Abstract
Five Escherichia coli type 1 pilus mutants that had point mutations in fimH, the gene encoding the type 1 pilus adhesin FimH, were characterized. FimH is a minor component of type 1 pili that is required for the pili to bind and agglutinate guinea pig erythrocytes in a mannose-inhibitable manner. Point mutations were located by DNA sequencing and deletion mapping. All mutations mapped within the signal sequence or in the first 28% of the predicted mature protein. All mutations were missense mutations except for one, a frameshift lesion that was predicted to cause the loss of approximately 60% of the mature FimH protein. Bacterial agglutination tests with polyclonal antiserum raised to a LacZ-FimH fusion protein failed to confirm that parental amounts of FimH cross-reacting material were expressed in four of the five mutants. The remaining mutant, a temperature-sensitive (ts) fimH mutant that agglutinated guinea pig erythrocytes after growth at 31°C but not at 42°C, reacted with antiserum at both temperatures in a manner similar to the parent. Consequently, this mutant was chosen for further study. Temperature shift experiments revealed that new FimH biosynthesis was required for the phenotypic change. Guinea pig erythrocyte and mouse macrophage binding experiments using the ts mutant grown at the restrictive and permissive temperatures revealed that whereas erythrocyte binding was reduced to a level comparable to that of a fimH insertion mutant at the restrictive temperature, mouse peritoneal macrophages were bound with parental efficiency at both the permissive and restrictive temperatures. Also, macrophage binding by the ts mutant was insensitive to mannose inhibition after growth at 42°C but sensitive after growth at 31°C. The ts mutant thus binds macrophages with one receptor specificity at 31°C and another at 42°C.
Type 1 pili are filamentous proteinaceous appendages produced by several members of the Enterobacteriaceae. In Escherichia coli, type 1 pili have been studied extensively with regard to their genetics, biosynthesis, and ability to bind mannose-containing receptor molecules on a variety of eucaryotic cells (reviewed in reference 29). Although the pili are made principally of a single protein monomer, the product of the fimA gene, several minor protein components are also incorporated (12, 34). These are most often found at the ends of pili and are organized into fibrillar structures (15). One of the minor components, the product of the fimH gene (FimH), binds directly to the receptor (18). Whereas the specificity of the interaction of the fimH product can be influenced by other fimbrial components (23), several studies have linked certain naturally occurring fimH allelic types to the specificity of receptor binding (43, 46) and the strength of receptor binding (45). Additional experiments have suggested that some fimH allelic differences can contribute to tissue tropism (35, 43, 44). Gene fusion experiments have indicated that FimH binding capacity resides in the amino one-third to one-half of the protein (17, 48). However, point mutations in various parts of the coding region can effect a change in specificity for particular types of ligands (46). This is consistent with what is found with other bacterial adhesins (4). FimH has been used to express foreign antigens by inserting heterologous gene segments into the fimH gene (33) and used on its own as an effective immunogen in preventing experimental urinary tract infections in mice (20).
We have been particularly interested in the factors influencing the specificity of FimH binding and how FimH affects proper pilus structure (reviewed in 29). Some years ago, we isolated a number of fimH point mutants (13). These mutants were isolated following enrichment for individuals that formed pellicles when grown in static broth (a property associated with FimH) in the presence of a mannose analogue (α-methyl-mannoside) that normally inhibits pellicle formation. All mutants isolated in this manner were defective in their ability to bind guinea pig erythrocytes. Additionally, several of the mutants produced pili with altered morphology.
In this report, we further characterize five of the fimH mutants isolated in the previous study. These five mutants represent all allelic classes of the 11 mutants initially isolated (13). In particular, we concentrate upon one of the mutants that was conditionally defective in erythrocyte agglutination, showing that erythrocyte and macrophage binding are differentially affected by the mutation. The results indicate that FimH-mediated attachment to different types of eucaryotic cells can occur through different mechanisms.
MATERIALS AND METHODS
Bacteria, bacteriophage, plasmids, and media.
The bacterial strains, all E. coli K-12 derivatives, bacteriophage, and plasmids used in this investigation are listed in Table 1. Media consisted of L agar and L broth (28), λ-broth (28), tetrazolium agar (42), and MinA broth and agar (28). Antibiotics were added as previously described (30) unless otherwise noted.
TABLE 1.
E. coli strains, bacteriophage, and plasmids used in this study
| Strain, phage, or plasmid | Description | Source or reference |
|---|---|---|
| E. coli | ||
| JM101 | Δ(lac-proAB) supE thi/F′ lacIqZΔM15 traD36 proAB | 27 |
| EC901 | leu-6 argE3 proA2 lacY1 his-4 thi-1 galK2 ara-14 xyl-5 srl-31 hsdR4 recA13 srl::Tn10 | 38 |
| ORN103 | thr-1 leu-6 thi-1 Δ(argF-lac)U169 xyl-7 ara-13 mtl-2 gal-6 rspL tonA2 minA minB Δ(fimEACDFGH) | 31 |
| ORN172 | thr-1 leuB thi-1 Δ(argF-lac)U169 xyl-7 ara-13 mtl-2 gal-6 rpsL tonA2 supE44 Δ(fimBEACDFGH)::kan pilG1 | 50 |
| ORN206 | JM101 except Δ(fimBEACDFGH) and recA13 Kanr | P1 transduction from ORN172, then EC901, then ORN103 |
| ORN155 | ORN115 except (Tn5 inserted adjacent [3′] to fimH) | 13 |
| ORN160 | ORN155 except fimH241 | 13 |
| ORN162 | ORN155 except fimH204 (has identical lesion to fimH236) | 13 |
| ORN163 | ORN155 except fimH218 (has identical lesion to fimH236) | 13 |
| ORN164 | ORN155 except fimH236 | 13 |
| ORN165 | ORN155 except fimH244 | 13 |
| ORN157 | ORN155 except fimH205(ts) | 13 |
| ORN158 | ORN155 except fimH208 | 13 |
| ORN115 | thr-1 leuB thi-1 Δ(argF-lac)U169 malA1 xyl-7 ara-13 mtl-2 gal-6 rpsL fhuA2 supE44 pilG1 | 47 |
| ORN133 | Same as ORN115 except fimH′-kan Mal− | 26 |
| ORN183 | Same as ORN157 except Mal+ | 13 |
| ORN175 | Same as ORN115 except Mal+ | 16 |
| ORN204 | Same as ORN133 except Mal+ | 11 |
| Bacteriophage P1 | vir | Laboratory collection |
| Plasmids | ||
| pBR322 | ColE1, Apr Tcr | 3 |
| pACYC184 | P15A, Cmr Tcr | 5 |
| pUR288 | lacZ fusion vector Apr | 37 |
| pSH2 | pACYC184 fimBEAICDFGH Cmr | 10 |
| pORN118 | pSH2 except PstI site in fimH changed to XhoI Cmr | 25 |
| pORN123 | pBR322 ΔPvuII Apr | 32 |
| pORN148 | pORN123 fimD′ fimF fimG fimH Apr | 36 |
| pORN303 | pUR288 with lacZ-fimH fusion | This study |
| pORN304 | Bal31 fimH deletion mutant of pORN118; terminal 22 codons removed | This study |
| pORN305 | Bal31 fimH deletion mutant of pORN118; terminal 82 codons removed | This study |
| pORN306a | KpnI-SalI deletion mutant of pSH2; terminal 276 codons of fimH removed | This study |
| pORN307b | PvuII-SalI deletion mutant of pSH2; entire fimH gene deleted | This study |
| pORN142 | ColE1, fimH with adjacent Tn5 insertion | 13 |
| pORN241 | pORN142 except fimH241 allele | 13 |
| pORN236 | pORN142 except fimH236 allele | 13 |
| pORN204 | pORN142 except fimH204 allele | 13 |
| pORN218 | pORN142 except fimH218 allele | 13 |
| pORN244 | pORN142 except fimH244 allele | 13 |
| pORN205 | pORN142 except fimH205 allele | 13 |
| pORN208 | pORN142 except fimH208 allele | 13 |
The KpnI site in fimH was first converted to an XhoI site by partial digestion of the pSH2 plasmid with KpnI (pSH2 has two KpnI sites), followed by end filling with DNA polymerase I and XhoI linker ligation as described by Orndorff and Falkow (31).
