Abstract
The gut microbiome influences host physiology and pathophysiology through a number of pathways, one of which is microbial production of chemical metabolites which interact with host signaling pathways. Short chain fatty acids (SCFAs) are a class of gut microbial metabolites known to activate multiple signaling pathways in the host. Growing evidence indicates that the gut microbiome is linked to blood pressure, that SCFAs modulate blood pressure regulation, and that delivery of exogenous SCFAs lowers blood pressure. Given that hypertension is a key risk factor for cardiovascular disease, the examination of novel contributors to blood pressure regulation has the potential to lead to novel approaches or treatments. Thus, this review will discuss SCFAs with a focus on their host G protein-coupled receptors including G protein-coupled receptor 41 (GPR41), GPR43, and GPR109A, as well as olfactory receptor 78 (OLFR78) and OLFR558. This includes a discussion of the ligand profiles, G protein coupling, and tissue distribution of each receptor. We will also review phenotypes relevant to blood pressure regulation which have been reported to date for Gpr41, Gpr43, Gpr109a, and Olfr78 KO mice. In addition, we will consider how SCFA signaling influences physiology at baseline, and, how SCFA signaling may contribute to blood pressure regulation in settings of hypertension. In sum, this review will integrate current knowledge regarding how SCFAs and their receptors regulate blood pressure.
Keywords: Gpr41, Gpr43, Gpr109a, Olfr78, blood pressure, SCFAs
Introduction
Hypertension is a major risk factor for cardiovascular disease and stroke, as well as a hallmark of obesity, diabetes and metabolic syndrome1–3. Successful hypertension prevention and treatment are key to decreasing the risk of those diseases. Based on the American Heart Association’s definition of hypertension (systolic blood pressure (SBP)⩾130 mmHg, diastolic blood pressure (DBP)⩾80 mmHg), it is estimated that 47% of US adults, approximately 121.5 million adults4, are hypertensive (52% for males and 43% for females1,5). For those <65 years of age hypertension is more prevalent in males; the opposite is true for those who are ⩾65 years of age1. Hypertension incidence also varies by race, with black men and black women both having a cumulative incidence of 76% by age 55, as compared to 54% for white men and 40% for white women6. Compounding these issues, 39% of US adults with hypertension are unaware that they are hypertensive1.
Growing evidence indicates that the gut microbiota influences a wide variety of traits in the host organisms, including atherosclerosis7,8, irritable bowel syndrome9,10, immune disorders11–13, and kidney disease14–17. A primary way that microbes affect the host is via the production of metabolites that are absorbed into the host bloodstream, where they can activate signaling pathways to alter physiology. A particularly well-studied class of gut microbial metabolites are short chain fatty acids (SCFAs), which are simple straight-chain carboxylic acid molecules. In recent years, multiple groups have connected SCFAs to blood pressure regulation, and, to hypertension18–23. Here, we will review our current understanding of SCFAs, their receptors, and their effects on physiology.
Short Chain Fatty Acids (SCFAs)
Short chain fatty acids (SCFAs) are metabolites produced by microbial fermentation of dietary carbohydrates, and are perhaps the most well-studied of the gut microbial metabolites24. The term ‘SCFAs’ commonly refers to the three most abundant species of SCFAs, which are all 2-4 carbon straight-chain compounds (acetate, propionate, and butyrate) - this is how we will use the term in this review. However, SCFAs can also include shorter (formate) or longer (valerate) compounds, as well as branched compounds (isobutyrate, isovalerate, and 2-methylbutanoate).
Indigestible complex carbohydrates in the large intestine undergo fermentation by anaerobic microbes, resulting in the production of metabolites including SCFAs25. The most abundant butyrate-producing bacteria in the colon belongs to the Clostridia clusters IV and XIVa, which includes species related to Firmicutes, Eubacterium and Roseburia26–28. Acetate and propionate are primarily produced by Bacteriodetes phyla29,30.
As referenced above, gut microbiota produce SCFAs as a byproduct of metabolism; in fact, the concentration of SCFAs in the colonic lumen is ~100mM31. The ratio of acetate:propionate:butyrate in the colon is approximately 60:20:2032,33. SCFAs transverse from the colonic lumen to the blood by monocarboxylate transporters34, as well as by diffusion35. SCFA levels within the circulating blood of the host are much lower than in the colonic lumen, albeit still substantial. For example, acetate, the most abundant of the SCFAs, is typically 50-500 μM in circulating plasma32,36–38. Plasma acetate levels, as well as the ratio of acetate:propionate:butyrate, varies based on diet25,39, but acetate is consistently the most abundant (ratios have been reported to vary from 40:26:34 to 78:17:539,40).
As previously stated, SCFAs are produced by the gut microbiota in quantities that significantly contribute to host circulating levels. Notably, host metabolism can also produce SCFAs, but it is unclear whether host SCFA production meaningfully contributes to host plasma levels. A study by Perry, et al found that plasma acetate, as well as plasma butyrate and propionate, are nearly undetectable in germ-free mice (that is, mice which entirely lack microbiota)37. However, a study from Kimura, et al reported that plasma acetate only decreases by ~40% in germ-free animals41. Furthermore, a recent report indicated that mice treated with antibiotics had dramatically suppressed SCFAs in the colonic lumen, but no change in plasma SCFAs42. Thus, although there is agreement that gut microbial metabolism contributes to circulating SCFAs, the precise contribution of gut vs host metabolism remains to be fully understood.
Cell Biology of SCFA Receptors
SCFAs circulating in the blood can alter signaling pathways via several different classes of host proteins, most notably histone deacetylases (HDACs) and G protein-coupled receptors (GPCRs). HDACs remove acetyl groups from histones, increasing the affinity between histones and DNA to repress transcription43. There are four classes of human HDACs based on their conserved sequences to yeast HDACs: I, II (including IIA and IIB), III, and IV43 . SCFAs, primarily butyrate, are prominent inhibitors of HDAC activity44,45. For example, in rats with cardiac hypertrophy, treatment with sodium butyrate was beneficial in reducing LV wall thickness, cardiomyocyte diameters, and hypertrophic indices46. It was suggested these effects were mediated by decreased mRNA expression of HDAC subtypes46. In another report, mice treated with SCFAs (acetate, butyrate, and propionate) showed reduced mRNA expression of HDAC subtypes in kidney tissue, and were protected against folic-acid nephropathy47. Results of this study suggest SCFA-mediated HDAC inhibition contributes to the pathophysiology of acute kidney injury.
SCFAs can also act as ligands and bind to host microbial metabolite-sensing GPCRs, and this is the primary focus of this review. GPCRs are cell surface receptors containing seven transmembrane alpha helices and an extracellular amino terminus48. Given their highly conserved sequences and structure, GPCRs are widely studied for their ability to sense a variety of stimuli49. To date, GPCRs that have been identified to bind SCFAs include: GPR41, GPR43, GPR109A, OR51E2, and OR51E150–53; the expression of these GPCRs in the top 10 single cell types is shown in Figure 1.
Figure 1. Top 10 single cell types expressing (A) Gpr43, (B) Gpr41, (C) Gpr109a, (D) Or51e2 (mouse ortholog: Olfr78), and (E) Or51e1 (mouse ortholog: Olfr558).
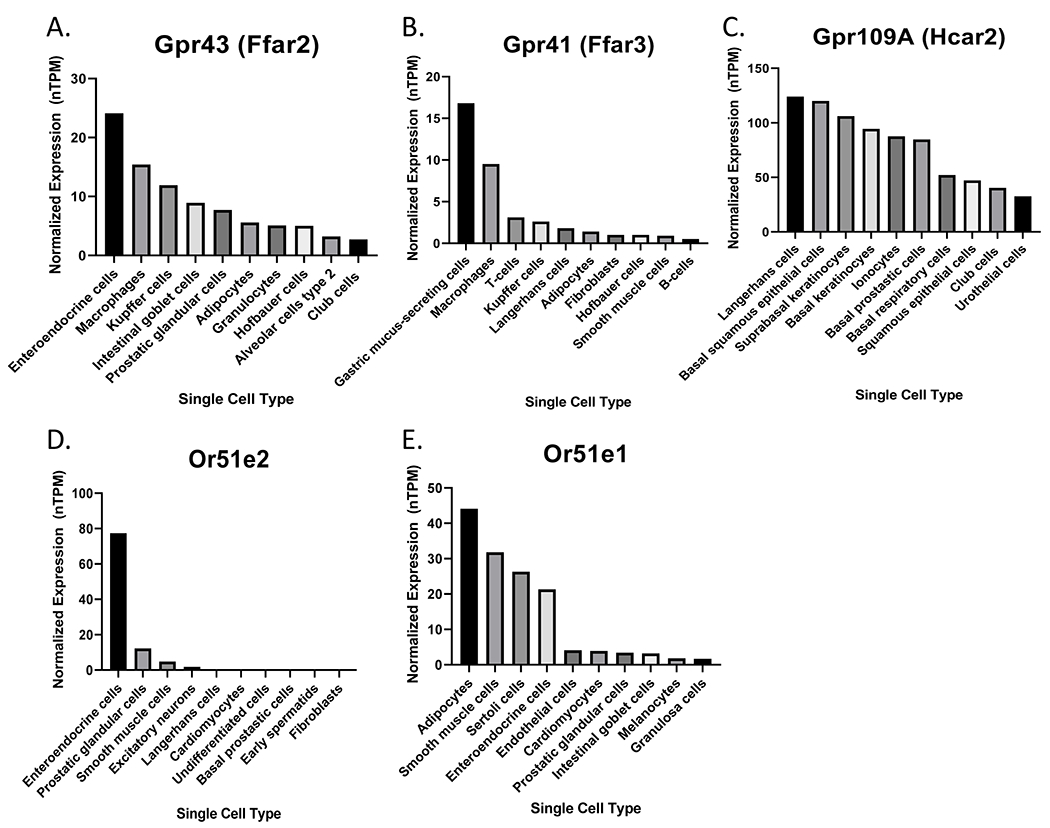
Values are reported in normalized transcripts per million (nTPM)134. Data available from Human Protein Atlas (v21.1.proteinatlas.org)135–139.
