Abstract
A growing number of familial Mediterranean fever (FMF) patients in Israel do not have a single country of origin for all four grandparents. We aimed to predict the Mediterranean fever gene (MEFV) variant most likely to be found for an individual FMF patient, by a machine learning approach. This study was conducted at the Sheba Medical Center, a referral center for FMF in Israel. All Jewish referrals included in this study carried an FMF associated variant in MEFV as shown by genetic testing performed between 2001 and 2017. We introduced the term ‘origin score’ to capture the dose and different combinations of the grandparents’ origin. A machine learning approach was used to analyze the data. In a total of 1781 referrals included in this study, the p.Met694Val variant was the most common, and the variants p.Glu148Gln and p.Val726Ala second and third most common, respectively. Of 26 countries of origin analyzed, those that increased the likelihood of a referral to carry specific variants were identified in North Africa for p.Met694Val, Europe for p.Val726Ala, and west Asia for p.Glu148Gln. Fourteen of the studied countries did not show a highly probable variant. Based on our results, it is possible to describe an association between modern day origins of the three most common MEFV variant types and a geographical region. A strong geographic association could arise from positive selection of a specific MEFV variant conferring resistance to endemic infectious agents.
Subject terms: Genetics, Immunology
Introduction
Familial Mediterranean fever (FMF) is the most common syndrome in the group of hereditary auto-inflammatory diseases1. It is an autosomal recessive disease that mainly associates with variants in the MEFV gene, located on chromosome 16. MEFV encodes the pyrin protein, which is important for the inflammatory response to infectious agents2. More than 300 variants of the MEFV gene have been identified in Infevers https://fmf.igh.cnrs.fr/ISSAID/infevers/search.php?n=1 (Infevers: an online database for autoinflammatory mutations. Copyright. Available at https://infevers.umai-montpellier.fr/ Accessed at (02/2022)3–6). The five most common variants (p.Met694Val, p.Val726Ala, p.Met694Ile, p.Met680Ile and p.Glu148Gln) account for the vast majority of cases7,8. The prevalence of FMF is highest among ethnic inhabitants of the Mediterranean basin with a carrier rate of up to 1 in 4 in certain populations. In recent years, the disease has been reported in ethnically heterogeneous patients around the globe9–13. Israel is considered an endemic area for FMF14,15. Its current population has diverse origins in the Jewish diaspora including Europe, northern Africa, and Asia. There is a correlation between the p.Met694Val variant and Jewish Moroccan ethnicity as well as with a severe disease phenotype16,17. However, associations between other countries of origin and FMF variants have been only partially established18. Such knowledge is important to understand the epidemiology of FMF. Here we use a novel approach, based on a machine learning algorithm, to predict the mutation type carried by a patient based on the countries of origin of his/her parents or grandparents.
Subjects and methods
Setting and patient selection
This study was conducted at The Chaim Sheba Medical Center in Tel Hashomer, Israel, which is a referral center for genetic testing and evaluation of FMF patients. First, we collected data on all referrals to our center for genetic analysis by their primary physician following a clinical suspicion for FMF between 2001 and 2017. All referrals negative for variants in MEFV were excluded. Since mixed origins mainly characterize Jewish patients only Jewish referrals were included in our study group. Data regarding the gender and the specific variant of each referral was extracted from medical records. This research was approved by Sheba Medical Center institutional ethics committee. All methods were performed in accordance with the relevant guidelines and regulations.
Genetic analysis of MEFV
For the genetic analysis, DNA was extracted from 100 µl of blood taken from the referral using a Puregene kit (Gentra Inc.) and was screened for five known variants in MEFV, LRG190t1:c.2080A > G p.(Met694Val), c.2177 T > C p.Val726Ala, c.422G > C, p.Glu148Gln, c.20420G > A or c.2040G > C p.Met680Ile, and c.2082G > A p.Met694Ile, using a commercial kit (Gamidagen) or polymerase chain reaction (PCR) amplification and restriction enzyme analysis19.