The PvuII site in fimH was first converted to an XhoI site by partial digestion of the pSH2 plasmid with PvuII (pSH2 has five PvuII sites) and ligation of XhoI linkers as described by Orndorff and Falkow (31).
Genetic techniques.
Transformation with plasmid DNA followed the method described by Lederberg and Cohen (21). P1 transduction followed the method described by Miller (28).
Recombinant DNA techniques.
Restriction endonuclease cleavage, plasmid and chromosomal DNA isolation, fragment purification, end filling, ligation, and subcloning were performed as previously described (36). DNA sequencing was performed on double-stranded plasmid DNA as previously described (50) using 20-oligonucleotide primer pairs bracketing various regions of the fimH and lacZ genes. PCR product sequencing of chromosomal and plasmid DNA was carried out as described by Russell and Orndorff (36). Bal31 exonuclease digestions were performed as described by Maniatis et al. (24).
Construction and induction of the lacZ-fimH fusion, and isolation of the fusion protein.
A lacZ-fimH fusion was created by KpnI-PstI digestion of pORN148 followed by end filling and ligation to BamHI-digested, end-filled plasmid pUR288. This created plasmid pORN303. The fusion product contained approximately 92% of the mature FimH product. Pilot experiments revealed that the most efficient production of fusion protein was obtained by diluting an overnight culture of the strain containing the fusion plasmid (ORN206/pORN303) 1:50 in fresh warm L-broth containing ampicillin (100 μg/ml) and 5 mM isopropyl-β-d-thiogalactopyranoside (IPTG), followed by 4 h of growth with shaking at 37°C. After this period, bacteria were harvested by centrifugation (7,500 × g for 10 min), and the pellet was resuspended in 1× protein sample buffer (19) and boiled for 5 min. After a brief centrifugation to remove the insoluble material, various amounts of the supernatant were subjected to electrophoresis on a 10% discontinuous polyacrylamide gel (19). The fusion protein band was detected following Coomassie brilliant blue staining at the position expected from the predicted size of the fusion protein induced by IPTG. For preparative gels (prepared as above except 1.5-mm thick), the approximate location and amount of the fusion protein band were determined by staining marker strips containing high-molecular-weight protein standards (Bio-Rad) with Coomassie brilliant blue stain.
Immunological methods.
Rabbit polyclonal antiserum against the LacZ-FimH fusion protein was raised by injecting a New Zealand White rabbit (ca. 2 kg) with a macerated acrylamide gel slice that contained approximately 100 μg of a LacZ-FimH fusion protein in complete Freund's adjuvant. Booster doses of 100 μg of fusion protein in incomplete Freund's adjuvant were administered approximately every 2 weeks with blood drawn beginning after the fourth boost and every 2 weeks thereafter, for a total of five bleedings. Antiserum was stored at −20°C, and aliquots were ammonium sulfate precipitated (7) and concentrated approximately fivefold in phosphate-buffered saline (PBS) prior to use.
Immunological and functional detection of FimH.
The presence of FimH was determined immunologically by bacterial agglutination reactions performed in 96-well round-bottomed microtiter plates. Bacteria from overnight cultures were isolated by brief centrifugation (1 to 2 min in a microcentrifuge) and concentrated two- to fivefold in PBS (approximately 4 × 109 to 10 × 109 cells per ml) depending upon the experiment. Twenty-five microliters of antiserum was serially twofold diluted in microtiter wells, and 25 μl of the bacterial suspension was added to each well. Microtiter plates were incubated at room temperature for 1 h and then refrigerated (4°C) until the negative control wells (containing a fimH insertion mutant) had settled (typically 24 to 48 h).
FimH function was assayed by the ability of E. coli to agglutinate guinea pig erythrocytes. Agglutination tests were conducted in 96-well round-bottomed microtiter plates in which overnight cultures, concentrated twofold, were serially twofold diluted and the contents of each well were mixed by adding 25 μl of a 4% suspension of fresh guinea pig erythrocytes. Incubation proceeded as above until erythrocytes in the negative control wells (containing the fimH insertion mutant as above) had settled (typically 15 to 24 h).
Recombination mapping of fimH mutations.
Transformants that contained a chromosomal fimH mutant allele received plasmids containing deletion derivatives of fimH via transduction. Transductants (800 to 1,000 colonies) were recovered from agar plates using a cotton swab. The material from each swab was expressed into approximately 1.0 ml of PBS, and 0.3 ml was added to a microcentrifuge tube containing 100 μl of settled fresh guinea pig erythrocytes. The bacteria and erythrocytes were mixed by inversion and incubated (to allow binding) for 10 min. The red blood cells were isolated by centrifugation for approximately 1 s in a microcentrifuge, and the supernatant was aspirated. The pellets were resuspended and washed with 1.0 ml of PBS five more times. The final pellet was resuspended in 0.5 ml of distilled water (to lyse the erythrocytes), and 2.0 ml of L-broth with chloramphenicol was added. This mixture was incubated overnight with shaking at 37°C to expand the population that remained bound to the erythrocytes. The following day, red blood cell debris was removed from 1.0 ml of the culture by a 2-s microcentrifugation step, and the bacteria in the supernatant were subsequently isolated by centrifugation for 2 min. The isolated bacteria were washed in 1.0 ml of PBS and resuspended in 0.75 ml of PBS. Fifty microliters of settled guinea pig erythrocytes was then added. The subsequent incubation, washing, and enrichment were repeated an additional two times. After the last outgrowth, cultures were streaked onto L-agar plates, and 20 individual colonies were scored for their ability to agglutinate erythrocytes.
Erythrocyte binding assay.
