Abstract
Alzheimer's disease (AD) is a progressive disease, and the number of AD patients is increasing every year as the population ages. One of the pathophysiological mechanisms of AD is thought to be the effect of metabolomic abnormalities. There have been several studies of metabolomic abnormalities of AD, and new biomarkers are being investigated. Metabolomic studies have been attracting attention, and the aim of this study was to identify metabolomic biomarkers associated with AD and mild cognitive impairment (MCI). Of the 927 participants in the Nakayama Study conducted in Iyo City, Ehime Prefecture, 106 were selected for this study as Control (n = 40), MCI (n = 26), and AD (n = 40) groups, matched by age and sex. Metabolomic comparisons were made across the three groups. Then, correlations between metabolites and clinical symptoms were examined. The blood mRNA levels of the ornithine metabolic enzymes were also measured. Of the plasma metabolites, significant differences were found in ornithine, uracil, and lysine. Ornithine was significantly decreased in the AD group compared to the Control and MCI groups (Control vs. AD: 97.2 vs. 77.4; P = 0.01, MCI vs. AD: 92.5 vs. 77.4; P = 0.02). Uracil and lysine were also significantly decreased in the AD group compared to the Control group (uracil, Control vs. AD: 272 vs. 235; P = 0.04, lysine, Control vs. AD: 208 vs. 176; P = 0.03). In the total sample, the MMSE score was significantly correlated with lysine, ornithine, thymine, and uracil. The Barthel index score was significantly correlated with lysine. The instrumental activities of daily living (IADL) score were significantly correlated with lysine, betaine, creatine, and thymine. In the ornithine metabolism pathway, the spermine synthase mRNA level was significantly decreased in AD. Ornithine was decreased, and mRNA expressions related to its metabolism were changed in the AD group compared to the Control and MCI groups, suggesting an association between abnormal ornithine metabolism and AD. Increased betaine and decreased methionine may also have the potential to serve as markers of higher IADL in elderly persons. Plasma metabolites may be useful for predicting the progression of AD.
Subject terms: Neuroscience, Biomarkers, Medical research, Neurology
Introduction
Alzheimer’s disease (AD) is a progressive disease, and the number of patients is increasing every year as the population ages. A variety of factors including genetic, epigenetic, and environmental factors, as well as aging, are thought to contribute to the disease, but none have as yet been clearly identified yet. Physical risk factors such as type 2 diabetes, obesity, hypertension, and hyperlipidemia are thought to potentially increase the risk of AD1. These risk factors are thought to be related to metabolic changes. Therefore, the study of blood metabolomics may show some underlying mechanisms associated with the pathophysiology of AD and may uncover new biomarkers and early therapeutic targets for AD2,3.
Blood-based biomarkers for AD are thought to reflect the pathology of the brain and are easier to collect than those in cerebrospinal fluid4,5. Metabolomics has attracted much attention6,7. Capillary electrophoresis-time-of-flight mass spectrometry (CE-TOFMS) is one of the comprehensive, quantitative, and high-throughput analytical tools used for metabolomics8,9. To date, CE-TOFMS has identified abnormal blood metabolites in patients with various neuropsychiatric diseases such as schizophrenia10, bipolar disorder11, major depressive disorder12, and autism spectrum disorder13. Preceding blood metabolomic studies of AD showed that the changes in some amino acids and polyamines, such as ornithine, might reflect the pathologies involved14,15. However, these changes in AD have not been consistently confirmed, and few studies have reported on mild cognitive impairment (MCI).
Thus, the purpose of this study was to profile metabolites in the blood of MCI, AD, and age-sex-matched cognitively normal controls (Control). Then, the correlations among clinical characteristics including cognitive function and activities of daily living (ADL) and the plasma levels of metabolites were examined. Finally, gene expression analyses of enzymes in candidate metabolic pathways were performed.
Material and methods
Subjects from the Nakayama Study
Nakayama Town is a rural community in Iyo City, Ehime Prefecture, Japan, with a population of 2655. The Nakayama Study was a population-based complete enumeration survey including all residents over 65 years of age in Nakayama Town from January 2017 through April 2018. Of the 1,512 residents, 927 were qualified for the study16,17, 40 subjects who met the criteria for AD and 26 who met the criteria for aMCI were selected out of 927 people, and then 40 healthy controls were randomly selected from remaining 861 subjects to be age and sex matched to the 40 AD subjects. All subjects or their families signed written, informed consent forms approved by the institutional ethics committees of Ehime University Graduate School of Medicine (Approval Number: 1610004). This study was performed in accordance with the Declaration of Helsinki. Basically, blood sampling was drawn after 2 h after eating, but 2 patients had their blood drawn before eating and 10 participant s had their blood drawn less than 2 h after eating. A self-administered questionnaire was used to collect data on education, smoking and drinking history, daily physical activity level, medical history, and medication use16,17. After data collection, the data were checked by trained interviewers. Height and weight were measured with light clothing, and the body mass index (BMI) was calculated from the measured values. Head magnetic resonance imaging (MRI) and the Mini Mental State Examination (MMSE) were also performed. All AD patients met the diagnostic criteria according to the National Institute on Aging/Alzheimer’s Association18, and a diagnosis of amnestic MCI was made for patients who satisfied the following criteria: (1) normal general cognitive function, with a Mini-Mental State Examination (MMSE) score ≥ 23; (2) on the delayed recall test of the Wechsler Memory Scale-Revised, the cutoff scores for any cognitive impairment were selected according to education status (≤ 8 points for 16 years of education, ≤ 4 points for 8–15 years, and ≤ 2 points for 0–7 years); (3) neuropsychiatric examination showing absence of dementia or depression as determined by geriatric neuropsychiatrists according to the Diagnostic and Statistical Manual of Mental Disorders, 3rd edition, revised (DSM-III R) (American Psychiatric Association, 1997) criteria; (4) no ADL impairment; and (5) hippocampal and parietal lobe atrophy without apparent cerebrovascular disease on brain MRI.