GPR41 (free fatty acid receptor 3, FFAR3) and GPR43 (free fatty acid receptor 2, FFAR2) have been identified in smooth muscle cells, adipose, spleen, pancreatic tissues, and immune cells53–59, where they play significant roles in host health and disease. The most potent agonists for GPR41 and GPR43 are acetate, propionate, and butyrate53,60,61. Studies conflict as to whether beta-hydroxybutyrate (βHB), a ketone body, acts as an agonist or antagonist for GPR4162–66. A 2011 study reported that βHB is an antagonist for GPR4162. However, a 2013 study demonstrated that βHB is a GPR41 dose-dependent agonist, and were unable to find evidence of antagonism66. In our view, the data showing that βHB is an agonist for GPR41 is quite strong; however, it is important to note that βHB is used experimentally as both an antagonist64,65 and an agonist63, resulting in some confusion in the literature.
Previously reported ligand profiles and EC50’s for GPR41 and GPR43 vary by both assay and species tested, but, acetate, propionate and butyrate are detected in the micromolar range67,68 GPR41’s reported EC50 for acetate, propionate, and butyrate are: 393-1072μM, 6-127μM, and 33-158μM, respectively. Reported EC50’s for GPR43 for acetate, propionate, and butyrate are: 35-431μM, 14-290μM, and 28-371μM, respectively67,68. In addition, these receptors do have additional (weaker) ligands67,68. As stated above, circulating levels of acetate range from 50-500 μM in circulating plasma32,36–38, and thus it is likely that physiological changes in acetate may alter GPR41 and/or GPR43 signaling. SCFAs which bind to these two receptors activate cell signaling through interactions with G proteins coupled to the receptor.
Additional reports find that GPR109A (alternatively referred to as HCAR2 or NIACR1) is also a metabolite-sensing receptor, albeit not as extensively studied as the previously mentioned SCFA receptors59,69–71. GPR109A recognizes both niacin and butyrate, but binds butyrate with relatively low affinity69. Indeed, it has been demonstrated in GTPyS assays that niacin activates GPR109A with an EC50 ~ 250nM70, although concentrations of butyrate needed to activate the receptor are within the millimolar range, at ~1 mmol/L59. Similar to GPR41 and GPR43, GPR109A signals by activating the Gi/o protein, thus inhibiting adenylyl cyclase53,59,69. Studies have demonstrated that GPR109A is primarily expressed in immune cells50 and the intestinal lumen52, and expression of this receptor in the presence of its ligands plays a role in regulating host immune function and metabolism.
Olfactory receptors (OR) constitute the largest group of GPCRs, and recent findings demonstrate that two receptors within this family, OLFR78 and OLFR558, respond to SCFAs50,72,73. Of note, a new unified nomenclature has been proposed for olfactory receptor naming across species, and has been adapted for some but not all species74. In this article we will use the widely used root ‘Olfr’ nomenclature for murine receptors. Although olfactory receptors comprise the largest gene family in the genome75, only three olfactory receptors have 1:1 orthology among placental mammals76. OLFR78 (human ortholog: OR51E2) and OLFR558 (human ortholog: OR51E1) are two of these well-conserved receptors. OLFR78 is activated by acetate with an EC50 of 2.01mM - 2.35mM, and propionate with an EC50 of 0.63-0.92mM50,73,77,78. OLFR558 is activated by butyrate with an EC50 of ~0.2 mM (when co-transfected with the Golf protein)73,79.
Importantly, the ligand profile of OLFR78 is very similar to its human ortholog OR51E280, and the ligand profile of OFLR558 is very similar to its human ortholog OR51E173,79, with both OLFR558 and OR51E1 having additional (structurally related) ligands. Some groups81,82 have reported that beta-ionone (β-ionone) and/or androstenone derivatives also act as ligands for OR51E2. There are also reports that lactate is a partial agonist for OLFR7877,83,84; however, lactate does not activate OR51E283,85. Both OLFR78 and OLFR558 activation increases cAMP production, and in olfactory tissues, this happens via coupling to the olfactory G protein (Golf)86. In vitro, OLFR78 (and other ORs) likely can also couple to Gs; for both Gs and Golf, OR activation leads to increases in cAMP. Importantly, it was recently demonstrated that OLFR78 (and OR51E2) heterodimerize with GPR132, and that the heteromer receptor is sensitive to lactate85. The idea that some olfactory receptors dimerize to other GPCRs to influence cell signaling pathways is a noteworthy development in OR research.
SCFAs and Blood Pressure Regulation
Numerous studies have found an association between gut microbes and hypertension. For example, there are shifts in the gut microbiota of hypertensive subjects (‘dysbiosis’) in mice, in rats, and in humans5,87–89. In addition, transplanting microbes from a hypertensive subject into germ-free mice (which do not have any native microbes) can transfer the hypertensive phenotype90. Intriguingly, another study has shown that a high salt diet (a known risk factor for hypertension) depletes a specific strain of gut microbes, and depleting this strain can prevent salt-sensitive hypertension87. SCFA transporters also link SCFAs to blood pressure regulation. Yang, et al. found that decreased expression of colonic Slc5a8 (which can transport butyrate) is associated with more butyrate in the cecal content but less butyrate in the circulation in the spontaneously hypertensive rat (SHR)91.
Changes in SCFAs have also been reported in Dahl rats. One study reported that dietary salt is associated with increased fecal levels of acetate, propionate, and isobutyrate (but not butyrate) in Dahl rats92. Another study reported that dietary salt did not alter plasma acetate, valerate or butyrate93; however, it is not surprising that fecal levels may change without altering plasma levels. On the other hand, it was reported that fecal transplants can alter plasma SCFAs in Dahl salt-sensitive rats94. Together, it seems that the connections between gut microbes, SCFAs, and blood pressure are not always clear. In fact, several groups have attempted to measure blood pressure in animals without any gut microbes (germ-free models), but results have varied with some reporting decreased blood pressure95, a trend toward increased blood pressure96, or no change in blood pressure97. Although some of these differences may be due to the difficulties of working with germ-free models without compromising their germ-free status (to date, telemetry blood pressure measurements have not been performed in germ-free animals), it is also possible that differing findings may be due to the fact that germ-free mice are ‘missing’ not just one signaling pathway, but a multitude of pathways (due to the absence of all gut microbial metabolites), and likewise, that altering fecal or plasma levels of a SCFA can affect multiple receptors. Thus, in the following sections, we will focus on specific effects which can be assigned to specific SCFAs and/or specific GPCRs, in the hopes of beginning to disentangle these physiologically important yet interconnected and complex signaling pathways.
SCFAs and vasodilation
A key regulator of blood pressure is vasoconstriction and vasodilation of blood vessels5. In 1928, acetate was first reported as a vasodilator which drops BP due to vasodilation98. Acetate was used as a pH buffer in dialysis solutions until the 1980s, when it was recognized that acetate was contributing to hypotension in patients via a vasodilatory effect99–102. Numerous studies have now documented dose-dependent vasodilation of acetate, propionate, and butyrate which promote hypotension20,22,23,80,103. This effect is markedly attenuated after denudation of the epithelium103. Butyrate is also reported to dilate phenylephrine pre-constricted rat mesenteric and gracilis muscle arteries ex vivo64.
As a result of these vasodilatory actions, acetate, propionate, and butyrate have blood pressure lowering effects. When delivered acutely (for example, via i.v. or i.p.), SCFAs cause acute hypotension which develops in seconds and recovers over minutes80,104. Chronic intake of acetate105, butyrate106,107, or propionate19 also lowers blood pressure. Consistent with these findings, SCFAs lower BP even without prebiotic sources in the diet108. Recent evidence indicates that Dendrobium officinale ultrafine powder (DOFP) alters intestinal flora, increases fecal and serum SCFAs, and enhances vascular endothelial vasodilation function in metabolic hypertension109. This effect was blocked by the eNOS inhibitor L-NAME109. In humans, it was reported that higher levels of plasma but not fecal butyrate was positively associated with ambulatory arterial stiffness index (AASI)110. Although no association between bacterial α and β diversity and AASI was found, two main bacteria taxa were associated with human AASI: Lactobacillus spp. and Clostridium app110. However, the role of these two bacteria taxa in human arterial stiffness needs to be determined. Of note, there are no published studies showing that SCFAs can lower blood pressure in humans as an intervention (however, there is a promising preprint in this area111).