Overnight cultures of fimH point mutants and positive and negative control strains (described below) were harvested by microcentrifugation, resuspended in PBS, and mixed 1:1 by volume to give a final concentration of approximately 5 × 106 cells/ml. Sixty microliters of each mixture to be tested was added to an Eppendorf tube. Ten microliters was removed, diluted, and titered on maltose-tetrazolium plates to obtain the ratio of the two strains. To the bacteria remaining in the tube, 0.1 ml of settled guinea pig erythrocytes was added. The resulting suspension was then gently mixed and incubated at room temperature for 10 min. Adherent bacteria were removed by a brief (approximately 3 s) centrifugation to pellet erythrocytes and erythrocyte-bound bacteria. A portion of the supernatant was diluted and plated, and the resulting ratio was compared to the starting ratio.
In each assay, a fimH point mutant was mixed with a fimH insertion mutant (strain ORN204). The insertion mutant provided a negative control within each assay. To ensure that the changes in ratio accurately reflected differences in binding ability, pilot experiments included mixtures of two parental strains and two fimH insertion mutants. The strain combinations used in this case were ORN115 with ORN175 and ORN133 with ORN204. These strains were also mixed in parent-insertion mutant pairs. The strains in all mixtures were distinguished by their different maltose utilization phenotype on maltose-tetrazolium agar (refer to Table 1). Normalization of mutant binding values was done in part to reduce the effects of artifactual variability between assays (e.g., erythrocyte age and concentration).
Macrophage binding assay.
Resident (unelicited) peritoneal macrophages from male BALB/c mice 8 to 12 weeks of age were used in these experiments. Macrophages were harvested, delivered into 48-well cluster culture plates, and incubated overnight as described previously (11). Macrophages (approximately 1 × 105 to 2 × 105 cells/well in 0.5 ml of tissue culture medium) were exposed to approximately 106 E. coli (grown overnight in λ-broth, and harvested and mixed pairwise as described [11]) that were added in 25 μl of PBS for 10 min at 37°C. After incubation, wells were washed four times (each wash was with 0.5 ml of PBS). After the final wash, 0.5 ml of PBS containing 0.1% Triton X-100 was added to each well to lyse the macrophages. Approximately 5 min after the Triton X-100 additions, the contents of the wells were diluted and plated. The exposure of bacteria to Triton X-100 had no effect on bacterial viability. Control wells that contained no macrophages were used to assess nonspecific binding of bacteria. In no instance was the level of binding appreciable (>10% of that of macrophage-containing wells). When α-methyl-mannoside (αmm) was added to inhibit E. coli binding, a small volume of a 1.0 M αmm stock solution (in PBS) was added to a final concentration of 50 mM. Control wells had an equivalent volume of PBS added. PBS solutions used to remove unbound E. coli also contained 50 mM αmm.
In each assay (typically performed at least in duplicate), the strain mixtures were the same as those indicated for the erythrocyte binding assay just described. That is, binding efficiency was determined relative to a fimH insertion mutant. In order to assess the degree of macrophage binding inhibition effected by αmm, comparisons between parent and point mutant were normalized to a fimH insertion mutant (whose binding is insensitive to αmm) by using mixtures containing the strain to be assayed and an insertion mutant. This normalization controlled for well-to-well variation in macrophage number and allowed internal calibration of the degree of αmm binding inhibition.
Statistical and DNA sequence analysis methods.
Standard deviation of the mean was calculated with the Microsoft Excel STDEV function. Standard error was calculated as the standard deviation divided by the square root of the number of experiments. The significance of mean differences was determined by Student's t test. Both tests were provided by the Microsoft Excel version 4 statistics package. Statistically significant differences were defined as a P of <0.05. Genetics Computer Group (version 7) SIGCLEAVE analysis using the cleavage rules of von Heijne (49) was used to assess possible alterations in the FimH signal cleavage site due to signal sequence mutations in fimH.
Nucleotide sequence accession numbers.
The sequences of the five representative fimH mutant alleles have been deposited in GenBank with accession numbers as follows: fimH241, AF154925; fimH236, AF154926; fimH244, AF154927; fimH205, AF154928; and fimH208, AF154929.
RESULTS
Mapping point lesions in fimH.
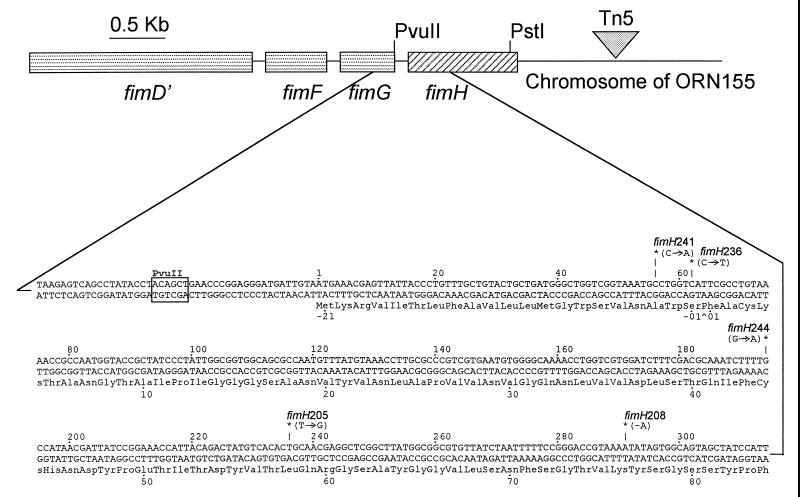
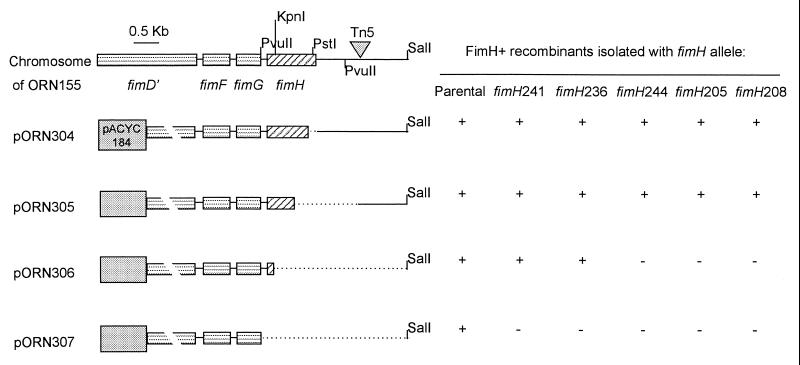
DNA sequencing of the plasmid-borne fimH alleles used to create the chromosomal mutants described by Harris et al. (13) provided the sequence of both strands of the entire fimH gene. The sequence of 11 alleles revealed that 5 were unique, each having a single lesion at the sites marked in Fig. 1. (The area sequenced included the fimH coding region plus a minimum of 20 bp on either side.) Sequencing of PCR amplicons from the chromosomal fimH alleles in all 11 strains revealed that they contained the same lesion as the plasmid-borne allele. (The entire gene was not sequenced in these cases [data not shown].) As a separate test that the lesions sequenced were responsible for the hemagglutination-negative phenotype, recombination mapping was carried out. This mapping involved enrichment for hemagglutination-positive individuals that were produced as a result of recombination between a chromosomal fimH allele having one of the point mutations and a set of in vitro-generated deletion derivatives of fimH residing on plasmids. The results of this mapping procedure (Fig. 2) were in good agreement with the lesion location indicated by DNA sequencing. Also, since the mutant alleles were not resequenced in their entirety after their introduction into the chromosome via homologous recombination (13), this mapping procedure indicated that unappreciated sequence differences in or around the chromosomal fimH gene were not responsible for the mutant phenotype.