Quantitative metabolome analysis
Blood sampling was performed during the participants’ visits for the Nakayama Study. Samples were sent from Ehime University to Tokushima University, where each 50-µL sample was mixed with 450 µL of methanol containing internal standards (10 µM) and vortexed. Chloroform (500 µL) and Milli-Q water (200 µL) were added, mixed thoroughly, then centrifuged (2300×g, 4 °C, 5 min). To remove macromolecules, 375 μL of the aqueous layer were filtered through a 5-kDa cutoff filter (EMD Millipore, Billerica, MA, USA). The filtrate was lyophilized and dissolved in 50 µL of Milli-Q water containing the reference compound before mass spectrometry analysis.
Metabolome measurements were conducted at Tokushima University. Plasma metabolite profiling and a mixture of 55 standard metabolites (50 µM each, HMT, Tsuruoka, Japan) were analyzed using a capillary electrophoresis electrospray ionization time-of-flight mass spectrometry (CE-ESI-TOFMS) system (Agilent 7100 CE - 6230 TOFMS, Agilent Technologies, Palo Alto, CA, USA). 55 cation mode metabolites with standard were measured. Then, the 35 metabolites were selected by counts (> 1000) and signal-to-noise ratio (> 3.0) of the peak area from the extracted ion chromatogram.
Blood sample for mRNA expression
Total RNA was isolated from whole peripheral blood of the same participant in the metabolomic study using PaxGene Blood RNA Systems tubes (BD, Tokyo, Japan) according to the standard protocol. The RNA concentration and purity measured by NanoDrop-1000 (Thermo Fisher Scientific, Yokohama, Japan) showed a 260/280 ratio between 1.8 and 2.0. Then, cDNA was synthesized using RNA (1.0 μg) with the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). The specific TaqMan probes (Integrated DNA Technologies, Iowa, USA) were Hs.PT.39a.22214836 for glyceraldehyde-3-phosphate dehydrogenase (GAPDH), Hs.PT.56a.491276 for argininosuccinate lyase (ASL), Hs.PT.58.14740388 for nitric oxide synthase2 (NOS2), Hs.PT.56a.2920438 for argininosuccinate synthase1 (ASS1), Hs.PT.58.22990692.g for ornithine transcarbamylase (OTC), Hs.PT.56a.20779559 for arginase1 (ARG1), Hs.PT.58.2742899 for antizyme inhibitor 1 (AZIN1), Hs.PT.58.1971913 for ornithine decarboxylase antizyme1 (OAZ1), Hs.PT.58.26956824 for ornithine decarboxylase antizyme2 (OAZ2), Hs.PT.58.4341481 for ornithine decarboxylase antizyme3 (OAZ3), Hs.PT.58.27029915 for ornithine decarboxylase 1 (ODC1), and Hs.PT.58.15157347 for spermine synthase (SMS). Our previous studies have consistently identified GAPDH as a suitable housekeeping gene for blood gene expression analysis using the Paxgene blood RNA system19,20. The final volume of reactions was 10 µL with TaqMan Universal Master Mix (Applied Biosystems). Expression levels were examined in duplicate with a specific sample as a reference. The ΔΔCt method using StepOne software (Applied Biosystems) was used to measure the relative expression levels.
Data analysis
Descriptive statistics were calculated for the Control, MCI, and AD groups. Normality of distributions was checked using the Shapiro–Wilk test. The mean difference among three groups was tested by one-way analysis of variance with Bonferroni’s multiple comparisons test or the Kruskal–Wallis test with Bonferroni’s multiple comparisons test according to normality. Spearman’s correlation test was used to determine the correlations among clinical parameters and each metabolite or mRNA level. All analyses were carried out using IBM® SPSS® Version 26. A two-tailed P < 0.05 was considered significant in all analyses.
Results
Participants’ characteristics
Study participants’ characteristics including age, sex, height, weight, BMI, MMSE, education, Barthel Index (BI), instrumental activities of daily living (IADL), geriatric depression scale 15 (GDS15), and medical history are presented in Table 1. In the table of subject characteristics (Table 1), patients with inadequate history taking were included: one subject with no Barthel Index, one subject with inadequate history taking in Diabetes, Hyperlipidemia, Drinking, and Smoking, respectively. The Control, MCI, and AD groups were selected so that they would be matched in sex, height, and weight. The MMSE scores were significantly decreased (Control vs. MCI vs. AD: 28.4 vs. 25.3 vs. 18.4, P < 0.01). The majority of the participants had finished junior high school. The BI was significantly lower in the AD group compared to the MCI and Control groups (Control vs. AD: 98.3 vs 93.6, P = 0.02, MCI vs. AD: 98.3 vs 93.6, P < 0.01). The same results were obtained for IADL (Control vs. AD: 11.5 vs 6.00, P < 0.01, MCI vs. AD: 11.2 vs. 6.00, P < 0.01). Of all participants, 37 (35.2%) had a history of drinking and 21 (20.0%) of smoking. The rates of hypertension, diabetes mellitus, and hyperlipidemia were 75 (70.8%), 19 (18.1%), and 34 (32.4%), respectively. Although the number with hyperlipidemia was significantly higher in the Control group compared to the AD group (20 vs, 6; P < 0.01). In the Participant characteristics (Blood test), significant results were obtained for ALT, but additional tests did not yield significant results (Table 2).
Table 1.
Participant characteristics. One (AD) had no information on diabetes, hyperlipidemia, drinking and smoking.