Two recent studies reported that that acute delivery of SCFAs not only decreases mean arterial pressure (MAP), but simultaneously suppresses heart rate (HR)64,104, and these studies suggest that SCFAs lowers HR via modulating sympathetic tone104. Acetate is also reported to affect cardiac function104. Propionate is shown to induce cardiac protection by significantly attenuating cardiac hypertrophy, fibrosis, vascular dysfunction, and hypertension in angiotensin II infused hypertension mice and apolipoprotein E KO mice models19. Butyrate is shown to lower BP and decrease HR after administration into the colon in rat. However, the hypotensive effect was dramatically decreased by subphrenic vagotomy and pretreatment with βHB (used as an antagonist of Gpr41/43 in this study, but also reported as an agonist by other groups – see “Cell Biology of SCFA Receptors” section above)64. Finally, SCFAs are reported to have protective effects on endothelial dysfunction induced by angiotensin II in rat aortic endothelial cells and rat aortas112.
Effects of SCFAs on blood pressure regulation via GPCRs
In this section we will review current literature113–117 regarding the roles of specific SCFA GPCRs in blood pressure regulation (Figure 2).
Figure 2. Ligands and cardiovascular phenotypes of SCFAs GPCR receptors, as well as sites of expression which have been suggested to contribute to these phenotypes.
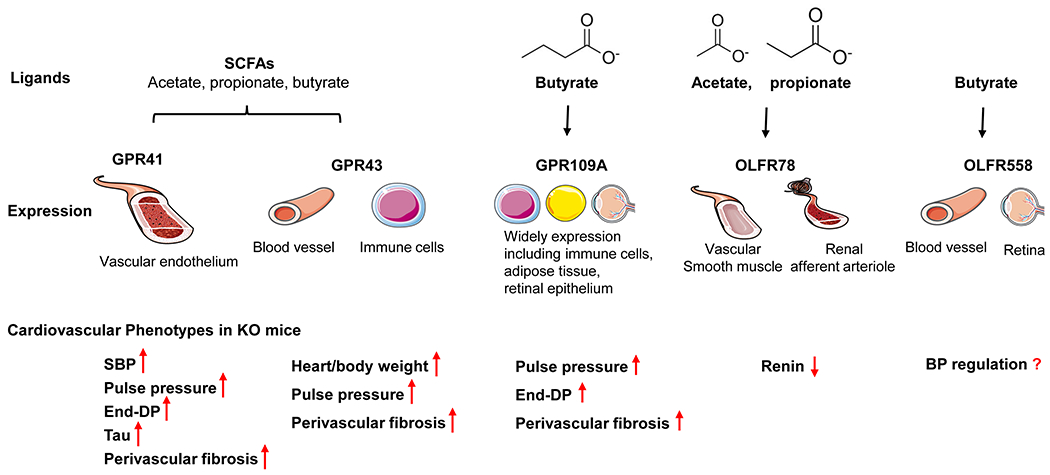
This figure summarizes current knowledge regarding the ligands and sites of expression for GPR41, GPR43, GPR109A, OLFR78, and OLFR558. In addition, cardiovascular phenotypes in KO mice are outlined. Gpr41 KO mice exhibit increased SBP103, with higher pulse pressure, end-diastolic pressure (DP), tau, and perivascular fibrosis108. Gpr43 KO mice have an increased heart/body weight, pulse pressure, and perivascular fibrosis108. Gpr109a KO mice are reported to have a higher pulse pressure, end-DP, as well as perivascular fibrosis108. Olfr78 KO mice have a lower plasma renin activity39,49. Images in this figure are from Servier Medical Art (smart.servier.com).
GPR41 is expressed in the vascular endothelium118, and, in the autonomic and sensory ganglia in mouse and human62,119 indicating that it is well-positioned to modulate vascular tone in response to SCFAs. To date, two studies have examined blood pressure in Gpr41 whole-animal KO mice, both with similar findings: Natarajan et al103 and Kaye et al108. Using blood pressure telemetry, Natarajan et al reported that whole-animal Gpr41 KO have increased systolic pressure and increased pulse pressure, as well as an increased heart weight/body weight ratio103. Although Kaye, et al did not publish systolic pressure data, they did report increased pulse pressure as measured by cardiac catherisation108; given that increased pulse pressure is indicative of isolated systolic hypertension, it seems likely that systolic pressure was also elevated in these animals. Unlike the Natarajan study, Kaye, et al did not observe an increase in the heart weight/body weight ratio. However, Natarajan et al specify that heart weight/body weight was increased in 6-month old KOs103 (Kaye, et al do not report the age of mice that were examined). If Kaye, et al examined younger mice this could explain the lack of significance in this measurement (and it does appear that Kaye, et al see a trend toward an increase)108. Finally, Kaye, et al reported that Gpr41 KO have an increase in end-diastolic pressure, Tau, and perivascular fibrosis; these parameters were not examined in the Natarajan, et al study.
GPR43 is highly expressed in immune cells120, and has also been reported in blood vessels50. Gpr43 KO mice have increased heart to body weight and higher perivascular fibrosis, but without a difference in BP as measured via cardiac catheterisation108. Nakai, et al. provided the first human evidence that Gpr43 is differentially modulated in essential hypertension: they found that hypertensive subjects had lower levels of Gpr43 expression in immune cells121. It was also reported that Gpr41 and Gpr43 expression in circulating immune cells is negatively associated with human arterial stiffness; presumably, decreased Gpr41 and Gpr43 decreases the response to BP-lowering SCFAs such as butyrate110. These data suggest that GPR43 signaling deficiency helps to drive the pro-inflammatory phenotype in hypertensive subjects121. Intriguingly, Ang et al found that GPR41 and GPR43 can heterodimerize in monocytes and macrophages, and that the heterodimer exhibited increased Ca2+ signaling and β-arrestin-2 recruitment. The enhanced signaling was attenuated by antagonizing GPR43, inhibiting Gαq inhibition (YM254890), or inhibiting Gαi122. The possibility of GPR41-GPR43 heterodimers adds an additional level of complexity to studies of GPR41 and GPR43.
GPR109A is widely expressed in white and brown adipose tissue, keratinocytes, various immune cells, and likely in microglia123. Regarding blood pressure regulation, GPR109A in the rostral ventrolateral medulla (RVLM) plays a role in central blood pressure control, where activation by its ligand nicotinic acid (NA) leads to Ca2+-dependent L-glutamate release, subsequently increasing neuronal oxidative stress and sympathetic activity123.
Gpr109a KO mice were reported to have elevated end-diastolic pressure and pulse pressure, as well as perivascular fibrosis. However, Gpr109a KO mice demonstrated no differences in BP compared to WT mice at baseline (measured via cardiac catheterisation)108. Double KO of Gpr43/Gpr109a have significantly larger heart to body weight ratios, and increased end-diastolic pressure and pulse pressure108, but without changes in BP at baseline (as measured by tail-cuff)108. Gpr109a is also expressed in the retinal epithelium where it plays a role in the inflammatory pathway of diabetic retinopathy124. Gpr109a expression is upregulated in diabetic retinopathy, suggesting that increased Gpr109a expression (and elevation of its ligand βHB) may help fight inflammation124. Notably, Gpr109a has relatively high expression in immune cells (as does Gpr43), and SCFA GPCR expression in this cell type may also be quite important in blood pressure regulation. For example, immune cell infiltration in renal tissue125 and perivascular adipose tissue126 is required for the progression of hypertension. Immune cells are an important component of blood pressure regulation125,126, where they ultimately alter blood pressure by altering parameters such as peripheral vascular resistance, cardiac output, or renal sodium absorption126. An important effect of immune cell infiltration in the vasculature is the vascular remodeling, including changes in lumen diameter and in media-to-lumen ratio127. Tissue-specific knockout models will be required to definitely determine whether the origin of a change in vascular function is due to expression of a SCFA GPCR in an immune cell, or, in vascular cells.
OLFR78 is expressed in vascular smooth muscle cells in a variety of organs; intriguingly, it is only found in a subset of the smooth muscle cells of each vessel50. OLFR78 also localizes to renal afferent arterioles, where it has been shown to impact renin release50. Specifically, it was reported that isolated renal glomeruli with attached juxtaglomerular apparati (JGA) from Olfr78 WT mice release renin in response to propionate, but this response is absent in JGA/glomeruli from Olfr78 whole-body KO mice50. Consistent with this, it was shown that Olfr78 KO mice have lower plasma renin activity36,50. Recently, it was reported that both Olfr78 whole-animal KO mice and Olfr78fl/fl Renin-Cre KO mice have decreased renin protein levels associated with glomeruli36. Despite these changes in renin, however, Olfr78 KO mice are normotensive when blood pressure is measured via telemetry36. As the most closely related olfactory receptor of Olfr78, Olfr558 is widely expressed in many tissues including kidney and heart73,128. Although a role for OLFR558 in blood pressure regulation has not been reported, Olfr558 expression is found in vascular smooth muscle cells in kidney, heart128, and in the retina129, suggesting that Olfr558 may play a role in blood pressure regulation.