FIG. 1.
Diagram of the fimH gene (diagonally striped rectangle) in the chromosome of strain ORN155. The entire fim gene cluster (fimBEACDEFGH) is located at approximately 98 min on the E. coli genetic map (more precise coordinates can be obtained from GenBank, accession no. AE000502, and reference 2). All genes in the cluster are transcribed left to right. The Tn5 insertion adjacent to fimH is not drawn to scale. Approximately one-third of the 5′ end of the fimH gene is expanded to show the sequence. Nucleotide numbering follows the convention in GenBank. Amino acids are numbered, with positive numbers denoting residues in the mature protein. Negative numbers indicate the amino acids in the signal sequence. The caret (^) below the sequence denotes the signal processing site (12). Mutation sites are designated by asterisks (∗) and allele number. Arrows show the base change involved. A minus sign indicates a deletion. Amino acid changes accompanying the mutations and the phenotypes conferred by the lesions are summarized in Table 2.
FIG. 2.
Mapping of mutations shown in Fig. 1 by recombination. Mutants with chromosomal fimH alleles shown on the right side of the figure were incubated with the plasmids shown at the left. Recombinants capable of hemagglutination were recovered following enrichment as described in the text. The fimH gene is denoted by the diagonally striped rectangle. The dotted line indicates the region deleted. Specific deletions (codons deleted) from the 3′ end of FimH are listed in Table 1. Deletions were created by removing either the restriction fragments indicated or by using Bal31 exonuclease digestions starting from the XhoI site in pORN118 as described in the text.
Variety of phenotypes displayed by fimH point mutants.
Whereas all fimH mutants failed to agglutinate guinea pig erythrocytes, certain fimH point mutants displayed additional properties (summarized in Table 2). Two of these properties involved the generation of aberrant pilus morphologies (class II and class III), and a third involved a conditional agglutination phenotype, all of which were initially noted by Harris et al. (13). Class II mutants (represented here by strain ORN164) had longer than normal pili. Class III mutants (represented here by strain ORN158) had much longer and very sparse pili. The lesion associated with the class II phenotype predicted a change in the last amino acid at the signal sequence site from serine to leucine. The class III mutant had a frameshift lesion predicted to generate a truncated FimH product approximately 40% of the normal size. In addition to the aberrantly fimbriated mutants, a mutant with a conditional (temperature sensitive [ts]) erythrocyte agglutination phenotype was isolated (Table 2). This mutant (strain ORN157, carrying the fimH205 allele) had morphologically similar pili at both the permissive and restrictive temperatures but failed to agglutinate erythrocytes at the restrictive temperature (13).
TABLE 2.
Properties of fimH mutants
| Lesion locationa (nt) | No. of isolatesb | Morphologic classc | Representative fimH allele (strain no.) | Type of lesion, amino acid change if missense (amino acid position)d | Phenotype or other relevant properties |
|---|---|---|---|---|---|
| 56 | 5 | I | fimH241 (ORN160) | Missense, Ala (−3)→asp | Amino acid change in signal sequence near processing site |
| 62 | 3e | II | fimH236 (ORN164) | Missense, Ser (−1)→Leu | Amino acid change in signal sequence at processing site |
| 194 | 1 | I | fimH244 (ORN165) | Missense, Cys (44)→Tyr | Amino acid change in first of four cysteine residues |
| 236 | 1 | I | fimH205 (ORN157) | Missense, Leu (58)→Arg | Amino acid change produces a conditional erythrocyte agglutination phenotype |
| 291 | 1f | III | fimH208 (ORN158) | Frameshift, deletion of 1 nucleotide | Frameshift lesion predicts a truncated protein approximately 40% of normal size |
Nucleotide (nt) residue altered. Refer to Fig. 1 for coordinates. The parental fimH allele is identical to that of the fully sequenced E. coli K-12 strain MG1655, referenced in the text.
Number of isolates refers to the number of independently isolated fimH mutants having the same mutation by DNA sequencing and recombination analysis. Five identical mutant alleles were found at position 56 (fimH241, fimH229, fimH222, fimH247, and fimH246 [13]). Three identical alleles were found at position 62 (fimH336, fimH204, and fimH218 [13]).
Morphologic class refers to the classes observed by Harris et al. (13). Class I mutants have normal-appearing pili, class II mutants have longer than normal pili, and class III mutants have fewer and longer pili. See text for additional details.
See Fig. 1 for base pair changes, lesion location, and amino acid position.
Originally, only one mutant having the representative allele (fimH236) was scored as being a class II mutant (13). Retrospective analysis of available electron micrographs revealed that at least one of the two additional mutants was simply overlooked in the original screen (data not shown).
In addition to strain ORN158, one additional mutant was scored as being a class III mutant in our original phenotypic screening (13). Upon examination of Southern blots using DNA encoding fimH as a probe, and PCR sequencing of the chromosomal fimH allele in this mutant, it was concluded that there had been an aberrant introduction of this allele into the chromosome (our unpublished observations).
Is the defect in the fimH products one of localization or function?
In the initial characterization of fimH point mutants (13), it was inferred that the fimH product was being expressed and properly localized because the mutants formed pellicles in static broth, a property eliminated by fimH insertion mutations (13). However, in these tests, we could not conclude that parental amounts of mutant FimH were being expressed because the level of FimH expression needed for pellicle formation could have been lower than that needed for erythrocyte agglutination.
In order to provide a measurement of the fimH product that was not linked to function, we produced and employed polyclonal rabbit antiserum raised against a lacZ-fimH translational fusion. The reactivity of four of the five mutants was distinguishably less than that of the parental strain (data not shown). Only the ts mutant having the fimH205 allele showed reactivity indistinguishable from the parent's (see next section). The decreased reactivity of the four mutants made further functional comparisons to the parent problematical, because we could not conclusively state the reason for the reduced antiserum reactivity (e.g., poor cross-reactivity, reduced FimH expression, or exposure in the pilus fiber). For these reasons, further characterization of the binding properties of these mutants was curtailed, and results focused on the fimH205 mutant.
FimH expression in the ts mutant at the restrictive and permissive temperatures.
