Graphical abstract
Keywords: COVID-19, Antiviral drug, Mβ-CD, Inclusion complex, SARS-CoV-2 (3CLPro) inhibitor, In silico
Abstract
During the current outbreak of the novel coronavirus disease 2019 (COVID-19), researchers have examined several antiviral drugs with the potential to inhibit the proliferation of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The antiviral drug acyclovir (AVR), which is used to treat COVID-19, in complex with methyl-β-cyclodextrin (Mβ-CD) was examined in the solution and solid phases. UV–visible and fluorescence spectroscopic analyses confirmed that the guest (AVR) was included inside the host (Mβ-CD) cavity. A solid inclusion complex of AVR was prepared by co-precipitation, physical mixing, kneading, and bath sonication methods at a 1:1 ratio of Mβ-CD:AVR. The prepared Mβ-CD:AVR inclusion complex was characterized using Fourier transform infrared spectroscopy (FTIR), X-ray powder diffraction (XRD), thermogravimetric analysis (TGA), differential scanning calorimetry (DSC), and scanning electron microscopy (SEM) analysis. Phase solubility studies indicated the Mβ-CD:AVR inclusion complex exhibited a higher stability constant and linear enhancement in AVR solubility with increasing Mβ-CD concentrations. In silico analysis of the Mβ-CD/AVR inclusion complex confirmed that AVR drugs show potential as inhibitors of SARS-CoV-2 3C-like protease (3CLpro) receptors. Results obtained using the PatchDock and FireDock servers indicated that the most favorable docking ligand was Mβ-CD:AVR, which interacted with SARS-CoV-2 (3CLPro) protease inhibitors with high geometric shape complementarity scores (2522 and 5872) and atomic contact energy (-313.77 and −214.70 kcal mol−1). Our results suggest that the Mβ-CD/AVR inclusion complex inhibits the main protease of SARS-CoV-2, although further wet-lab experiments are needed to verify these findings.
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the cause of the coronavirus disease 2019 (COVID-19) outbreak, which was first detected in December 2019 [1]. This disease has resulted in serious challenges to international health security and created global health and socioeconomic problems. Fever, myalgia, cough, dyspnea, and weariness are symptoms of this illness. Since COVID-19-specific medicines and vaccines are less available, the mortality of the disease is high. Various medications available have been used to treat COVID-19, and studies aimed at the development of treatments, particularly vaccines, are underway. In addition to vaccination, antiviral medications may be useful for treating COVID-19 [2]. Viruses contain two types of non-structural proteins: papain-like (PLpro) and 3-chymotrypsin-like (3CLpro) proteases. Viruses also contain RNA polymerase and helicase [3]. Both of these proteases (PLpro and 3CLpro) are involved in virus transcription and replication, with 3CLpro thought to play the most important role in viral replication. The crystal structure of COVID-19 3CLpro (PDB ID:7DJR) is publicly available in the Protein Data Bank (PDB). Chymotrypsin-like proteases play a critical role in the life cycle of viruses and are stable during COVID-19. SARS-CoV-2 3C-like protease (3CLpro) receptors were recently demonstrated to be viable therapeutic targets [4]. Drug repurposing, also known as reframing, can be used to control the spread of dangerous microorganisms that seriously threaten human health. Although FDA-approved antiviral drugs are known to be safe for use in humans, their effectiveness against SARS-CoV-2 must be evaluated [5]. The effectiveness of these drugs in preventing or reducing the severity of COVID-19 symptoms remains unclear. Therefore, studies are urgently needed to identify drug candidates targeting different SARS-CoV-2 proteins to improve therapies for COVID-19.
Antiviral drugs are commonly used to treat viral infections [6]. As most antivirals target the viral replication mechanism within organisms, these drugs must be both safe and efficacious to prevent harm to the host and eradicate the infection. Acyclovir (AVR) is a well-known antiviral medication used to treat herpes simplex virus infections [7]. Neither parenteral nor oral administration of currently available AVR formulations can result in sufficient AVR concentrations in target areas. The ineffectiveness of oral AVR therapy is related to its low permeability and limited water solubility [8], [9]. Thus, various methods have been developed to enable oral administration of AVR to ensure delivery, which can result in systemic side effects such as acute renal failure and neurotoxicity. The physicochemical properties of antivirals, such as their solubility, stability, and permeability, influence the administration route. Excipients can be used to improve the dispersion and absorption of drugs and minimize the antiviral dose required for effective therapy. Increasing dissolution, which enhances bioavailability, can improve the therapeutic efficacy of water-insoluble drugs [10].
A potential approach for improving apparent solubility and dissolution rates, and thus the bioavailability of poorly soluble drugs, involves the formation of inclusion complexes [11]. Drug complexation with cyclodextrins (CDs) can modify the physicochemical properties of a drug to increase its dissolution and bioavailability [12]. CDs contain cavities that can encapsulate hydrophobic groups to generate inclusion complexes without affecting the guest molecule structure or chemical characteristics. CDs are biocompatible, do not elicit an immunological response, and exhibit low toxicity. Commercially viable formulations for oral, parenteral, nasal, pulmonary, and cutaneous drug delivery have been prepared as inclusion complexes containing bioactive chemicals [13]. Our research groups have been involved in the last decade focusing mainly on the study of nanomaterials, drug complexation characteristics, and their usefulness in sensing, pharmaceutical, and other applications [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31]. In addition, to treat COVID-19, CDs have been utilized to encapsulate putative antiviral interactions at various stages of the viral lifecycle [32], [33], [34], [35], [36]. β-CD can be modified by adding different groups to form a range of derivatives with increased water solubility and lower toxicity compared to the parent CD. Owing to its high water solubility and entrapment rate, methyl β-CD (Mβ-CD) has received considerable attention among these derivatives [37]. Methylation of the CD ring makes the inner surface of the torus less steric and more hydrophobic, which can improve entrapment efficiency [38]. Complexation of AVR with β-CD was previously demonstrated and suggested as a feasible option for improving dissolution characteristics [39], [40], [41]. However, the effects of Mβ-CD on the water solubility and inclusion of AVR have not been investigated.
This study was conducted to systematically investigate the formation of an inclusion complex of AVR and Mβ-CD to enhance its solubility and dissolution. The inclusion complex of Mβ-CD:AVR in a 1:1 stoichiometric ratio was characterized, and the interaction of the Mβ-CD:AVR complex with SARS-CoV-2 (3CLPro) protease inhibitors was investigated in molecular docking simulations.
2. Materials and methods
2.1. Materials
All chemical reagents used in the experiments were of analytical grade. Mβ-CD (average molecular weight = 1310, ≥ 98% purity) and AVR (average molecular weight = 225) were from Sigma-Aldrich (St. Louis, MO, USA). Deionized water with resistivity > 10MΩ /cm and pH of Phosphate-buffered saline 7.4 (sterile-filtered, suitable for cell culture) was used. Deionized water was purified using a Milli-Q water purification system (Millipore, Billerica, MA, USA).
2.2. Preparation of Mβ-CD:AVR inclusion complex
Mβ-CD:AVR inclusion complexes were prepared using co-precipitation, physical mixing, kneading, and bath sonication methods. Co-precipitation (CP) was performed using equimolar solutions of AVR in ethanol as the organic phase and Mβ-CD in distilled water for the binary system. Briefly, 1 g of Mβ-CD was dissolved in 30 mL of water, and 0.1681 g of AVR was dissolved in 20 mL of ethanol solution. The AVR organic phase was added to the aqueous phase of Mβ-CD with continuous stirring on a magnetic stirrer (30 °C, 24 h, 300 rpm). In addition, equimolar quantities of Mβ-CD and AVR were ground in a mortar for 15 min to obtain a physical mixture (PM), followed by the addition of water:ethanol (1:1 v/v) and trituration to obtain a homogenous paste for 15 min, with drying at 45 °C in an oven to obtain the kneading method (KM). Finally, an Mβ-CD and AVR mixture in equimolar ratios was subjected to bath sonication (BS) for 24 h. The resulting CP and BS solutions were filtered through a 0.45-μm membrane filter; the filtrate was stored at −80 °C for 48 h and used to prepare the Mβ-CD:AVR inclusion complex.