We can also gain insight into how SCFA GPCRs function by considering how SCFA GPCR expression is altered by various stimuli. For example, a study by Nakai, et al assayed the expression of GPR41, GPR43 and GPR109A in immune cells of humans with and without hypertension using qPCR130. While they did not observe changes in GPR41 or GPR109A expression, they found a decrease in GPR43 expression in hypertensive subjects. GPR43 expression was strongly negatively correlated with blood pressure, and this association was significant even after adjusting for age, body mass index, and sex. Interestingly, this same study found that hypertensive subjects had increased plasma acetate and butyrate, which was counter to what the authors had expected. Based on their own previous work105,108, they presumed that higher levels of acetate and butyrate would lower blood pressure, not raise it. Intriguingly, they propose that increased levels of SCFAs may be ineffective in these patients due to the concomitant downregulation of GPR43. A separate study by Weber, et al131 examined the renal expression of Gpr41, Gpr43 and Olfr78 in a mouse model of hypertension (the Angiotensin II-infusion model). They found a decrease in Gpr41 and Gpr43 expression but an increase in Olfr78 expression by qPCR. They also used commercially available antibodies to examine protein level changes; although to our knowledge these antibodies have not been validated, they saw similar changes by western blot. Finally, a study by Brunskill132, et al used microarray to identify “genes that confer the identity of the renin cell” and found not only that Olfr558 is enriched in renin cells, but that the expression of Olfr558 is upregulated in mice treated with captopril (an angiotensin converting enzyme inhibitor).
Future Directions and Challenges
SCFA signaling is complex, and this complexity presents a clear challenge. First, there are multiple SCFAs; in fact, in addition to the three major SCFAs—acetate, propionate, and butyrate - SCFAs can induce shorter or longer, as well as branched compounds. Second, these SCFAs activate multiple receptors including but not limited to GPR41, GPR43, GPR109A, OLFR78, and OLFR558, some of which are found in multiple tissues, and some of which can heterodimerize. Each SCFA can activate multiple receptors, and each receptor can be activated by one or more than one SCFA. Moreover, multiple bacteria can produce each SCFA. Apparently, this pathway is more of a ‘web’ than a linear pathway. Thus, understanding the role of each individual member of this web (each SCFA and each host protein) is a major challenge.
To meet this challenge, we suggest that future studies which reduce aspects of this complexity will provide valuable clues: for example, tissue-specific KOs of a SCFA receptor, and/or, dosing animals with one SCFA at a time to understand the role of each SCFA. KO studies for SCFA receptors should also consider whether there may be compensatory changes in the remaining receptors which could influence interpretation of the results, and SCFA levels should be routinely measured for studies examining SCFA GPCRs to rule out (or rule in?) changes in SCFAs themselves. We must then use this information to inform and understand our interpretation of data from the more complex, intact system.
Looking ahead, this area holds great promise given that SCFA levels can be altered via dietary interventions. Thus, a nutritional therapeutic strategy – for example, the use of prebiotics and/or probiotics4,133 - could be developed to prevent or attenuate hypertension. In future studies, it will be important to better understand the functional roles of these pathways in both normotension and hypertension, and to further explore how we may be able to strategically target SCFA signaling pathways to promote health.
Conclusion
Both SCFAs and SCFA GPCRs are known to influence blood pressure regulation, but the involvement of multiple SCFAs and multiple SCFA receptors makes this area of research complex. However, studies using both acute and chronic SCFA treatment as well as studies in KO animals have begun to unravel this complexity.
Supplementary Material
Sources of funding
We are grateful for support from the National Science Foundation Graduate Research Fellowship (DGE-1746891to BM), the NIDDK Diabetic Complications Consortium (RRID:SCR_001415m www.diacomp.org), grants DK076169 and DK 115255 (subaward to JLP), and R56DK107726 (to JLP).
Footnotes
Disclosures
The authors have no competing interests to declare.
Reference List
- 1.Virani SS, Alonso A, Aparicio HJ, Benjamin EJ, Bittencourt MS, Callaway CW, Carson AP, Chamberlain AM, Cheng S, Delling FN, et al. Heart Disease and Stroke Statistics-2021 Update: A Report From the American Heart Association. Circulation. 2021;143:e254–e743. doi: 10.1161/CIR.0000000000000950 [DOI] [PubMed] [Google Scholar]
- 2.Safar ME, Balkau B, Lange C, Protogerou AD, Czernichow S, Blacher J, Levy BI, Smulyan H. Hypertension and Vascular Dynamics in Men and Women With Metabolic Syndrome. J Am Coll Cardiol. 2013;61:12–19. doi: 10.1016/j.jacc.2012.01.088 [DOI] [PubMed] [Google Scholar]
- 3.Jones DW, Whelton PK, Allen N, Clark D, Gidding SS, Muntner P, Nesbitt S, Mitchell NS, Townsend R, Falkner B, et al. Management of Stage 1 Hypertension in Adults With a Low 10-Year Risk for Cardiovascular Disease: Filling a Guidance Gap A Scientific Statement From the American Heart Association. Hypertension. 2021;77:E58–E67. doi: 10.1161/Hyp.0000000000000195 [DOI] [PubMed] [Google Scholar]
- 4.Kang Y, Cai Y. Gut microbiota and hypertension: From pathogenesis to new therapeutic strategies. Clin Res Hepatol Gastroenterol. 2018;42:110–117. doi: 10.1016/j.clinre.2017.09.006 [DOI] [PubMed] [Google Scholar]
- 5.Duttaroy AK. Role of Gut Microbiota and Their Metabolites on Atherosclerosis, Hypertension and Human Blood Platelet Function: A Review. Nutrients. 2021;13. doi: ARTN 144 10.3390/nu13010144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas SJ, Booth JN 3rd, Dai C, Li X, Allen N, Calhoun D, Carson AP, Gidding S, Lewis CE, Shikany JM, et al. Cumulative Incidence of Hypertension by 55 Years of Age in Blacks and Whites: The CARDIA Study. J Am Heart Assoc. 2018;7. doi: 10.1161/JAHA.117.007988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung YM, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: nature09922 [pii]; 10.1038/nature09922 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Z, Bergeron N, Levison BS, Li XS, Chiu S, Jia X, Koeth RA, Li L, Wu Y, Tang WHW, et al. Impact of chronic dietary red meat, white meat, or non-meat protein on trimethylamine N-oxide metabolism and renal excretion in healthy men and women. Eur Heart J. 2019;40:583–594. doi: 10.1093/eurheartj/ehy799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dahlqvist G, Piessevaux H. Irritable bowel syndrome: the role of the intestinal microbiota, pathogenesis and therapeutic targets. Acta Gastroenterol Belg. 2011;74:375–380. [PubMed] [Google Scholar]
- 10.Goto Y, Kurashima Y, Kiyono H. The gut microbiota and inflammatory bowel disease. Curr Opin Rheumatol. 2015;27:388–396. doi: 10.1097/BOR.0000000000000192 [DOI] [PubMed] [Google Scholar]
- 11.Berer K, Mues M, Koutrolos M, Rasbi ZA, Boziki M, Johner C, Wekerle H, Krishnamoorthy G. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature. 2011;479:538–541. doi: 10.1038/nature10554 [DOI] [PubMed] [Google Scholar]
- 12.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: nature08530 [pii]; 10.1038/nature08530 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alemao CA, Budden KF, Gomez HM, Rehman SF, Marshall JE, Shukla SD, Donovan C, Forster S, Yang IA, Keely S, et al. Impact of diet and the bacterial microbiome on the mucous barrier and immune disorders. Allergy. 2020. doi: 10.1111/all.14548 [DOI] [PubMed] [Google Scholar]
