-TFIIA subcomplex is capable of binding to TATA box sequences in nucleosomal DNA in vitro (57) and a cryo-EM structure of TBP-TFIIA-nucleosome was recently resolved (22) (Figure 6F). To investigate if this chromatin feature could be identified in cells, fragMaps from TBP DFF-ChIP data from HFF cells (19) were created. This approach allows the visualization of the distribution of DNA fragments of various lengths (y-axis) protected from DFF at each position ± 350 bp surrounding a MaxTSS (x-axis). The maximum average value in the window sets the darkest value in the heatmap. A total of 11 561 truQuant TSRs were identified in HFF cells from PRO-Cap data (43). Two fragMaps were created, for truQuant TSRs containing a TBP-motif in the –36 to –19 region upstream of the MaxTSS (n = 308) and for the truQuant TSRs without the TBP motif (n = 11,253). The two common features most readily apparent are the PIC (∼75 bp fragments) and the PIC/+1 nucleosome (∼230 bp fragments) (Figure 6F). Interestingly, unique to the TBP-motif containing truQuant TSRs are TBP containing ∼45 bp fragments and ∼160–170 bp fragments containing TBP and a nucleosome (Figure 6F). SLNF11, NRIP3 and GADD45G are examples of TBP-motif containing genes with some of the highest TBP/nucleosome feature counts (Figure 6F). Of the 9253 TRSs that overlap between the HFF and HAP1 data sets, the 253 TSRs that contain a TBP binding motif are significantly more TBP-dependent than the rest (Supplementary Figure S6C; P < 0.0001, Mann–Whitney test).
Having identified four TBP containing features, we were interested in determining how they partitioned across the 308 TSRs containing the TBP motif. This was accomplished by quantifying the amount of each feature for each TSR using the feature parameters specified in Supplementary Figure S6D and calculating the Pearson's correlation coefficient for each feature with all others and with the sum of all TBP containing complexes (Supplementary Figure S6D). As anticipated, the TBP feature had a moderate positive correlation (r = 0.5) with the PIC. The TBP feature also has a moderate positive correlation (r = 0.49) with the TBP/nucleosome. However, almost no correlation (r = 0.08) between PIC and TBP/nucleosome was found. That is, at TBP motif containing loci, TBP will ultimately associate with either a PIC or with a nucleosome, but not with both. Because the size of the TBP/nucleosome is the same as a Pol II abutted to a nucleosome, we verified that the TBP/nucleosome feature was absent in a Pol II (Ser5P) fragMap (19) for the same 308 truQuant TSRs and for the rest of the TSRs (Supplementary Figure S6E). Altogether, these data show that TBP motif containing truQuant TSRs can exhibit a TBP/nucleosome feature and this complex cannot co-exist with PIC to any significant degree (Supplementary Figure S6F).
Ribosomal protein genes (RPGs)
Finally, we turned our attention to the 77 RPGs that are generally highly expressed and have a special initiator that lacks purines. The average base distribution around the MaxTSS for the RPGs is dominated by the RPG initiator from –4 to +5 (Supplementary Figure S6G). Interestingly, there is a G rich region from +20 to +35 reminiscent of the TAF1 dependent promoters (see Figure 5B) except for those promoters the region starts farther upstream at about +5. A little less than half of the RPG promoters have a TA rich region from –32 to –25 that might interact with TBP although two studies in Drosophila (58,59) have concluded that RPGs utilize the TBP like protein TRF2 (TBPL1 in humans) instead of TBP. The human RPGs have a wide variation in TBP-dependency from 1 to 8, but otherwise fairly normal and uniform dependencies on TFIIB, TAF1 and XPB (avg. dependencies of 5.3, 1.8 and 1.4 respectively) (Supplementary Figure S6H). Because of the TBP dependency of the human RPGs, fragMaps were generated from the 2000 bp region surrounding the MaxTSS of the 77 RPGs in HFFs (Supplementary Figure S6I). Pol II (F12 antibody to the large Pol II subunit) and Pol II Ser5P DFF-ChIP data clearly demonstrated that RPG promoters have the standard paused Pol II (free pause ∼50 bp fragments and nucleosome abutted ∼180 bp fragments) as well as PICs (∼70 bp fragments) and nucleosome abutted PICs (∼230 bp fragments). TBP DFF-ChIP confirmed that PICs were prominent features with some TBP/nuc complexes (∼175 bp fragments) and TBP upstream protection (∼45 bp fragments). H3K4me3 DFF-ChIP revealed highly positioned +1 nucleosomes. The relatively high TBP dependency and the dominant TBP containing PIC over the RPG promoters suggests that TBP plays a major role in expression of human RPGs. However, we cannot rule out a function for TRF2.
DISCUSSION
In this study, we report the effects of selective, acute depletion of TBP, TAF1, TFIIB and XPB on transcription in human cells. Depletions were robust, with less than 10% of the factors remaining after a 2 h treatment. Effects were quantified for the All TSRs promoter set (n = 72 095), which includes major and minor promoters driving active genes in the sense and divergent direction and active enhancers, and for the truQuant TSRs set, which includes the major promoter for each active gene (n = 10 273). Initiation was quantified using PRO-Seq 5′ end data from promoter-proximally paused Pol II. Gene body transcription for the truQuant set was quantified using PRO-Seq 3′ ends downstream of the main pause region up to the main CPS after blocklisting other promoters in that region. Remarkably, for both promoter sets only TFIIB depletion caused an average ∼10-fold reduction on initiation by Pol II equivalent to the level of factor depletion. The average dependencies for TBP, TAF1, and XPB were only 1.6-, 2.0- and 1.7-fold respectively for the larger All TSRs set. Though the average dependency for these three factors was relatively modest across both promoter sets, values for individual promoters varied over a broad range, particularly for TBP.