2.3. Characterizations
Ultraviolet–visible (UV–Vis) and fluorescence spectral measurements were performed using a UV 3220 spectrometer (Optizen, Redmond, WA, USA) and F-2700 spectrofluorometer (Hitachi, Tokyo, Japan). Fourier-transform infrared spectroscopy (FTIR) was performed using an FTIR spectrophotometer (Perkin Elmer, Waltham, MA, USA) at a scanning range of 4000–400 cm−1. The powder X-ray diffractometry (XRD) patterns for AVR, Mβ-CD, and Mβ-CD:AVR inclusion complexes were obtained using an X-ray diffractometer (PANalytical X’Pert Philips, MRD model; Malvern Panalytical, Malvern, UK) at a voltage and current of 40 kV and 30 mA, respectively (wavelength: 1.5405 Å scanning speed; 5° min−1, range; 10–80°). Thermogravimetric analysis (TGA) and differential scanning calorimetry (DSC) analysis of β-CD and the Mβ-CD:AVR inclusion complex (temperature range: 25–400 °C and heating rate: 10 °C min−1 under nitrogen) was performed using a TGA-Q5000 thermal analyzer (TA Instruments, New Castle, DE, USA) and DSC Q200 (TA Instruments). The surface morphologies of AVR, Mβ-CD, and Mβ-CD:AVR inclusion complexes were examined by scanning electron microscopy (SEM) with a FESEM Leo Supra 50VP (Carl Zeiss SMT, Oberkochen, Germany) microscope at an accelerating voltage of 200 kV. All the samples were immobilized on double-sided carbon tape without further coating as the extent of sample preparation.
2.4. Phase solubility studies
The apparent stability constant and complexation efficacy of Mβ-CD:AVR inclusion complex formation was determined as described by Higuchi and Connors [42]. The effect of Mβ-CD on the solubility of AVR was investigated by adding excess AVR to aqueous solutions containing increasing amounts of Mβ-CD (2–12 mM). To remove undissolved particulates, the solutions were sonicated in an ultrasonic bath for 1 h, incubated at 25 °C, and filtered through a 0.45 μm filter. After reaching equilibrium, the amount of AVR in the samples was measured using UV–Vis spectrophotometry. The Mβ-CD concentration versus AVR concentration was plotted to yield a phase solubility diagram. Three parallel samples were evaluated for each sample. The slopes of the phase solubility diagrams and intrinsic solubility were used to compute the stability constant values for the Mβ-CD:AVR inclusion complex (equation (1)). Complexation efficiency was evaluated to determine the optimum conditions for Mβ-CD:AVR inclusion, as it reflects the ability of Mβ-CD to AVR and is independent of intrinsic AVR drug solubility (equation (2)).
| (1) |
| (2) |
2.5. Determination of encapsulation efficiency and drug content
The total amount of free or unincorporated AVR entrapped in the Mβ-CD:AVR inclusion complex was determined by UV–Vis spectrophotometry at 256 nm. Briefly, 0.16 g of prepared Mβ-CD:AVR inclusion complexes were dissolved in 25 mL of phosphate-buffered saline at pH 7.4 and sonicated for 30 min; the encapsulated amount of AVR was calculated at 256 nm. Before analysis, the formulations were agitated at 4000 rpm for 30 min to separate the active component from any undissolved Mβ-CD in the Mβ-CD:AVR solution. Equation (3) was used to determine the AVR drug content (DC) and encapsulation efficiency (EE):
| (3) |
| (4) |
where AVRexperimental and AVRtheroretical are the experimental (absorption measurements) and theoretical (solid complex product) amounts of AVR in the Mβ-CD:AVR inclusion complexes.
2.6. Molecular modeling
The PatchDock and FireDock servers (https://bioinfo3d.cs.tau.ac.il/PatchDock/) (https://bioinfo3d.cs.tau.ac.it/FireDock/) were used for molecular docking of the binary (Mβ-CD:AVR) and ternary (Mβ-CD:AVR /SARS-CoV-2 (3CLPro; PDB ID:7DJR) complexes. The chemical structures of AVR and Mβ-CD were retrieved from the Cambridge database, manually drawn with ChemDraw Professional 16.0 and Gaussian 9.0, and then optimized with Chem3D 16.0. (CambridgeSoft, PerkinElmer). PDB format was used to save the optimized structures. The SARS-CoV-2 (3CLPro) X-ray crystallographic structure (PDB ID:7DJR) was obtained from the PBD. Using the refined 3D structure of Mβ-CD (Mβ-CD:AVR) as the receptor and optimized 3D structure of AVR and 7DJR as the ligands, molecular docking was performed using the PatchDock and FireDock servers. Many docked files at different locations were generated in PatchDock. FireDock was used to further examine the docked files by displaying the binding energy for each file. For the UCSF chimera, docked files with the lowest global energy were chosen for further analysis. Molecular docking was performed to determine the most advantageous confirmation of the molecular representations of AVR. Mβ-CD, Mβ-CD:AVR, and Mβ-CD:AVR/SARS-CoV-2 (3CLPro) were produced using UCSF-Chimera 1.8.1 (https://www.cgl.ucsf.edu/chimera) to validate the experimental findings. Finally, using the default scoring mechanism of the PatchDock and FireDock servers, the configuration with the highest docking score and lowest binding affinity (kcal/mol) was identified. Each computation yielded the most advantageous Mβ-CD:AVR and Mβ-CD:AVR/SARS-CoV-2 (3CLPro) combinations, which were further examined.
2.7. Statistical analysis
The statistical analysis was performed using OriginLab (OriginPro 8.5) Software. All the data were expressed as the mean ± standard deviation (SD). The comparison values were performed with a one-way of variance (ANOVA) test and results were considered statistically significant if the p-value < 0.05.
3. Results and discussion
3.1. Optical properties
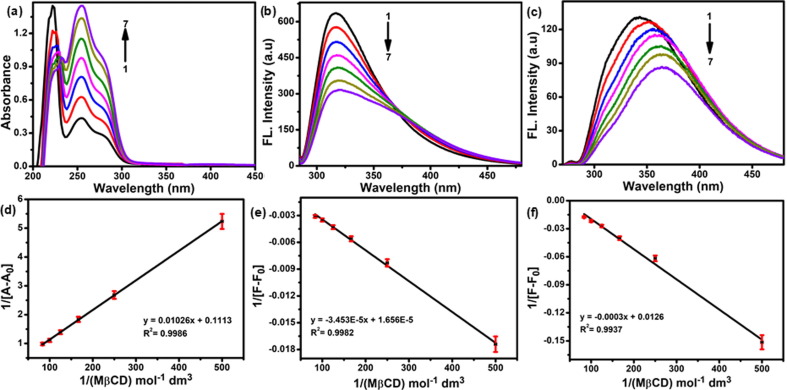
The UV–Vis and fluorescence spectral properties of AVR in the presence of Mβ-CD were determined. Fig. 1 (a) shows the absorption spectra of AVR (2.25 × 10−5 M) in the presence of Mβ-CD at concentrations of 0–12 mM (pH 7.4) in an aqueous solution. The absorption spectra of AVR showed significant absorption maxima and shoulder peaks at 222, 256, and 280 nm (π–π* transition) [43]. As the quantity of Mβ-CD increased, the absorbance of AVR increased, but the absorption region was slightly red-shifted in the Mβ-CD:AVR inclusion complex. The solubility of AVR in Mβ-CD and reduction of the guest molecule were likely responsible for these results, and the spectral alterations indicate strong interactions between AVR and Mβ-CD [44]. Additionally, increased absorbance of Mβ-CD solutions has been linked to improved drug solubility via hydrophobic interactions in the Mβ-CD cavity. Fluorescence spectroscopy was performed to investigate AVR inclusion in Mβ-CD. Fig. 1(b,c) shows the fluorescence spectra of AVR at different concentrations of Mβ-CD (0–12 mM) with excitation wavelengths of 256 and 280 nm and emission maxima of 316 and 342 nm. With the continuous addition of Mβ-CD, the fluorescence intensity of AVR decreased at a wavelength of 256 nm without any marginal shift; at a wavelength of 280 nm, the emission intensity declined, although there was a substantial redshift [45]. The decrease in fluorescence intensity was linked to AVR partitions in the highly hydrophobic Mβ-CD cavity, suggesting the formation of an Mβ-CD:AVR inclusion complex. Mβ-CD inclusion complexation typically increases fluorescence intensity but was shown to be reduced in a few cases [45].