Computational analysis
Origin score
In genetic studies, it is usually straightforward to investigate the association between country of origin and variants using mathematical tools such as Bayes rule. However, given the ancestral diversity of the Israeli Jewish population, the subjects referred to our center often do not have a single country of origin. It was therefore necessary to construct a model using machine learning in order to perform statistical analysis. We included in the analysis countries from which at least 15 referrals originated. Based on this threshold, the data used for the analysis included 26 countries (out of 48 reported to be countries of origin for parents or grandparents by patients in the cohort). First, data on referrals and countries of origin were tabulated in a matrix with a row representing a subject and a column representing a possible country of origin. We then calculated an “Origin Score” in the following way: In each cell we stored the fraction of the subject’s origin from each country. For example, if a referral has two grandparents from Algeria, one from Morocco, and one from Iraq, then the values of the corresponding cells will be 0.5, 0.25, and 0.25, respectively, and cells corresponding to all other countries will be assigned a value of 0. If information about the country of origin of one of the grandparents was missing, it was assumed that both grandparents from that side had the same country of origin. Subjects without information on at least one grandparent from each side were excluded from the analysis. Based on the method described above, we calculated for every country, the sum of origin scores of subjects with any level of ancestry from that country. The sum of origin scores per country is presented in Fig. S1.
Machine learning approach
The logic behind our novel machine learning based approach is that the level at which we are able to predict if a person has a specific variant based on his/her origin is an indication of the strength of the correlation between the origin and the variant. Clearly, the stronger the association between a given country of origin and a specific variant the more accurate is the prediction. For the machine learning approach, we used the logistic regression module "scikit-learn” in Python 2.720. Logistic regression is a linear model used to measure the relationship between the categorical dependent variable and one or more independent variables by estimating probabilities describing the possible outcomes based on the logistic function. In our study, we used the countries of origin as the independent variables and attempted to predict the specific variant as a categorical dependent variable (i.e., if the person has or does not have the specific variant). The performance of the prediction was evaluated by the area under the curve (AUC) measure, which shows the deviation of the performance from a random prediction, which has an AUC value of 0.5 while a perfect prediction has a value of 1. We validated our model using tenfold cross validation (dividing the data randomly each time to 90% for training and 10% for testing) and by bootstrapping (where subsets were resampled with replacement 1000 times and patients that were not included in the sample were used as the test dataset). For each prediction, we averaged each vector coefficients and used the result to identify origins that are positively and negatively associated with certain variants. Since our analysis revealed that the p.Met694Ile and p.Met680Ile variants were very rare in our study sample, we excluded these variants from the subsequent analysis. Therefore, we included in the final analysis only the three most common variant types: p.Met694Val, p.Val726Ala and p.Glu148Gln. The data used for the prediction of the country of origin included only patients that carry a single type of mutation, either homozygous or heterozygous. Compound heterozygotes were not included since we did not have enough data for each compound heterozygous pair.
Selection of country groups
We combined the different countries of origin into groups that contained four countries each, covering four possible origins per patient. We formed a group from the four countries that were ranked highest in their association with each variant and another group with the four countries that were ranked as the least associated.
Ethics committee
The study has been approved by the appropriate ethics committee. Informed consent was waived by the ethics institutional review board (IRB) – Sheba Medical Center (SMC-9763-12).
Results
A total of 1842 referrals for MEFV genetic testing had at least one MEFV gene variant. After excluding 61 subjects with uncommon variants, we included 1781 subjects (52% females) in our analysis (Table 1). The number of subjects detected with each MEFV variant is presented in Fig. S2. The p.Met694Val variant was found in 72% of the referrals. Out of the 1286 referrals that carry p.Met694Val, 18% (240) had another variant; these referrals were compound heterozygous for either p.Glu148Gln, p.Val726Ala, p.Met680Ile, or p.Met694Ile. The prevalence of referrals with the p.Glu148Gln and p.Val726Ala variants were 23% (419 subjects) and 19% (342 subjects), respectively. Of the referrals that carried p.Glu148Gln, 35% (150) were compound heterozygous either with p.Met694Val, or p.Val726Ala. Of those with the p.Val726Ala variant, 42% (145) were compound heterozygous.