| Control | MCI | AD | P | |
|---|---|---|---|---|
| 40 | 26 | 40 | ||
| Age (SD) years | 81.8 ± 2.26 | 82.4 ± 5.49 | 83.8 ± 4.79 | 0.14 |
| Female (%) | 26/40 (65.0) | 17/26 (65.4) | 26/40 (65.0) | 0.99 |
| Height (SD) cm | 150 ± 8.10 | 149 ± 11.1 | 146 ± 7.87 | 0.15 |
| Weight (SD) kg | 54.1 ± 9.36 | 50.1 ± 8.68 | 49.0 ± 9.55 | 0.04 |
| BMI (SD) kg/m2 | 24.0 ± 2.87 | 22.5 ± 2.91 | 22.9 ± 3.49 | 0.14 |
| MMSE (SD) | 28.4 ± 1.51 | 25.3 ± 1.66 | 18.4 ± 3.72 | < 0.01 |
| Education (%) | ||||
| ≤ Finished primary school | 1/40 (2.5) | 3/26 (11.5) | 6/40 (15.0) | |
| ≤ Finished junior high school | 25/40 (62.5) | 12/26 (46.2) | 26/40 (65.0) | |
| ≤ Finished high school | 13/40 (32.5) | 10/26 (38.5) | 8/40 (20.0) | |
| ≤ Finished college, university | 1/40 (2.5) | 1/26 (3.8) | 0/40 (0.0) | |
| Barthel Index (SD) | 98.3 ± 5.13 | 98.3 ± 8.83 | 93.6 ± 10.8 | < 0.01 |
| IADL (SD) | 11.5 ± 2.22 | 11.2 ± 3.25 | 6.00 ± 4.82 | < 0.01 |
| GDS15 (SD) | 2.45 ± 2.01 | 3.08 ± 2.24 | 3.33 ± 2.97 | 0.45 |
| Medical history (%) | ||||
| Hypertension (%) | 28/40 (70.0) | 18/26 (69.2) | 31/40 (77.5) | 0.68 |
| Diabetes (%) | 9/40 (22.5) | 3/26 (11.5) | 7/39 (17.9) | 0.53 |
| Hyperlipidemia (%) | 20/40 (50.0) | 9/26 (34.6) | 6/39 (15.4) | < 0.01 |
| Drinking (%) | 16/40 (40.0) | 10/26 (38.5) | 11/39 (28.2) | 0.51 |
| Smoking (%) | 8/40 (20.0) | 5/26 (19.2) | 8/39 (20.5) | 0.99 |
BMI body mass index, MMSE Mini Mental State Examination, IADL instrumental activities of daily living, GDS15 geriatric depression scale 15.
Table 2.
Participant characteristics (Blood test).
| Control (N = 40) | MCI (N = 26) | AD (N = 40) | P | |
|---|---|---|---|---|
| WBC (× 103/μL) | 6.08 ± 1.50 | 6.31 ± 3.94 | 5.84 ± 1.31 | 0.64 |
| RBC (× 106/μL) | 4.32 ± 0.41 | 4.27 ± 0.65 | 4.24 ± 0.40 | 0.74 |
| Hb (g/dL) | 13.3 ± 1.19 | 13.0 ± 1.62 | 13.1 ± 1.32 | 0.69 |
| Ht (%) | 41.3 ± 3.55 | 40.7 ± 5.16 | 40.6 ± 3.89 | 0.71 |
| MCV (fL) | 95.6 ± 3.55 | 95.9 ± 7.06 | 96.0 ± 5.09 | 0.96 |
| MCH (pg) | 30.7 ± 1.34 | 30.7 ± 2.75 | 30.9 ± 2.03 | 0.93 |
| MCHC (%) | 32.1 ± 0.73 | 32.0 ± 1.13 | 32.2 ± 0.85 | 0.84 |
| PLT (× 104/μL) | 23.3 ± 3.83 | 26.4 ± 20.2 | 23.2 ± 6.27 | 0.80 |
| TP (g/dL) | 7.35 ± 0.40 | 7.23 ± 0.46 | 7.24 ± 0.38 | 0.39 |
| ALB (g/dL) | 4.19 ± 0.30 | 4.08 ± 0.35 | 4.12 ± 0.31 | 0.39 |
| Total-Bil (mg/dL) | 0.77 ± 0.28 | 0.69 ± 0.23 | 0.70 ± 0.25 | 0.40 |
| AST (U/L) | 24.8 ± 5.96 | 26.2 ± 9.19 | 26.9 ± 14.1 | 0.73 |
| ALT (U/L) | 17.7 ± 7.34 | 18.5 ± 11.1 | 18.6 ± 29.0 | 0.04 |
| ALP (U/L) | 243 ± 75.4 | 264 ± 77.8 | 284 ± 201 | 0.32 |
| LDH (U/L) | 223 ± 33.6 | 237 ± 56.6 | 228 ± 31.9 | 0.76 |
| γ-GT (U/L) | 22.4 ± 11.5 | 21.6 ± 10.8 | 34.9 ± 78.7 | 0.80 |
| CPK (U/L) | 117 ± 43.5 | 121 ± 71.0 | 126 ± 92.3 | 0.66 |
| Tchol (mg/dL) | 194 ± 27.3 | 193 ± 38.0 | 192 ± 31.2 | 1.00 |
| LDL (mg/dL) | 108 ± 21.7 | 110 ± 28.3 | 110 ± 28.5 | 0.88 |
| HDL (mg/dL) | 58.2 ± 15.7 | 60.0 ± 15.9 | 57.1 ± 15.0 | 0.76 |
| HbA1c (NGSP, %) | 5.91 ± 0.68 | 5.66 ± 0.37 | 5.96 ± 1.00 | 0.44 |
| GA (%) | 16.3 ± 3.10 | 15.8 ± 1.96 | 17.4 ± 4.92 | 0.48 |
| BUN (mg/dL) | 17.8 ± 3.93 | 17.2 ± 5.90 | 17.0 ± 4.98 | 0.38 |
| Cr (mg/dL) | 0.72 ± 0.19 | 0.74 ± 0.37 | 0.74 ± 0.23 | 0.80 |
| eGFR (mL/min/1.73 m2) | 80.7 ± 11.3 | 81.5 ± 15.3 | 77.6 ± 14.1 | 0.66 |
| UA (mg/dL) | 4.82 ± 0.81 | 4.62 ± 1.32 | 4.70 ± 1.37 | 0.79 |
| Na (mEq/L) | 141 ± 2.05 | 141 ± 2.47 | 141 ± 2.49 | 0.40 |
| K (mEq/L) | 4.94 ± 0.70 | 5.06 ± 0.74 | 4.99 ± 0.45 | 0.76 |
| Cl (mEq/L) | 103 ± 2.21 | 103 ± 3.23 | 103 ± 2.93 | 0.67 |
| TSH (μIU/mL) | 2.97 ± 1.89 | 2.89 ± 2.21 | 2.37 ± 1.27 | 0.35 |
| FT4 (ng/dL) | 1.14 ± 0.19 | 1.07 ± 0.16 | 1.11 ± 0.13 | 0.49 |
| CRP (mg/dL) | 0.12 ± 0.14 | 0.07 ± 0.10 | 0.10 ± 0.12 | 0.14 |
Comparison of plasma metabolites in the Control, MCI, and AD groups
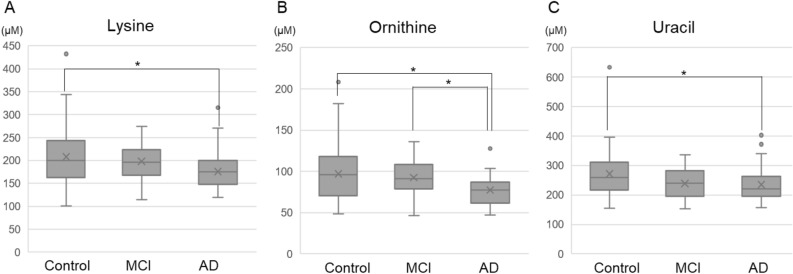
In the plasma metabolite comparisons, 35 metabolites were examined. Significant differences were found only in ornithine, uracil, and lysine (Table 3, Fig. 1). For ornithine, there was a significant difference between the Control and AD groups and between the MCI and AD groups (Control vs. AD: 97.2 µM vs. 77.4 µM; P = 0.01, MCI vs. AD: 92.5 µM vs. 77.4 µM; P = 0.02). For uracil and lysine, there were significant differences between the Control and AD groups (uracil, Control vs. AD: 272 µM vs. 235 µM; P = 0.04; Lysine, Control vs. AD: 208 µM vs. 176 µM; P = 0.03).