- 14.Vaziri ND. CKD impairs barrier function and alters microbial flora of the intestine: a major link to inflammation and uremic toxicity. Curr Opin Nephrol Hypertens. 2012;21:587–592. doi: 10.1097/MNH.0b013e328358c8d5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noel S, Martina-Lingua MN, Bandapalle S, Pluznick J, Hamad AR, Peterson DA, Rabb H. Intestinal microbiota-kidney cross talk in acute kidney injury and chronic kidney disease. Nephron Clin Pract. 2014;127:139–143. doi: 10.1159/000363209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li YJ, Chen X, Kwan TK, Loh YW, Singer J, Liu Y, Ma J, Tan J, Macia L, Mackay CR, et al. Dietary Fiber Protects against Diabetic Nephropathy through Short-Chain Fatty Acid-Mediated Activation of G Protein-Coupled Receptors GPR43 and GPR109A. J Am Soc Nephrol. 2020. doi: 10.1681/ASN.2019101029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gong J, Noel S, Pluznick JL, Hamad ARA, Rabb H. Gut Microbiota-Kidney Cross-Talk in Acute Kidney Injury. Semin Nephrol. 2019;39:107–116. doi: 10.1016/j.semnephrol.2018.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adnan S, Nelson JW, Ajami NJ, Venna VR, Petrosino JF, Bryan RM, Durgan DJ. Alterations in the gut microbiota can elicit hypertension in rats. Physiol Genomics. 2016:physiolgenomics 00081 02016. doi: 10.1152/physiolgenomics.00081.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartolomaeus H, Balogh A, Yakoub M, Homann S, Marko L, Hoges S, Tsvetkov D, Krannich A, Wundersitz S, Avery EG, et al. Short-Chain Fatty Acid Propionate Protects From Hypertensive Cardiovascular Damage. Circulation. 2019;139:1407–1421. doi: 10.1161/CIRCULATIONAHA.118.036652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knock G, Psaroudakis D, Abbot S, Aaronson PI. Propionate-induced relaxation in rat mesenteric arteries: a role for endothelium-derived hyperpolarising factor. J Physiol. 2002;538:879–890. doi: PHY_13105 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marques FZ, Nelson EM, Chu PY, Horlock D, Fiedler A, Ziemann M, Tan JK, Kuruppu S, Rajapakse NW, El-Osta A, et al. High Fibre Diet and Acetate Supplementation Change the Gut Microbiota and Prevent the Development of Hypertension and Heart Failure in DOCA-Salt Hypertensive Mice. Circulation. 2016. doi: 10.1161/CIRCULATIONAHA.116.024545 [DOI] [PubMed] [Google Scholar]
- 22.Mortensen FV, Nielsen H, Mulvany MJ, Hessov I. Short chain fatty acids dilate isolated human colonic resistance arteries. Gut. 1990;31:1391–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nutting CW, Islam S, Daugirdas JT. Vasorelaxant effects of short chain fatty acid salts in rat caudal artery. Am J Physiol. 1991;261:H561–H567. [DOI] [PubMed] [Google Scholar]
- 24.Nogal A, Valdes AM, Menni C. The role of short-chain fatty acids in the interplay between gut microbiota and diet in cardio-metabolic health. Gut Microbes. 2021;13:1–24. doi: 10.1080/19490976.2021.1897212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud DJ, Bakker BM. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res. 2013;54:2325–2340. doi: 10.1194/jlr.R036012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pryde SE, Duncan SH, Hold GL, Stewart CS, Flint HJ. The microbiology of butyrate formation in the human colon. FEMS Microbiol Lett. 2002;217:133–139. doi: 10.1111/j.1574-6968.2002.tb11467.x [DOI] [PubMed] [Google Scholar]
- 27.Sanna S, van Zuydam NR, Mahajan A, Kurilshikov A, Vich Vila A, Vosa U, Mujagic Z, Masclee AAM, Jonkers D, Oosting M, et al. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat Genet. 2019;51:600–605. doi: 10.1038/s41588-019-0350-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levy M, Thaiss CA, Elinav E. Metabolites: messengers between the microbiota and the immune system. Genes Dev. 2016;30:1589–1597. doi: 10.1101/gad.284091.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng W, Ao H, Peng C. Gut Microbiota, Short-Chain Fatty Acids, and Herbal Medicines. Front Pharmacol. 2018;9:1354. doi: 10.3389/fphar.2018.01354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reichardt N, Vollmer M, Holtrop G, Farquharson FM, Wefers D, Bunzel M, Duncan SH, Drew JE, Williams LM, Milligan G, et al. Specific substrate-driven changes in human faecal microbiota composition contrast with functional redundancy in short-chain fatty acid production. ISME J. 2018;12:610–622. doi: 10.1038/ismej.2017.196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bugaut M. Occurrence, absorption and metabolism of short chain fatty acids in the digestive tract of mammals. Comp Biochem Physiol B. 1987;86:439–472. doi: 10.1016/0305-0491(87)90433-0 [DOI] [PubMed] [Google Scholar]
- 32.Muller M, Hernandez MAG, Goossens GH, Reijnders D, Holst JJ, Jocken JWE, van Eijk H, Canfora EE, Blaak EE. Circulating but not faecal short-chain fatty acids are related to insulin sensitivity, lipolysis and GLP-1 concentrations in humans. Sci Rep. 2019;9:12515. doi: 10.1038/s41598-019-48775-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takagi R, Sasaki K, Sasaki D, Fukuda I, Tanaka K, Yoshida K, Kondo A, Osawa R. A Single-Batch Fermentation System to Simulate Human Colonic Microbiota for High-Throughput Evaluation of Prebiotics. PLoS One. 2016;11:e0160533. doi: 10.1371/journal.pone.0160533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coady MJ, Chang MH, Charron FM, Plata C, Wallendorff B, Sah JF, Markowitz SD, Romero MF, Lapointe JY. The human tumour suppressor gene SLC5A8 expresses a Na+-monocarboxylate cotransporter. J Physiol. 2004;557:719–731. doi: 10.1113/jphysiol.2004.063859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Charney AN, Micic L, Egnor RW. Nonionic diffusion of short-chain fatty acids across rat colon. Am J Physiol. 1998;274:G518–524. doi: 10.1152/ajpgi.1998.274.3.G518 [DOI] [PubMed] [Google Scholar]
- 36.Poll BG, Xu J, Gupta K, Shubitowski TB, Pluznick JL. Olfactory receptor 78 modulates renin but not baseline blood pressure. Physiol Rep. 2021;9:e15017. doi: 10.14814/phy2.15017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perry RJ, Peng L, Barry NA, Cline GW, Zhang D, Cardone RL, Petersen KF, Kibbey RG, Goodman AL, Shulman GI. Acetate mediates a microbiome-brain-beta-cell axis to promote metabolic syndrome. Nature. 2016;534:213–217. doi: 10.1038/nature18309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shubitowski TB, Poll BG, Natarajan N, Pluznick JL. Short-chain fatty acid delivery: assessing exogenous administration of the microbiome metabolite acetate in mice. Physiol Rep. 2019;7:e14005. doi: 10.14814/phy2.14005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levrat MA, Remesy C, Demigne C. High propionic acid fermentations and mineral accumulation in the cecum of rats adapted to different levels of inulin. J Nutr. 1991;121:1730–1737. [DOI] [PubMed] [Google Scholar]
- 40.den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud DJ, Bakker BM. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. Journal of Lipid Research. 2013;54:2325–2340. doi: 10.1194/jlr.R036012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kimura I, Miyamoto J, Ohue-Kitano R, Watanabe K, Yamada T, Onuki M, Aoki R, Isobe Y, Kashihara D, Inoue D, et al. Maternal gut microbiota in pregnancy influences offspring metabolic phenotype in mice. Science. 2020;367. doi: 10.1126/science.aaw8429 [DOI] [PubMed] [Google Scholar]
- 42.Osada Y, Nakagawa S, Ishibe K, Takao S, Shimazaki A, Itohara K, Imai S, Yonezawa A, Nakagawa T, Matsubara K. Antibiotic-induced microbiome depletion alters renal glucose metabolism and exacerbates renal injury after ischemia-reperfusion injury in mice. Am J Physiol Renal Physiol. 2021;321:F455–F465. doi: 10.1152/ajprenal.00111.2021 [DOI] [PubMed] [Google Scholar]
- 43.Park SY, Kim JS. A short guide to histone deacetylases including recent progress on class II enzymes. Exp Mol Med. 2020;52:204–212. doi: 10.1038/s12276-020-0382-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boffa LC, Vidali G, Mann RS, Allfrey VG. Suppression of histone deacetylation in vivo and in vitro by sodium butyrate. J Biol Chem. 1978;253:3364–3366. [PubMed] [Google Scholar]
- 45.Vidali G, Boffa LC, Bradbury EM, Allfrey VG. Butyrate suppression of histone deacetylation leads to accumulation of multiacetylated forms of histones H3 and H4 and increased DNase I sensitivity of the associated DNA sequences. Proc Natl Acad Sci U S A. 1978;75:2239–2243. doi: 10.1073/pnas.75.5.2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patel BM. Sodium Butyrate Controls Cardiac Hypertrophy in Experimental Models of Rats. Cardiovasc Toxicol. 2018;18:1–8. doi: 10.1007/s12012-017-9406-2 [DOI] [PubMed] [Google Scholar]