TFIIB had the highest dependency even though only about half of it was found associated with chromatin, which could have lessened its dependence as the depleted factor was replaced from the free pool. TFIIB is unique among the GTFs because it interacts Pol II near the active site (60). It is not only required for Pol II transcription in vitro with the full GTF set, it is also required for transcription on pre-melted TATA box templates with only pure Pol II and TBP (61). Point mutations in TFIIB have been reported which affect TSS selection (62) and the efficiency of early elongation (63). This last point is particularly interesting since the most TFIIB-dependent promoters have a distinctive initially transcribed region where the nascent hybrid would be especially stable due to high GC content (Figure 5B). Alternatively, the initially transcribed region for the most TFIIB dependent promoters has a significant enrichment for C residues. Because CTP is the limiting NTP in the cells (64), transcription would be slower which would favor abortive initiation. It is possible that TFIIB helps to overcome this abortive initiation by stabilizing the short hybrid. Interestingly, TFIIB dependence had an inverse correlation with the amount of PIC assembled (Figure 6E).
Both TBP and TAF1 are key constituents of TFIID which is generally thought to be recruited to all Pol II promoters as the first step in the formation of a PIC. We found that only a small fraction of promoters (<10%) were highly TBP dependent. Those promoters are distinguished by the presence of a TBP-specific motif about 30 bp upstream of the TSS. One potential explanation for the low number of TBP dependent genes is the existence of two TBP related factors TRF2 (TBPL1) and TRF3 (TBPL2) that might substitute for TBP. However, TRF2 functions primarily in germ line cells and is not required for embryonic development (65,66). TRF2 does not contain the crucial residues used by TPB to recognize the TATA element and cannot substitute for TBP in TFIID (67). We cannot rule out a significant function for TRF2 in HAP1 cells, but it should not be considered a potential substitute for TBP. The TRF3 gene is not expressed in HAP1 cells based on our PRO-Seq data. The footprint of TFIID on promoter DNA in vitro (4), the general sequence signature of Pol II promoters (18), and detailed functional analyses of several TATA-less promoters all emphasize that promoter recognition likely involves important contacts downstream of the TSS, particularly from about +25 to +35. Structural work reinforces earlier observations that TAF1 is primarily responsible for recognition of these G-rich downstream elements (DSEs). Consistent with this, we find that the most TAF1 dependent promoters are enriched in G residues from the TSS to ∼+35, which includes the previously described DSEs (Figure 5B). Recent studies demonstrate that PICs assembled in vitro with TFIID on fully TATA-less promoters contain TBP interacting with DNA about 30 bp upstream of the expected TSS (3). This agrees with the idea that TBP is generally present in PICs even in the absence of any high-affinity TBP binding site.
Dependency on XPB was subtle, but XPB depletion did have a negative effect on almost all Pol II promoters. The role of the ATPase activity of XPB in Pol II transcription has been difficult to resolve. The covalent inhibitor triptolide blocks Pol II initiation in vitro (68) and in vivo if added at high enough concentration for extended periods (69). However, depletion of XPB in cells did not affect Pol II transcription and also eliminated the negative effect of triptolide (10). A recent study demonstrated that template DNA within a complete Pol II PIC is already partly distorted, poised to become fully melted (70). Thus, while XPB in PICs may not be generally required for initiation, covalently blocking its ATPase activity could ultimately interfere with template melting or promoter clearance. The promoters most dependent on XPB have a distinctive sequence composition, GC rich around –30 and G-poor over the DSEs, indicating that these promoters should be the least likely to support the assembly of a stable PIC (Figure 5B). This was exactly what was found when looking at PIC heatmaps in which XPB dependence was inversely correlated with PIC levels (Figure 6E). This suggests that XPB, like TFIIB, is most important when a stable PIC is least likely to assemble.
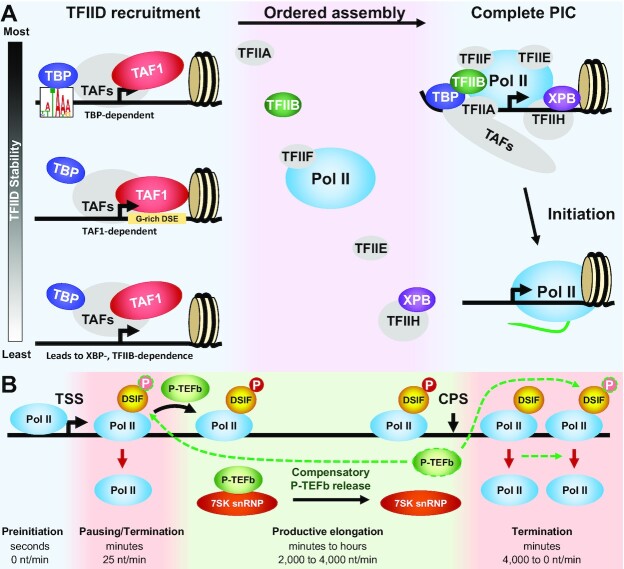
As shown diagrammatically on the left in Figure 7A, two very broad promoter classes can be envisioned which primarily rely either on TBP-template interactions or TAF-template interactions. Only TBP dependent promoters also support a sub-PIC complex centered on the TBP-specific motif (Figure 6F, S6E), emphasizing how central TBP-template interactions are for this promoter class. TBP dependent promoters are less likely to have directly abutted +1 nucleosomes, while such contact is more likely for the TAF1-dependent promoters (Figures 5C, 6E). This is consistent with the possibility that PIC assembly could be facilitated by TAF interactions with adjacent H3K4me3-modified nucleosomes. Our TBP DFF-ChIP analysis also revealed that the TBP/nucleosome is a common feature in loci with TBP-specific motifs and high levels of TBP (Figure 6E, S6E). A direct interaction of TBP with a nucleosome has only previously been demonstrated in vitro (22,57). The TBP/nucleosome is incompatible with conventional PIC formation. The ability of TBP to interact directly with the nucleosome may partially explain why TBP-specific motif containing promoters are driven by TFIID in vivo even though transcription of such promoters in vitro requires only TBP. The downstream TAF-DNA interactions could prevent inactivation of the promoter by nucleosome encroachment to interact with TBP.