Fig. 1.
(a) UV–Visible and (b, c) fluorescence spectra of AVR (2.25 × 10-5 M) at pH 7.4 at various concentrations of Mβ-CD: (1) 0, (2) 0.002 M, (3) 0.004 M, (4) 0.006 M, (5) 0.008 M, (6) 0.010 M, and (7) 0.012 M and (d–f) Benesi–Hildebrand plots of 1/[A − A0] and 1/[F − F0] vs. 1/[Mβ-CD].
The 1:1 M ratio and binding constant (K) of the AVR and Mβ-CD inclusion complexes were calculated using the Benesi–Hildebrand equations (5), (6).
Absorption.
| (5) |
Fluorescence.
| (6) |
The plots of 1/A-A0 and 1/F-F0 vs. 1/[Mβ-CD] for AVR in Fig. 1(d–e) showed an excellent linear correlation (0.9986, 0.9982, and 0.9937), indicating the establishment of a 1:1 inclusion complex. The 'K' values of 96.59, 58.65, and 91.14 M−1 were much greater than the intercept and slope values of the Benesi–Hildebrand plot, possibly because of the improved solubility of AVR and greater Mβ-CD:AVR inclusion complex formation.
3.2. FTIR spectroscopy
FTIR is useful for studying the interactions responsible for the formation of the Mβ-CD:AVR inclusion complex as host and guest molecules. The FTIR spectra of Mβ-CD, AVR, and the Mβ-CD:AVR inclusion complex prepared using CP, PM, KM, and BS are shown in Fig. 2 a. The FTIR spectrum of Mβ-CD showed a large O—H stretching peak at 3392 cm−1, C—H stretching peak at 2928 cm−1, and C—O—) stretching peaks at 1179, 1085, and 1038 cm−1 compatible with the bonds of ether and hydroxyl groups, methoxy (O-CH3) group stretching peak at 2863 cm−1, and crystallized water peak at 1641 cm−1, respectively [46]. The FTIR spectrum of pure AVR exhibited N—H stretching vibrations at 3442 cm−1, O—H stretching vibrations at 3149 cm−1, O—H stretching at 2687 cm−1, C O stretching at 1698 cm−1, and N—H bending at 1631 cm−1 [47]. Formation of the Mβ-CD:AVR complex was suggested by the peak variation and a decrease in the absorption band strength in the inclusion of complex spectra. The FTIR spectrum of CP showed that all absorption bands were of the same strength, revealing little interaction between AVR and Mβ-CD. Noticeable characteristic peaks of AVR were observed at approximately 1200–1600 cm−1, but the strength of the absorption bands in the range 3000–3450 cm−1 was lower for the inclusion complex, suggesting the formation of Mβ-CD:AVR. The FTIR spectra of Mβ-CD:AVR complexes (PM and KM) significantly differed from those of Mβ-CD and AVR complexes, showing a larger and wider peak in the region of the —COOH group at approximately 1698 cm−1, as well as an increase in C O stretching due to addition of —COOH groups. All other diffraction peaks in this region were altered in AVR drug formulations using the two types of preparation procedures (PM and KM), indicating that AVR and Mβ-CD have significant effects. The AVR peaks at 3442 cm−1 (N—H stretching) and 3149 cm−1 (O—H stretching) were present in all compositions; however, the FTIR peaks of AVR at 2500–2000 cm−1 were eliminated in the BS compositions, suggesting the formation of Mβ-CD:AVR inclusion complexes. Differences in the FTIR spectra of the PM, KM, and BS samples, such as peak shifts or intensity reductions up to nearly a complete absence, showed varying degrees of interaction and amorphization in the Mβ-CD:AVR inclusion complex.
Fig. 2.
(a) FTIR spectra and (b) XRD pattern of Mβ-CD, AVR, Mβ-CD:AVR inclusion complex by co-precipitation (CP), physical mixture (PM), kneading method (KM), and bath sonication (BS) method.
3.3. X-ray powder diffraction
The degree of crystallinity of AVR was determined by comparing the typical peaks and their intensities, and the Mβ-CD:AVR inclusion complex was examined. The XRD patterns of Mβ-CD, AVR, and Mβ-CD:AVR inclusion complex prepared using the CP, PM, KM, and BS methods are shown in Fig. 2b. The XRD pattern of Mβ-CD was characterized by only two distinctive peaks at diffraction angles of 2θ = 11.1° and 18.2°, indicating that the excipient was in an amorphous [46]. AVR showed a high degree of crystallinity based on the height of sharp peaks at 10.3°, 12.9°, 15.8°, 18.9°, 20.8°, 23.7°, 26.1°, and 29.2° [39]. The XRD pattern of the CP of the Mβ-CD:AVR inclusion complex showed multiple peaks at 11.3°, 18.5°, 19.3°, 23.0°, 23.7°, 25.0°, and 28.4°, indicating the crystalline nature of AVR. In the cases of PM (10.3°, 11.8°, 18.2°, 23.8°, 26.0°, and 29.1°) and KM (11.5°, 18.7°, 23.9°, and 28.7°), the XRD spectra appeared as the superposition of the single components. Both diffraction patterns showed decreased peak strengths, the absence of peaks, and loss of crystallinity, likely because of the amorphous nature of Mβ-CD. The FTIR data supports these results. In contrast, XRD examination of the BS-prepared sample demonstrated that the Mβ-CD:AVR inclusion complex was completely amorphous, with no crystalline characteristics in the diffraction peaks. Furthermore, once the AVR molecule was placed in the Mβ-CD cavity, it did not form a crystalline structure by interacting with other AVR molecules. As no distinct diffraction angles were observed, the final product was amorphous, which agrees with the DSC results.
3.4. Thermal analysis
DSC and TGA can be used to assess variations in the physical and chemical properties of materials and determine the formation of Mβ-CD:AVR inclusion complexes (Fig. 3 ). The DSC curve of Mβ-CD exhibited a typical broad effect at 50 °C, corresponding to the loss of water molecules, as shown in Fig. 3a [48]. Another peak in an endothermic position was observed at a higher temperature of 365 °C, indicating Mβ-CD breakdown. Owing to the moisture loss, the AVR thermogram showed an exothermic peak at 105 °C and a sudden intense peak at 260 °C, corresponding to the melting point [9]. However, both peaks emerged and significantly shifted to the Mβ-CD endothermic peak in the CP of the Mβ-CD:AVR inclusion complex, and the decreased intensity of the peak indicated partial complexation and/or association between AVR and Mβ-CD. The disappearance of the Mβ-CD peak supported that AVR was uniformly dispersed in the complex and that the PM of the Mβ-CD:AVR inclusion complex was rather amorphous [49]. The KM method for the Mβ-CD:AVR inclusion complex DSC thermogram revealed a strong endothermic peak at 350 °C and broad peaks at 150 °C. As a result, the endothermic peaks of AVR and Mβ-CD were significantly altered in the KM sample. An endothermic signal was consistent with AVR melting on the DSC thermogram of the BS sample for the Mβ-CD:AVR inclusion complex did not appear at approximately 260 °C, demonstrating that AVR was dispersed in the Mβ-CD cavity. The AVR interaction in the Mβ-CD inclusion complex structure was confirmed based on this behavior. An AVR peak was observed in the CP, PM, and KM samples of the Mβ-CD:AVR inclusion complex. AVR shifted from a crystalline to an amorphous, which agrees with the XRD results.
Fig. 3.
(a) Differential scanning calorimetry and (b) thermogravimetric analysis of Mβ-CD, AVR, Mβ-CD:AVR inclusion complex by co-precipitation (CP), physical mixture (PM), kneading method (KM), and bath sonication (BS) method.
