Abstract
A gram-negative bacterium, Sphingomonas sp. strain A1, isolated as a producer of alginate lyase, has a characteristic cell envelope structure and forms a mouth-like pit on its surface. The pit is produced only when the cells have to incorporate and assimilate alginate. An alginate uptake-deficient mutant was derived from cells of strain A1. One open reading frame, algS (1,089 bp), exhibiting homology to the bacterial ATP-binding domain of an ABC transporter, was cloned as a fragment complementing the mutation. algS was followed by two open reading frames, algM1 (972 bp) and algM2 (879 bp), which exhibit homology with the transmembrane permeases of ABC transporters. Disruption of algS of strain A1 resulted in the failure to incorporate alginate and to form a pit. Hexahistidine-tagged AlgS protein (AlgSHis6) overexpressed in Escherichia coli and purified by Ni2+ affinity column chromatography showed ATPase activity. Based on these results, we propose the occurrence of a novel pit-dependent ABC transporter system that allows the uptake of macromolecules.
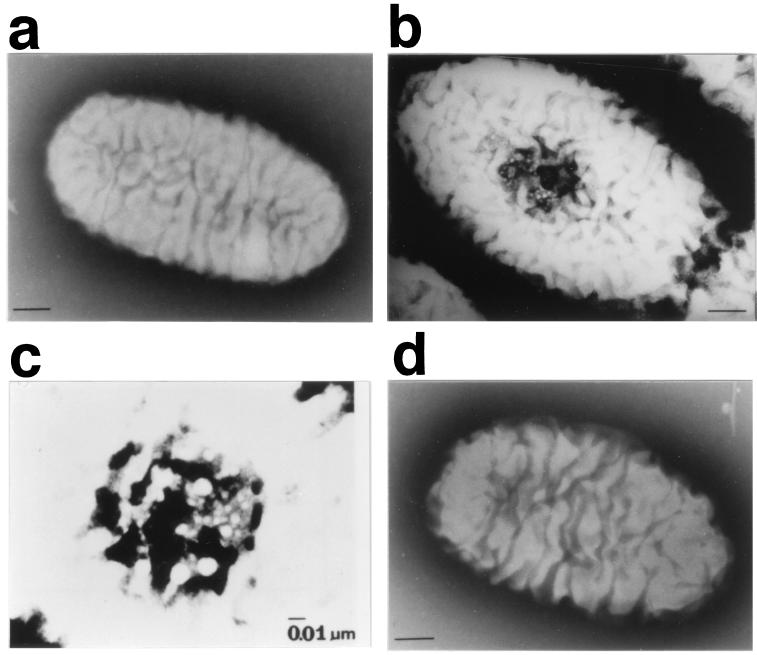
Alginate is a heteropolysaccharide consisting of α-l-mannuronate and its 5′-epimer, α-l-guluronate, and is produced by brown seaweeds and certain bacteria. Other than its utilization in the medical, chemical, and food areas, this biopolymer has been widely associated with chronic Pseudomonas aeruginosa infection in the lungs of cystic fibrosis patients (5). A bacterium, Sphingomonas sp. strain A1, was isolated from soil as a potent producer of alginate lyase catalyzing the depolymerization of alginate (24). The cells of strain A1 are covered by many large plaits (Fig. 1a), and a mouth-like pit (0.02 to 0.1 μm in diameter) is formed on their surface when they are grown in a medium containing alginate as the sole carbon source (10, 11) (Fig. 1b and c). The pit formed in the presence of alginate disappears when the cells are transferred to a medium without alginate (10, 11). In the presence of alginate, a specific region of the cell surface and its neighborhood is intensely stained with an agent that interacts with mucopolysaccarides (alginate), and thin sections of cells show an irregular site where the cell membrane sinks deeply into the cytosol (10, 11).
FIG. 1.
Cell surface structures of strain A1 cells grown in the absence (a) and presence (b and c) of alginate (HAY medium) and that of an A1dS disruptant (d) grown in the presence of alginate (HAY medium). (a, b, and d) Bar, 0.1 μm. Panel c is reproduced from reference 10 with permission of the publisher. The particles in the pit are alginate gel.