Figure 7.
Effects of GTF depletion on initiation and elongation. (A) Phases of Pol II initiation. TFIID is recruited by core promoter elements and/or a positioned +1 nucleosome and through interactions with other TFs. TBP-dependent promoters have the most stable recruitment due to TBP interaction with its binding motif which is likely quickly stabilized by TFIIA. TAF1-dependency relies in part on TAF1 interaction with a G-rich downstream element and a positioned +1 nucleosome. XBP- and TFIIB-dependence occurs on the least stable interactions that require rapid initiation before loss of TFIID. After TFIID recruitment, the other GTFs associate to complete the PIC which then initiates. (B) RNA polymerase II elongation control with a depiction of the effect of compensatory P-TEFb release from the 7SK snRNP. Lifetimes and elongation rates for the four phases of the transcription cycle are indicated. P-TEFb dependent phosphorylation of DSIF normally occurs during the transition into productive elongation and is reversed downstream of the CPS. Termination occurs during promoter-proximal pausing and downstream of the CPS. Reduction of productive elongation leads to compensatory release of P-TEFb from the snRNP which we hypothesize leads to enhanced phosphorylation of DSIF to both increase gene body transcription and to delay termination downstream of the CPS. Green dashed outline and arrows indicate activity of released P-TEFb.
Since the large majority of promoters lack TBP-specific motifs, PIC assembly at these promoters should rely on the overall interaction of TFIID with DNA, particularly TAF1 with the DSEs. However, dependency on TAF1 is typically only about 2-fold. In this context it is important to note earlier observations that TFIID-promoter interactions can be independent of both TBP and TAF1. Tora and colleagues have shown that a subcomplex of TFIID lacking both TBP and TAF1 can be purified and crucially can support transcription in vitro, even on a TATA promoter (23). More recent studies reported that a stable TFIID core depends particularly on TAF4 and not on TBP or TAF1. This is particularly relevant because TAF4 can provide specific promoter contacts upstream of the TSS (6). PIC assembly in the absence of either TBP or TAF1 might therefore be supported by subcomplexes of TFIID. As noted above, promoters that are most dependent on either TFIIB or XPB seem to lack the canonical TFIID promoter elements (Figure 5B). For these promoters, factors bound upstream of ∼-40 could interact with Mediator to directly recruit Pol II. Pol II in turn can interact with TFIIH and TFIIB, providing the necessary minimal machinery to support transcript initiation in the absence of both TBP and TAF1.
Comparison of our results with previous studies in which GTFs were knocked down illuminates some similarities and discrepancies. Fant et al. (71) rapidly depleted TAF1 using TRIM-Away in HCT116 cells and reported PRO-Seq data demonstrating that paused transcripts increased globally while gene body transcripts were relatively constant. This finding along with in vitro assays led to the conclusion that TFIID is required for pausing. These results are not consistent with our observations that TBP or TAF1 depletion leads to a decrease in paused Pol II due to inhibition of initiation and not to a downstream displacement of pause locations (Figure 1C). The basis for this discrepancy is not clear, but it is worth noting that the dataset normalization in Fant et al. (71) did not rely on spike-ins and the depletion of TAF1 in that study also caused an even higher level of depletion of TAF2. Another study depleted TFIIB from HeLa cells with siRNA. Based on microarray analysis, these authors found that TFIIB was dispensable for transcription of many human promoters but was essential for herpes simplex virus-1 gene transcription and replication (72). Most cells survived the 80 h treatment with siRNA. The relative distributions of mRNA levels were not dramatically affected in that study but effects on the absolute level of mRNAs were not determined. TBP knockout in mice was lethal at the embryonic blastocyst stage, but prior to that stage Pol I and Pol III transcription was dramatically reduced while Pol II transcription remained robust as determined by nuclear run on assays (73). This is similar to our finding with Pol II. We did not observe strong reduction in Pol I and some Pol III transcription but we would stress that our knockdowns lasted only 2 h. Teves et al. (74) found that depletion of TBP during mitosis using a drug induced degron system (6 h treatment) showed only small effects on Pol II as measured by ChRO-Seq in asynchronous mESCs. As we also found, there were no changes observed for Pol I, but Pol III transcription was dramatically affected. Petrenko et al. (75) used anchor away to deplete many GTFs including TBP, TFIIB and TAF1 in budding yeast and concluded based on ChIP-Seq results that all of these depletions broadly affected transcription. That study was focused on PIC formation and not transcription and used a method that can lead to loss of function of entire complexes such as TFIID even though only one subunit is targeted (76). We note that direct comparisons of transcription factor function between yeast and mammals should be interpreted with caution.
We find that after digesting nuclei with DFF, PICs can be readily recovered with an antibody to TBP (Figure 6F, S6E) (19). This is somewhat surprising, since a complete PIC should immediately support initiation and therefore PICs would to be expected to be transient and difficult to detect. In the conventional PIC assembly pathway, TFIIH is the last GTF to associate, suggesting the possibility that complexes with PIC dimensions recovered with a TBP antibody could have resulted from incomplete TFIIH loading. However, PICs are also effectively recovered with an antibody to the Pol II CTD phosphorylated at Ser5 (19). Such complexes must have contained Cdk7 and presumably also XPB. In the final steps of in vitro PIC assembly, the downstream template associations of the TAFs are displaced by XPB (Figure 1A). We speculate that nascent PICs could be trapped in a transcriptionally inactive state because of failure to complete this final stage of PIC assembly.
Supporting the idea that at least some PICs are relatively stably bound, we found that depletion of TBP led to an increase in TSRs that are nearby highly TBP-dependent TSRs driven either by Pol II or Pol III (Figure 3). Presumably, the loss of TBP destabilizes the PIC and allows the transcriptional machinery to utilize other TSRs that were buried under the TBP containing complexes over the TBP-dependent TSR. These complexes can include nucleosomes that are associated with the PIC or directly with TBP (19). While reduction of TBP-dependent PICs can be easily imagined to occur after depletion of TBP, the loss of neighboring nucleosomes downstream for Pol II or upstream for Pol III must be more complicated. The simplest explanation is that Pol II and Pol III PICs stabilize the nearby nucleosomes which are usually close packed (19) unlike in bulk chromatin. Loss of the PIC’s influence on stability could allow remodeling and access of the nearby TSRs to the transcription machinery.