TGA was performed to evaluate the thermal stability of Mβ-CD, AVR, and the Mβ-CD:AVR inclusion complex for the CP, PM, KM, and BS solid inclusion complexes scanned between 20 °C and 400 °C at 10 °C/min (Fig. 3b). When guest molecules are placed in the CD cavities, their melting, boiling, or sublimation points frequently change or disappear. The weight of Mβ-CD remained constant as the temperature was increased to 300 °C. At 340–390 °C, noticeable weight loss of Mβ-CD occurred, demonstrating degradation of the Mβ-CD molecules [50]. The initial step at 50–200 °C reflected the loss of water molecules in the hydrated crystals of AVR powder, and the second stage at 50–200 °C was related to the loss of water molecules in the hydrated crystals of AVR powder. Degradation of the side chain from the guanosine rings causes a second weight loss at 295 °C, resulting in non-bonded guanosine [51]. The first melting step in the CP of the Mβ-CD:AVR inclusion complex began at 244 °C and persisted till 287 °C. The melting temperature was related to an initial weight loss of 22.5%. The second, larger section started at 341 °C and increased to 390 °C, resulting in weight loss of 38.4% at the decomposition temperature. There were two weight loss phases in the Mβ-CD:AVR inclusion complex prepared by PM because of Mβ-CD evaporation (below 84 °C) and AVR disintegration (241 °C). The TGA thermograms showed that the major degradation phase of Mβ-CD changed from 281 °C to 380 °C, with a lower weight loss ratio (45.8%) than pristine Mβ-CD. The TGA curves of KM and BS of Mβ-CD:AVR inclusion complexes revealed three different stages of mass loss, which was attributable to water evaporation below 100 °C and AVR evaporation at 170–240 °C and 147–237 °C. At 364 °C and 369 °C, the Mβ-CD:AVR inclusion complexes prepared by KM and BS showed a moderate and slow reduction, indicating that the inclusion structure was well-protected for AVR. The decomposition of Mβ-CD was assigned to the second stage at approximately 375 °C, which was supported by the Mβ-CD curve, with roughly 70% and 72% of sharp mass losses in both samples (KM and BS) including Mβ-CD [51]. Mβ-CD:AVR inclusion complex BS was substantially more thermally stable compared to the Mβ-CD:AVR inclusion complexes prepared using CP, PM, and KM. Thus, compared to pure AVR, thermal evaporation of AVR in the Mβ-CD:AVR inclusion complex occurred at a higher temperature, indicating that an inclusion complex of AVR and Mβ-CD was successfully developed. These findings agree with those of the characterization results, indicating that AVR was present in the Mβ-CD cavity.
3.5. Morphology
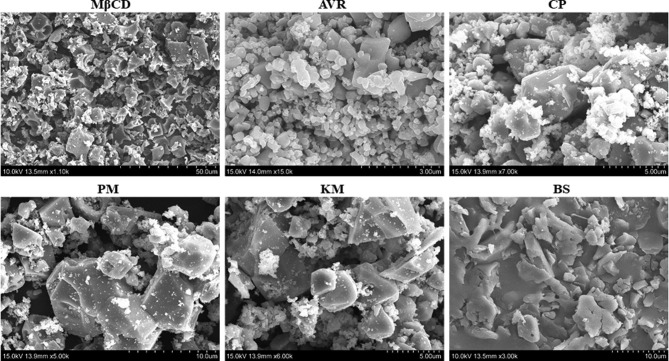
Fig. 4 shows the surface morphology of Mβ-CD, AVR, and Mβ-CD:AVR inclusion complexes prepared by CP, PM, KM, and BS analyzed using SEM. Mβ-CD is composed of amorphous spherical particles, whereas AVR is composed of crystals of variable form and size [52], [53]. The distinctive AVR crystals, mixed with the Mβ-CD spherical particles or adhering to their surface, were visible in the SEM image of the CP-prepared Mβ-CD:AVR inclusion complex system, revealing the presence of the crystalline AVR drug. This indicates that in the solid state, the two species did not interact. The morphologies of the PM and KM particles of the Mβ-CD:AVR inclusion complexes showed considerable deformation and aspect, indicating the presence of a new solid phase. These findings suggest that the crystalline structure of the systems was changed or the presence of novel solid inclusion complexes. However, the original shape of AVR and Mβ-CD disappeared in the SEM image of the BS-prepared Mβ-CD:AVR inclusion complex system because of the irregular amorphous particles. These modifications indicate that the inclusion complex had been successfully formed, which agrees with the results of XRD and TGA. The BS method is among the most reliable methods for creating inclusion compounds and is widely used in food and medicine applications.
Fig. 4.
Scanning electron microscopy analysis of Mβ-CD, AVR, Mβ-CD:AVR inclusion complex by co-precipitation (CP), physical mixture (PM), kneading method (KM), and bath sonication (BS) method.
3.6. Phase solubility studies
Phase solubility analysis was performed by plotting the quantity of AVR against the concentration of Mβ-CD. As an inclusion complex formed between AVR and Mβ-CD, the aqueous solubility of AVR gradually increased in a concentration-dependent manner as a factor of the Mβ-CD concentration, as observed in the phase solubility diagram drawn using UV–Vis measurements (Fig. 5 ). Hydrophobic interactions between AVR and Mβ-CD may be the underlying mechanism of this effect. According to Higuchi and Connors [42], the phase solubility diagram can be classified as a type AL. The formation of an Mβ-CD:AVR inclusion complex between AVR and the Mβ-CD complexing agent was associated with enhanced AVR solubility in aqueous Mβ-CD solutions. Furthermore, in the tested concentration range, greater values were observed for the regression coefficient (R2 = 0.9961), slope (0.12), and intercept (0.45). Formation of a 1:1 Mβ-CD:AVR inclusion complex as indicated by the linear curve obtained under the experimental conditions. Previous studies showed that AVR tends to form 1:1 inclusion complexes with HPβ-CD [9] and β-CD [41], [53], [54]. The Mβ-CD:AVR inclusion complex had apparent stability constant (Ks) of 75.7 M−1 and a complexation efficiency of 1.36.
Fig. 5.
Phase solubility diagrams of Mβ-CD:AVR inclusion complex systems (n = 3).
3.7. Encapsulation efficiency and drug content
The two most significant metrics for evaluating the quality of an inclusion complex are drug loading content and encapsulation efficiency. The ability of Mβ-CD to encapsulate AVR is directly reflected by the encapsulation efficiency. Based on the UV–Vis spectrophotometer measurements, the encapsulation efficiency (EE) and drug content (DC) were determined (Table 1 ). The different Mβ-CD:AVR inclusion complex preparation methods substantially impacted the encapsulation efficiency and drug; these values for AVR in the Mβ-CD:AVR inclusion complex (CP, PM, KM, and BS) were approximately 64.3 ± 2.11, 58.7 ± 1.83, 74.8 ± 0.98, and 89.5 ± 2.35%, respectively. In addition, the drug content of the Mβ-CD:AVR inclusion complexes (CP, PM, KM, and BS) ranged from 3.72 ± 0.11% to 5.84 ± 0.02%. Of the four different Mβ-CD:AVR inclusion complexes, the BS method for preparing the Mβ-CD:AVR inclusion complex was more efficient than the other methods. AVR was efficiently loaded into Mβ-CD, which has a hydrophilic outer surface and can readily encapsulate AVR to form the Mβ-CD:AVR inclusion complex [55]. Thus, the Mβ-CD:AVR inclusion complex showed the best effect at a molar ratio of 1:1, likely because the hydrophobic cavity of Mβ-CD has sufficient space to encapsulate and interact with AVR through hydrogen bonding and van der Waals forces.
Table 1.
Encapsulation efficiency (EE) and drug content (DC) of Mβ-CD:AVR inclusion complexes prepared by different methods (n = 3).
| Mβ-CD:AVR inclusion complex | CP | PM | KM | BS |
|---|---|---|---|---|
| EE% | 64.3 ± 2.11 | 58.7 ± 1.83 | 74.8 ± 0.98 | 89.5 ± 2.35 |
| DC% | 4.82 ± 0.01 | 3.72 ± 0.11 | 5.63 ± 0.12 | 5.84 ± 0.02 |
3.8. Molecular docking
To support rational drug design, molecular docking techniques can be utilized to determine the binding affinities of various ligands for the target protein structure [56]. We performed docking studies to examine the interaction between AVR and the Mβ-CD cavity. We found that AVR interacts in a 1:1 ratio with Mβ-CD, with optimum stability constant and complexation efficiency. We chose the top five complex docking molecules (Mβ-CD:AVR inclusion complex and Mβ-CD:AVR interaction with SARS-CoV-2 (3CLPro) protease inhibitors) based on the integrated docking scores for bond analysis of structures obtained from PatchDock and FireDock (Table 2 and Fig. 6, Fig. 7 ) [57], [58].