Table 1.
Prevalence of common MEFV variants in Jewish FMF referrals to genetic testing between 2001 and 2017 in one referral center.
| Variant, zygosity status | Patient number by gender | Total | |
|---|---|---|---|
| Female | Male | ||
| p.Met694Val, (HTZ) | 421 | 403 | 824 |
| p.Glu148Gln, (HTZ) | 142 | 119 | 261 |
| p.Met694Val, (HMZ) | 122 | 100 | 222 |
| p.Val726Ala, (HTZ) | 88 | 72 | 160 |
| p.Met694Val and p.Glu148Gln, Compound heterozygous E148Q | 58 | 63 | 121 |
| p.Met694Val and p.Val726Ala, Compound heterozygous | 49 | 67 | 116 |
| p.Val726Ala, (HMZ) | 16 | 21 | 37 |
| p.Val726Ala and p.Glu148Gln, apparent compound heterozygous | 15 | 14 | 29 |
| p.Glu148Gln, (HMZ) | 6 | 2 | 8 |
| p.Met694Val and p.Met680Ile, Compound heterozygous | 2 | 2 | |
| p.Met694Val and p.Met694Ile, Compound heterozygous | 1 | 1 | |
| Total | 920 | 861 | 1781 |
HMZ homozygote, HTZ heterozygote.
The results presented here are based on 10-Fold cross-validation (see Methods). Similar results were achieved using bootstrap resampling with replacement 1000 times (See supplementary Fig. S3).
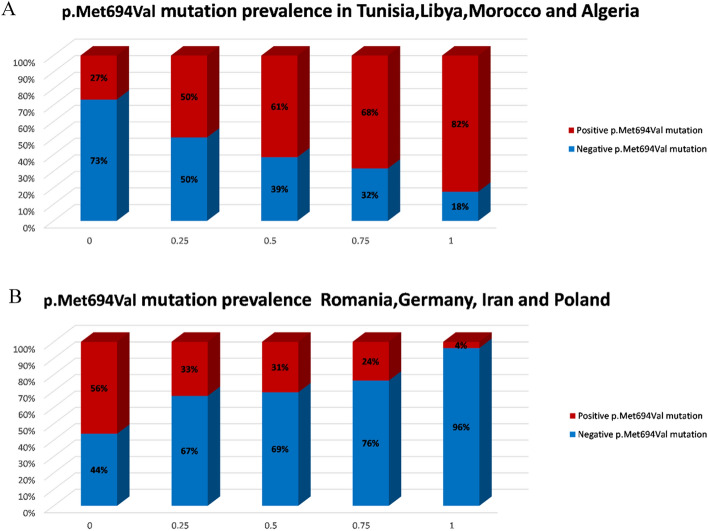
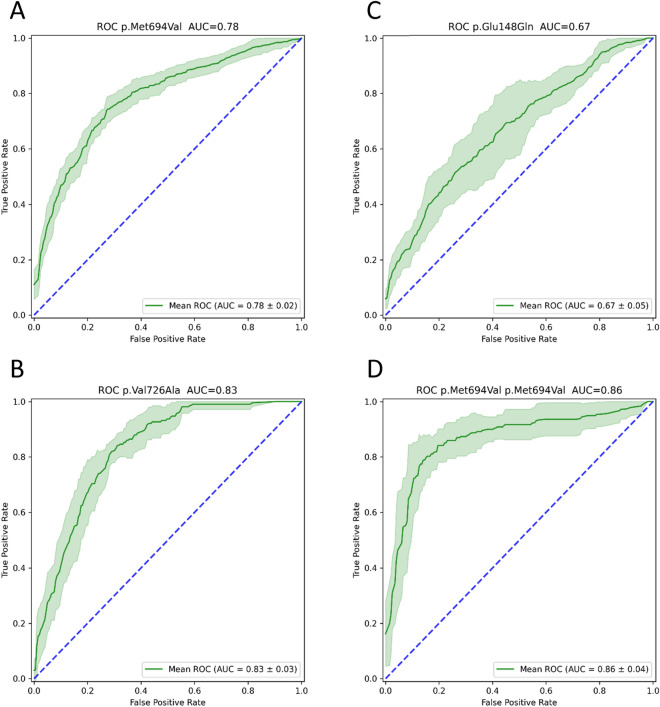
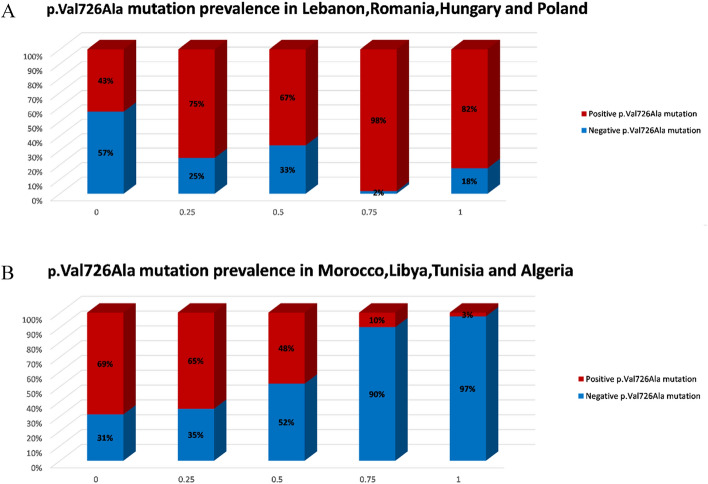
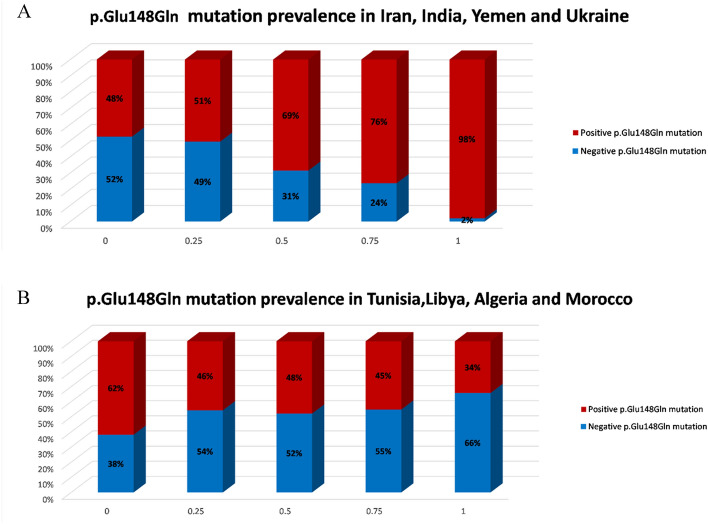