Sequence-specific TFs play critical roles in promoting transcription initiation by recruitment of the PIC, chromatin remodelers, and coactivators (77,78). Ubiquitously expressed TFs include the well-studied SP1, YY1, ETS, NRF1 and NF-Y which have been found in promoter regions (79). Binding sites for these factors were enriched near the TSSs for a substantial fraction of the promoters in the All TSRs set (Figure 6A). The SP1, NRF1 and NF-Y sites were found primarily upstream of TSSs. These sites showed a 10-bp periodic spacing over some portion of the upstream region, consistent with the possibility that particular orientations of the factor along the face of DNA favor interactions with the core transcriptional machinery. The NF-Y sites in particular were primarily located upstream of the usual position of TBP-specific motifs, consistent with the fact that NF-Y sites are enriched for the most TBP-dependent promoters (Figure 6B). YY1 sites are most likely to occur at or just downstream of the TSS, a strikingly different location from the other factor sites. This suggests that YY1 is not functioning as a stimulatory factor for conventional PIC assembly but instead as a direct participant in the transcription complex. That possibility is consistent with earlier reports that YY1 is able to drive transcription in vitro from a supercoiled template in the absence of TBP and TFIIH (80). Furthermore, TFIIB relieves YY1 transcriptional repression of vitamin D receptor-mediated transcriptional response (81). YY1 motifs are correlated with TSR strength but anti-correlated with dependence on the GTFs, particularly TFIIB (Figure 6D). That is, YY1 sites are most often found at/near the TSSs of relatively strong promoters that nevertheless lack other motifs that could direct transcript initiation. YY1-dependent promoters could represent one class of promoters that do not function primarily through assembly of a conventional PIC.
One of the major findings of our study was the blunted effect of GTF depletion on Pol II transcription of gene bodies. The decrease in promoter-proximally paused transcripts caused by reduced efficiency of initiation was much greater than that seen in gene bodies (Figure 4D). This likely resulted from higher levels of P-TEFb activity, because any reduction in productive elongation in cells leads to release of P-TEFb from the 7SK snRNP (31), presumably to compensate for the decrease (Figure 7B). In turn this leads to an additional decrease in the paused Pol II, an effect most easily seen for the genes with the lowest initial level of productive elongation (Supplementary Figure S4B). It is likely that the increased dependency calculated from reduction of paused Pol II due to increased P-TEFb was relatively minor because on average only 10% of paused Pol II complexes enter productive elongation and the rest terminate with a half-life of seconds to minutes (69,82,83). A similar compensatory effect by P-TEFb on pause release was recently hypothesized following depletion of the Med 14 Mediator subunit (84).
The dramatic alteration in behavior of Pol II downstream of the CPS seen after depletion of TFIIB and to a lesser extent TAF1 (Figure 4F, Supplementary Figure S4A) may also be explained by the increase in active P-TEFb. TFIIB depletion led to an increase in the level of nascent transcripts downstream of the CPS and a dramatic increase in the average distance of Pol II downstream from the CPS. This is likely caused by a reduction in termination efficiency. In yeast, TFIIB has been implicated in Pol II termination downstream of CPSs and in linking the 5′ and 3′ ends of genes (85,86). Additionally, TFIIB interaction with Pol II blocks the RNA exit channel (87), suggesting a possible direct effect on termination. In yeast and mammals, TFIIB has been found to localize at 5′ and 3′ ends of genes where it interacts with cleavage and polyadenylation factors (88–91). However, the Fisher lab has reported that P-TEFb can regulate the exonuclease activity of Xrn2 (92) which drives termination. Recent studies have uncovered a mechanism in which regulation of DSIF phosphorylation by P-TEFb and the opposing function of phosphatases PP1 and perhaps PP4 are involved in the termination process in yeast (93) and mammals (94). Based on these last observations, we hypothesize that the effect of TFIIB depletion on termination is due to increased P-TEFb activity triggered in compensation for reduced gene body transcription. In support of this idea, rampant runaway transcription downstream of CPSs has been reported as a consequence of knockdown of 7SK leading to excess P-TEFb activity in cells (95,96). Additional support for this model can be seen in Figure 4, where depletion of TAF1 leads to both release of P-TEFb from the 7SK snRNP and a termination defect similar to but more modest than the defect caused by depletion of TFIIB. The fact that depletion of TFIIB and TAF1 have effects on termination at 3′ end of genes that are proportional to their effects on initiation favors a common mechanism involving P-TEFb rather than on the specific properties of the individual GTFs.
TBP is also required for transcription driven by RNA polymerases I and III (47). We did not detect major changes in transcription of preribosomal 45S rDNA loci (average dependency of 1.5) as a result of depletion of TBP. This could be due to much more stable binding of TBP to the Pol I promoter (55). Dominant TBP containing features directly upstream of the 45S rRNA TSS and at other sites upstream and downstream of the 45S gene have been visualized with DFF-ChIP (49). Among the Pol III transcription units, only tRNA genes were severely affected upon reduction of TBP levels (average dependency ∼10). Other Pol III targets including the snRNA genes U6, 7SK and 7SL were unperturbed following TBP depletion (average dependency of 1.1). Neither Pol I nor Pol III activity had any significant dependency on the other factors. The type II class tRNA promoters contain a TATA-like sequence upstream of the TSS while the U6 and 7SK type III promoters contain both a TATA-like sequence and upstream proximal (PSE) and distal (DSE) sequence elements (97). These additional regulatory sequences for the type III promoters are recognized by multiprotein complexes that may support effective Pol III recruitment and transcription initiation (98) even when TBP levels are greatly reduced. The U1–U5, U11 and U12 snRNA genes which are transcribed by Pol II were strongly dependent on TBP and TFIIB but not TAF1. This agrees with previous results showing that a different TBP-TAF complex that does not include TAF1 is formed at these loci (48).