Table 2.
Computed using PatchDock and FireDock servers scores of the top 5 docked models of Mβ-CD:AVR inclusion complex (a) and Mβ-CD:AVR inclusion complex interaction with SARS-CoV-2 (3CLPro) protease inhibitors (b).
| (a) |
Mβ-CD:AVR |
||||||
|---|---|---|---|---|---|---|---|
| Patchdock server |
FireDock server |
||||||
| S. No. | Score | Area (Å2) | ACE kal/mol | Global Energy kal/mol | Attractive VdW kal/mol | Repulsive VdW kal/mol | ACE kal/mol |
| 1 | 2522 | 330.10 | −313.77 | −41.51 | −15.99 | 5.28 | −14.07 |
| 2 | 2520 | 333.90 | −296.01 | −41.23 | −14.49 | 4.79 | −14.67 |
| 3 | 2496 | 304.60 | −301.74 | −41.03 | −14.70 | 5.36 | −14.71 |
| 4 | 2470 | 329.80 | −299.80 | −40.76 | −15.37 | 4.63 | −13.91 |
| 5 | 2462 | 337.00 | −313.37 | −40.74 | −15.39 | 6.76 | −15.08 |
| (b) | Mβ-CD:AVR/SARS-CoV-2 (3CLPro) protease inhibitors | ||||||
| 1 | 5872 | 796.30 | −214.70 | −21.67 | −18.49 | 8.85 | −7.29 |
| 2 | 5768 | 679.90 | 100.86 | −13.56 | −16.86 | 5.22 | −2.94 |
| 3 | 5714 | 693.50 | 94.42 | −10.44 | −11.30 | 3.51 | −3.23 |
| 4 | 5702 | 675.60 | −210.64 | −9.61 | −7.04 | 2.65 | −5.56 |
| 5 | 5634 | 705.20 | −147.33 | −5.01 | −13.09 | 3.79 | 0.57 |
Fig. 6.
Schematic representation of the energetically most favorable complexes. Docking studies of the chemical and 3D structure of AVR, Mβ-CD, and Mβ-CD:AVR inclusion complex with stick and sphere arrangements. AVR, acyclovir; Mβ-CD, methyl β-cyclodextrin.
Fig. 7.
Schematic representation of the energetically most favorable complexes. Docking studies of Mβ-CD:AVR complex interaction with SARS-CoV-2 (3CLPro) protease inhibitors. Protein is shown in cartoon representation. AVR, acyclovir; Mβ-CD, methyl β-cyclodextrin.
The PatchDock geometric shape complementarity score, the approximate interface area size of the complex, and atomic contact energy were 2522, 330.10 Å, and − 313.77 kcal/mol, respectively, when docked with the Mβ-CD:AVR inclusion complex (Fig. 6). Furthermore, the FireDock global (binding) energy of the solution, attractive and repulsive van der Waals forces to the global binding energy, and atomic contact energy were − 41.51, −15.99, 5.28, and − 14.07 kcal/mol, respectively. The optimal docked structure showed a value of − 41.51 kcal/mol (this value is related to the free binding energy; an increased negative value indicates more free binding energy) [59], [60], [61], [62]. Fig. 6 shows that the Mβ-CD:AVR inclusion complex has stronger binding and was advantageous because of the hydrophobic characteristics of the Mβ-CD cavity, which contains more methyl groups than other β-CDs. As expected, complexation with Mβ-CD had a substantial solubilizing effect on AVR, revealing agreement between the computational simulations and experimental results. The ligands were evaluated using a molecular docking model until minimum energy was reached. The active site amino acid residues are examined in molecular docking studies on receptor-ligand interactions [63].
Inhibiting the viral 3CLpro in the treatment of COVID-19 would benefit from the strong binding affinity that anti-bacterial drugs, anti-hypertensive drugs, and natural substances with antiviral capabilities have exhibited to 3CLpro. In the battle against COVID-19, cyclodextrin (Mβ-CD) has also been employed to encapsulate potential antiviral targeting at various viral lifecycle stages. From these observations, we successfully docked the Mβ-CD:AVR inclusion complex with SARS-CoV-2 (3CLPro) protease inhibitors (PDB ID:7DJR) to predict the selected binding site along with the favored orientation of the antiviral drug Mβ-CD:AVR inclusion complex inside SARS-CoV-2 (3CLPro) protease inhibitors [64]. AVR structure-based best-docked ligand conformation was assessed to look at the precise chemical interaction in the receptors active pocket [65], [66]. The energetically favorable conformation of the docked poses revealed that the Mβ-CD:AVR inclusion complex bound to the protease inhibitors (Fig. 7). The Mβ-CD:AVR/SARS-CoV-2 (3CLPro) protease inhibitors showed the highest docking score (5872) and lowest atomic contact energy (−214 kcal/mol) in the PatchDock calculations. In addition, the lowest global energy, attractive van der Waals forces, and repulsive van der Waals forces to the global binding energy, and atomic contact energy were − 21.67, −18.49, 8.85, and − 7.29 kcal/mol in the FireDock calculations. The Mβ-CD:AVR inclusion complex also showed hydrogen interactions with active residues and bound to the active cavity of the SARS-CoV-2 main protease at the phenylalanine core residues, whereas fewer interactions were observed at another protein site [67], [68], [69], [70], [71], [72], [73]. The results in Table 2 show that the atomic contact energy was lower than that of the Mβ-CD:AVR inclusion complex, indicating that the Mβ-CD:AVR inclusion complex is preferentially bound to SARS-CoV-2 (3CLPro) protease inhibitors. Therefore, the Mβ-CD:AVR inclusion complex may function as an inhibitor of the SARS-CoV-2 (3CLPro) protease receptor.
4. Conclusions
A series of antiviral peptides may be useful for developing new therapeutic options for SARS-CoV-2. The Mβ-CD:AVR inclusion complex was predicted to strongly inhibit SARS-CoV-2 in silico computational studies with remarkable docking scores and binding energies. The Mβ-CD:AVR inclusion complex exhibited different physicochemical characteristics from free AVR and Mβ-CD. The association constant of the Mβ-CD:AVR inclusion complexes were determined, and a 1:1 stoichiometry was confirmed. Inclusion complexes and AVR, Mβ-CD, or their different preparation mixtures showed differences in their molecular structures and physicochemical characteristics. Moreover, the stability and morphology of AVR encapsulated in Mβ-CD were determined, and the increased solubility of AVR in the complex was confirmed. The Mβ-CD:AVR inclusion complex is bound strongly to the active sites of the protein target of SARS-CoV-2 (3CLPro) protease inhibitors, with predicted the highest docking score at 5872, atomic contact energy of − 214 kcal/mol, and global binding energy of − 21.67 kcal/mol. In conclusion, this Mβ-CD:AVR inclusion complex against the 3CLPro receptor of SARS-CoV-2 should be considered as a promising platform that could be prolonged to other antiviral drugs with the aim of effective AVR drug formulation and antiviral treatments.