Table 3.
Mean values of plasma amino acids.
| Control (N = 40) | MCI (N = 26) | AD (N = 40) | P | |
|---|---|---|---|---|
| Alanine (µM) | 509 ± 90.0 | 504 ± 112 | 495 ± 109 | 0.82 |
| Arginine (µM) | 107 ± 25.0 | 105 ± 28.4 | 103 ± 26.7 | 0.81 |
| Asparagine (µM) | 59.7 ± 9.93 | 58.9 ± 12.0 | 56.9 ± 11.6 | 0.51 |
| Aspartic acid (µM) | 30.1 ± 8.72 | 30.4 ± 9.67 | 28.4 ± 14.8 | 0.11 |
| Cysteine (µM) | 10.3 ± 2.47 | 9.90 ± 2.72 | 9.99 ± 2.56 | 0.83 |
| Glutamine (µM) | 752 ± 91.6 | 725 ± 119 | 746 ± 109 | 0.29 |
| Glutamic acid (µM) | 113 ± 35.3 | 110 ± 49.1 | 104 ± 49.2 | 0.22 |
| Glycine (µM) | 288 ± 63.8 | 320 ± 115 | 287 ± 73.1 | 0.24 |
| Histidine (µM) | 88.3 ± 15.2 | 82.6 ± 15.1 | 80.8 ± 13.2 | 0.06 |
| Isoleucine (µM) | 63.6 ± 23.1 | 64.9 ± 17.5 | 59.6 ± 20.3 | 0.37 |
| Leucine (µM) | 118 ± 38.1 | 122 ± 29.6 | 110 ± 29.9 | 0.24 |
| Lysine (µM) | 208 ± 62.9 | 198 ± 45.6 | 176 ± 40.5 | 0.02 |
| Methionine (µM) | 23.0 ± 7.33 | 23.8 ± 7.01 | 24.8 ± 6.88 | 0.32 |
| Phenylalanine (µM) | 82.2 ± 14.5 | 78.0 ± 14.1 | 76.0 ± 16.6 | 0.13 |
| Proline (µM) | 233 ± 89.6 | 203 ± 47.8 | 223 ± 75.0 | 0.51 |
| Serine (µM) | 153 ± 30.1 | 156 ± 28.4 | 147 ± 32.1 | 0.36 |
| Threonine (µM) | 145 ± 34.3 | 130 ± 34.5 | 134 ± 37.2 | 0.19 |
| Tryptophan (µM) | 60.2 ± 13.0 | 57.9 ± 14.7 | 56.2 ± 11.9 | 0.40 |
| Tyrosine (µM) | 81.1 ± 18.6 | 75.4 ± 15.1 | 75.3 ± 23.8 | 0.23 |
| Valine (µM) | 243 ± 51.7 | 250 ± 48.4 | 244 ± 52.7 | 0.83 |
| Anthranilic acid (µM) | 1.12 ± 0.42 | 0.95 ± 0.23 | 0.99 ± 0.28 | 0.25 |
| Betaine (µM) | 72.1 ± 20.9 | 72.7 ± 18.1 | 68.3 ± 20.3 | 0.33 |
| Choline (µM) | 33.2 ± 6.75 | 33.3 ± 7.23 | 31.7 ± 6.68 | 0.45 |
| Citrulline (µM) | 40.6 ± 13.2 | 40.1 ± 17.5 | 37.5 ± 9.19 | 0.75 |
| Creatine (µM) | 49.9 ± 29.5 | 50.0 ± 25.9 | 52.5 ± 33.8 | 0.95 |
| Creatinine (µM) | 71.7 ± 17.5 | 71.9 ± 40.0 | 67.9 ± 23.5 | 0.21 |
| GABA (µM) | 4.23 ± 4.33 | 4.42 ± 10.3 | 3.38 ± 4.92 | 0.14 |
| Hydroxyproline (µM) | 13.6 ± 5.15 | 13.2 ± 5.80 | 13.2 ± 6.24 | 0.71 |
| Hypoxanthine (µM) | 16.2 ± 4.98 | 14.3 ± 4.63 | 15.0 ± 5.90 | 0.11 |
| Kynurenine (µM) | 2.22 ± 0.64 | 2.05 ± 0.63 | 1.97 ± 0.60 | 0.19 |
| N,N-Dimethylglycine (µM) | 4.37 ± 1.14 | 4.36 ± 1.26 | 4.62 ± 1.36 | 0.59 |
| Ornithine (µM) | 97.2 ± 34.1 | 92.5 ± 20.5 | 77.4 ± 17.7 | < 0.01 |
| Thymine (µM) | 68.0 ± 22.2 | 71.4 ± 20.6 | 78.8 ± 30.0 | 0.07 |
| Uracil (µM) | 272 ± 82.9 | 239 ± 49.4 | 235 ± 59.1 | 0.04 |
| Uridine (µM) | 14.2 ± 4.25 | 14.5 ± 6.76 | 14.3 ± 5.47 | 0.95 |
Figure 1.