For the utilization of polysaccharides and other macromolecules, microbes usually degrade them by means of extracellular enzymes and then take the depolymerized low-molecular-weight (LMW) products up through their membranes. However, alginate lyase is exclusively localized in the cytoplasm, and the periplasmic and membrane fractions contain no alginate lyase activity (25). Furthermore, no extracellular alginate-depolymerizing activities have been detected in concentrated culture fluid of strain A1 (24, 25), when assayed by measuring the changes in viscosity and absorbance at 235 nm (for alginate lyase) or analysis of the depolymerization profile by thin-layer chromatography (data not shown). The amino acid sequence of alginate lyase purified from strain A1 was 53 amino acids shorter in the N-terminal region than that expected from the open reading frame (ORF) of the aly gene. However, this was not thought to be due to the presence of a signal peptide, because little alginate lyase activity has been detected in the periplasmic fraction and the mature enzyme is generated from the precursor (26). These results strongly suggest the occurrence of a direct uptake system for alginate in cells of strain A1. Here, we report the molecular basis of the macromolecule (alginate) uptake system mediated by a novel pit-dependent ABC transporter.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. Sphingomonas sp. strain A1 was isolated from a ditch as a producer of alginate lyase (24). Escherichia coli DH5α was used as the host for cosmid and plasmid propagation, HB101 harboring plasmid pRK2013 (Kmr) (CLONTECH) was used as helper cells for triparental mating, and BL21(DE3)pLysS was used as the host for protein expression using the pET3a or pET14b vector (Novagen).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Sphingomonas sp. | ||
| A1 | Wild type | 23 |
| AL-L | Alginate uptake-deficient mutant | This study |
| A1dS | algS::Kmr | This study |
| E. coli | ||
| DH5α | General cloning host strain | TOYOBO |
| HB101 | General cloning host strain | CLONTECH |
| BL21(DE3)pLysS | General protein expression strain | Novagen |
| Plasmids | ||
| pKS13 | Tetrcos Mob+ RK2 replicon | 14 |
| pKTY320 | Ampr Cmr Mob+ p15A replicon | 14 |
| pRK2013 | Kmr Tra+ ColE1 replicon | CLONTECH |
| pET3a | E. coli expression vector, Ampr | Novagen |
| pET14b | E. coli expression vector, Ampr | Novagen |
| pBluescriptII SK+ | E. coli cloning vector, Ampr | Stratagene |
| pBE11 | 23-kb Sau3AI fragment of strain A1 genomic DNA in pKS13 | This study |
| pALC1 | 2.8-kb PstI fragment in pKS13 | This study |
| pALC2 | 6-kb EcoRI fragment in pKS13 | This study |
| pAS3 | algS in pET3a | This study |
| pAS4 | algS in pET14b | This study |
Mutant isolation.
Cells of strain A1 grown aerobically at 30°C in an AYA medium [0.2% sodium alginate, 0.05% yeast extract, 0.1% (NH4)2SO4, 0.05% MgSO4 · 7H2O, 0.1% KH2PO4, 0.16% Na2HPO4 (pH 7.2)] were mutagenized with N-methyl-N′-nitro-N-nitrosoguanidine (1) and then spread on agar (1.5%) plates of AYS medium (0.2% sodium alginate, 0.01% yeast extract, and salts as above [pH 7.2]). After 2 days of incubation at 30°C, small colonies that formed on the plates were selected, and one of them was designated AL-L.
Alginate lyase activity was determined as described in reference 10. Briefly, cell extracts were incubated with 0.1% sodium alginate in 50 mM Tris-HCl (pH 7.2), and then the increase in absorbance at 235 nm was monitored. One unit of enzyme activity is defined as the amount of enzyme that releases 5.7 nmol of the product per min at 30°C. Alginate in culture fluids was enzymatically determined by using purified alginate lyase III (9) as described before (15). Cell growth in HAY medium (1.0% sodium alginate, 0.5% yeast extract, and salts as above [pH 7.2]) was turbidimetrically measured at 600 nm.
DNA cloning and related experiments.
A gene library of strain A1 was constructed as follows. Genomic DNA was partially digested with Sau3AI and then fractionated on an agarose gel. The DNA fragments (15 to 25 kb in size) recovered with Gene Clean II (BIO 101) were ligated into the BamHI site in a broad-host-range cosmid vector, pKS13 (Tetr) (14), packaged in vitro into lambda phage particles, and then used to infect E. coli DH5α. The library was used to transform the AL-L cells through triparental mating in the presence of helper plasmid pRK2013 (14). Transconjugants were selected on AY (0.5% sodium alginate, 0.02% yeast extract, and salts [pH 7.2]) plates containing tetracycline (20 μg/ml) and polymyxin B (5 μg/ml). E. coli DH5α and HB101 were sensitive to polymyxin B, while the cells of strain A1 were resistant to this antibiotic.
DNA sequencing was carried out by the dideoxy chain termination method using plasmid pBluescriptII SK+ (Stratagene). The nucleotide sequence was analyzed using DNASIS software (Hitachi Co., Ltd.).
Antibody preparation and Western blot analysis.
Plasmid pAS3 was constructed by introducing the PCR-amplified fragment of algS into the NdeI and BamHI sites of the pET3a vector. The PCR primers used were 5′-CATATGGTAGCAAGCGTCAGCATT-3′ and 5′-GGATCCTCAGTAAATAGAGGCCTG-3′. E. coli BL21(DE3)pLysS harboring pAS3 was grown at 37°C until the culture reached an A600 of 0.5. IPTG was then added to a final concentration of 1 mM, and the culture was continued for a further 2 h. AlgS was purified as inclusion bodies, solubilized stepwise with urea from 8 to 0 M, and then used for antibody production. Cytoplasmic and membrane fractions of strain A1 cells grown in an AY medium were prepared by the method described in reference 12, and the membrane fraction was solubilized with 1% octyl glucoside.
algS gene disruption.
A 1.2-kb kanamycin resistance gene from pUC4K (Pharmacia) was inserted into the EcoRV site of a 2.8-kb PstI fragment containing algS, ligated into vector pKTY320 (Ampr) (14), and then transconjugated into cells of strain A1 through triparental mating. Small colonies on the AY plates containing kanamycin (50 μg/ml) and polymyxin B (5 μg/ml) were checked by restreaking on AY plates containing ampicillin (25 μg/ml), and gene disruptants (Kmr and Amps) generated via homologous recombination were selected.