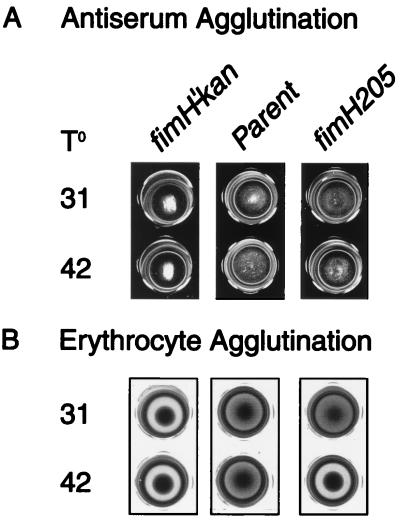
Bacterial agglutination of ORN157 (carrying the ts fimH205 allele) using dilutions of FimH-specific antiserum revealed no noticeable difference between the parent and mutant in terms of antibody reactivity after growth at both the permissive and restrictive temperatures (summarized in Fig. 3A). In contrast, erythrocyte agglutination effected by the mutant was decidedly reduced (Fig. 3B).
FIG. 3.
Microtiter agglutination reactions of strain ORN157 bearing the fimH205 allele compared to the parental strain (ORN155) and strain ORN133, a fimH insertion mutant (fimH′-kan). (A) Bacterial agglutination of mutants grown at the temperatures indicated, 31 or 42°C, concentrated twofold from overnight cultures, and incubated with FimH antiserum (1:4 dilution) as described in the text. (B) Microtiter guinea pig erythrocyte agglutination by the same strains obtained under the same growth condition. Incubation conditions are described in the text.
Role of temperature in FimH-mediated erythrocyte agglutination in fimH205 mutants.
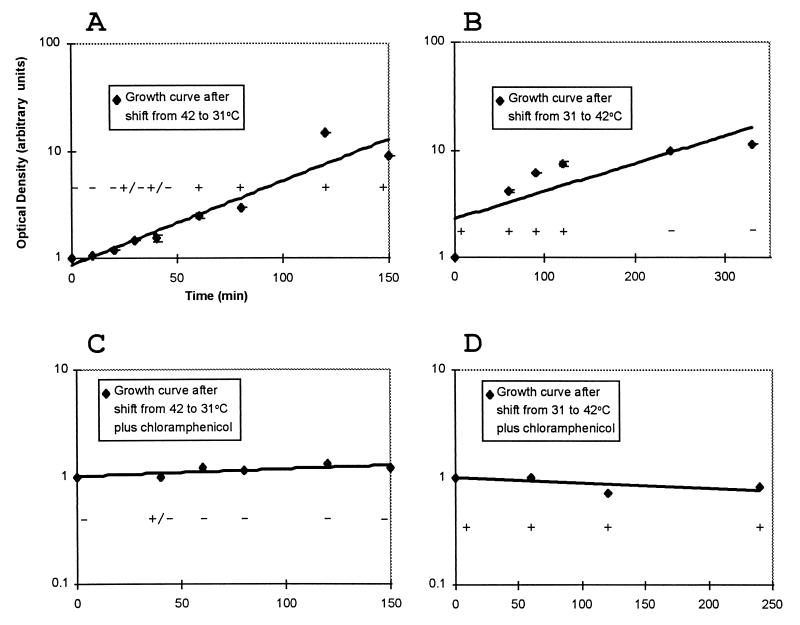
Two ways that a temperature shift could effect a change in FimH function are (i) via a spontaneous (instantaneous) conformational change in existing FimH molecules and (ii) via a conformational change in nascent or newly synthesized fimH product (requiring time for new synthesis). We tested these two possibilities by examining the kinetics with which the hemagglutination phenotype (Hag) changed after a temperature shift in the presence and absence of sufficient chloramphenicol to halt protein synthesis. Our results (Fig. 4) revealed that the change from Hag+ to Hag− (and vice versa) was not instantaneous and required new protein synthesis. Thus, it appeared that a temperature shift could not induce a structural change in existing FimH molecules.
FIG. 4.
Kinetics of the appearance of a hemagglutination-positive or -negative phenotype after a shift in temperature from 31 to 42°C and vice versa. (A and B) Emergence and disappearance, respectively, of the hemagglutination phenotype following a temperature shift. (C and D) Same shifts but in the presence of chloramphenicol (20 μg/ml). Diamonds represent points on growth curves taken from at least two experiments. Optical density at 600 nm values were normalized to the starting value in each experiment. Consequently, the units are arbitrary. The line shown through the points is a trend line drawn from a logarithmic regression analysis of the points. Hemagglutination was scored as a positive (+), weakly positive (+/−), or no hemagglutination (−) depending on the strength of the agglutination reaction. To test hemagglutination, samples were removed at the times indicated, diluted (or concentrated) to a constant optical density, and tested for hemagglutination of guinea pig erythrocytes as described in the text. Error bars represent standard error of the mean.
Erythrocyte and macrophage binding by fimH205 mutants.
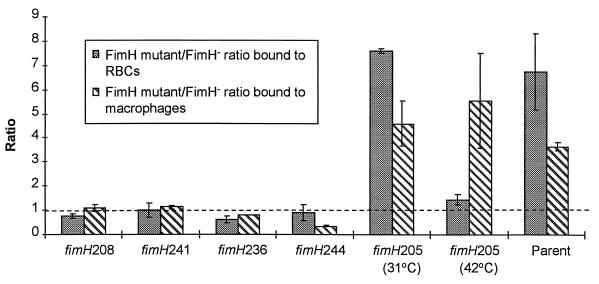
Guinea pig erythrocyte binding was greatly influenced by temperature in the fimH205 mutant: fimH205 mutants bound erythrocytes with statistically the same effectiveness as a fimH insertion mutant at 42°C but had the binding effectiveness of the parental strain at 31°C (Fig. 5). In contrast, the ability of the fimH205 mutant to bind to macrophages was statistically the same as that of the parent at both 31 and 42°C (Fig. 5). For comparison, the results of this binding assay with the four other fimH mutants (the ones that did not agglutinate in FimH antiserum at parental levels) are shown. These four mutants bound with the same effectiveness (statistically) as the fimH insertion mutant.
FIG. 5.
Comparison of the binding ability of mutants having the fimH lesions indicated relative to a fimH insertion mutant (fimH′-kan strain ORN133, denoted as FimH− in the legend box) and compared to the parental (ORN155) strain for their ability to bind guinea pig erythrocytes (RBCs) and resident BALB/c peritoneal macrophages. All mutant strains were grown at 37°C except for the fimH205 mutant, which was grown at the temperatures indicated. Parental binding values (ratio of the parental strain to the fimH insertion mutant; far right-hand bars were not significantly affected by the growth temperatures employed (31, 37, or 42°C), and for economy, a single average value is shown. Vertical bars represent the standard error of the mean. The dashed line indicates a 1:1 ratio between the test strain and the fimH insertion mutant. At least two experiments were averaged.
Macrophage binding by the ts mutant grown at the restrictive temperature was mannose insensitive.