Origination in Tunisia, Libya, Morocco, and Algeria was positively associated with the p.Met694Val variant, roughly 70% referrals of Moroccan decent carried this variant. Origination in Romania, Germany, Iran and Poland reduced the chance of carrying the p.Met694Val variant (Fig. 1A, B). The performance of this prediction is demonstrated by the AUC of 0.78 (Fig. 2A). Moreover, by multivariate logistic regression analysis we demonstrated that Libya, Tunisia, Morocco and Algeria as countries of origin contributed the most to the probability that a referral would be homozygous for p.Met694Val variant with even higher degree of certainty (AUC = 0.86; Fig. 2D). Referrals with the p.Val726Ala variant had a high probability of originating from Lebanon, Romania, Hungary, or Poland (Fig. 3A). Ancestors from Morocco, Libya, Tunisia, and Algeria reduced the likelihood of this variant (Fig. 3B) with an AUC of 0.83 (Fig. 2B). Iran, India, Yemen and Ukraine are the origins that contribute the most to the existence of p.Glu148Gln variant (Fig. 4A), whereas Tunisia, Libya, Algeria and Morocco had an opposite impact (Fig. 4B) with an AUC of 0.67 (Fig. 2C). Fourteen of the studied countries did not show strong association with a single MEFV variant.