Results reported here with GTF knockdown and DFF-ChIP suggest that conventional models of assembly of the Pol II transcriptional machinery at promoters are over-simplified. Loss of TBP has little effect outside of TBP-specific motif containing promoters and loss TAF1 causes surprisingly modest reduction of transcription at promoters lacking the non-TBP-specific motif. The roles of subcomplexes of TFIID and the +1 nucleosome in PIC assembly should be considered, as well as the possibility that Pol II recruitment by upstream factors provides promoter function at some locations in the absence of canonical consensus elements near the TSS. It will be important to determine the full repertoire of mechanisms that control assembly and stability of functional Pol II transcription complexes at promoters.
DATA AVAILABILITY
Raw and processed PRO-Seq data for this manuscript can be obtained from GEO GSE194153 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE194153). Previously published PRO-Cap and DFF-Seq datasets of HFFs can also be obtained from GEO with identifications GSE113394 and GSE185763, respectively.
Supplementary Material
Contributor Information
Juan F Santana, Department of Biochemistry and Molecular Biology, The University of Iowa, Iowa City, IA 52242, USA.
Geoffrey S Collins, Department of Biochemistry and Molecular Biology, The University of Iowa, Iowa City, IA 52242, USA.
Mrutyunjaya Parida, Department of Biochemistry and Molecular Biology, The University of Iowa, Iowa City, IA 52242, USA.
Donal S Luse, Department of Cardiovascular and Metabolic Sciences, Lerner Research Institute, Cleveland Clinic, Cleveland, OH 44195, USA.
David H Price, Department of Biochemistry and Molecular Biology, The University of Iowa, Iowa City, IA 52242, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of General Medical Sciences [GM126908 to D.H.P. and GM121428 to D.S.L.]. Funding for open access charge: NIH [GM121428 and GM126908].
Conflict of interest statement. None declared.
REFERENCES
- 1. Luse D.S. The RNA polymerase II preinitiation complex. Through what pathway is the complex assembled?. Transcription. 2014; 5:e27050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Burley S.K., Roeder R.G.. Biochemistry and structural biology of transcription factor IID (TFIID). Annu. Rev. Biochem. 1996; 65:769–799. [DOI] [PubMed] [Google Scholar]
- 3. Chen X., Qi Y., Wu Z., Wang X., Li J., Zhao D., Hou H., Li Y., Yu Z., Liu W.et al.. Structural insights into preinitiation complex assembly on core promoters. Science. 2021; 372: [DOI] [PubMed] [Google Scholar]
- 4. Louder R.K., He Y., Lopez-Blanco J.R., Fang J., Chacon P., Nogales E.. Structure of promoter-bound TFIID and model of human pre-initiation complex assembly. Nature. 2016; 531:604–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Weinzierl R.O., Dynlacht B.D., Tjian R.. Largest subunit of drosophila transcription factor IID directs assembly of a complex containing TBP and a coactivator. Nature. 1993; 362:511–517. [DOI] [PubMed] [Google Scholar]
- 6. Patel A.B., Louder R.K., Greber B.J., Grunberg S., Luo J., Fang J., Liu Y., Ranish J., Hahn S., Nogales E.. Structure of human TFIID and mechanism of TBP loading onto promoter DNA. Science. 2018; 362:aau8872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lagrange T., Kapanidis A.N., Tang H., Reinberg D., Ebright R.H.. New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB. Genes. Dev. 1998; 12:34–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Deng W., Roberts S.G.. A core promoter element downstream of the TATA box that is recognized by TFIIB. Genes. Dev. 2005; 19:2418–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tirode F., Busso D., Coin F., Egly J.M.. Reconstitution of the transcription factor TFIIH: assignment of functions for the three enzymatic subunits, XPB, XPD, and cdk7. Mol. Cell. 1999; 3:87–95. [DOI] [PubMed] [Google Scholar]
- 10. Alekseev S., Nagy Z., Sandoz J., Weiss A., Egly J.M., Le May N., Coin F.. Transcription without XPB establishes a unified helicase-independent mechanism of promoter opening in eukaryotic gene expression. Mol Cell. 2017; 65:504–514. [DOI] [PubMed] [Google Scholar]
- 11. Yella V.R., Bansal M.. DNA structural features of eukaryotic TATA-containing and TATA-less promoters. FEBS Open Bio. 2017; 7:324–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gershenzon N.I., Ioshikhes I.P.. Synergy of human pol II core promoter elements revealed by statistical sequence analysis. Bioinformatics. 2005; 21:1295–1300. [DOI] [PubMed] [Google Scholar]
- 13. Kuras L., Kosa P., Mencia M., Struhl K.. TAF-Containing and TAF-independent forms of transcriptionally active TBP in vivo. Science. 2000; 288:1244–1248. [DOI] [PubMed] [Google Scholar]
- 14. Pugh B.F., Tjian R.. Transcription from a TATA-less promoter requires a multisubunit TFIID complex. Genes. Dev. 1991; 5:1935–1945. [DOI] [PubMed] [Google Scholar]
- 15. Petrenko N., Struhl K.. Comparison of transcriptional initiation by RNA polymerase II across eukaryotic species. Elife. 2021; 10:e67964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vo Ngoc L., Huang C.Y., Cassidy C.J., Medrano C., Kadonaga J.T.. Identification of the human DPR core promoter element using machine learning. Nature. 2020; 585:459–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kutach A.K., Kadonaga J.T.. The downstream promoter element DPE appears to be as widely used as the TATA box in Drosophila core promoters. Mol. Cell Biol. 2000; 20:4754–4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Luse D.S., Parida M., Spector B.M., Nilson K.A., Price D.H.. A unified view of the sequence and functional organization of the human RNA polymerase II promoter. Nucleic Acids Res. 2020; 48:7767–7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Spector B.M., Parida M., Li M., Ball C.B., Meier J.L., Luse D.S., Price D.H.. Differences in RNA polymerase II complexes and their interactions with surrounding chromatin on human and cytomegalovirus genomes. Nat. Commun. 