Electron microscopy.
Cells of strain A1 or a disruptant were aerobically grown at 30°C in HAY medium. Transmission electron microscopy was performed as described in reference 10.
Overexpression and purification of AlgSHis6.
The above-described PCR fragment of algS was inserted into pET14b (Novagen) to add a hexahistidine tag at the N terminus of AlgS, the resulting plasmid being designated pAS4. E. coli BL21(DE3)pLysS harboring pAS4 was grown at 30°C until the culture reached an A600 of 0.5, and then IPTG was added to a final concentration of 1 mM. After cultivation for 12 h at 16°C, about 1 g (wet weight) of cells from 400 ml of culture was harvested by centrifugation, washed with 50 ml of 50 mM Tris-HCl (pH 7.6), and then resuspended in 10 ml of 50 mM Tris-HCl (pH 7.6)–0.1 mM EDTA–0.1 mM phenylmethylsulfonyl fluoride–5 mM ATP. The cells were ultrasonically treated at 9 kHz for 20 min at 0°C, and the resulting supernatant was applied to a 5-ml Ni2+ affinity column (Pharmacia). After washing the column with 50 ml of buffer A (50 mM Tris-HCl [pH 7.6], 0.1 mM EDTA, 20% glycerol, 5 mM ATP) containing 10 mM imidazole and 1% Triton X-100 (wash I) and 50 ml of buffer A containing 100 mM imidazole (wash II), AlgSHis6 was eluted with 10 ml of buffer A containing 500 mM imidazole and then dialyzed against buffer A at 4°C overnight. Protein concentrations were determined using Bradford reagent (Sigma), with bovine serum albumin as a standard.
ATPase activity.
The reaction was initiated by the addition of 50 μl of 5 mM ATP and 5 mM MgCl2 in 50 mM Tris-HCl (pH 8.0) to 50 μl of an AlgSHis6 solution in 50 mM Tris-HCl (pH 8.0)–5 mM ATP–0.1 mM EDTA–20% glycerol. After incubation at 37°C for an appropriate time, the reaction was stopped by the addition of 0.1 ml of 12% sodium dodecyl sulfate (SDS). The amount of inorganic phosphoric acid (Pi) liberated was determined colorimetrically (4). The initial velocity was determined from at least three time points.
Chemicals.
Sodium alginate from an edible seaweed, Eisenia bicyclis (average molecular weight, 25,700), was purchased from Nacalai Tesque Co., Ltd., Kyoto, Japan. LMW alginate (average molecular weight, 1,000) was prepared by digestion of sodium alginate with purified alginate lyase III and purified by centrifugal filtration with Ultrafree-MC (Millipore).
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper have been deposited in the DDBJ/EMBL/GenBank nucleotide sequence databases under accession number AB011415.
RESULTS
Characterization of mutant strain AL-L.
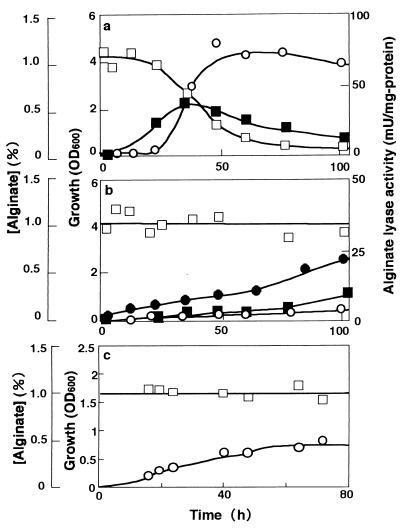
Unless otherwise specified, high-molecular-weight alginate (average molecular weight, 25,700) was used as the carbon source throughout the study. Wild-type strain A1 showed growth on HAY medium by assimilating alginate and producing alginate lyase (Fig. 2a). To obtain mutants unable to use alginate for growth, cells of strain A1 were mutated, and AL-L was isolated as one such mutant. The growth of AL-L on HAY medium was barely supported by yeast extract included in the medium, and alginate in the medium remained undigested. The alginate lyase activity of AL-L was extremely low, although it was detectable (Fig. 2b). However, when LMW alginate (average molecular weight, 1,000) was included in the HAY medium in place of alginate, AL-L cells resumed growth (Fig. 2b) with the formation of alginate lyase, the level of growth being comparable to that of strain A1 cultured similarly (data not shown). These results indicated that AL-L has a mutation in the alginate uptake system and not in alginate metabolism.
FIG. 2.
Growth of strains A1 (a), AL-L (b), and A1dS (c) on alginate. Cells were grown in HAY medium (1 liter) containing alginate (1%) except that the cells of A1dS were cultured in 6 ml of the same medium. ●, growth of AL-L in HAY medium with LMW alginate. ○, growth on alginate; ■, alginate lyase activity of alginate-grown cells; □, alginate concentration in medium.
Complementation of the mutation in AL-L.