Figure 1.
Prevalence of the countries of origin most indicative for the p.Met694Val variant: (A) Tunisia, Libya, Morocco, and Algeria are the countries of origin most positively associated with the p.Met694Val variant. (B) Romania, Germany, Iran and Poland are the countries of origin most negatively associated with the p.Met694Val variant. Red and blue bars indicate patients who carry and do not carry the p.Met694Val variant, respectively. The x-axis indicates the amount of grandparents from the four countries (0 being none, 1 being all four).
Figure 2.
ROC of multivariate logistic regression variant prediction. The figure represents the performance of the multivariate logistic regression model that uses 27 features (26 countries and sex of the patient) to predict whether a patient: (A) carries the variant p.Met694Val, (B) carries the variant p.Val726Ala, (C) carries the variant p.Glu148Gln, and (D) is homozygous for p.Met694Val.
Figure 3.
Prevalence of the countries of origin most indicative for the p.Val726Ala variant: (A) Lebanon, Romania, Hungary, and Poland are the countries of origin most positively associated with the p.Val726Ala variant. (B) Morocco, Libya, Tunisia, and Algeria are the countries of origin most negatively associated with the p.Val726Ala variant. Red and blue bars indicate patients who carry and do not carry the p.Val726Ala variant, respectively. The x-axis indicates the amount of grandparents from the four countries (0 being none, 1 being all four).
Figure 4.
Prevalence of the countries of origin most indicative for the p.Glu148Gln variant: Countries that were found to be the most positively (A) and negatively (B) associated origins for a patient who carries a positive p.Glu148Gln variant. Red and blue bars represent the patients who carry and do not carry the p.Glu148Gln variant, respectively. The X-axis indicates the amount of grandparents from the four countries (0 being none, 1 being all four).
Discussion
Many types of variants in the MEFV gene are associated with FMF. The five most commonly identified mutation types have been denoted as the founder mutations7,8. An association between ethnicity and the type of variant has been suggested, but a clear connection has not been established17. This study was conducted in order to demonstrate such link: We sought to demonstrate that a patient's variant could be predicted based on his/her family origins. Given the ancestral diversity of the large study population and the fact that the subjects rarely had a single country of origin, we constructed a model using machine learning in order to perform statistical analysis. We introduced an origin score to quantify the ethnic complexity of each individual.
Analysis of the study population, which included 1781 Israeli referrals for FMF testing mainly due to FMF suspicion, allowed us to extract reliable results despite the ethnic diversity of the study population. Our results showed that the p.Met694Val variant was the most prevalent among the study population, identified in 72% of studied subjects. Referrals whose parents or grandparents came to Israel from Tunisia, Libya, Algeria, or Morocco were most likely to carry this specific variant. The second most common mutation, p.Glu148Gln, was observed in 23% of the cohort; the most common countries of origin were Iran, Yemen, India, and Ukraine. The p.Val726Ala variant was the third most common, found in 19% of subjects. The countries of origin that mostly contributed to the existence of this variant are Lebanon, Romania, Hungary, and Poland. Notably, an inverse relationship exists between the p.Met694Val variant and p.Val726Ala with regards to country of origin. Countries that are most correlated with p.Met694Val are those least likely to be predictive of p.Val726Ala and vice versa.