2022; 13:2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cianfrocco M.A., Kassavetis G.A., Grob P., Fang J., Juven-Gershon T., Kadonaga J.T., Nogales E.. Human TFIID binds to core promoter DNA in a reorganized structural state. Cell. 2013; 152:120–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Patel A.B., Greber B.J., Nogales E.. Recent insights into the structure of TFIID, its assembly, and its binding to core promoter. Curr. Opin. Struct. Biol. 2020; 61:17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang H., Xiong L., Cramer P.. Structures and implications of TBP-nucleosome complexes. Proc. Natl. Acad. Sci. U.S.A. 2021; 118:e2108859118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wieczorek E., Brand M., Jacq X., Tora L.. Function of TAF(II)-containing complex without TBP in transcription by RNA polymerase iI. Nature. 1998; 393:187–191. [DOI] [PubMed] [Google Scholar]
- 24. Wright K.J., Marr M.T., Tjian R.. TAF4 nucleates a core subcomplex of TFIID and mediates activated transcription from a TATA-less promoter. Proc. Natl. Acad. Sci. U. S. A. 2006; 103:12347–12352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Basehoar A.D., Zanton S.J., Pugh B.F.. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004; 116:699–709. [DOI] [PubMed] [Google Scholar]
- 26. Fischer V., Schumacher K., Tora L., Devys D. Global role for coactivator complexes in RNA polymerase II transcription. Transcription. 2019; 10:29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. van Nuland R., Schram A.W., van Schaik F.M., Jansen P.W., Vermeulen M., Marc Timmers H.T.. Multivalent engagement of TFIID to nucleosomes. PLoS One. 2013; 8:e73495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lauberth S.M., Nakayama T., Wu X., Ferris A.L., Tang Z., Hughes S.H., Roeder R.G.. H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell. 2013; 152:1021–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dollinger R., Gilmour D.S.. Regulation of promoter proximal pausing of RNA polymerase II in metazoans. J. Mol. Biol. 2021; 433:166897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Peterlin B.M., Price D.H.. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell. 2006; 23:297–305. [DOI] [PubMed] [Google Scholar]
- 31. Peterlin B.M., Brogie J.E., Price D.H.. 7SK snRNA: a noncoding RNA that plays a major role in regulating eukaryotic transcription. Wiley Interdiscip. Rev. RNA. 2012; 3:92–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nabet B., Ferguson F.M., Seong B.K.A., Kuljanin M., Leggett A.L., Mohardt M.L., Robichaud A., Conway A.S., Buckley D.L., Mancias J.D.et al.. Rapid and direct control of target protein levels with VHL-recruiting dTAG molecules. Nat. Commun. 2020; 11:4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ball C.B., Nilson K.A., Price D.H.. Use of the nuclear walk-on methodology to determine sites of RNA polymerase II initiation and pausing and quantify nascent RNAs in cells. Methods. 2019; 159-160:165–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nilson K.A., Lawson C.K., Mullen N.J., Ball C.B., Spector B.M., Meier J.L., Price D.H.. Oxidative stress rapidly stabilizes promoter-proximal paused pol II across the human genome. Nucleic Acids Res. 2017; 45:11088–11105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sluder A.E., Price D.H., Greenleaf A.L.. An activity necessary for in vitro transcription is a DNase inhibitor. Biochimie. 1987; 69:1199–1205. [DOI] [PubMed] [Google Scholar]
- 36. Ball C.B., Parida M., Santana J.F., Spector B.M., Suarez G.A., Price D.H.. Nuclear export restricts gdown1 to a mitotic function. Nucleic Acids Res. 2022; 50:1908–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li M., Ball C.B., Collins G., Hu Q., Luse D.S., Price D.H., Meier J.L.. Human cytomegalovirus IE2 drives transcription initiation from a select subset of late infection viral promoters by host RNA polymerase II. PLoS Pathog. 2020; 16:e1008402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bailey T.L., Elkan C.. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell Syst. Mol. Biol. 1994; 2:28–36. [PubMed] [Google Scholar]
- 41. Bailey T.L., Machanick P.. Inferring direct DNA binding from chip-seq. Nucleic Acids Res. 2012; 40:e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Crooks G.E., Hon G., Chandonia J.M., Brenner S.E.. WebLogo: a sequence logo generator. Genome Res. 2004; 14:1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Parida M., Nilson K.A., Li M., Ball C.B., Fuchs H.A., Lawson C.K., Luse D.S., Meier J.L., Price D.H.. Nucleotide resolution comparison of transcription of human cytomegalovirus and host genomes reveals universal use of RNA polymerase II elongation control driven by dissimilar core promoter elements. Mbio. 2019; 10:e02047-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Anandapadamanaban M., Andresen C., Helander S., Ohyama Y., Siponen M.I., Lundstrom P., Kokubo T., Ikura M., Moche M., Sunnerhagen M.. High-resolution structure of TBP with TAF1 reveals anchoring patterns in transcriptional regulation. Nat. Struct. Mol. Biol. 2013; 20:1008–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kwak H., Fuda N.J., Core L.J., Lis J.T.. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science. 2013; 339:950–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shi W., Zhou W.. Frequency distribution of TATA box and extension sequences on human promoters. BMC Bioinformatics. 2006; 7(Suppl. 4):S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cormack B.P., Struhl K.. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell. 1992; 69:685–696. [DOI] [PubMed] [Google Scholar]