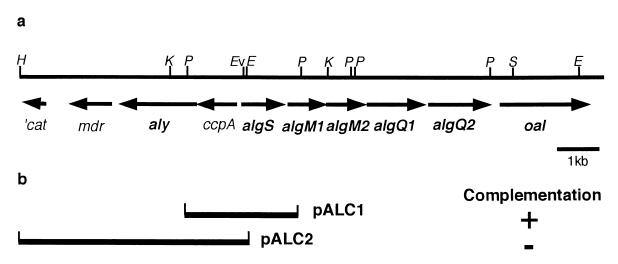
Eighteen clones that were able to complement the defect in AL-L and allowed it to assimilate alginate were obtained from a gene library of strain A1. Recombinant cosmid DNAs isolated from these clones were classified into four groups with respect to the restriction patterns, and all of them were confirmed to contain a common genomic DNA fragment. The growth and alginate assimilation of one strain (AL-LC) containing a recombinant plasmid were comparable to those of strain A1 (data not shown). The recombinant plasmid (pBE11) in AL-LC contained a 23-kb DNA insert.
By using a vector, pKS13, two derivative plasmids (pALC1 and pALC2) were constructed with the restriction fragments derived from the 23-kb insert cloned into AL-LC and then transconjugated to AL-L. Plasmid pALC1 enabled the mutant cells to grow well on HAY medium, but pALC2 failed to complement the mutation (Fig. 3b).
FIG. 3.
(a) Physical map of the 15-kb genomic fragment of strain A1 contained in pBE11. H, HindIII; K, KpnI; P, PstI; Ev, EcoRV; E, EcoRI; S, SphI. (b) Complementation testing of AL-L. Plasmids pALC1 and pALC2 were constructed by inserting the 2.8-kb PstI and 6-kb HindIII-EcoRI fragments, respectively, into pKS13, followed by introduction into AL-L. Growth (+) was determined on AY medium.
Nucleotide sequence of the cloned DNA fragment and analysis.
The nucleotide sequence of the 15-kb fragment cloned on pBE11 was analyzed (Fig. 3a) (accession number AB011415). Examination of the sequence revealed one possible ORF in a common region in pALC1 and pALC2; this ORF was designated algS. Five possible ORFs, designated algM1, algM2, algQ1, algQ2, and oal (W. Hashimoto, O. Miyake, K. Momma, S. Kawai, and K. Murata, submitted for publication), were coded downstream of algS. About 1 kb upstream from algS, the alginate lyase gene, aly (26), was found in the opposite orientation, and there were also some ORFs encoding amino acid sequences homologous to those of catalase from E. coli (p13029, cat) (partial), multidrug resistance protein from Bacillus subtilis (p39843, mdr), and catabolite control protein from Bacillus megaterium (p46828, ccpA).
The ORF of algS consisted of 1,089 nucleotides, encoding 363 amino acids with a calculated molecular size of 40 kDa. The deduced amino acid sequence exhibited homology to those of many proteins belonging to the superfamily of ATP-binding domains of ABC (ATP-binding cassette) transporters. The AlgS sequence was 52% and 48% identical to those of UgpC (20) and MalK (6) of E. coli, respectively. The Walker motifs (3, 22) were conserved. Hydropathy analyses suggested that AlgS is a soluble protein.
The algM1 ORF of 972 nucleotides coded for a protein of 324 amino acids with a molecular size of 37 kDa. The algM2 ORF (879 nucleotides) coded for a protein of 293 amino acids with a molecular size of 33 kDa. The deduced amino acid sequences of AlgM1 and AlgM2 exhibited 40 and 30% identity to those of the transmembrane proteins LPLB (p39128) and LPLC (p39129) from Bacillus subtilis, respectively; however, the functions of LPLB and LPLC have not been reported. AlgM1 and AlgM2 also showed homology to the membrane domains of bacterial sugar ABC transporters. AlgM1 was 26% identical to UgpA (20), and AlgM2 was 24% identical to UgpE (20). The hydropathy profiles indicated that AlgM1 and AlgM2 each contain six putative transmembrane helices. A sequence matching the consensus permease EAA motif (EAA---G---------I-LP) (13) was also found in both proteins.
The next two putative ORFs, algQ1 (1,578 nucleotides) and algQ2 (1,548 nucleotides), coded for proteins of 526 amino acids (60 kDa) and 516 amino acids (60 kDa), respectively. The amino acid sequences of AlgQ1 and AlgQ2 showed 74% identity to each other. The two proteins were partially homologous to a lipoprotein, LPLA (p37966), from B. subtilis (8) (22%).
The −35 and −10 consensus sequences were found upstream of the sequence of algS, and a strong stem-loop structure was found between the stop codon of algQ2 and the start codon of oal. The distances between the algS, algM1, algM2, algQ1, and algQ2 genes were short, being 94, 56, 78, and 146 bp, respectively, and a promoter-like sequence was absent in each space. Therefore, the algS, algM1, algM2, algQ1, and algQ2 genes appear to constitute an operon.
Three point mutations were found in algS on sequence analysis of AL-L genomic DNA. Cytosine481 was mutated to thymine, causing a change of Pro161 in Walker motif B of AlgS to Ser. Thymine620 was mutated to cytosine (Val207 to Ala), and adenine881 was mutated to guanine (Glu294 to Gly).
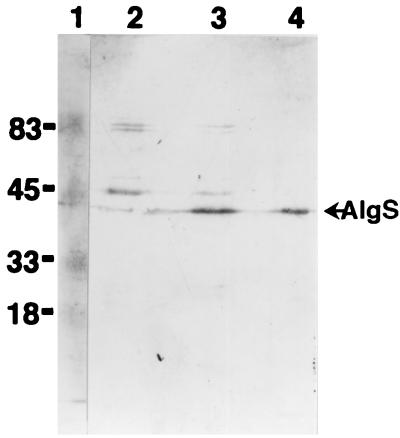
Western blotting analysis of AlgS.