Comparisons of mean plasma amino acid values for (A) lysine, (B) ornithine, and (C) uracil. *P < 0.05.
Relationships between metabolite relative concentrations and clinical symptoms
The relationships between the concentrations of metabolites and clinical symptoms evaluated by the MMSE, BI, IADL, and GDS15 were examined. In total samples, the MMSE score was significantly correlated with lysine (r = 0.23, P < 0.05), ornithine (r = 0.28, P < 0.01) and thymine (r = − 0.22, P < 0.05). The BI was significantly correlated with lysine (r = 0.21, P < 0.05). The IADL score was significantly correlated with lysine (r = 0.20, P < 0.05), betaine (r = 0.34, P < 0.01), creatine (r = − 0.25, P < 0.05), and thymine (r = − 0.27, P < 0.01). The GDS15 score was correlated with creatinine (r = − 0.23, P < 0.05).
Comparison of mRNA expression levels of metabolic enzymes
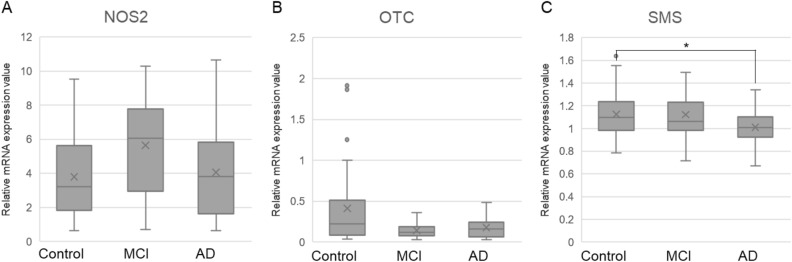
The mRNA expression levels of the metabolic enzymes in ornithine pathways were measured (Fig. 2). SMS, NOS2, and OTC mRNA levels were significantly different among the three groups (Table 4). On post hoc analysis, the SMS mRNA level was significantly lower in the AD group than in the Control group (Control vs. AD: 1.13 vs 1.01; P = 0.04). The NOS2 mRNA level was higher in the MCI group than in the Control group (Control vs. MCI: 3.80 vs 5.66; P = 0.06), whereas the OTC mRNA level was lower in the MCI group compared to the Control group (Control vs. MCI: 0.41 vs 0.14; P = 0.06).
Figure 2.
Comparisons of mRNA expression levels of metabolism enzymes for (A) NOS2, (B) OTC, and (C) SMS. *P < 0.05.
Table 4.
Relative mRNA expression levels of metabolism enzymes.
| N | Control | N | MCI | N | AD | P | |
|---|---|---|---|---|---|---|---|
| ASL | 37 | 1.39 ± 0.21 | 24 | 1.47 ± 0.24 | 40 | 1.35 ± 0.34 | 0.26 |
| NOS2 | 33 | 3.80 ± 2.51 | 23 | 5.66 ± 2.85 | 32 | 4.06 ± 2.91 | 0.04 |
| ASS1 | 37 | 0.48 ± 0.28 | 24 | 0.32 ± 0.10 | 37 | 0.36 ± 0.14 | 0.14 |
| OTC | 35 | 0.41 ± 0.49 | 25 | 0.14 ± 0.09 | 34 | 0.18 ± 0.13 | 0.04 |
| ARG1 | 39 | 0.42 ± 0.27 | 26 | 0.41 ± 0.27 | 39 | 0.40 ± 0.28 | 0.83 |
| AZIN1 | 37 | 1.17 ± 0.15 | 26 | 1.21 ± 0.19 | 38 | 1.18 ± 0.19 | 0.56 |
| OAZ1 | 37 | 0.83 ± 0.35 | 24 | 0.80 ± 0.40 | 38 | 0.82 ± 0.42 | 0.64 |
| OAZ2 | 37 | 1.29 ± 0.26 | 25 | 1.35 ± 0.32 | 38 | 1.25 ± 0.33 | 0.46 |
| OAZ3 | 39 | 1.34 ± 0.65 | 25 | 1.63 ± 0.73 | 39 | 1.53 ± 0.69 | 0.17 |
| ODC1 | 39 | 1.45 ± 0.46 | 25 | 1.47 ± 0.61 | 39 | 1.26 ± 0.39 | 0.12 |
| SMS | 38 | 1.13 ± 0.20 | 25 | 1.12 ± 0.21 | 38 | 1.01 ± 0.14 | 0.01 |
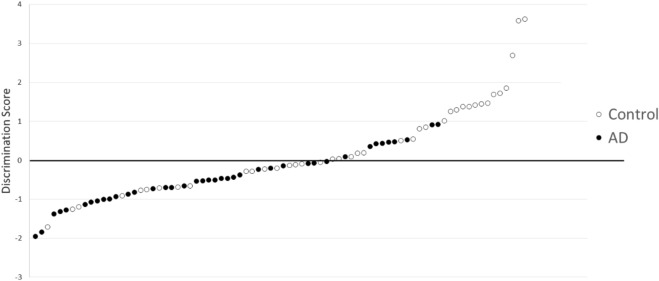
Discriminant analysis of the AD and Control groups
Discriminant analysis was conducted with the levels of ornithine, lysine, and uracil (Wilks lambda = 0.82, P < 0.01). The discrimination score was calculated for each sample as follows:
The analysis demonstrated sensitivity and specificity of 77.5% and 57.5%, respectively (Fig. 3).
Figure 3.
Discrimination score for three metabolites (○: Control, ●: AD). The distribution of scores, with sensitivity and specificity of 77.5% and 57.5%, respectively, in differentiating between healthy controls and AD patients.