- 48. Zaborowska J., Taylor A., Roeder R.G., Murphy S.. A novel TBP-TAF complex on RNA polymerase II-transcribed snRNA genes. Transcription. 2012; 3:92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ball C.B., Parida M., Li M., Spector B.M., Suarez G.A., Meier J.L., Price D.H.. Human cytomegalovirus infection elicits global changes in host transcription by RNA polymerases I, II, and III. Viruses. 2022; 14:779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Marshall N.F., Price D.H.. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J Biol Chem. 1995; 270:12335–12338. [DOI] [PubMed] [Google Scholar]
- 51. Biglione S., Byers S.A., Price J.P., Nguyen V.T., Bensaude O., Price D.H., Maury W.. Inhibition of HIV-1 replication by P-TEFb inhibitors DRB, seliciclib and flavopiridol correlates with release of free P-TEFb from the large, inactive form of the complex. Retrovirology. 2007; 4:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mayer A., di Iulio J., Maleri S., Eser U., Vierstra J., Reynolds A., Sandstrom R., Stamatoyannopoulos J.A., Churchman L.S.. Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell. 2015; 161:541–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ball C.B., Li M., Parida M., Hu Q., Ince D., Collins G.S., Meier J.L., Price D.H.. Human cytomegalovirus IE2 both activates and represses initiation and modulates elongation in a context-dependent manner. Mbio. 2022; e0033722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Scruggs B.S., Gilchrist D.A., Nechaev S., Muse G.W., Burkholder A., Fargo D.C., Adelman K.. Bidirectional transcription arises from two distinct hubs of transcription factor binding and active chromatin. Mol Cell. 2015; 58:1101–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hasegawa Y., Struhl K.. Promoter-specific dynamics of TATA-binding protein association with the human genome. Genome Res. 2019; 29:1939–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ravarani C.N., Chalancon G., Breker M., de Groot N.S., Babu M.M.. Affinity and competition for TBP are molecular determinants of gene expression noise. Nat. Commun. 2016; 7:10417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Godde J.S., Nakatani Y., Wolffe A.P.. The amino-terminal tails of the core histones and the translational position of the TATA box determine TBP/TFIIA association with nucleosomal DNA. Nucleic Acids Res. 1995; 23:4557–4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang Y.L., Duttke S.H., Chen K., Johnston J., Kassavetis G.A., Zeitlinger J., Kadonaga J.T.. TRF2, but not TBP, mediates the transcription of ribosomal protein genes. Genes Dev. 2014; 28:1550–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Baumann D.G., Gilmour D.S.. A sequence-specific core promoter-binding transcription factor recruits TRF2 to coordinately transcribe ribosomal protein genes. Nucleic Acids Res. 2017; 45:10481–10491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sainsbury S., Niesser J., Cramer P.. Structure and function of the initially transcribing RNA polymerase II-TFIIB complex. Nature. 2013; 493:437–440. [DOI] [PubMed] [Google Scholar]
- 61. Pan G., Greenblatt J.. Initiation of transcription by RNA polymerase II is limited by melting of the promoter DNA in the region immediately upstream of the initiation site. J. Biol. Chem. 1994; 269:30101–30104. [PubMed] [Google Scholar]
- 62. Fairley J.A., Evans R., Hawkes N.A., Roberts S.G.. Core promoter-dependent TFIIB conformation and a role for TFIIB conformation in transcription start site selection. Mol. Cell. Biol. 2002; 22:6697–6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Pal M., Ponticelli A.S., Luse D.S.. The role of the transcription bubble and TFIIB in promoter clearance by RNA polymerase iI. Mol. Cell. 2005; 19:101–110. [DOI] [PubMed] [Google Scholar]
- 64. Traut T.W. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 1994; 140:1–22. [DOI] [PubMed] [Google Scholar]
- 65. Martianov I., Fimia G.M., Dierich A., Parvinen M., Sassone-Corsi P., Davidson I.. Late arrest of spermiogenesis and germ cell apoptosis in mice lacking the TBP-like TLF/TRF2 gene. Mol. Cell. 2001; 7:509–515. [DOI] [PubMed] [Google Scholar]
- 66. Martianov I., Velt A., Davidson G., Choukrallah M.A., Davidson I.. TRF2 is recruited to the pre-initiation complex as a testis-specific subunit of TFIIA/ALF to promote haploid cell gene expression. Sci. Rep. 2016; 6:32069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Dantonel J.C., Wurtz J.M., Poch O., Moras D., Tora L.. The TBP-like factor: an alternative transcription factor in metazoa?. Trends Biochem. Sci. 1999; 24:335–339. [DOI] [PubMed] [Google Scholar]
- 68. Titov D.V., Gilman B., He Q.L., Bhat S., Low W.K., Dang Y., Smeaton M., Demain A.L., Miller P.S., Kugel J.F.et al.. XPB, a subunit of TFIIH, is a target of the natural product triptolide. Nat. Chem. Biol. 2011; 7:182–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Price D.H. Transient pausing by RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:4810–4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Dienemann C., Schwalb B., Schilbach S., Cramer P.. Promoter distortion and opening in the RNA polymerase II cleft. Mol. Cell. 2019; 73:97–106. [DOI] [PubMed] [Google Scholar]
- 71. Fant C.B., Levandowski C.B., Gupta K., Maas Z.L., Moir J., Rubin J.D., Sawyer A., Esbin M.N., Rimel J.K., Luyties O.et al.. TFIID enables RNA polymerase II promoter-proximal pausing. Mol. Cell. 2020; 78:785–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gelev V., Zabolotny J.M., Lange M., Hiromura M., Yoo S.W., Orlando J.S., Kushnir A., Horikoshi N., Paquet E., Bachvarov D.et al.. A new paradigm for transcription factor TFIIB functionality. Sci Rep. 2014; 4:3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Martianov I., Viville S., Davidson I.. RNA polymerase II transcription in murine cells lacking the TATA binding protein. Science. 2002; 298:1036–1039. [DOI] [PubMed] [Google Scholar]