The expression of AlgS in strain A1 was detected using antiserum for AlgS (Fig. 4). AlgS was mainly detected in the membrane fraction of strain A1, with the expected apparent molecular size of 40 kDa.
FIG. 4.
Western blot analysis of AlgS. Lane 1, size markers (in kilodaltons); lane 2, cytoplasmic fraction of strain A1; lane 3, membrane fraction of strain A1; lane 4, purified AlgS expressed in E. coli. Proteins were loaded at 8 μg for lanes 2 and 3 and 0.5 μg for lane 4.
Gene disruptant of algS.
Three algS disruptants (Kmr Amps) were obtained from among 130 Kmr colonies, one of which was designated A1dS. Gene disruption was confirmed by Southern hybridization, PCR, sequence analysis, and complementation with plasmid pALC1 (data not shown). AldS did not assimilate alginate in HAY medium (Fig. 2c). As in the case of AL-L (Fig. 2b), the limited growth observed was at the expense of the yeast extract in the medium. Electron microscopy analysis revealed that the mutant cells grown on HAY medium did not show the formation of a pit on their cell surface (Fig. 1d). These results indicated that AlgS is a component of the alginate incorporation machinery and has the ability to regulate the formation of a pit.
ATPase activity of purified AlgSHis6.
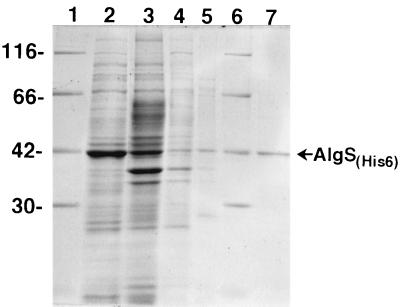
To facilitate the purification of AlgS, a hexahistidine tag was added to the N terminus of AlgS, and E. coli cells transformed with pAS4 were induced with IPTG at the late exponential phase of growth. About half of the AlgSHis6 protein was expressed in a soluble form when the cells were grown at a lower temperature (16°C), and AlgSHis6 was purified with a single Ni2+ affinity column to yield a greater than 95% pure preparation (Fig. 5). The purified AlgSHis6 was reactive with an anti-AlgS antibody (data not shown).
FIG. 5.
Purification of AlgSHis6: SDS-PAGE of fractions obtained at various stages of purification, stained with Coomassie blue. Lanes 1 and 6, size markers (in kilodaltons); lane 2, soluble fraction; lane 3, column flowthrough; lane 4, wash I; lane 5, wash II; lane 7, imidazole eluate. Proteins were loaded at 5.8 μg for lanes 2, 3, and 4 and 1 μg for lane 7. For lane 5, the same volume of the protein solution as in lane 4 was loaded.
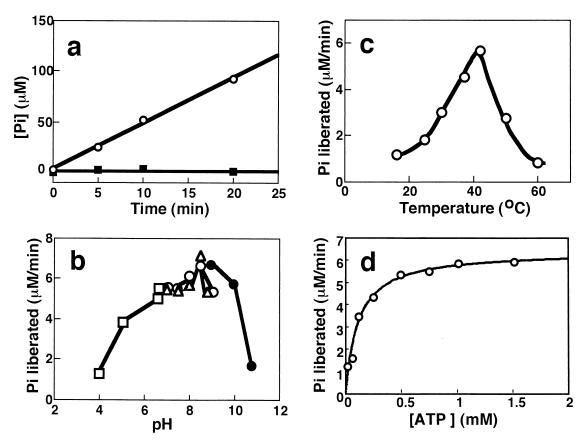
Since AlgS seemed to be an ATP binding protein of ABC transporters, the ATPase activity of AlgSHis6 was examined by determining the liberation of ADP (by thin-layer chromatography; data not shown) and Pi from ATP. AlgSHis6 hydrolyzed ATP in the presence of Mg2+ (Fig. 6a), Mn2+, Co2+, and Ca2+. The enzyme was most active at pH 8.5 (Fig. 6b) and at 42°C (Fig. 6c). AlgS was stable at 42°C for 30 min at pH 8.0, and 50% of the activity was lost after 30 min of incubation at 55°C.
FIG. 6.
Properties of the ATPase activity of AlgSHis6. (a) Linearity of the ATPase reaction at pH 8.0 and 37°C. ○, with 2.5 mM MgCl2; ■, without MgCl2. The final concentration of AlgSHis6 was 0.5 mg/ml. (b) pH dependence at 37°C. Fifty microliters of 100 mM buffers of various pHs containing 10 mM ATP and 5 mM MgCl2 was added to 50 μl of an AlgSHis6 (1.2 mg/ml) solution dialyzed against 5 mM Tris-HCl (pH 8.0)–0.1 mM EDTA–20% glycerol. □, sodium acetate; ○, Tris-HCl; ▵, sodium HEPES; ●, sodium CAPSO. (c) Temperature dependence at pH 8.0 (HEPES). The final concentration of AlgSHis6 was 0.5 mg/ml. (d) Dependence on the ATP concentration at pH 8.0 and 37°C. Fifty microliters of various amounts of an ATP and MgCl2 (molar ratio, 2:1) mixture solution was added to 50 μl of an AlgSHis6 solution (0.9 mg/ml) including 0.05 mM EDTA and 20% glycerol. The solid line is the theoretical curve for Km = 0.11 mM and Vmax = 6.42 μM/min.