Discussion
In the present study, ornithine, uracil, and lysine were decreased in AD patients compared to healthy controls. The decrease of ornithine may be consistent with the previous study showing that ornithine was decreased in the CSF of AD patients, and that the polyamine pathway (and metabolites of the tryptophan-kynurenine pathway) was also downregulated21. In addition, it has been reported that decreased ornithine is associated with lower cognitive function in the general population22. As for ornithine metabolism pathways (Fig. 4), it has been shown that metabolites of arginine and polyamine are altered in the blood of MCI and AD patients15. However, AD patients have been reported to have decreased lipid metabolites such as HDL subfractions and DHA, and increased glutamine and ornithine levels22. Knockout mice that lack an ornithine degradation system develop fatal hyperammonemia, and ornithine is considered to be an essential amino acid for the urea cycle23. Similarly, in humans, ornithine accumulation has been shown to cause hyperornithinemia-hyperammonemia-homocitrullinuria syndrome. At present, the cause of this disorder is not clear, but clinically, it is believed to cause mental retardation22. The present results indicated that ornithine and polyamine pathways may be associated with cognitive function in AD. There has been no report showing significant changes in uracil and lysine in AD, and further studies of their involvement in the pathophysiology of AD are needed.
Figure 4.
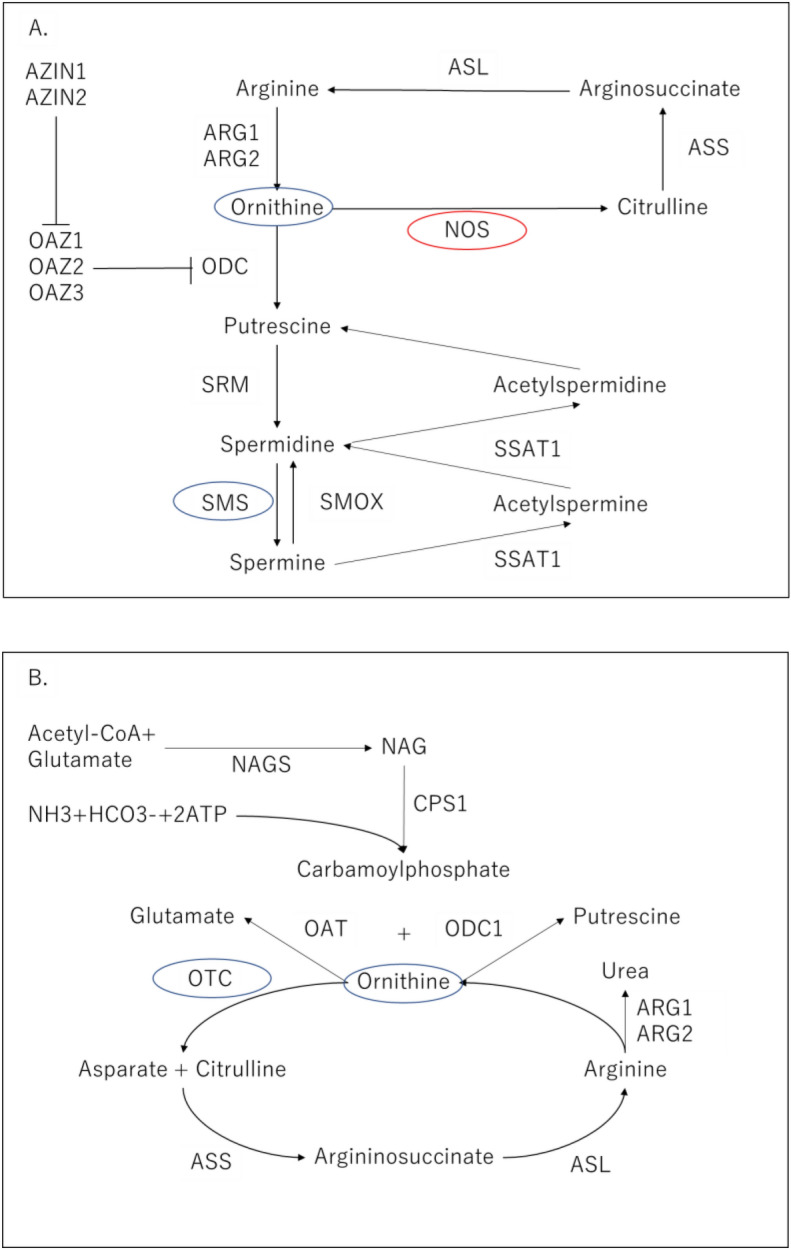
Polyamine pathway and urea cycle. (A) The polyamine pathway in mammalian cells (33,586,680 Fig. 1) ARG1: arginase1, ARG2 arginase2, ODC ornithine decarboxylase, SRM spermidine synthase, SMS spermine synthase, SMOX spermine oxidase, AZIN1 antizyme inhibitor 1, OAZ1 ornithine decarboxylase antizyme 1, SSAT1 spermidine/spermine N1-acetyltransferase 1, NOS nitric oxide synthase, ASL argininosuccinate lyase, ASS argininosuccinate synthase. (B) Urea cycle (31,978,055 Fig. 1) NAG N-acetyl glutamate, NAGS N-acetylglutamate synthase, CPS1 carbamoyl phosphate synthetase I, OAT ornithine aminotransferase, ornithine aminotransferase, ODC1 ornithine decarboxylase 1, OTC ornithine transcarbamylase.
Interestingly, there was a significant positive correlation between betaine and IADL. Betaine is known to act against various biological stresses and serve as a substrate in the betaine-homocysteine S-methyltransferase (BHMT) reaction, converting homocysteine to methionine24. Betaine intake is known to improve cognitive impairment in both AD mice25 and AD patients26. Thus, high betaine levels may be associated with higher ADL in elderly persons.
We hypothesized that the decreased ornithine level might be due to an aberrant polyamine pathway or urea cycle (Fig. 4). Thus, mRNA levels of enzymes in the polyamine pathway and urea cycle were examined. In the polyamine pathway, the SMS mRNA level was significantly decreased in AD, and the NOS2 mRNA level was increased in MCI. In the urea cycle, the OTC mRNA level was decreased in MCI. There may be some correlations between the ornithine concentration and mRNAs related to the amino acid, but direct relationships among them could not be found.
The discriminant analysis between the AD and Control groups using the concentrations of 3 metabolites (ornithine, lysine, and uracil) demonstrated sensitivity and specificity of 77.5% and 57.5%, respectively. The relatively low specificity suggested that only three metabolites had not enough potential to use in the clinical setting for differentiating AD from healthy subjects, although there were significant relationships among these three metabolites and AD. Considering AD and MCI due to AD-related cognitive changes as a continuous spectrum, the intermediate discrimination score of the MCI group is reasonable (mean ± SD, AD − 0.53 ± 0.69 vs. MCI 0.02 ± 0.93 vs. Control 0.40 ± 1.22). Emerging evidence has suggested that biomarkers detecting progression of AD are needed for more personalized treatments, and blood metabolomics are among such novel candidates27,28.