- 47.Liu Y, Li YJ, Loh YW, Singer J, Zhu W, Macia L, Mackay CR, Wang W, Chadban SJ, Wu H. Fiber Derived Microbial Metabolites Prevent Acute Kidney Injury Through G-Protein Coupled Receptors and HDAC Inhibition. Front Cell Dev Biol. 2021;9:648639. doi: 10.3389/fcell.2021.648639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Latorraca NR, Venkatakrishnan AJ, Dror RO. GPCR Dynamics: Structures in Motion. Chem Rev. 2017;117:139–155. doi: 10.1021/acs.chemrev.6b00177 [DOI] [PubMed] [Google Scholar]
- 49.Basith S, Cui M, Macalino SJY, Park J, Clavio NAB, Kang S, Choi S. Exploring G Protein-Coupled Receptors (GPCRs) Ligand Space via Cheminformatics Approaches: Impact on Rational Drug Design. Front Pharmacol. 2018;9:128. doi: 10.3389/fphar.2018.00128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pluznick JL, Protzko RJ, Gevorgyan H, Peterlin Z, Sipos A, Han J, Brunet I, Wan LX, Rey F, Wang T, et al. Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proc Natl Acad Sci U S A. 2013;110:4410–4415. doi: 10.1073/pnas.1215927110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Priyadarshini M, Kotlo KU, Dudeja PK, Layden BT. Role of Short Chain Fatty Acid Receptors in Intestinal Physiology and Pathophysiology. Compr Physiol. 2018;8:1091–1115. doi: 10.1002/cphy.c170050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun M, Wu W, Liu Z, Cong Y. Microbiota metabolite short chain fatty acids, GPCR, and inflammatory bowel diseases. J Gastroenterol. 2017;52:1–8. doi: 10.1007/s00535-016-1242-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van der Hee B, Wells JM. Microbial Regulation of Host Physiology by Short-chain Fatty Acids. Trends Microbiol. 2021;29:700–712. doi: 10.1016/j.tim.2021.02.001 [DOI] [PubMed] [Google Scholar]
- 54.Tazoe H, Otomo Y, Karaki S, Kato I, Fukami Y, Terasaki M, Kuwahara A. Expression of short-chain fatty acid receptor GPR41 in the human colon. Biomed Res. 2009;30:149–156. doi: 10.2220/biomedres.30.149 [DOI] [PubMed] [Google Scholar]
- 55.Xiong Y, Miyamoto N, Shibata K, Valasek MA, Motoike T, Kedzierski RM, Yanagisawa M. Short-chain fatty acids stimulate leptin production in adipocytes through the G protein-coupled receptor GPR41. Proc Natl Acad Sci U S A. 2004;101:1045–1050. doi: 10.1073/pnas.2637002100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang W, Xiao Y, Huang X, Chen F, Sun M, Bilotta AJ, Xu L, Lu Y, Yao S, Zhao Q, et al. Microbiota Metabolite Short-Chain Fatty Acids Facilitate Mucosal Adjuvant Activity of Cholera Toxin through GPR43. J Immunol. 2019;203:282–292. doi: 10.4049/jimmunol.1801068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Veprik A, Laufer D, Weiss S, Rubins N, Walker MD. GPR41 modulates insulin secretion and gene expression in pancreatic beta-cells and modifies metabolic homeostasis in fed and fasting states. FASEB J. 2016;30:3860–3869. doi: 10.1096/fj.201500030R [DOI] [PubMed] [Google Scholar]
- 59.Pluznick JL. Gut microbiota in renal physiology: focus on short-chain fatty acids and their receptors. Kidney Int. 2016;90:1191–1198. doi: 10.1016/j.kint.2016.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brown AJ, Goldsworthy SM, Barnes AA, Eilert MM, Tcheang L, Daniels D, Muir AI, Wigglesworth MJ, Kinghorn I, Fraser NJ, et al. The Orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J Biol Chem. 2003;278:11312–11319. doi: 10.1074/jbc.M211609200 [DOI] [PubMed] [Google Scholar]
- 61.Le Poul E, Loison C, Struyf S, Springael JY, Lannoy V, Decobecq ME, Brezillon S, Dupriez V, Vassart G, Van Damme J, et al. Functional characterization of human receptors for short chain fatty acids and their role in polymorphonuclear cell activation. J Biol Chem. 2003;278:25481–25489. doi: 10.1074/jbc.M301403200 [DOI] [PubMed] [Google Scholar]
- 62.Kimura I, Inoue D, Maeda T, Hara T, Ichimura A, Miyauchi S, Kobayashi M, Hirasawa A, Tsujimoto G. Short-chain fatty acids and ketones directly regulate sympathetic nervous system via G protein-coupled receptor 41 (GPR41). Proc Natl Acad Sci U S A. 2011;108:8030–8035. doi: 10.1073/pnas.1016088108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nooromid M, Chen EB, Xiong L, Shapiro K, Jiang Q, Demsas F, Eskandari M, Priyadarshini M, Chang EB, Layden BT, et al. Microbe-Derived Butyrate and Its Receptor, Free Fatty Acid Receptor 3, But Not Free Fatty Acid Receptor 2, Mitigate Neointimal Hyperplasia Susceptibility After Arterial Injury. J Am Heart Assoc. 2020;9:e016235. doi: 10.1161/JAHA.120.016235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Onyszkiewicz M, Gawrys-Kopczynska M, Konopelski P, Aleksandrowicz M, Sawicka A, Kozniewska E, Samborowska E, Ufnal M. Butyric acid, a gut bacteria metabolite, lowers arterial blood pressure via colon-vagus nerve signaling and GPR41/43 receptors. Pflugers Arch. 2019;471:1441–1453. doi: 10.1007/s00424-019-02322-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Priyadarshini M, Layden BT. FFAR3 modulates insulin secretion and global gene expression in mouse islets. Islets. 2015;7:e1045182. doi: 10.1080/19382014.2015.1045182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Won YJ, Lu VB, Puhl HL 3rd, Ikeda SR. beta-Hydroxybutyrate modulates N-type calcium channels in rat sympathetic neurons by acting as an agonist for the G-protein-coupled receptor FFA3. J Neurosci. 2013;33:19314–19325. doi: 10.1523/JNEUROSCI.3102-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Le Poul E, Loison C, Struyf S, Springael JY, Lannoy V, Decobecq ME, Brezillon S, Dupriez V, Vassart G, Van Damme J, et al. Functional characterization of human receptors for short chain fatty acids and their role in polymorphonuclear cell activation. J Biol Chem. 2003;278:25481–25489. doi: 10.1074/jbc.M301403200 [DOI] [PubMed] [Google Scholar]
- 68.Brown AJ, Goldsworthy SM, Barnes AA, Eilert MM, Tcheang L, Daniels D, Muir AI, Wigglesworth MJ, Kinghorn I, Fraser NJ, et al. The orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J Biol Chem. 2003;278:11312–11319. doi: 10.1074/jbc.M211609200 [DOI] [PubMed] [Google Scholar]
- 69.Thangaraju M, Cresci GA, Liu K, Ananth S, Gnanaprakasam JP, Browning DD, Mellinger JD, Smith SB, Digby GJ, Lambert NA, et al. GPR109A is a G-protein-coupled receptor for the bacterial fermentation product butyrate and functions as a tumor suppressor in colon. Cancer Res. 2009;69:2826–2832. doi: 10.1158/0008-5472.CAN-08-4466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gharbaoui T, Skinner PJ, Shin YJ, Averbuj C, Jung JK, Johnson BR, Duong T, Decaire M, Uy J, Cherrier MC, et al. Agonist lead identification for the high affinity niacin receptor GPR109a. Bioorg Med Chem Lett. 2007;17:4914–4919. doi: 10.1016/j.bmcl.2007.06.028 [DOI] [PubMed] [Google Scholar]
- 71.Snelson M, Tan SM, Higgins GC, Lindblom RSJ, Coughlan MT. Exploring the role of the metabolite-sensing receptor GPR109a in diabetic nephropathy. Am J Physiol Renal Physiol. 2020;318:F835–F842. doi: 10.1152/ajprenal.00505.2019 [DOI] [PubMed] [Google Scholar]
- 72.Spehr M, Munger SD. Olfactory receptors: G protein-coupled receptors and beyond. J Neurochem. 2009;109:1570–1583. doi: 10.1111/j.1471-4159.2009.06085.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kuhns VLH, Sanchez J, Sarver DC, Khalil Z, Rajkumar P, Marr KA, Pluznick JL. Characterizing novel olfactory receptors expressed in the murine renal cortex. Am J Physiol-Renal. 2019;317:F172–F186. doi: 10.1152/ajprenal.00624.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Olender T, Jones TEM, Bruford E, Lancet D. A unified nomenclature for vertebrate olfactory receptors. BMC Evol Biol. 2020;20:42. doi: 10.1186/s12862-020-01607-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Malnic B, Godfrey PA, Buck LB. The human olfactory receptor gene family. Proc Natl Acad Sci U S A. 2004;101:2584–2589. doi: 10.1073/pnas.0307882100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Niimura Y, Matsui A, Touhara K. Extreme expansion of the olfactory receptor gene repertoire in African elephants and evolutionary dynamics of orthologous gene groups in 13 placental mammals. Genome Res. 2014;24:1485–1496. doi: 10.1101/gr.169532.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chang AJ, Ortega FE, Riegler J, Madison DV, Krasnow MA. Oxygen regulation of breathing through an olfactory receptor activated by lactate. Nature. 