GTP was utilized with 82% efficiency compared to ATP, and CTP (74%), TTP (62%), and UTP (31%) were also utilized with lower efficiency than ATP. The Km values for ATP were determined to be 0.11 mM (Fig. 6d) and 0.68 mM at pHs 8.0 and 7.0 (data not shown), respectively, at 37°C. The ATP homologues 5′-adenylyl-imidodiphosphate, adenylyl(β,γ-methylene)-diphosphonate, and adenosine 5′-O-(3-thio)triphosphate significantly inhibited the ATPase activity of AlgSHis6.
DISCUSSION
AL-L, isolated as a mutant incapable of assimilating alginate, was thought to have damage in the gene for either alginate uptake or alginate metabolism. However, extremely low alginate lyase activity was detected in AL-L cells grown in a medium containing alginate, and AL-L was able to grow in a medium containing LMW alginate (Fig. 2b), thereby producing a substantial amount of alginate lyase. Furthermore, pALC2 carrying the alginate lyase gene (aly) could not complement the mutation. These results indicated that AL-L has a mutation in the transport of alginate but not in the alginate lyase gene. The low activity of alginate lyase in AL-L cells grown in the presence of alginate could be explained by the failure of alginate incorporation, since alginate lyase formation is strictly dependent on the presence of alginate (24).
The mutation in AL-L was complemented by pALC1 (Fig. 3b). pALC1 encodes the full lengths of ccpA and algS. However, pALC2, which encodes the full length of ccpA, did not restore the growth of AL-L. These observations imply that the mutation was in algS. Indeed, the mutation was confirmed to be in algS by sequence analysis of AL-L genomic DNA. Disruptant analysis also indicated that AlgS is a prerequisite for the incorporation of alginate, because disruptant A1dS could not grow on alginate as a carbon source (Fig. 2c).
The AlgS protein showed homology with other bacterial ABC transporters responsible for the import of sugars. The archetypal ABC transporter, which has been well studied as to the transport of maltose in E. coli and histidine in Salmonella enterica serovar Typhimurium (2, 18), exists as a membrane-bound complex composed of two ATP-binding cassette domains (ATPase) and two transmembrane domains (permease). Two ORFs, algM1 and algM2, were coded downstream of algS, and the deduced amino acid sequences of AlgM1 and AlgM2 showed homology with the membrane permease domains of bacterial ABC transporters. AlgS, AlgM1, and AlgM2 showed homology with UgpC, UgpA, and UgpE, respectively, which are members of the ABC transporter family involved in the uptake of glycerol 3-phosphate (20). These results suggest that the alginate transporter of strain A1 belongs to the ABC transporters. Because the soluble AlgS protein was detected in the membrane fraction, the alginate transporter may be assumed to exist as a membrane-bound complex in strain A1 cells.
In the bacterial ABC transporter systems, the substrates are transported across the cytoplasmic membrane concomitant with the hydrolysis of ATP by the ABC transporter (2, 18). The purified ATP-binding cassette domain, HisP, of the histidine transport system and MalK of the maltose transport system in S. enterica serovar Typhimurium have been reported to exhibit ATPase activity so far (19, 23), although the activity is low compared with that of the ABC transporter complex. Similarly to HisP and MalK, AlgSHis6 catalyzed the liberation of ADP and Pi from ATP and is thought to be an ATPase supplying the energy for the transport of alginate.
Bacterial ABC transporters involved in nutrient uptake have been reported to show affinity toward LMW substrates, and to the best of our knowledge, no reports have been published so far on high-molecular-weight substrates like alginate. LMW substrates to be transported are first bound to soluble receptors (periplasmic substrate-binding proteins) and then transferred to an ABC transporter. The gene for the binding protein is generally found in the neighborhood of genes encoding the ATP-binding domain or permease domain. As for the alginate uptake system, no ORFs that encode close homologs of sugar-binding proteins were found upstream or downstream of the algS, algM1, and algM2 genes. However, the amino acid sequences of AlgQ1 and AlgQ2 exhibit low homology with that of lipoprotein LPLA, which is a distant homolog of the E. coli maltose-binding protein MalE. It has been reported that substrate-binding proteins in gram-positive bacteria are often lipoproteins tethered to the cell surface (7). Therefore, AlgQ1 and AlgQ2 may act as substrate-binding proteins in this ABC transporter system. AlgQ1 and AlgQ2 did not have a clear signal peptide sequence or the consensus sequence of the E. coli lipoprotein (7).
Alginate transport and assimilation in strain A1 are thought to be controlled simultaneously, since transporter genes algS, algM1, algM2, the alginate lyase gene aly, and the gene of oligoalginate lyase, oal, which degrades the reaction products of alginate lyase (W. Hashimoto, O. Miyake, K. Momma, S. Kawai, and K. Murata, submitted for publication), have been shown to form a cluster (Fig. 3a). The alginate lyase is an inducible enzyme, and the expression of aly may be controlled by ccpA adjacent to aly. Since a putative promoter and terminator were found in front of algS and in back of algQ2, respectively, the genes from algS to algQ2 may constitute an operon.