- 74. Teves S.S., An L., Bhargava-Shah A., Xie L., Darzacq X., Tjian R.. A stable mode of bookmarking by TBP recruits RNA polymerase II to mitotic chromosomes. Elife. 2018; 7:e35621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Petrenko N., Jin Y., Dong L., Wong K.H., Struhl K.. Requirements for RNA polymerase II preinitiation complex formation in vivo. Elife. 2019; 8:e43654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Gallego O., Specht T., Brach T., Kumar A., Gavin A.C., Kaksonen M.. Detection and characterization of protein interactions in vivo by a simple live-cell imaging method. PLoS One. 2013; 8:e62195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Frontini M., Imbriano C., diSilvio A., Bell B., Bogni A., Romier C., Moras D., Tora L., Davidson I., Mantovani R.. NF-Y recruitment of TFIID, multiple interactions with histone fold TAF(II)s. J. Biol. Chem. 2002; 277:5841–5848. [DOI] [PubMed] [Google Scholar]
- 78. Borggrefe T., Yue X.. Interactions between subunits of the mediator complex with gene-specific transcription factors. Semin. Cell Dev. Biol. 2011; 22:759–768. [DOI] [PubMed] [Google Scholar]
- 79. Benner C., Konovalov S., Mackintosh C., Hutt K.R., Stunnenberg R., Garcia-Bassets I.. Decoding a signature-based model of transcription cofactor recruitment dictated by cardinal cis-regulatory elements in proximal promoter regions. PLoS Genet. 2013; 9:e1003906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Usheva A., Shenk T.. TATA-binding protein-independent initiation: YY1, TFIIB, and RNA polymerase II direct basal transcription on supercoiled template DNA. Cell. 1994; 76:1115–1121. [DOI] [PubMed] [Google Scholar]
- 81. Raval-Pandya M., Dhawan P., Barletta F., Christakos S.. YY1 represses vitamin D receptor-mediated 25-hydroxyvitamin D(3)24-hydroxylase transcription: relief of repression by CREB-binding protein. Mol. Endocrinol. 2001; 15:1035–1046. [DOI] [PubMed] [Google Scholar]
- 82. Steurer B., Janssens R.C., Geverts B., Geijer M.E., Wienholz F., Theil A.F., Chang J., Dealy S., Pothof J., van Cappellen W.A.et al.. Live-cell analysis of endogenous GFP-RPB1 uncovers rapid turnover of initiating and promoter-paused RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:E4368–E4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Shao R., Kumar B., Lidschreiber K., Lidschreiber M., Cramer P., Elsasser S.J.. Distinct transcription kinetics of pluripotent cell states. Mol. Syst. Biol. 2022; 18:e10407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Jaeger M.G., Schwalb B., Mackowiak S.D., Velychko T., Hanzl A., Imrichova H., Brand M., Agerer B., Chorn S., Nabet B.et al.. Selective mediator dependence of cell-type-specifying transcription. Nat. Genet. 2020; 52:719–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. O’Brien M.J., Ansari A.. Beyond the canonical role of TFIIB in eukaryotic transcription. Curr. Genet. 2022; 68:61–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Allepuz-Fuster P., O’Brien M.J., Gonzalez-Polo N., Pereira B., Dhoondia Z., Ansari A., Calvo O.. RNA polymerase II plays an active role in the formation of gene loops through the rpb4 subunit. Nucleic Acids Res. 2019; 47:8975–8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kostrewa D., Zeller M.E., Armache K.J., Seizl M., Leike K., Thomm M., Cramer P.. RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature. 2009; 462:323–330. [DOI] [PubMed] [Google Scholar]
- 88. Doris S.M., Chuang J., Viktorovskaya O., Murawska M., Spatt D., Churchman L.S., Winston F.. Spt6 is required for the fidelity of promoter selection. Mol. Cell. 2018; 72:687-699 e686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Singh B.N., Hampsey M.. A transcription-independent role for TFIIB in gene looping. Mol. Cell. 2007; 27:806–816. [DOI] [PubMed] [Google Scholar]
- 90. Wang Y., Fairley J.A., Roberts S.G.. Phosphorylation of TFIIB links transcription initiation and termination. Curr. Biol. 2010; 20:548–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Eaton J.D., Francis L., Davidson L., West S.. A unified allosteric/torpedo mechanism for transcriptional termination on human protein-coding genes. Genes Dev. 2020; 34:132–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Sanso M., Levin R.S., Lipp J.J., Wang V.Y., Greifenberg A.K., Quezada E.M., Ali A., Ghosh A., Larochelle S., Rana T.M.et al.. P-TEFb regulation of transcription termination factor xrn2 revealed by a chemical genetic screen for cdk9 substrates. Genes Dev. 2016; 30:117–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Parua P.K., Booth G.T., Sanso M., Benjamin B., Tanny J.C., Lis J.T., Fisher R.P.. A cdk9-pp1 switch regulates the elongation-termination transition of RNA polymerase II. Nature. 2018; 558:460–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Parua P.K., Kalan S., Benjamin B., Sanso M., Fisher R.P.. Distinct Cdk9-phosphatase switches act at the beginning and end of elongation by RNA polymerase II. Nat. Commun. 2020; 11:4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Castelo-Branco G., Amaral P.P., Engstrom P.G., Robson S.C., Marques S.C., Bertone P., Kouzarides T.. The non-coding snRNA 7SK controls transcriptional termination, poising, and bidirectionality in embryonic stem cells. Genome Biol. 2013; 14:R98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Guo J., Li T., Price D.H.. Runaway transcription. Genome Biol. 2013; 14:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Dieci G., Fiorino G., Castelnuovo M., Teichmann M., Pagano A.. The expanding RNA polymerase III transcriptome. Trends Genet. 2007; 23:614–622. [DOI] [PubMed] [Google Scholar]
- 98. Tatosyan K.A., Stasenko D.V., Koval A.P., Gogolevskaya I.K., Kramerov D.A.. TATA-like boxes in RNA polymerase III promoters: requirements for nucleotide sequences. Int. J. Mol. Sci. 2020; 21:3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed PRO-Seq data for this manuscript can be obtained from GEO GSE194153 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE194153). Previously published PRO-Cap and DFF-Seq datasets of HFFs can also be obtained from GEO with identifications GSE113394 and GSE185763, respectively.