All in all, the machine learning approach identified a single highly probable MEFV variant in 12 of the origins studied. The same was not the case for FMF referrals of other origins including Iraqi-Jews despite their high origin score, suggesting that at least two MEFV variants are probable in those origins. Based on the obtained results we deduce that the common MEFV variants in the Israeli Jewish population of our time have origins in a different geographical area: p.Met694Val in North Africa, p.Val726Ala in Europe and p.Glu148Gln in Asia. In general, the machine learning results are consistent with already established variants frequencies in North African, and Ashkenazi Jews9, yet they add a larger geographic scope and a better, country-wise perspective. For instance, our study identified Lebanon, a longtime residence of a small and relatively isolated Sephardi community, as the fourth country predicting Val726Ala, a variant considered to be of Ashkenazi origin (Ashkenazi allele frequency (AF) = 0.04, GnomAD21, https://gnomad.broadinstitute.org) (Table 2), perhaps as a consequence of random genetic drift. It is also intriguing that Ukraine, a residence of Ashkenazi Jewry, was found to be an origin of the p.Glu148Gln variant, along with a distant cluster of Asian countries. This finding could arise from the ethnic composition Ukraine immigrants to Israel, which includes mixed families of Ashkenazi and non-Jewish origins22. The geographical pattern of the p.Met694Val and p.Val726Ala variants observed in our study does not extend to the non-Jewish Caucasian populations of Europe (AF = 0.0009, gnomAD21) nor to the non-Jewish North-African population23,24, consistent with genetic drift, randomly occurring in small and isolated populations of the Jewish diaspora9, undergoing an evolution-based positive selection. Indeed a plague endemic could pose a rapid selection for MEFV variants introduced to Middle Eastern-derived populations early on25. Specifically the p.Met694Val and p.Val726Ala variants were shown to impede the evasion of Yersinia pestis detection by the intracellular pathogen sensing system, which is mediated by the pyrin inflammasome26. Leukocytes from asymptomatic carriers mounted higher IL-1β levels in response to Y. pestis in In-vitro studies25, Emphasizing the selective advantage of MEFV heterozygotes.
Table 2.
Frequency of the p.Met694Val , p.Val726Ala and p.glu148Gln variants in Jewish and corresponding geographic non-Jewish populations.
| Ethnicity (healthy subjects) | Variant frequency p.Met694Val | Variant frequency p. Val726Ala | Variant frequency p. Glu148Gln | References |
|---|---|---|---|---|
| North African Jews in Israel (n = 100) | 0.08 | 0 | 0.05 | Stoffman et al.29 |
| Ashkenazi Jews in Israel (n = 3472) | 0 | 0.0425 | 0.05818 | GnomAD V.3.1.221 |
| Iranian Jews in Israel (n = 100) | 0 | 0 | 0.025 | Stoffman et al.29 |
| Yemen Jews in Israel (n = 36) | 0.0138 | 0 | 0.0276 | Feld et al.30 |
| European—Non-Finish (n = 68,010) | 0.0001764 | 0.0007205 | 0.01266 | GnomAD V.3.1.221 |
| South Asians (n = 4828) | 0 | 0 | 0.2981 | GnomAD V.3.1.221 |
Our study also identified an Asian origin for the p.Glu148Gln variant in the Israeli FMF referrals. An Asian origin is in agreement with the high frequency of this variant in the non-Jewish south and east Asian populations (AF = 0.298 and 0.280, respectively, GnomAD21), (Table 2). This observation may be rooted in the early settlement of the Asian Jewish diaspora and its admixture with the local population27. Considering that FMF morbidity is scarce in south Asian countries, the clinical significance of the p.Glu148Gln variant is uncertain, and it’s inclusion among the variants associated with FMF needs to be carefully discussed. A recent study showed a 17-fold increased penetrance of FMF in compound heterozygotes carrying both the p.Glu148Gln and p.Met694Val variants, over heterozygotes carrying the p. Met694Val variant alone, in north African Israeli-Jews28. This suggests that the p.Glu148Gln might be considered pathogenic in certain ethnicities.
The assignment of a certain variant to a particular origin might be compromised by two confounders: first, the study cohort mainly comprised symptomatic referrals, which may underrepresents low penetrance variants such as p.Val726Ala and p.Glu148Gln. However, the studied population concords with those served by practitioners, and therefore our results answer and are appropriate for medical needs. Second, the exclusion of compound heterozygous subjects could somewhat skew the results. However, the distribution of the excluded mutations is comparable to their distribution in the populations affected by this step. Therefore, its impact on the results is minimal.