2015;527:240–244. doi: 10.1038/nature15721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Saito H, Chi Q, Zhuang H, Matsunami H, Mainland JD. Odor coding by a Mammalian receptor repertoire. Sci Signal. 2009;2:ra9. doi: 10.1126/scisignal.2000016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xu J, Pluznick JL. Key Amino Acids Alter Activity and Trafficking of a Well-conserved Olfactory Receptor. Am J Physiol Cell Physiol. 2022. doi: 10.1152/ajpcell.00440.2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pluznick JL, Protzko RJ, Gevorgyan H, Peterlin Z, Sipos A, Han J, Brunet I, Wan LX, Rey F, Wang T, et al. Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proc Natl Acad Sci U S A. 2013;110:4410–4415. doi: 1215927110 [pii]; 10.1073/pnas.1215927110 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Neuhaus EM, Zhang W, Gelis L, Deng Y, Noldus J, Hatt H. Activation of an olfactory receptor inhibits proliferation of prostate cancer cells. J Biol Chem. 2009;284:16218–16225. doi: 10.1074/jbc.M109.012096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Abaffy T, Bain JR, Muehlbauer MJ, Spasojevic I, Lodha S, Bruguera E, O’Neal SK, Kim SY, Matsunami H. A Testosterone Metabolite 19-Hydroxyandrostenedione Induces Neuroendocrine Trans-Differentiation of Prostate Cancer Cells via an Ectopic Olfactory Receptor. Front Oncol. 2018;8:162. doi: 10.3389/fonc.2018.00162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Aisenberg WH, Huang J, Zhu W, Rajkumar P, Cruz R, Santhanam L, Natarajan N, Yong HM, De Santiago B, Oh JJ, et al. Defining an olfactory receptor function in airway smooth muscle cells. Sci Rep. 2016;6:38231. doi: 10.1038/srep38231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhou T, Chien MS, Kaleem S, Matsunami H. Single cell transcriptome analysis of mouse carotid body glomus cells. J Physiol. 2016;594:4225–4251. doi: 10.1113/JP271936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vadevoo SMP, Gunassekaran GR, Lee C, Lee N, Lee J, Chae S, Park JY, Koo J, Lee B. The macrophage odorant receptor Olfr78 mediates the lactate-induced M2 phenotype of tumor-associated macrophages. Proc Natl Acad Sci U S A. 2021;118. doi: 10.1073/pnas.2102434118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Belluscio L, Gold GH, Nemes A, Axel R. Mice deficient in G(olf) are anosmic. Neuron. 1998;20:69–81. doi: 10.1016/s0896-6273(00)80435-3 [DOI] [PubMed] [Google Scholar]
- 87.Wilck N, Matus MG, Kearney SM, Olesen SW, Forslund K, Bartolomaeus H, Haase S, Mahler A, Balogh A, Marko L, et al. Salt-responsive gut commensal modulates TH17 axis and disease. Nature. 2017;551:585–589. doi: 10.1038/nature24628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mell B, Jala VR, Mathew AV, Byun J, Waghulde H, Zhang Y, Haribabu B, Vijay-Kumar M, Pennathur S, Joe B. Evidence for a link between Gut Microbiota and Hypertension in the Dahl rat model. Physiol Genomics. 2015;47:187–197. doi: physiolgenomics.00136.2014 [pii]; 10.1152/physiolgenomics.00136.2014 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang T, Santisteban MM, Rodriguez V, Li E, Ahmari N, Carvajal JM, Zadeh M, Gong MH, Qi YF, Zubcevic J, et al. Gut Dysbiosis Is Linked to Hypertension. Hypertension. 2015;65:1331–1340. doi: 10.1161/Hypertensionaha.115.05315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li J, Zhao F, Wang Y, Chen J, Tao J, Tian G, Wu S, Liu W, Cui Q, Geng B, et al. Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome. 2017;5:14. doi: 10.1186/s40168-016-0222-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yang T, Magee KL, Colon-Perez LM, Larkin R, Liao YS, Balazic E, Cowart JR, Arocha R, Redler T, Febo M, et al. Impaired butyrate absorption in the proximal colon, low serum butyrate and diminished central effects of butyrate on blood pressure in spontaneously hypertensive rats. Acta Physiol. 2019;226. doi: ARTN e13256 10.1111/apha.13256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bier A, Braun T, Khasbab R, Di Segni A, Grossman E, Haberman Y, Leibowitz A. A High Salt Diet Modulates the Gut Microbiota and Short Chain Fatty Acids Production in a Salt-Sensitive Hypertension Rat Model. Nutrients. 2018;10. doi: 10.3390/nu10091154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chakraborty S, Galla S, Cheng X, Yeo JY, Mell B, Singh V, Yeoh B, Saha P, Mathew AV, Vijay-Kumar M, et al. Salt-Responsive Metabolite, beta-Hydroxybutyrate, Attenuates Hypertension. Cell Rep. 2018;25:677–689 e674. doi: 10.1016/j.celrep.2018.09.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mell B, Jala VR, Mathew AV, Byun J, Waghulde H, Zhang YJ, Haribabu B, Vijay-Kumar M, Pennathur S, Joe B. Evidence for a link between gut microbiota and hypertension in the Dahl rat. Physiol Genomics. 2015;47:187–197. doi: 10.1152/physiolgenomics.00136.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Joe B, McCarthy CG, Edwards JM, Cheng X, Chakraborty S, Yang T, Golonka RM, Mell B, Yeo JY, Bearss NR, et al. Microbiota Introduced to Germ-Free Rats Restores Vascular Contractility and Blood Pressure. Hypertension. 2020:HYPERTENSIONAHA12015939. doi: 10.1161/HYPERTENSIONAHA.120.15939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Karbach SH, Schonfelder T, Brandao I, Wilms E, Hormann N, Jackel S, Schuler R, Finger S, Knorr M, Lagrange J, et al. Gut Microbiota Promote Angiotensin II-Induced Arterial Hypertension and Vascular Dysfunction. J Am Heart Assoc. 2016;5. doi: 10.1161/JAHA.116.003698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gordon HA, Wostmann BS, Bruckner-Kardoss E. Effects of Microbial Flora on Cardiac Output and Other Elements of Blood Circulation. Proc Soc Exp Biol Med. 1963;114:301–304. [DOI] [PubMed] [Google Scholar]
- 98.Bauer W, Richards DW. A Vaso-Dilator Action of Acetates. J Physiol-London. 1928;66:371–378. doi: DOI 10.1113/jphysiol.1928.sp002534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Keshaviah PR. The role of acetate in the etiology of symptomatic hypotension. Artif Organs. 1982;6:378–387. [DOI] [PubMed] [Google Scholar]
- 100.Kirkendol PL, Devia CJ, Bower JD, Holbert RD. A comparison of the cardiovascular effects of sodium acetate, sodium bicarbonate and other potential sources of fixed base in hemodialysate solutions. Trans Am Soc Artif Intern Organs. 1977;23:399–405. [DOI] [PubMed] [Google Scholar]
- 101.Pagel MD, Ahmad S, Vizzo JE, Scribner BH. Acetate and bicarbonate fluctuations and acetate intolerance during dialysis. Kidney Int. 1982;21:513–518. [DOI] [PubMed] [Google Scholar]
- 102.Hakim RM, Pontzer MA, Tilton D, Lazarus JM, Gottlieb MN. Effects of acetate and bicarbonate dialysate in stable chronic dialysis patients. Kidney Int. 1985;28:535–540. doi: 10.1038/ki.1985.161 [DOI] [PubMed] [Google Scholar]
- 103.Natarajan N, Hori D, Flavahan S, Steppan J, Flavahan NA, Berkowitz DE, Pluznick JL. Microbial short chain fatty acid metabolites lower blood pressure via endothelial G protein-coupled receptor 41. Physiol Genomics. 2016;48:826–834. doi: 10.1152/physiolgenomics.00089.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Poll BG, Xu JJ, Jun S, Sanchez J, Zaidman NA, He XJ, Lester L, Berkowitz DE, Paolocci N, Gao WD, et al. Acetate, a Short-Chain Fatty Acid, Acutely Lowers Heart Rate and Cardiac Contractility Along with Blood Pressure. J Pharmacol Exp Ther. 2021;377:39–50. doi: 10.1124/jpet.120.000187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Marques FZ, Nelson E, Chu PY, Horlock D, Fiedler A, Ziemann M, Tan JK, Kuruppu S, Rajapakse NW, El-Osta A, et al. High-Fiber Diet and Acetate Supplementation Change the Gut Microbiota and Prevent the Development of Hypertension and Heart Failure in Hypertensive Mice. Circulation. 2017;135:964–977. doi: 10.1161/CIRCULATIONAHA.116.024545 [DOI] [PubMed] [Google Scholar]
- 106.Wang L, Zhu Q, Lu A, Liu X, Zhang L, Xu C, Liu X, Li H, Yang T. Sodium butyrate suppresses angiotensin II-induced hypertension by inhibition of renal (pro)renin receptor and intrarenal renin-angiotensin system. J Hypertens. 2017;35:1899–1908. doi: 10.1097/HJH.0000000000001378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kim S, Goel R, Kumar A, Qi Y, Lobaton G, Hosaka K, Mohammed M, Handberg EM, Richards EM, Pepine CJ, et al. Imbalance of gut microbiome and intestinal epithelial barrier dysfunction in patients with high blood pressure. Clin Sci (Lond). 2018;132:701–718. doi: 10.1042/CS20180087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kaye DM, Shihata WA, Jama HA, Tsyganov K, Ziemann M, Kiriazis H, Horlock D, Vijay A, Giam B, Vinh A, et al. Deficiency of Prebiotic Fiber and Insufficient Signaling Through Gut Metabolite-Sensing Receptors Leads to Cardiovascular Disease. Circulation. 2020;141:1393–1403. doi: 10.1161/CIRCULATIONAHA.119.043081 [DOI] [PubMed] [Google Scholar]