This work’s strength was that it was a population-based, complete enumeration study, but it had some limitations. Since few studies have examined the relationships between metabolomics and neurodegenerative diseases, further studies are warranted to validate the current findings in other cohorts, both cross-sectionally and longitudinally. Further research is needed to diagnose other neurodegenerative diseases such as dementia with Lewy bodies. The significance level of 5% for metabolites was not adjusted for multiple testing, which may lead to type I error. Moreover, the sample size was relatively small. Due to the limited power of this study, the negative findings should not be interpreted as showing no associations.
Conclusion
Ornithine was decreased and mRNA expressions related to its metabolism were changed in the AD group compared to the Control and MCI groups, suggesting an association between abnormal ornithine metabolism and AD. Increased betaine and decreased methionine may have the potential to serve as markers of better IADL in elderly persons. Our results suggest that plasma metabolomics may be useful for predicting the progression of AD.
Supplementary Information
Acknowledgements
The authors would like to thank Ms. Chiemi Onishi for technical assistance. This study was supported by the Japan Agency for Medical Research and Development (JP18dk0307076, JP20dk0207025 and JP20dk0307076). This work was partially supported by a Health and Labour Science Research Grant from the Japanese Ministry of Health, Labour and Welfare and a Grant-in-Aid for Scientific Research from the Japanese Ministry of Education, Culture, Sports, Science and Technology, JSPS KAKENHI Grant Numbers 18H02752, 18K07564, 19K08075 and 20K16628.
Author contributions
T.O. and J.I. wrote the main manuscript text. T.O., Y.Y., T.N. and K.M. conducted the experiments. A.T., H.S. and T.M. collected the samples and managed the clinical data. T.M., S.N., J.I., A.T., T.O. and S.U. planed and organized the research. All authors reviewed the manuscript.
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-19670-y.
References
- 1.Yu JT, Xu W, Tan CC, Andrieu S, Suckling J, Evangelou E, Pan A, Zhang C, Jia J, Feng L, Kua EH, Wang YJ, Wang HF, Tan MS, Li JQ, Hou XH, Wan Y, Tan L, Mok V, Tan L, Dong Q, Touchon J, Gauthier S, Aisen PS, Vellas B. Evidence-based prevention of Alzheimer's disease: systematic review and meta-analysis of 243 observational prospective studies and 153 randomised controlled trials. J. Neurol. Neurosurg. Psychiatry. 2020;91:1201–1209. doi: 10.1136/jnnp-2019-321913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang YY, Sun YP, Luo YM, Peng DH, Li X, Yang BY, Wang QH, Kuang HX. Biomarkers for the clinical diagnosis of Alzheimer's disease: Metabolomics analysis of brain tissue and blood. Front. Pharmacol. 2021;12:700587. doi: 10.3389/fphar.2021.700587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reveglia P, Paolillo C, Ferretti G, De Carlo A, Angiolillo A, Nasso R, Caputo M, Matrone C, Di Costanzo A, Corso G. Challenges in LC-MS-based metabolomics for Alzheimer's disease early detection: targeted approaches versus untargeted approaches. Metabolomics. 2021;17:78. doi: 10.1007/s11306-021-01828-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ochi S, Iga JI, Funahashi Y, Yoshino Y, Yamazaki K, Kumon H, Mori H, Ozaki Y, Mori T, Ueno SI. Identifying blood transcriptome biomarkers of Alzheimer's disease using transgenic mice. Mol. Neurobiol. 2020;57:4941–4951. doi: 10.1007/s12035-020-02058-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teunissen CE, Verberk IMW, Thijssen EH, Vermunt L, Hansson O, Zetterberg H, van der Flier WM, Mielke MM, Del Campo M. Blood-based biomarkers for Alzheimer's disease: Towards clinical implementation. Lancet Neurol. 2022;21:66–77. doi: 10.1016/S1474-4422(21)00361-6. [DOI] [PubMed] [Google Scholar]
- 6.Hurtado MO, Kohler I, de Lange EC. Next-generation biomarker discovery in Alzheimer's disease using metabolomics—From animal to human studies. Bioanalysis. 2018;10:1525–1546. doi: 10.4155/bio-2018-0135. [DOI] [PubMed] [Google Scholar]
- 7.Fleszar MG, Wisniewski J, Zboch M, Diakowska D, Gamian A, Krzystek-Korpacka M. Targeted metabolomic analysis of nitric oxide/L-arginine pathway metabolites in dementia: Association with pathology, severity, and structural brain changes. Sci. Rep. 2019;9:13764. doi: 10.1038/s41598-019-50205-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soga T, Igarashi K, Ito C, Mizobuchi K, Zimmermann HP, Tomita M. Metabolomic profiling of anionic metabolites by capillary electrophoresis mass spectrometry. Anal. Chem. 2009;81:6165–6174. doi: 10.1021/ac900675k. [DOI] [PubMed] [Google Scholar]
- 9.Soga T, Ohashi Y, Ueno Y, Naraoka H, Tomita M, Nishioka T. Quantitative metabolome analysis using capillary electrophoresis mass spectrometry. J. Proteome Res. 2003;2:488–494. doi: 10.1021/pr034020m. [DOI] [PubMed] [Google Scholar]