Supplementary Information
Author contributions
O.A., R.B., R.U., A.L. and S.K. contributed to the study conception and design. Material preparation, data collection and analysis were performed by A.L., A.S., S.D.. The first draft of the manuscript was written by A.S., D.S. and S.K. and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.”
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Data availability
Upon request—From Prof. Shay.
Code availability
Mentioned in the method section.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Orit Adato, Ronen Brenner, Ron Unger and Shaye Kivity
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-19538-1.
References
- 1.Ciccarelli F, De Martinis M, Ginaldi L. An update on autoinflammatory diseases. Curr. Med. Chem. 2014;21:261–269. doi: 10.2174/09298673113206660303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schnappauf O, Chae JJ, Kastner DL, Aksentijevich I. The Pyrin inflammasome in health and disease. Front. Immunol. 2019;10:1745. doi: 10.3389/fimmu.2019.01745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Menthière CS, et al. INFEVERS: the Registry for FMF and hereditary inflammatory disorders mutations. Nucleic Acids Res. 2003;31:282–285. doi: 10.1093/nar/gkg031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Touitou I, et al. Infevers: an evolving mutation database for auto-inflammatory syndromes. Hum. Mutat. 2004;24:194–198. doi: 10.1002/humu.20080. [DOI] [PubMed] [Google Scholar]
- 5.Milhavet F, et al. The infevers autoinflammatory mutation online registry: update with new genes and functions. Hum. Mutat. 2008;29:803–808. doi: 10.1002/humu.20720. [DOI] [PubMed] [Google Scholar]
- 6.Van Gijn ME, et al. New workflow for classification of genetic variants’ pathogenicity applied to hereditary recurrent fevers by the International Study Group for Systemic Autoinflammatory Diseases (INSAID) J. Med. Genet. 2018;55:530–537. doi: 10.1136/jmedgenet-2017-105216. [DOI] [PubMed] [Google Scholar]
- 7.Drenth JPH, Van Der Meer JWM. The inflammasome: a linebacker of innate defense. N. Engl. J. Med. 2006;355:730–732. doi: 10.1056/NEJMcibr063500. [DOI] [PubMed] [Google Scholar]
- 8.Kucuk, A. et al. Familial mediterranean fever. Acta Medica (Hradec Králové)57, 97–104 (2014). [DOI] [PubMed]
- 9.Ben-Chetrit E, Touitou I. Familial mediterranean fever in the world. Arthritis Care Res. 2009;61:1447–1453. doi: 10.1002/art.24458. [DOI] [PubMed] [Google Scholar]
- 10.Migita, K. et al. Familial Mediterranean fever in Japan. Med. (United States)91, 337–343 (2012). [DOI] [PubMed]
- 11.Jesus AA, et al. Hereditary autoinflammatory syndromes: a Brazilian Multicenter Study. J. Clin. Immunol. 2012;32:922–932. doi: 10.1007/s10875-012-9688-x. [DOI] [PubMed] [Google Scholar]
- 12.Majeed HA, et al. The spectrum of familial mediterranean fever gene mutations in arabs: report of a large Series. Semin. Arthritis Rheum. 2005;34:813–818. doi: 10.1016/j.semarthrit.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 13.Esmaeili M, Bonyadi M, Rafeey M, Sakha K, Somi MH. Common MEFV mutation analysis in Iranian Azeri Turkish patients with familial mediterranean fever. Semin. Arthritis Rheum. 2008;37:334–338. doi: 10.1016/j.semarthrit.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 14.Brenner R, et al. Familial Mediterranean fever and incidence of cancer. Arthritis Rheumatol. 2018;70:127–133. doi: 10.1002/art.40344. [DOI] [PubMed] [Google Scholar]