The disruptant A1dS failed to form a pit on its cell surface even in the presence of alginate. A1dM1 also failed to form a pit under the same conditions and did not grow on alginate in HAY medium (K. Momma, Y. Mishima, W. Hashimoto, and K. Murata, unpublished data). Although the intrinsic function of AlgS and AlgM1 in pit formation is not yet clear, it is possible that pit formation in the presence or absence of alginate is regulated by AlgS and/or AlgM1.
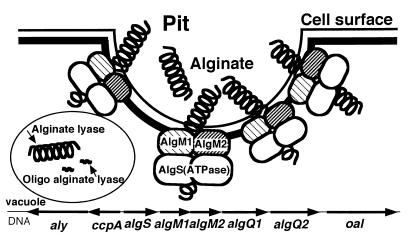
The molecular machinery that we found in strain A1 is different from the typical prokaryotic ABC transporter systems in that (i) the system allows the incorporation of macromolecules and (ii) the system is linked with a pit formed on the cell surface. Based on these observations, we can postulate a novel model for the macromolecule (alginate) uptake mechanism (Fig. 7). Briefly, alginate in the external medium is concentrated in the pit and then transported into the cytosol by an ABC transporter consisting of AlgS, AlgM1, and AlgM2. AlgS generates energy through ATP hydrolysis and transfers it to permease proteins, AlgM1 and AlgM2.
FIG. 7.
Putative model for the pit-dependent macromolecule incorporation system in strain A1. A macromolecule (alginate) present in the medium is first concentrated in the pit and then incorporated through the actions of AlgM1, AlgM2, and AlgS. Alginate is depolymerized by alginate lyase and oligoalginate lyase.
Gram-negative bacteria contain stereospecific solute transport systems in their inner membranes and nonstereoselective porins in their outer membranes that allow the passage of small ions, nutrients, and metabolic products across these lipid bilayer structures (16, 17, 21). Macromolecules such as proteins and polysaccharides are excreted out of cells by means of specially equipped export machineries. However, no data have been published regarding the direct import of macromolecules such as alginate. We could, for the first time, show the presence of a cleverly equipped novel system for the uptake of macromolecules in bacteria. Recently, pectin was also found to be directly transported into cells of strain A1 (W. Hashimoto, S. Mori, K. Momma, and K. Murata, unpublished data). Some interesting questions remain. How is pit formation controlled by the presence or absence of macromolecule substrates? How are macromolecules concentrated in the pit? How does alginate cross the outer membrane? Is the direct macromolecule uptake system energetically more efficient than the transport of depolymerized LMW products? Investigations on these issues may open the way for studying unexplored uptake machineries for macromolecules such as proteins, DNAs, and polysaccharides.
ACKNOWLEDGMENTS
We are grateful to Masamichi Takagi of the University of Tokyo for the kind gift of plasmids pKS13 and pKTY320.
This work was supported in part by grants-in-aid from the Ministry of Education, Science, Sports and Culture of Japan (10556017, 10145229, 11132237, 11460039, and 09876026 to K. Murata).
Keiko Momma and Wataru Hashimoto contributed equally to this work.
ADDENDUM
We have changed the name for ORF algH to algS to avoid confusion, because algH has already been used in other literature concerning alginate metabolism.
REFERENCES
- 1.Adelberg E A, Mandel M, Chen G C C. Optimal conditions for mutagenesis by N-methyl-N′-nitro-N-nitrosoguanidine in Escherichia coli K12. Biochem Biophys Res Commun. 1965;18:788–795. [Google Scholar]
- 2.Ames G F-L. Bacterial periplasmic transport systems: structure, mechanism, and evolution. Annu Rev Biochem. 1986;55:397–425. doi: 10.1146/annurev.bi.55.070186.002145. [DOI] [PubMed] [Google Scholar]
- 3.Bianchet M A, Ko Y H, Amzel L M, Pedersen P L. Modeling of nucleotide binding domains of ABC transporter proteins based on a F1-ATPase/recA topology: structural model of the nucleotide binding domains of the cystic fibrosis transmembrane conductance regulator (CFTR) J Bioenerg Biomembr. 1997;29:503–524. doi: 10.1023/a:1022443209010. [DOI] [PubMed] [Google Scholar]
- 4.Chifflet S, Torriglia A, Chiesa R, Tolosa S. A method for the determination of inorganic phosphate in the presence of labile organic phosphate and high concentrations of protein: application to lens ATPases. Anal Biochem. 1988;168:1–4. doi: 10.1016/0003-2697(88)90002-4. [DOI] [PubMed] [Google Scholar]
- 5.Gecesa P, Russell N J, editors. Pseudomonas infection and alginate. London, United Kingdom: Chapman & Hall; 1990. [Google Scholar]