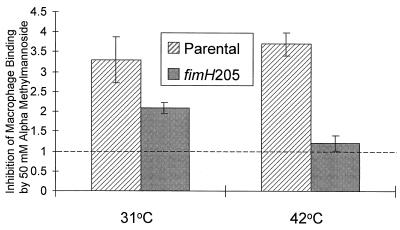
The ability of the nonmetabolizable mannose analog αmm to inhibit macrophage binding was assessed with a fimH205 mutant (ORN183) grown at 31 and 42°C. The degree to which the addition of 50 mM αmm inhibited binding was compared with binding by the parental strain (Fig. 6). Binding inhibition was quantitated by normalizing the reduction in binding effected by αmm on the parent and mutant relative to a fimH insertion mutant present as an internal control in all assays. For the parental strain, αmm inhibited binding approximately 3.5-fold regardless of the temperature (31, 37, and 42°C tested). For the mutant, however, αmm failed to change the binding significantly in 42°C-grown cells. After growth at 31°C, αmm inhibited macrophage binding of the mutant to a degree that was statistically the same as inhibition in the parent.
FIG. 6.
Degree of inhibition of macrophage binding by αmm in the fimH205 mutant grown at the restrictive and permissive temperatures. Macrophage binding was carried out in microtiter wells containing either the parental strain (ORN115) or a fimH205 mutant strain (ORN183) each mixed with a fimH′-kan insertion mutant strain (ORN204) as described in the text. The degree to which αmm inhibited binding of the parent or fimH205 mutant (ordinate values) was calculated from the change in ratio of these strains normalized to the change exhibited by the fimH′-kan insertion mutant. A value of 1 (dashed line) defines the value expected if there was no effect of αmm addition. Results presented are the averages of three separate experiments, each performed in duplicate. Error bars denote standard error of the mean.
DISCUSSION
The results reported herein define the location of lesions and phenotypic properties of five fimH point mutants isolated after site-directed mutagenesis (13). Although 11 independently isolated fimH mutants were initially identified, pilot experiments revealed that some had identical fimH lesions. The five described here represent one of each allelic type isolated. One mutant with a novel conditional erythrocyte binding phenotype was further defined as having an altered binding specificity after growth under permissive and restrictive conditions.
The fimH mutants examined here were obtained by enriching for individuals that formed pellicles in static broth (a property associated with FimH) in the presence of αmm (13). αmm is a mannose analogue that inhibits pellicle formation as well as erythrocyte agglutination by type 1 pili. Since pellicle formation is eliminated in fimH insertion mutants (13), we expected that most, if not all, of the fimH mutants would express parental levels of FimH and that the lesions would thus define regions of FimH required for erythrocyte binding. However, in the present study, bacterial agglutination tests with polyclonal antiserum raised against a LacZ-FimH fusion protein did not confirm that parental amounts of FimH were being produced in four of the five mutants. Whereas it is possible that some of these mutants have lesions that define areas of FimH responsible for erythrocyte binding and are poorly reactive with antibody as a consequence, we could not rule out the possibility that their failure to agglutinate (or bind to) erythrocytes was due entirely to inadequate FimH exposure or expression.
Two of the four mutants that did not express normal amounts of FimH-cross-reacting material (those with the fimH236 and fimH208 alleles) displayed signs of defective pilus biogenesis (13). We assume that their phenotypes result from the aberrant routing of the defective products in the pilus biogenesis process. In the case of the fimH236 mutation, the lesion lies in an area of the gene encoding the signal sequence (12), and the altered amino acid would thus not be part of the mature protein. However, the fimH236 lesion may result in an alteration in the site at which the signal is cleaved (49), creating a defect in the amino-terminal portion of the mature protein. The nature of the fimH208 lesion (a frameshift lesion approximately one-third of the way into the gene produced a garbled sequence of 33 amino acids before terminating, leaving a product approximately 40% of normal size, 33% of which was garbled) produces a mutant with the most dramatically altered pilus morphologic phenotype (13). Such mutations may affect the ability of the product to properly interact with the FimC chaperone protein (14) or with minor pilus components needed for pilus assembly (15, 36, 39). The recently acquired crystalline structure of the FimC-FimH complex should prove helpful (6) in this regard.
By far the most straightforward mutant to characterize was the ts fimH205 mutant. This mutant had parental levels of FimH and had a very pronounced phenotype. The lesion defining this allele converted a leucine to an arginine approximately one-fifth of the way through the mature FimH protein (amino acid position 58). The location and possible importance of this particular amino acid have been previously noted in studies by Sokurenko et al. (45, 46), who examined naturally occurring fimH alleles. In one of their studies, the fimH allele from E. coli K-12 strain CSH50 had arginine at the 58 position (GenBank accession no. A36976). This allele conferred the ability to bind to periodate-treated fibronectin in a mannose-inhibitable fashion. In fact, only one additional amino acid change in the CSH50 fimH allele, at position 201 (a histidine in place of a threonine), kept the two alleles from being identical. (Our parental fimH allele was identical to that of E. coli K-12 strain MG1655 that has been completely sequenced [2]; GenBank accession number U14003.) Sokurenko et al. (46) commented on the similarity of the fimH205 mutants to carry out protein-protein interactions in pellicle formation and the ability of the CSH50 FimH to bind to periodate-treated fibronectin. It was thus reassuring to find, in the present study, that the two mutants have one very specific genotypic feature in common. However, in the report of Sokurenko et al. (46), no mention was made of a conditional nature of fibronectin binding. Also, the binding reported was subject to inhibition by mannose. (The protein-protein binding conferred by the fimH205 allele in forming a pellicle [13] and in binding to macrophages was insensitive to mannose inhibition.) A recent report by Pouttu et al. (35) has indicated that the collagen binding ability of FimH is related to an amino acid change at residue 62. Whether this binding was mediated through protein-protein interactions in collagen was not specifically addressed. In any case, it appears that residues 58 to 62 of the FimH protein are important for determining receptor binding specificity. In addition to the above, it has been previously reported that linker insertion mutagenesis that changed residue 56 abolished mannose-sensitive binding (41).
In the present study, the nature of the ts defect in products from the fimH205 allele was pointed out in experiments that measured phenotypic lag after a temperature shift. Since new protein synthesis was required to effect a change in phenotype after a temperature shift, presynthesized FimH evidently could not undergo a conformational change to assume the new activity. Rather, a new (or nascent) FimH molecule was needed to assume the new configuration responsible for the altered activity. Also, the differences in phenotypic lag when shifting from the permissive to the nonpermissive temperature and vice versa were consistent with the idea that comparatively few pili (containing functional FimH products) were sufficient for hemagglutination. That is, acquisition of the hemagglutination-positive phenotype after a temperature shift was much more rapid than the loss of the hemagglutinating ability after a reverse shift.
Of most interest to us was the finding that the ts mutant carrying the fimH205 allele had different eucaryotic cell binding specificities when grown at the permissive and restrictive temperatures. At the restrictive growth temperature, the fimH205 mutant, while not binding to erythrocytes, still bound to macrophages through a FimH-specific interaction that was insensitive to mannose inhibition. The most likely explanation for this phenotype involves the earlier observation (10) that at the restrictive growth temperature, this mutant forms pellicles that appear to involve FimH-FimH interactions that are insensitive to mannose inhibition. This protein-protein type of interaction may be employed here in binding to macrophages. That is, macrophages may have a protein(s) on their surface capable of interacting with the mutant version of FimH. This explanation implies that erythrocytes lack such a protein(s). In related experiments by Sokurenko et al. (43), a correlation was made between the degree of affinity of FimH products (from naturally occurring fimH alleles) for monomannose and their affinity for cultured uroepithelial cells. It would thus appear that FimH has the potential to display a number of binding specificities.