CRediT authorship contribution statement
Sonaimuthu Mohandoss: Conceptualization, Methodology, Writing – original draft. Ramaraj Sukanya: Methodology, Investigation. Sivarasan Ganesan: Writing – review & editing. Fatemah H. Alkallas: Methodology, Investigation. Amira Ben Gouider Trabelsi: Conceptualization, Methodology, Investigation. Fedor V. Kusmartsev: Methodology, Investigation. Kuppu Sakthi Velu: Methodology, Investigation. Thambusamy Stalin: Conceptualization, Methodology, Investigation. Huang-Mu Lo: Validation, Writing – review & editing. Yong Rok Lee: Supervision, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (2021R1A2B5B02002436 and 2021R1F1A1061566). This research was funded by Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2022R223), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
References
- 1.Sharma A., Tiwari S., Deb M.K., Marty J.L. Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2): a global pandemic and treatment strategies. Int. J. Antimicrob. Agents. 2020;56 doi: 10.1016/j.ijantimicag.2020.106054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ng T.I., Correia I., Seagal J., Degoey D.A., Schrimpf M.R., Hardee D.J., Noey E.L., Kati W.M. Antiviral Drug Discovery for the Treatment of COVID-19 Infections. Viruses. 2022;14:1–27. doi: 10.3390/v14050961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vlachakis D., Papakonstantinou E., Mitsis T., Pierouli K., Diakou I., Chrousos G., Bacopoulou F. Molecular mechanisms of the novel coronavirus SARS-CoV-2 and potential anti-COVID19 pharmacological targets since the outbreak of the pandemic. Food Chem. Toxicol. 2020;146 doi: 10.1016/j.fct.2020.111805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prescott L. SARS-CoV-2 3CLpro whole human proteome cleavage prediction and enrichment/depletion analysis. Comput. Biol. Chem. 2022;98 doi: 10.1016/j.compbiolchem.2022.107671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mostafa A., Kandeil A., Elshaier Y.A.M.M., Kutkat O., Moatasim Y., Rashad A.A., Shehata M., Gomaa M.R., Mahrous N., Mahmoud S.H., Gaballah M., Abbas H., El Taweel A., Kayed A.E., Kamel M.N., El Sayes M., Mahmoud D.B., El-Shesheny R., Kayali G., Ali M.A. Fda-approved drugs with potent in vitro antiviral activity against severe acute respiratory syndrome coronavirus 2. Pharmaceuticals. 2020;13:1–24. doi: 10.3390/ph13120443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker V.S. ACYCLOVIR for SARS-CoV-2: An Old Drug with a New Therapeutic Purpose - An Observational Study. Int. J. Clin. Case rep. Rev. 2022;10(2) doi: 10.31579/2690-4861/199. In press. [DOI] [Google Scholar]
- 7.Donalisio M., Argenziano M., Rittà M., Bastiancich C., Civra A., Lembo D., Cavalli R. Acyclovir-loaded sulfobutyl ether-β-cyclodextrin decorated chitosan nanodroplets for the local treatment of HSV-2 infections. Int. J. Pharm. 2020;587 doi: 10.1016/j.ijpharm.2020.119676. [DOI] [PubMed] [Google Scholar]
- 8.Zielenkiewicz W., Koźbiał M., Golankiewicz B., Poznański J. Enhancement of aqueous solubility of tricyclic acyclovir derivatives by their complexation with hydroxypropyl-β-cyclodextrin. J. Therm. Anal. Calorim. 2010;101:555–560. doi: 10.1007/s10973-010-0847-0. [DOI] [Google Scholar]
- 9.Nair A.B., Attimarad M., Al-Dhubiab B.E., Wadhwa J., Harsha S., Ahmed M. Enhanced oral bioavailability of acyclovir by inclusion complex using hydroxypropyl-β-cyclodextrin. Drug Delivery. 2014;21:540–547. doi: 10.3109/10717544.2013.853213. [DOI] [PubMed] [Google Scholar]
- 10.Kalepu S., Nekkanti V. Insoluble drug delivery strategies: Review of recent advances and business prospects. Acta Pharmaceutica Sinica B. 2015;5:442–453. doi: 10.1016/j.apsb.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X., Zhang T., Lan Y., Wu B., Shi Z. Nanosuspensions Containing Oridonin/HP-β-Cyclodextrin Inclusion Complexes for Oral Bioavailability Enhancement via Improved Dissolution and Permeability. AAPS PharmSciTech. 2016;17:400–408. doi: 10.1208/s12249-015-0363-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saokham P., Muankaew C., Jansook P., Loftsson T. Solubility of cyclodextrins and drug/cyclodextrin complexes. Molecules. 2018;23:1–15. doi: 10.3390/molecules23051161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Real D.A., Bolaños K., Priotti J., Yutronic N., Kogan M.J., Sierpe R., Donoso-González O. Cyclodextrin-modified nanomaterials for drug delivery: Classification and advances in controlled release and bioavailability. Pharmaceutics. 2021;13 doi: 10.3390/pharmaceutics13122131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohandoss S., Maniyazagan M., Stalin T. A highly selective dual mode detection of Fe3+ ion sensing based on 1,5-dihydroxyanthraquinone in the presence of β-cyclodextrin. Mater. Sci. Eng., C. 2015;48:94–102. doi: 10.1016/j.msec.2014.11.060. [DOI] [PubMed] [Google Scholar]
- 15.Mohandoss S., Sivakamavalli J., Vaseeharan B., Stalin T. Fluorometric sensing of Pb2+ and CrO42- ions through host-guest inclusion for human lung cancer live cell imaging. RSC Adv. 2015;5:101802–101818. doi: 10.1039/c5ra17910f. [DOI] [Google Scholar]
- 16.Mohandoss S., Sivakamavalli J., Vaseeharan B., Stalin T. Host-guest molecular recognition based fluorescence On-Off-On chemosensor for nanomolar level detection of Cu2+ and Cr2O72- ions: Application in XNOR logic gate and human lung cancer living cell imaging, Sensors and Actuators, B. Chemical. 2016;234:300–315. doi: 10.1016/j.snb.2016.04.148. [DOI] [Google Scholar]
- 17.Maniyazagan M., Mohandoss S., Sivakumar K., Stalin T. N-phenyl-1-naphthylamine / b -cyclodextrin inclusion complex as a new fluorescent probe for rapid and visual detection of Pd 2 +, Spectrochimica Acta Part A : Molecular and Biomolecular. Spectroscopy. 2014;133:73–79. doi: 10.1016/j.saa.2014.04.183. [DOI] [PubMed] [Google Scholar]
- 18.Mohandoss S., Stalin T. Photochemical and computational studies of inclusion complexes between β-cyclodextrin and 1,2-dihydroxyanthraquinones. Photochem. Photobiol. Sci. 2017;16:476–488. doi: 10.1039/c6pp00285d. [DOI] [PubMed] [Google Scholar]
- 19.Mohandoss S., Stalin T. A new fluorescent PET sensor probe for Co2+ ion detection: computational, logic device and living cell imaging applications. RSC Adv. 2017;7:16581–16593. doi: 10.1039/c6ra27497h. [DOI] [Google Scholar]
- 20.Sonaimuthu M., Balakrishnan S.B., Kuppu S.V., Veerakanellore G.B., Thambusamy S. Spectral and proton transfer behavior of 1,4-dihydroxylanthraquinone in aqueous and confined media; molecular modelling strategy. J. Mol. Liq. 2018;259:186–198. doi: 10.1016/j.molliq.2018.03.042. [DOI] [Google Scholar]
- 21.Mohandoss S., Atchudan R., Edison T.N.J.I., Mishra K., Tamargo R.J.I., Palanisamy S., Yelithao K., You S.G., Lee Y.R. Rapid response and highly selective sensing of adenosine based on novel photoluminescent vanadium nanoclusters anchored on MoS2 nanosheets. Sens. Actuators, B. 2020;306 doi: 10.1016/j.snb.2019.127581. [DOI] [Google Scholar]
- 22.Sonaimuthu M., Nerthigan Y., Swaminathan N., Sharma N., Wu H.F. Photoluminescent hydrophilic cyclodextrin-stabilized cysteine-protected copper nanoclusters for detecting lysozyme. Anal. Bioanal. Chem. 2020;412:7141–7154. doi: 10.1007/s00216-020-02847-7. [DOI] [PubMed] [Google Scholar]