- 109.Li B, He X, Jin HY, Wang HY, Zhou FC, Zhang NY, Jie DY, Li LZ, Su J, Zheng X, et al. Beneficial effects of Dendrobium officinale on metabolic hypertensive rats by triggering the enteric-origin SCFA-GPCR43/41 pathway. Food Funct. 2021;12:5524–5538. doi: 10.1039/d0fo02890h [DOI] [PubMed] [Google Scholar]
- 110.Dinakis E, Nakai M, Gill PA, Yiallourou S, Sata Y, Muir J, Carrington M, Head GA, Kaye DM, Marques FZ. The Gut Microbiota and Their Metabolites in Human Arterial Stiffness. Heart Lung Circ. 2021;30:1716–1725. doi: 10.1016/j.hlc.2021.07.022 [DOI] [PubMed] [Google Scholar]
- 111.Jama HA, Rhys-Jones D, Nakai M, Yao CK, Climie RE, Sata Y, Anderson D, Creek DJ, Head GA, Kaye DM, et al. Gut microbial metabolites lower 24-hour systolic blood pressure in untreated essential hypertensive patients. medRxiv. 2022:2022.2006.2020.22276673. doi: 10.1101/2022.06.20.22276673 [DOI] [Google Scholar]
- 112.Robles-Vera I, Toral M, de la Visitacion N, Aguilera-Sanchez N, Redondo JM, Duarte J. Protective Effects of Short-Chain Fatty Acids on Endothelial Dysfunction Induced by Angiotensin II. Front Physiol. 2020;11:277. doi: 10.3389/fphys.2020.00277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Miyamoto J, Kasubuchi M, Nakajima A, Irie J, Itoh H, Kimura I. The role of short-chain fatty acid on blood pressure regulation. Curr Opin Nephrol Hy. 2016;25:379–383. doi: 10.1097/Mnh.0000000000000246 [DOI] [PubMed] [Google Scholar]
- 114.Yang F, Chen HW, Gao YH, An N, Li XY, Pan XD, Yang XY, Tian L, Sun JH, Xiong XJ, et al. Gut microbiota-derived short-chain fatty acids and hypertension: Mechanism and treatment. Biomed Pharmacother. 2020;130. doi: ARTN 110503 10.1016/j.biopha.2020.110503 [DOI] [PubMed] [Google Scholar]
- 115.Pluznick JL. Microbial Short-Chain Fatty Acids and Blood Pressure Regulation. Curr Hypertens Rep. 2017;19. doi: ARTN 25 10.1007/s11906-017-0722-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Natarajan N, Hori D, Flavahan S, Steppan J, Flavahan NA, Berkowitz DE, Pluznick JL. Microbial short chain fatty acid metabolites lower blood pressure via endothelial G-protein coupled receptor 41. Physiol Genomics. 2016:physiolgenomics 00089 02016. doi: 10.1152/physiolgenomics.00089.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kaye DM, Shihata W, Jama HA, Tsyganov K, Ziemann M, Kiriazis H, Horlock D, Vijay A, Giam B, Vinh A, et al. Deficiency of Prebiotic Fibre and Insufficient Signalling Through Gut Metabolite Sensing Receptors Leads to Cardiovascular Disease. Circulation. 2020. doi: 10.1161/CIRCULATIONAHA.119.043081 [DOI] [PubMed] [Google Scholar]
- 118.Natarajan N, Hori D, Flavahan S, Steppan J, Flavahan NA, Berkowitz DE, Pluznick JL. Microbial short chain fatty acid metabolites lower blood pressure via endothelial G protein-coupled receptor 41. Physiol Genomics. 2016;48:826–834. doi: 10.1152/physiolgenomics.00089.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nohr MK, Egerod KL, Christiansen SH, Gille A, Offermanns S, Schwartz TW, Moller M. Expression of the short chain fatty acid receptor GPR41/FFAR3 in autonomic and somatic sensory ganglia. Neuroscience. 2015;290:126–137. doi: 10.1016/j.neuroscience.2015.01.040 [DOI] [PubMed] [Google Scholar]
- 120.Muralitharan RR, Marques FZ. Diet-related gut microbial metabolites and sensing in hypertension. J Hum Hypertens. 2021;35:162–169. doi: 10.1038/s41371-020-0388-3 [DOI] [PubMed] [Google Scholar]
- 121.Nakai M, Ribeiro RV, Stevens BR, Gill P, Muralitharan RR, Yiallourou S, Muir J, Carrington M, Head GA, Kaye DM, et al. Essential Hypertension Is Associated With Changes in Gut Microbial Metabolic Pathways A Multisite Analysis of Ambulatory Blood Pressure. Hypertension. 2021;78:804–815. doi: 10.1161/Hypertensionaha.121.17288 [DOI] [PubMed] [Google Scholar]
- 122.Ang ZW, Xiong D, Wu M, Ding JL. FFAR2-FFAR3 receptor heteromerization modulates short-chain fatty acid sensing. Faseb J. 2018;32:289-+. doi: 10.1096/fj.201700252RR [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rezq S, Abdel-Rahman AA. Central GPR109A Activation Mediates Glutamate-Dependent Pressor Response in Conscious Rats. J Pharmacol Exp Ther. 2016;356:456–465. doi: 10.1124/jpet.115.229146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gambhir D, Ananth S, Veeranan-Karmegam R, Elangovan S, Hester S, Jennings E, Offermanns S, Nussbaum JJ, Smith SB, Thangaraju M, et al. GPR109A as an Anti-Inflammatory Receptor in Retinal Pigment Epithelial Cells and Its Relevance to Diabetic Retinopathy. Invest Ophth Vis Sci. 2012;53:2208–2217. doi: 10.1167/iovs.11-8447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rodriguez-Iturbe B, Pons H, Johnson RJ. Role of the Immune System in Hypertension. Physiol Rev. 2017;97:1127–1164. doi: 10.1152/physrev.00031.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Singh MV, Chapleau MW, Harwani SC, Abboud FM. The immune system and hypertension. Immunol Res. 2014;59:243–253. doi: 10.1007/s12026-014-8548-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mulvany MJ. Small artery remodelling in hypertension. Basic Clin Pharmacol Toxicol. 2012;110:49–55. doi: 10.1111/j.1742-7843.2011.00758.x [DOI] [PubMed] [Google Scholar]
- 128.Xu J, Choi R, Gupta K, Santhanam L, Pluznick J. Uncovering a sex-specific role for olfactory receptor 558 (Olfr558) in blood pressure regulation. In: FASEB Meeting. FASEB Journal; 2022. [Google Scholar]
- 129.Pronin A, Levay K, Velmeshev D, Faghihi M, Shestopalov VI, Slepak VZ. Expression of Olfactory Signaling Genes in the Eye. Plos One. 2014;9. doi: ARTN e96435 10.1371/journal.pone.0096435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Nakai M, Ribeiro RV, Stevens BR, Gill P, Muralitharan RR, Yiallourou S, Muir J, Carrington M, Head GA, Kaye DM, et al. Essential Hypertension Is Associated With Changes in Gut Microbial Metabolic Pathways: A Multisite Analysis of Ambulatory Blood Pressure. Hypertension. 2021:HYPERTENSIONAHA12117288. doi: 10.1161/HYPERTENSIONAHA.121.17288 [DOI] [PubMed] [Google Scholar]
- 131.Weber GJ, Foster J, Pushpakumar SB, Sen U. Altered microRNA regulation of short chain fatty acid receptors in the hypertensive kidney is normalized with hydrogen sulfide supplementation. Pharmacol Res. 2018;134:157–165. doi: 10.1016/j.phrs.2018.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Brunskill EW, Sequeira-Lopez ML, Pentz ES, Lin E, Yu J, Aronow BJ, Potter SS, Gomez RA. Genes that confer the identity of the renin cell. J Am Soc Nephrol. 2011;22:2213–2225. doi: 10.1681/ASN.2011040401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Marques FZ, Mackay CR, Kaye DM. Beyond gut feelings: how the gut microbiota regulates blood pressure. Nat Rev Cardiol. 2018;15:20–32. doi: 10.1038/nrcardio.2017.120 [DOI] [PubMed] [Google Scholar]
- 134.Karlsson M, Zhang C, Mear L, Zhong W, Digre A, Katona B, Sjostedt E, Butler L, Odeberg J, Dusart P, et al. A single-cell type transcriptomics map of human tissues. Sci Adv. 2021;7. doi: 10.1126/sciadv.abh2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Proteinatlas.org. 2022. Single cell type - FFAR3 - The Human Protein Atlas. [online] Available at: https://www.proteinatlas.org/ENSG00000185897-FFAR3/single+cell+type
- 136.Proteinatlas.org. 2022. Single cell type – FFAR2 - The Human Protein Atlas. [online] Available at: https://www.proteinatlas.org/ENSG00000126262-FFAR2/single+cell+type
- 137.Proteinatlas.org. 2022. Single cell type - HCAR2 - The Human Protein Atlas. [online] Available at: https://www.proteinatlas.org/ENSG00000182782-HCAR2/single+cell+type
- 138.Proteinatlas.org. 2022. Single cell type – OR51E2 - The Human Protein Atlas. [online] Available at: https://www.proteinatlas.org/ENSG00000167332-OR51E2/single+cell+type
- 139.Proteinatlas.org. 2022. Single cell type – OR51E1 - The Human Protein Atlas. [online] Available at: https://www.proteinatlas.org/ENSG00000180785-OR51E1/single+cell+type
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