Factors that determine FimH binding specificity have the potential to be exploited for processes that require conditional attachment and release of microorganisms from specific ligands. Already, the natural binding affinity of FimH for mannose has been used in combination with other binding domains to create chimeric adhesins (40). Another use for adhesin mutants with altered receptor specificities is in experiments examining the effects of bacterium-eucaryotic cell interactions. The effects of bacterial attachment on eucaryotic cell physiology are just beginning to be understood (1, 8, 22). The effects of specific types of attachment on eucaryotic cells are almost always deduced from experiments in which bacterial mutants that lack the attachment organelle (or the adhesive part of the organelle) are used as negative controls (9, 11, 16). In the case of type 1 pili, such mutants do not bind eucaryotic cells with an efficiency high enough to serve as a truly appropriate control (i.e., one that eliminates the effects of nonspecific binding rather than just the elimination of binding entirely). Attachment specificity mutants, such as the one isolated here, may allow better-controlled assays of the effect of receptor-ligand interactions to be determined.
ACKNOWLEDGMENTS
We thank Craig Altier for a critical reading of the manuscript and helpful suggestions.
This work was supported by grant AI 222223 from the Public Health Service and by the State of North Carolina.
REFERENCES
- 1.Baorto D M, Gao Z, Malaviya R, Dustin M L, van der Merwe A, Lublin D M, Abraham S N. Survival of FimH-expressing enterobacteria in macrophages relies on glycolipid traffic. Nature. 1997;389:636–639. doi: 10.1038/39376. [DOI] [PubMed] [Google Scholar]
- 2.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 3.Bolivar F. Construction and characterization of new cloning vehicles. III. Derivatives of plasmid pBR322 carrying unique EcoRI sites for selection of EcoRI-generated recombinant molecules. Gene. 1978;4:121–136. doi: 10.1016/0378-1119(78)90025-2. [DOI] [PubMed] [Google Scholar]
- 4.Carnoy C, Moseley S L. Mutational analysis of receptor binding mediated by the Dr family of Escherichia coli adhesins. Mol Microbiol. 1997;23:365–379. doi: 10.1046/j.1365-2958.1997.2231590.x. [DOI] [PubMed] [Google Scholar]
- 5.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived for the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1158. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choudhury D A, Thompson A, Stojanoff V, Langermann S, Pinkner J, Hultgren S J, Knight S D. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science. 1999;285:1061–1066. doi: 10.1126/science.285.5430.1061. [DOI] [PubMed] [Google Scholar]
- 7.Cooper H M, Paterson Y. Purification of immunoglobulin G fraction from antiserum, ascites fluid, or hybridoma supernatant. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology, supplement 37. New York, N.Y: John Wiley and Sons; 1997. p. 11.13.1. [Google Scholar]
- 8.Frankel G, Phillips A D, Rosenshine I, Dougan G, Kaper J B, Knutton S. Enteropathogenic and enterohemorrhagic Escherichia coli: more subversive elements. Mol Microbiol. 1998;30:911–921. doi: 10.1046/j.1365-2958.1998.01144.x. [DOI] [PubMed] [Google Scholar]
- 9.Godaly G, Fendeus B, Proudfoot A, Svensson M, Klemm P, Svanborg C. Role of fimbriae-mediated adherence for neutrophil migration across Escherichia coli-infected epithelial cell layers. Mol Microbiol. 1999;30:725–735. doi: 10.1046/j.1365-2958.1998.01104.x. [DOI] [PubMed] [Google Scholar]
- 10.Hagberg L, Hull R, Hull S, Falkow S, Freter R, Eden C S. Contribution of adhesion to bacterial persistence in the mouse urinary tract. Infect Immun. 1983;40:265–272. doi: 10.1128/iai.40.1.265-272.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamrick T S, Havell E A, Horton J R, Orndorff P E. Host and bacterial factors involved in innate ability of mouse macrophages to eliminate internalized unopsonized Escherichia coli. Infect Immun. 2000;68:125–132. doi: 10.1128/iai.68.1.125-132.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanson M S, Hempel J, Brinton C C., Jr Purification of the Escherichia coli type 1 pilin and minor pilus proteins and partial characterization of the adhesin protein. J Bacteriol. 1988;170:3350–3358. doi: 10.1128/jb.170.8.3350-3358.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris S L, Elliott D A, Blake M C, Must L M, Messenger M, Orndorff P E. Isolation and characterization of mutants with lesions affecting pellicle formation and erythrocyte agglutination by type 1 piliated Escherichia coli. J Bacteriol. 1990;172:6411–6418. doi: 10.1128/jb.172.11.6411-6418.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones C H, Pinkner J S, Nichols A V, Slonim L N, Abraham S N, Hultgren S J. FimC is a periplasmic PapD-like chaperone that directs assembly of type 1 pili in bacteria. Proc Natl Acad Sci USA. 1993;90:8397–8401. doi: 10.1073/pnas.90.18.8397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones C H, Pinkner J, Roth R, Heuser J, Nicholes A V, Abraham S N, Hultgren S J. FimH adhesin of type 1 pili is assembled into a fibrillar tip structure in the Enterobacteriaceae. Proc Natl Acad Sci USA. 1995;92:2081–2085. doi: 10.1073/pnas.92.6.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keith B R, Harris S L, Russell P W, Orndorff P E. Effect of type 1 piliation on in vitro killing of Escherichia coli by mouse peritoneal macrophages. Infect Immun. 1990;58:3448–3454. doi: 10.1128/iai.58.10.3448-3454.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knudsen T B, Klemm P. Probing the receptor recognition site of the FimH adhesin by fimbriae-displayed FimH-FocH hybrids. Microbiology. 1998;144:1919–1929. doi: 10.1099/00221287-144-7-1919. [DOI] [PubMed] [Google Scholar]
- 18.Krogfelt K A, Bergmans H, Klemm P. Direct evidence that the FimH protein is the mannose-specific adhesin of Escherichia coli type 1 fimbriae. Infect Immun. 1990;58:1995–1998. doi: 10.1128/iai.58.6.1995-1998.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Langermann S, Palaszynski S, Barnhart M, Auguste G, Pinkner J S, Burlein J, Barren P, Koenig S, Leath S, Jones C H, Hultgren S J. Prevention of mucosal Escherichia coli infection by FimH-adhesin-based systemic vaccination. Science. 1997;276:607–611. doi: 10.1126/science.276.5312.607. [DOI] [PubMed] [Google Scholar]
- 21.Lederberg E M, Cohen S N. Transformation of Salmonella typhimurium by plasmid deoxyribonucleic acid. J Bacteriol. 1974;119:1072–1074. doi: 10.1128/jb.119.3.1072-1074.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee C A. Type III secretion systems: machines to deliver bacterial proteins into eukaryotic cells? Trends Microbiol. 1997;5:148–156. doi: 10.1016/S0966-842X(97)01029-9. [DOI] [PubMed] [Google Scholar]