- 23.Mohandoss S., Palanisamy S., Priya V.V., Mohan S.K., Shim J.-J., Yelithao K., You S., Lee Y.R. Excitation-dependent multiple luminescence emission of nitrogen and sulfur co-doped carbon dots for cysteine sensing, bioimaging, and photoluminescent ink applications. Microchem. J. 2021;167 doi: 10.1016/j.microc.2021.106280. [DOI] [Google Scholar]
- 24.Mohandoss S., Palanisamy S., You S., Shim J., Lee Y.R. Rapid detection of silver ions based on luminescent carbon nanodots for multicolor patterning, smartphone sensors, and bioimaging applications. Anal. Methods. 2021:5719–5726. doi: 10.1039/d1ay01746b. [DOI] [PubMed] [Google Scholar]
- 25.Mohandoss S., Khanal H.D., Palanisamy S., You S., Shim J., Lee Y.R. Multiple heteroatom-doped photoluminescent carbon dots for ratiometric detection of Hg2+ ions in cell imaging and environmental applications. Anal. Methods. 2022:635–642. doi: 10.1039/d1ay02077c. [DOI] [PubMed] [Google Scholar]
- 26.Mohandoss S., Palanisamy S., You S., Rok Y. Synthesis of cyclodextrin functionalized photoluminescent metal nanoclusters for chemoselective Fe3+ ion detection in aqueous medium and its applications of paper sensors and cell imaging. J. Mol. Liq. 2022;356 doi: 10.1016/j.molliq.2022.118999. [DOI] [Google Scholar]
- 27.Umapathi R., Sonwal S., Ji M., Mohana G., Lee E., Jeon T., Kang S., Oh M., Suk Y. Colorimetric based on-site sensing strategies for the rapid detection of pesticides in agricultural foods : New horizons, perspectives, and challenges. Coord. Chem. Rev. 2021;446 doi: 10.1016/j.ccr.2021.214061. [DOI] [Google Scholar]
- 28.Umapathi R., Majid S., Sonwal S., Mohana G., Suk Y. Portable electrochemical sensing methodologies for on-site detection of pesticide residues in fruits and vegetables. Coord. Chem. Rev. 2022;453 doi: 10.1016/j.ccr.2021.214305. [DOI] [Google Scholar]
- 29.Umapathi R., Venkateswara C., Majid S., Mohana G., Kumar K., Oh M., Pil J., Suk Y. Recent advances in the use of graphitic carbon nitride-based composites for the electrochemical detection of hazardous contaminants. Coord. Chem. Rev. 2022;470 doi: 10.1016/j.ccr.2022.214708. [DOI] [Google Scholar]
- 30.Umapathi R., Park B., Sonwal S., Mohana G., Cho Y., Suk Y. Advances in optical-sensing strategies for the on-site detection of pesticides in agricultural foods. Trends Food Sci. Technol. 2022;119:69–89. doi: 10.1016/j.tifs.2021.11.018. [DOI] [Google Scholar]
- 31.Rani G.M., Umapathi R., Cho Y., Huh Y.S., Kim E., Park S. Sowing kernels for food safety : Importance of rapid on-site detction of pesticide residues in agricultural foods. 2022:1–11. doi: 10.1002/fft2.166. [DOI] [Google Scholar]
- 32.Braga S.S., Barbosa J.S., Santos N.E., El-Saleh F., Paz F.A.A. Cyclodextrins in antiviral therapeutics and vaccines. Pharmaceutics. 2021;13 doi: 10.3390/pharmaceutics13030409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jicsinszky L., Martina K., Cravotto G. Cyclodextrins in the antiviral therapy. J. Drug Delivery Sci. Technol. 2021;64 doi: 10.1016/j.jddst.2021.102589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garrido P.F., Calvelo M., Blanco-González A., Veleiro U., Suárez F., Conde D., Cabezón A., Piñeiro Á., Garcia-Fandino R. The Lord of the NanoRings: Cyclodextrins and the battle against SARS-CoV-2. Int. J. Pharm. 2020;588 doi: 10.1016/j.ijpharm.2020.119689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piñeiro Á., Pipkin J., Antle V., Garcia-Fandino R. Aggregation versus inclusion complexes to solubilize drugs with cyclodextrins. A case study using sulphobutylether-β-cyclodextrins and remdesivir. J. Mol. Liq. 2021;343 doi: 10.1016/j.molliq.2021.117588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dermawan D., Prabowo B.A., Rakhmadina C.A. In silico study of medicinal plants with cyclodextrin inclusion complex as the potential inhibitors against SARS-CoV-2 main protease (Mpro) and spike (S) receptor. Inf. Med. Unlocked. 2021;25 doi: 10.1016/j.imu.2021.100645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Santos P.S., Souza L.K.M., Araujo T.S.L., Medeiros J.V.R., Nunes S.C.C., Carvalho R.A., Pais A.C.C., Veiga F.J.B., Nunes L.C.C., Figueiras A. Methyl-β-cyclodextrin inclusion complex with β βcaryophyllene: Preparation, characterization, and improvement of pharmacological activitie. ACS Omega. 2017;2:9080–9094. doi: 10.1021/acsomega.7b01438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu B., Li W., Zhao J., Liu Y., Zhu X., Liang G. Physicochemical characterisation of the supramolecular structure of luteolin/cyclodextrin inclusion complex. Food Chem. 2013;141:940–945. doi: 10.1016/j.foodchem.2013.03.097. [DOI] [PubMed] [Google Scholar]
- 39.Plessing Rossel C., Sepúlveda Carreño J., Rodríguez-Baeza M., Bernabé Alderete J. Inclusion Complex of the Antiviral Drug Acyclovir with Cyclodextrin in Aqueous Solution and in Solid Phase. Quim. Nova. 2000;23:749–752. [Google Scholar]
- 40.Pedotti S., Pistarà V., Cannavà C., Carbone C., Cilurzo F., Corsaro A., Puglisi G., Ventura C.A. Synthesis and physico-chemical characterization of a β-cyclodextrin conjugate for sustained release of Acyclovir. Carbohydr. Polym. 2015;131:159–167. doi: 10.1016/j.carbpol.2015.05.071. [DOI] [PubMed] [Google Scholar]
- 41.Noviza D., Afrianto D., Halim A. The effect of milling process on acyclovir inclution complex with beta cyclodextrin. Journal of Chemical and Pharmaceutical Research. 2015;7:354–362. [Google Scholar]
- 42.alizadeh N., Poorbagher N. Host-guest inclusion complexes of sulfabenzamide with β- and methyl-β-cyclodextrins: Characterization, antioxidant activity and DFT calculation. J. Mol. Struct. 2022;1260 doi: 10.1016/j.molstruc.2022.132809. [DOI] [Google Scholar]
- 43.Luengo J., Aránguiz T., Sepúlveda J., Hernández L., Von Plessing C. Preliminary pharmacokinetic study of different preparations of acyclovir with β-cyclodextrin. J. Pharm. Sci. 2002;91:2593–2598. doi: 10.1002/jps.10245. [DOI] [PubMed] [Google Scholar]
- 44.Sbârcea L., Tănase I.M., Ledeți A., Cîrcioban D., Vlase G., Barvinschi P., Miclău M., Văruţ R.M., Suciu O., Ledeți I. Risperidone/randomly methylated β-cyclodextrin inclusion complex—compatibility study with pharmaceutical excipients. Molecules. 2021;26:1–17. doi: 10.3390/molecules26061690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y., Deng Z., Wang X., Shi Y., Lu Y., Fang S., Liang X. Formononetin/methyl-β-cyclodextrin inclusion complex incorporated into electrospun polyvinyl-alcohol nanofibers: Enhanced water solubility and oral fast-dissolving property. Int. J. Pharm. 2021;603 doi: 10.1016/j.ijpharm.2021.120696. [DOI] [PubMed] [Google Scholar]
- 46.Ribeiro A., Figueiras A., Santos D., Veiga F. Preparation and solid-state characterization of inclusion complexes formed between miconazole and methyl-β-cyclodextrin. AAPS PharmSciTech. 2008;9:1102–1109. doi: 10.1208/s12249-008-9143-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lembo D., Swaminathan S., Donalisio M., Civra A., Pastero L., Aquilano D., Vavia P., Trotta F., Cavalli R. Encapsulation of Acyclovir in new carboxylated cyclodextrin-based nanosponges improves the agent’s antiviral efficacy. Int. J. Pharm. 2013;443:262–272. doi: 10.1016/j.ijpharm.2012.12.031. [DOI] [PubMed] [Google Scholar]
- 48.Olga Cardoso A.F. Preparation and characterization of Trimethoprim inclusion complex with Methyl-β-Cyclodextrin and determination of its antimicrobial activity. Pharmaceutica Analytica Acta. 2015;06:6–10. doi: 10.4172/2153-2435.1000405. [DOI] [Google Scholar]