- 15.Zlotogora J. Autosomal recessive diseases among the Israeli Arabs. Hum. Genet. 2019;138:1117–1122. doi: 10.1007/s00439-019-02043-3. [DOI] [PubMed] [Google Scholar]
- 16.Moradian MM, et al. Comprehensive analysis of mutations in the MEFV gene reveal that the location and not the substitution type determines symptom severity in FMF. Mol. Genet. Genomic Med. 2017;5:742–750. doi: 10.1002/mgg3.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alghamdi M. Familial Mediterranean fever, review of the literature. Clin. Rheumatol. 2017;36:1707–1713. doi: 10.1007/s10067-017-3715-5. [DOI] [PubMed] [Google Scholar]
- 18.Özdemir FMA, Gülez N, Makay B. Evaluation of the international severity score for FMF (ISSF) scores in Turkish children diagnosed with FMF: a single-center experience. Clin. Rheumatol. 2021;40:3219–3225. doi: 10.1007/s10067-021-05652-4. [DOI] [PubMed] [Google Scholar]
- 19.Livneh A, et al. MEFV mutation analysis in patients suffering from amyloidosis of familial Mediterranean fever. Amyloid. 1999;6:1–6. doi: 10.3109/13506129908993281. [DOI] [PubMed] [Google Scholar]
- 20.Pedregosa FABIANPEDREGOSA, F. et al. Scikit-learn: machine learning in Python Gaël Varoquaux Bertrand Thirion Vincent Dubourg Alexandre Passos PEDREGOSA, VAROQUAUX, GRAMFORT ET AL. Matthieu Perrot. J. Mach. Learn. Res.12, 2825–2830 (2011).
- 21.Karczewski, K. J. et al. The mutational constraint spectrum quantified from variation in 141,456 humans, Genome Aggregation Database Consortium. Nature581, 19 (2020). [DOI] [PMC free article] [PubMed]
- 22.Reinharz, S., & DellaPergola, S. Jewish Intermarriage Around the World (Google eBook). 221 (2011).
- 23.Ait-Idir D, Djerdjouri B. Differential mutational profiles of familial Mediterranean fever in North Africa. Ann. Hum. Genet. 2020;84:423–430. doi: 10.1111/ahg.12404. [DOI] [PubMed] [Google Scholar]
- 24.Chaabouni HB, et al. MEFV mutations in Tunisian patients suffering from familial Mediterranean fever. Semin. Arthritis Rheum. 2007;36:397–401. doi: 10.1016/j.semarthrit.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 25.Park YH, et al. Ancient familial Mediterranean fever mutations in human pyrin and resistance to Yersinia pestis. Nat. Immunol. 2020;21:857–867. doi: 10.1038/s41590-020-0705-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chung LK, et al. The Yersinia Virulence Factor YopM Hijacks Host Kinases to Inhibit Type III Effector-Triggered Activation of the Pyrin Inflammasome. Cell Host Microbe. 2016;20:296–306. doi: 10.1016/j.chom.2016.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Behar, D. M. et al. Counting the founders: the matrilineal genetic ancestry of the Jewish Diaspora. PLoS One3, (2008). [DOI] [PMC free article] [PubMed]
- 28.Eyal, O., Shinar, Y., Pras, M. & Pras, E. Familial Mediterranean fever: Penetrance of the p.[Met694Val];[Glu148Gln] and p.[Met694Val];[=] genotypes. Hum. Mutat.41, 1866–1870 (2020). [DOI] [PubMed]
- 29.Stoffman N, et al. Higher than expected carrier rates for familial Mediterranean fever in various Jewish ethnic groups. Eur. J. Hum. Genet. 2000;8:307–310. doi: 10.1038/sj.ejhg.5200446. [DOI] [PubMed] [Google Scholar]
- 30.Feld O, Livneh A, Shinar Y, Berkun Y, Lidar M. MEFV mutation carriage in Israeli Jewish individuals from ethnicities with low risk for familial Mediterranean fever. J. Hum. Genet. 2009;54:369–371. doi: 10.1038/jhg.2009.33. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Upon request—From Prof. Shay.
Mentioned in the method section.