- 23.Madison B, Ofek I, Clegg S, Abraham S N. Type 1 fimbrial shafts of Escherichia coli and Klebsiella pneumoniae influence sugar-binding specificities of their FimH adhesins. Infect Immun. 1994;62:843–848. doi: 10.1128/iai.62.3.843-848.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maniatis T, Fritsch E R, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 25.Maurer L, Orndorff P E. A new locus, pilE, required for the binding of type 1 piliated Escherichia coli to erythrocytes. FEMS Microbiol Lett. 1985;30:59–66. [Google Scholar]
- 26.Maurer L M, Orndorff P E. Identification and characterization of genes determining receptor binding and pilus length of Escherichia coli type 1 pili. J Bacteriol. 1987;169:640–645. doi: 10.1128/jb.169.2.640-645.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Messing J, Crea R, Seeburg P H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981;9:309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 29.Orndorff P E. Escherichia coli type 1 pili. In: Miller V L, Kaper J B, Portnoy D A, Isberg R R, editors. Molecular genetics of bacterial pathogenesis. Washington, D.C.: American Society for Microbiology; 1994. pp. 91–111. [Google Scholar]
- 30.Orndorff P E, Falkow S. Organization and expression of genes responsible for type 1 piliation in Escherichia coli. J Bacteriol. 1984;159:736–744. doi: 10.1128/jb.159.2.736-744.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orndorff P E, Falkow S. Nucleotide sequence of pilA, the gene encoding the structural component of type 1 pili in Escherichia coli. J Bacteriol. 1985;162:454–457. doi: 10.1128/jb.162.1.454-457.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orndorff P E, Spears P A, Schauer D, Falkow S. Two modes of control of pilA, the gene encoding type 1 pilin in Escherichia coli. J Bacteriol. 1985;164:321–330. doi: 10.1128/jb.164.1.321-330.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pallesen L, Poulsen L K, Christiansen G, Klemm P. Chimeric FimH adhesin of type 1 fimbriae: a bacterial display system for heterologous sequences. Microbiology. 1995;141:2839–2848. doi: 10.1099/13500872-141-11-2839. [DOI] [PubMed] [Google Scholar]
- 34.Ponniah S, Endres R O, Hasty D L, Abraham S N. Fragmentation of Escherichia coli type 1 fimbriae exposes cryptic d-mannose-binding sites. J Bacteriol. 1991;173:4195–4202. doi: 10.1128/jb.173.13.4195-4202.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pouttu R, Puustinen T, Virkola R, Hacker J, Klemm P, Korhonen T K. Amino acid residue Ala-62 in the FimH fimbrial adhesin is critical for the adhesiveness of meningitis-associated Escherichia coli to collagens. Mol Microbiol. 1999;31:1747–1757. doi: 10.1046/j.1365-2958.1999.01311.x. [DOI] [PubMed] [Google Scholar]
- 36.Russell P W, Orndorff P E. Lesions in two Escherichia coli type 1 pilus genes after pilus number and length without affecting receptor binding. J Bacteriol. 1992;174:5923–5935. doi: 10.1128/jb.174.18.5923-5935.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruther U, Muller-Hill B. Easy identification of cDNA clones. EMBO J. 1983;2:1791–1794. doi: 10.1002/j.1460-2075.1983.tb01659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sato M, Staskawicz B J, Panopoulos N J, Peters S, Honma M. A host dependent hybrid plasmid suitable as a suicidal carrier for transposable elements. Plasmid. 1981;6:325–331. doi: 10.1016/0147-619x(81)90040-8. [DOI] [PubMed] [Google Scholar]
- 39.Saulino E T, Thanassi D G, Pinkner J S, Hultgren S J. Ramifications of kinetic partitioning on usher-mediated pilus biogenesis. EMBO J. 1998;17:2177–2185. doi: 10.1093/emboj/17.8.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schembri M A, Klemm P. Heterobinary adhesins based on the Escherichia coli FimH fimbrial protein. Appl Environ Microbiol. 1998;64:1628–1633. doi: 10.1128/aem.64.5.1628-1633.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schembri M A, Pallesen L, Connel H, Hasty D L, Klemm P. Linker insertion analysis of the FimH adhesin of type 1 fimbriae in an Escherichia coli fimH-null background. FEMS Microbiol Lett. 1996;137:257–263. doi: 10.1111/j.1574-6968.1996.tb08115.x. [DOI] [PubMed] [Google Scholar]
- 42.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 43.Sokurenko E V, Chesnokova V, Doyle R J, Hasty D L. Diversity of the Escherichia coli type 1 fimbrial lectin: differential binding to mannosides and uroepithelial cells. J Biol Chem. 1997;272:17880–17886. doi: 10.1074/jbc.272.28.17880. [DOI] [PubMed] [Google Scholar]
- 44.Sokurenko E V, Chesnokova V, Dykhuizen D E, Ofek I, Wu X R, Krogfelt K A, Struve C, Schembri M A, Hasty D L. Pathogenic adaptation of Escherichia coli by natural variation of the FimH adhesin. Proc Natl Acad Sci USA. 1998;95:8922–8926. doi: 10.1073/pnas.95.15.8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sokurenko E V, Courtney H S, Maslow J, Siitonen A, Hasty D L. Quantitative differences in adhesiveness of type 1 fimbriated Escherichia coli due to structural differences in fimH genes. J Bacteriol. 1995;177:3680–3686. doi: 10.1128/jb.177.13.3680-3686.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sokurenko E V, Courtney H S, Ohman D E, Klemm P, Hasty D L. FimH family of type 1 fimbrial adhesins: functional heterogeneity due to minor sequence variations among fimH genes. J Bacteriol. 1994;176:748–755. doi: 10.1128/jb.176.3.748-755.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spears P A, Schauer D, Orndorff P E. Metastable regulation of type 1 piliation in Escherichia coli and isolation and characterization of a phenotypically stable mutant. J Bacteriol. 1986;168:179–185. doi: 10.1128/jb.168.1.179-185.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thankavel K, Madison B, Ikeda T, Malaviya R, Shah A H, Arumugam P M, Abraham S N. Localization of a domain in the FimH adhesin of Escherichia coli type 1 fimbriae capable of receptor recognition and use of a domain-specific antibody to confer protection against experimental urinary tract infection. J Clin Invest. 1997;100:1123–1136. doi: 10.1172/JCI119623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.von Heijne G. Patterns of amino acids near signal sequence cleavage sites. Eur J Biochem. 1983;133:17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]
- 50.Woodall L D, Russell P W, Harris S L, Orndorff P E. Rapid, synchronous, and stable induction of type 1 piliation in Escherichia coli by using a chromosomal lacUV5 promoter. J Bacteriol. 1993;175:2770–2778. doi: 10.1128/jb.175.9.2770-2778.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]