- 49.De Freitas M.R., Rolim L.A., Soares M.F.D.L.R., Rolim-Neto P.J., De Albuquerque M.M., Soares-Sobrinho J.L. Inclusion complex of methyl-β-cyclodextrin and olanzapine as potential drug delivery system for schizophrenia. Carbohydr. Polym. 2012;89:1095–1100. doi: 10.1016/j.carbpol.2012.03.072. [DOI] [PubMed] [Google Scholar]
- 50.Li S., Yuan L., Zhang B., Zhou W., Wang X., Bai D. Photostability and antioxidant activity studies on the inclusion complexes of: Trans -polydatin with β-cyclodextrin and derivatives. RSC Adv. 2018;8:25941–25948. doi: 10.1039/c8ra04778b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Celebioglu A., Uyar T. Electrospun formulation of acyclovir/cyclodextrin nanofibers for fast-dissolving antiviral drug delivery. Mater. Sci. Eng., C. 2021;118 doi: 10.1016/j.msec.2020.111514. [DOI] [PubMed] [Google Scholar]
- 52.Figueiras A., Ribeiro L., Vieira M.T., Veiga F. Preparation and physicochemical characterization of omeprazole:methyl-beta- cyclodextrin inclusion complex in solid state. J. Incl. Phenom. Macrocycl. Chem. 2007;57:173–177. doi: 10.1007/s10847-006-9200-4. [DOI] [Google Scholar]
- 53.Maher M., Pandey M., Adhikari L., Semalty A., Semalty M. Effect of Hydrophilic Excipients on Cyclodextrin Complexes of Acyclovir in Improving Solubility, Dissolution and Permeability. Lett. Drug Des. Discovery. 2016;13:771–780. doi: 10.2174/1570180813666160517161058. [DOI] [Google Scholar]
- 54.Di Donato C., Iacovino R., Isernia C., Malgieri G., Varela-Garcia A., Concheiro A., Alvarez-Lorenzo C. Polypseudorotaxanes of pluronic® f127 with combinations of α-and β-cyclodextrins for topical formulation of acyclovir. Nanomaterials. 2020;10 doi: 10.3390/nano10040613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chulurks S., Jitapunkul K., Katanyutanon S., Toochinda P., Lawtrakul L. Stability enhancement and skin permeation application of nicotine by forming inclusion complex with β-cyclodextrin and methyl-β-cyclodextrin. Sci. Pharm. 2021;89 doi: 10.3390/scipharm89040043. [DOI] [Google Scholar]
- 56.Pinzi L., Rastelli G. Molecular docking: Shifting paradigms in drug discovery. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20184331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schneidman-Duhovny D., Inbar Y., Nussinov R., Wolfson H.J. PatchDock and SymmDock: Servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33:363–367. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mashiach E., Schneidman-Duhovny D., Andrusier N., Nussinov R., Wolfson H.J. FireDock: a web server for fast interaction refinement in molecular docking. Nucleic Acids Res. 2008;36:229–232. doi: 10.1093/nar/gkn186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mohandoss S., Atchudan R., Immanuel Edison T.N.J., Mandal T.K., Palanisamy S., You S.G., Napoleon A.A., Shim J.J., Lee Y.R. Enhanced solubility of guanosine by inclusion complexes with cyclodextrin derivatives: Preparation, characterization, and evaluation. Carbohydr. Polym. 2019;224 doi: 10.1016/j.carbpol.2019.115166. [DOI] [PubMed] [Google Scholar]
- 60.Mohandoss S., Atchudan R., Edison T.N.J.I., Mishra K., Tamargo R.J.I., Palanisamy S., Yelithao K., You S.G., Napoleon A.A., Lee Y.R. Enhancement of solubility, antibiofilm, and antioxidant activity of uridine by inclusion in β-cyclodextrin derivatives. J. Mol. Liq. 2020;306 doi: 10.1016/j.molliq.2020.112849. [DOI] [Google Scholar]
- 61.Mohandoss S., Edison T.N.J.I., Atchudan R., Palanisamy S., Prabhu N.M., Napoleon A.A., You S.G., Lee Y.R. Ultrasonic-assisted efficient synthesis of inclusion complexes of salsalate drug and β-cyclodextrin derivatives for potent biomedical applications. J. Mol. Liq. 2020;319 doi: 10.1016/j.molliq.2020.114358. [DOI] [Google Scholar]
- 62.Mohandoss S., Palanisamy S., You S.G., Shim J.J., Y. Rok Lee, Ultrasonication-assisted host–guest inclusion complexes of β-cyclodextrins and 5-hydroxytryptophan: Enhancement of water solubility, thermal stability, and in vitro anticancer activity. J. Mol. Liq. 2021;336 doi: 10.1016/j.molliq.2021.116172. [DOI] [Google Scholar]
- 63.Azam S.S., Abbasi S.W. Molecular docking studies for the identification of novel melatoninergic inhibitors for acetylserotonin-O-methyltransferase using different docking routines. Theor. Biol. Med. Modell. 2013;10:1–16. doi: 10.1186/1742-4682-10-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Deetanya P., Hengphasatporn K., Wilasluck P., Shigeta Y., Rungrotmongkol T., Wangkanont K. Interaction of 8-anilinonaphthalene-1-sulfonate with SARS-CoV-2 main protease and its application as a fluorescent probe for inhibitor identification, Computational and Structural. Biotechnol. J. 2021;19:3364–3371. doi: 10.1016/j.csbj.2021.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Choi R.H., Kim H.T. Density functional theory study of the dimer structure of amyloidogenic active sequence VQIVYK. Bull. Korean Chem. Soc. 2021;42(12):1579–1584. doi: 10.1002/bkcs.12416. [DOI] [Google Scholar]
- 66.Yu Q., Wu Y., Kang T., Choo J. Development of surface-enhanced Raman scattering-based immunoassay platforms using hollow Au nanostars for reliable SARS-CoV-2 diagnosis. Bull. Korean Chem. Soc. 2021;42(12):1699–1705. doi: 10.1002/bkcs.12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Baldwin E.T., Bhat T.N., Gulnik S., Liu B., Topol I.A., Kiso Y., Mimoto T., Mitsuya H., Erickson J.W. Structure of HIV-1 protease with KNI-272, a tight-binding transition-state analog containing allophenylnorstatine. Structure. 1995;3:581–590. doi: 10.1016/S0969-2126(01)00192-7. [DOI] [PubMed] [Google Scholar]
- 68.Rut W., Groborz K., Zhang L., Sun X., Zmudzinski M., Rolf Hilgenfeld M.D. Substrate specificity profiling of SARS-CoV-2 M pro protease provides basis for anti-COVID-19 drug design Institute of Biochemistry, Center for Structural and Cell Biology in Medicine, University of. BioRxiv. 2020 March 8. [Google Scholar]
- 69.Chuck C.P., Chong L.T., Chen C., Chow H.F., Wan D.C.C., Wong K.B. Profiling of substrate specificity of SARS-CoV 3CLpro. PLoS ONE. 2010;5:1–7. doi: 10.1371/journal.pone.0013197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Muramatsu T., Takemoto C., Kim Y.T., Wang H., Nishii W., Terada T., Shirouzu M., Yokoyama S. SARS-CoV 3CL protease cleaves its C-terminal autoprocessing site by novel subsite cooperativity. PNAS. 2016;113:12997–13002. doi: 10.1073/pnas.1601327113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sacco M.D., Ma C., Lagarias P., Gao A., Townsend J.A., Meng X., Dube P., Zhang X., Hu Y., Kitamura N., Hurst B., Tarbet B., Marty M.T., Kolocouris A., Xiang Y., Chen Y., Wang J. Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against Mpro and cathepsin L. Sci. Adv. 2020;6 doi: 10.1126/sciadv.abe0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shivanika C., Deepak Kumar S., Ragunathan V., Tiwari P., Sumitha A., Brindha Devi P. Molecular docking, validation, dynamics simulations, and pharmacokinetic prediction of natural compounds against the SARS-CoV-2 main-protease. J. Biomol. Struct. Dyn. 2022;40:585–611. doi: 10.1080/07391102.2020.1815584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hatmal M.M., Alshaer W., Al-Hatamleh M.A.I., Hatmal M., Smadi O., Taha M.O., Oweida A.J., Boer J.C., Mohamud R., Plebanski M. Comprehensive Structural and Molecular Comparison of Spike Proteins of SARS-CoV-2, SARS-CoV and MERS-CoV, and Their Interactions with ACE2. Cells. 2020;9 doi: 10.3390/cells9122638. [DOI] [PMC free article] [PubMed] [Google Scholar]