- 6.Gilson E, Nikaido H, Hofnung M. Sequence of the malK gene in E. coli K12. Nucleic Acids Res. 1982;10:7449–7458. doi: 10.1093/nar/10.22.7449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilson E, Alloing G, Schmidt T, Claverys J P, Dudler R, Hofnung M. Evidence for high affinity binding-protein dependent transport systems in gram-positive bacteria and in Mycoplasma. EMBO J. 1988;7:3971–3974. doi: 10.1002/j.1460-2075.1988.tb03284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gómez A, Ramón D, Sanz P. The Bacillus subtilis lipoprotein LplA causes cell lysis when expressed in Escherichia coli. Microbiology. 1994;140:1839–1845. doi: 10.1099/13500872-140-8-1839. [DOI] [PubMed] [Google Scholar]
- 9.Hisano T, Nishimura M, Yamashita T, Sakaguchi K, Takagi M, Imanaka T, Kimura A, Murata K. Production of bacterial alginate-specific lyase by recombinant Bacillus subtilis. J Ferment Bioeng. 1994;78:79–83. [Google Scholar]
- 10.Hisano T, Yonemoto Y, Yamashita T, Fukuda Y, Kimura A, Murata K. Direct uptake of alginate molecules through a pit on the bacterial cell surface: a novel mechanism for the uptake of macromolecules. J Ferment Bioeng. 1995;79:538–544. [Google Scholar]
- 11.Hisano T, Kimura N, Hashimoto W, Murata K. Pit structure on bacterial cell surface. Biochem Biophys Res Commun. 1996;220:979–982. doi: 10.1006/bbrc.1996.0526. [DOI] [PubMed] [Google Scholar]
- 12.Kawasaki S, Moriguchi R, Sekiya K, Nakai T, Ono E, Kume K, Kawahara K. The cell envelope structure of the lipopolysaccharide-lacking gram-negative bacterium Sphingomonas paucimobilis. J Bacteriol. 1994;176:284–290. doi: 10.1128/jb.176.2.284-290.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kerppola R E, Ames G F-L. Topology of the hydrophobic membrane-bound components of the histidine periplasmic permease. J Biol Chem. 1992;267:2329–2336. [PubMed] [Google Scholar]
- 14.Kimbara K, Hashimoto T, Fukuda M, Koana T, Takagi M, Oishi M, Yano K. Cloning and sequencing of two tandem genes involved in degradation of 2,3-dihydroxybiphenyl to benzoic acid in the polychlorinated biphenyl-degrading soil bacterium Pseudomonas sp. strain KKS102. J Bacteriol. 1989;171:2740–2747. doi: 10.1128/jb.171.5.2740-2747.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murata K, Yamaguchi H, Yonemoto Y, Sakaguchi K, Kimura A, Okayama K. Continuous depolymerization of alginate by a nonsupport bioreactor system containing flocculated bacterial cells. J Ferment Bioeng. 1992;73:172–174. [Google Scholar]
- 16.Nikaido H, Saier M H., Jr Transport proteins in bacteria: common themes in their design. Science. 1992;258:936–942. doi: 10.1126/science.1279804. [DOI] [PubMed] [Google Scholar]
- 17.Nikaido H. Porins and specific channels of bacterial outer membranes. Mol Microbiol. 1992;6:435–442. doi: 10.1111/j.1365-2958.1992.tb01487.x. [DOI] [PubMed] [Google Scholar]
- 18.Nikaido H, Hall J A. Overview of bacterial ABC transporters. Methods Enzymol. 1998;292:3–20. doi: 10.1016/s0076-6879(98)92003-1. [DOI] [PubMed] [Google Scholar]
- 19.Nikaido K, Liu P-Q, Ames G F-L. Purification and characterization of HisP, the ATP-binding subunit of a traffic ATPase (ABC transporter), the histidine permease of Salmonella typhimurium. J Biol Chem. 1997;272:27745–27752. doi: 10.1074/jbc.272.44.27745. [DOI] [PubMed] [Google Scholar]
- 20.Overduin P, Boos W, Tommassen J. Nucleotide sequence of the ugp genes of Escherichia coli K-12: homology to the maltose system. Mol Microbiol. 1988;2:767–775. doi: 10.1111/j.1365-2958.1988.tb00088.x. [DOI] [PubMed] [Google Scholar]
- 21.Saier M H., Jr Computer-aided analyses of transport protein sequences: gleaning evidence concerning function, structure, biogenesis, and evolution. Microbiol Rev. 1994;58:71–93. doi: 10.1128/mr.58.1.71-93.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walter C, Bentrup K H, Schneider E. Large scale purification, nucleotide binding properties, and ATPase activity of the MalK subunit of Salmonella typhimurium maltose transport complex. J Biol Chem. 1992;267:8863–8869. [PubMed] [Google Scholar]
- 24.Yonemoto Y, Murata K, Kimura A, Yamaguchi H, Okayama K. Bacterial alginate lyase: characterization of alginate lyase-producing bacteria and purification of the enzyme. J Ferment Bioeng. 1991;72:152–157. [Google Scholar]
- 25.Yonemoto Y, Yamaguchi H, Kimura A, Sakaguchi K, Okayama K, Murata K. Cloning of a gene for intracellular alginate lyase in a bacterium isolated from a ditch. J Ferment Bioeng. 1992;73:225–227. [Google Scholar]
- 26.Yonemoto Y, Tanaka H, Hisano T, Sakaguchi K, Abe S, Yamashita T, Kimura A, Murata K. Bacterial alginate lyase gene: nucleotide sequence and molecular route for generation of alginate lyase species. J Ferment Bioeng. 1993;75:336–342. [Google Scholar]