- 10.Koike S, Bundo M, Iwamoto K, Suga M, Kuwabara H, Ohashi Y, Shinoda K, Takano Y, Iwashiro N, Satomura Y, Nagai T, Natsubori T, Tada M, Yamasue H, Kasai K. A snapshot of plasma metabolites in first-episode schizophrenia: A capillary electrophoresis time-of-flight mass spectrometry study. Transl. Psychiatry. 2014;4:e379. doi: 10.1038/tp.2014.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshimi N, Futamura T, Kakumoto K, Salehi AM, Sellgren CM, Holmen-Larsson J, Jakobsson J, Palsson E, Landen M, Hashimoto K. Blood metabolomics analysis identifies abnormalities in the citric acid cycle, urea cycle, and amino acid metabolism in bipolar disorder. BBA Clin. 2016;5:151–158. doi: 10.1016/j.bbacli.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Umehara H, Numata S, Watanabe SY, Hatakeyama Y, Kinoshita M, Tomioka Y, Nakahara K, Nikawa T, Ohmori T. Altered KYN/TRP, Gln/Glu, and Met/methionine sulfoxide ratios in the blood plasma of medication-free patients with major depressive disorder. Sci. Rep. 2017;7:4855. doi: 10.1038/s41598-017-05121-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuwabara H, Yamasue H, Koike S, Inoue H, Kawakubo Y, Kuroda M, Takano Y, Iwashiro N, Natsubori T, Aoki Y, Kano Y, Kasai K. Altered metabolites in the plasma of autism spectrum disorder: A capillary electrophoresis time-of-flight mass spectroscopy study. PLoS ONE. 2013;8:e73814. doi: 10.1371/journal.pone.0073814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sriwichaiin S, Chattipakorn N, Chattipakorn SC. Metabolomic alterations in the blood and brain in association with Alzheimer's disease: Evidence from in vivo to clinical studies. J. Alzheimers Dis. 2021;84:23–50. doi: 10.3233/JAD-210737. [DOI] [PubMed] [Google Scholar]
- 15.Graham SF, Chevallier OP, Elliott CT, Holscher C, Johnston J, McGuinness B, Kehoe PG, Passmore AP, Green BD. Untargeted metabolomic analysis of human plasma indicates differentially affected polyamine and L-arginine metabolism in mild cognitive impairment subjects converting to Alzheimer's disease. PLoS ONE. 2015;10:e0119452. doi: 10.1371/journal.pone.0119452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ninomiya T, Nakaji S, Maeda T, Yamada M, Mimura M, Nakashima K, Mori T, Takebayashi M, Ohara T, Hata J, Kokubo Y, Uchida K, Taki Y, Kumagai S, Yonemoto K, Yoshida H, Muto K, Momozawa Y, Akiyama M, Kubo M, Ikeda M, Kanba S, Kiyohara Y, Group J-AS Study design and baseline characteristics of a population-based prospective cohort study of dementia in Japan: the Japan Prospective Studies Collaboration for Aging and Dementia (JPSC-AD) Environ. Health Prev. Med. 2020;25:64. doi: 10.1186/s12199-020-00903-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshino Y, Kumon H, Mori T, Yoshida T, Tachibana A, Shimizu H, Iga JI, Ueno SI. Effects of AGXT2 variants on blood pressure and blood sugar among 750 older Japanese subjects recruited by the complete enumeration survey method. BMC Genomics. 2021;22:287. doi: 10.1186/s12864-021-07612-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKhann GM, Knopman DS, Chertkow H, Hyman BT, Jack CR, Jr, Kawas CH, Klunk WE, Koroshetz WJ, Manly JJ, Mayeux R, Mohs RC, Morris JC, Rossor MN, Scheltens P, Carrillo MC, Thies B, Weintraub S, Phelps CH. The diagnosis of dementia due to Alzheimer's disease: Recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7:263–269. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watanabe SY, Iga J, Ishii K, Numata S, Shimodera S, Fujita H, Ohmori T. Biological tests for major depressive disorder that involve leukocyte gene expression assays. J. Psychiatr Res. 2015;66–67:1–6. doi: 10.1016/j.jpsychires.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 20.Watanabe S, Iga J, Nishi A, Numata S, Kinoshita M, Kikuchi K, Nakataki M, Ohmori T. Microarray analysis of global gene expression in leukocytes following lithium treatment. Hum. Psychopharmacol. 2014;29:190–198. doi: 10.1002/hup.2381. [DOI] [PubMed] [Google Scholar]
- 21.Muguruma Y, Tsutsui H, Noda T, Akatsu H, Inoue K. Widely targeted metabolomics of Alzheimer's disease postmortem cerebrospinal fluid based on 9-fluorenylmethyl chloroformate derivatized ultra-high performance liquid chromatography tandem mass spectrometry. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2018;1091:53–66. doi: 10.1016/j.jchromb.2018.05.031. [DOI] [PubMed] [Google Scholar]
- 22.van der Lee SJ, Teunissen CE, Pool R, Shipley MJ, Teumer A, Chouraki V, Melo van Lent D, Tynkkynen J, Fischer K, Hernesniemi J, Haller T, Singh-Manoux A, Verhoeven A, Willemsen G, de Leeuw FA, Wagner H, van Dongen J, Hertel J, Budde K, Willems van Dijk K, Weinhold L, Ikram MA, Pietzner M, Perola M, Wagner M, Friedrich N, Slagboom PE, Scheltens P, Yang Q, Gertzen RE, Egert S, Li S, Hankemeier T, van Beijsterveldt CEM, Vasan RS, Maier W, Peeters CFW, Jorgen Grabe H, Ramirez A, Seshadri S, Metspalu A, Kivimaki M, Salomaa V, Demirkan A, Boomsma DI, van der Flier WM, Amin N, van Duijn CM. Circulating metabolites and general cognitive ability and dementia: Evidence from 11 cohort studies. Alzheimers Dement. 2018;14:707–722. doi: 10.1016/j.jalz.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 23.Ma C, Hunt JB, Selenica MB, Sanneh A, Sandusky-Beltran LA, Watler M, Daas R, Kovalenko A, Liang H, Placides D, Cao C, Lin X, Orr MB, Zhang B, Gensel JC, Feola DJ, Gordon MN, Morgan D, Bickford PC, Lee DC. Arginase 1 insufficiency precipitates amyloid-beta deposition and hastens behavioral impairment in a mouse model of amyloidosis. Front. Immunol. 2020;11:582998. doi: 10.3389/fimmu.2020.582998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ueland PM. Choline and betaine in health and disease. J. Inherit. Metab. Dis. 2011;34:3–15. doi: 10.1007/s10545-010-9088-4. [DOI] [PubMed] [Google Scholar]
- 25.Ibi D, Hirashima K, Kojima Y, Sumiya K, Kondo S, Yamamoto M, Ando T, Hiramatsu M. Preventive effects of continuous betaine intake on cognitive impairment and aberrant gene expression in hippocampus of 3xTg mouse model of Alzheimer's disease. J. Alzheimers Dis. 2021;79:639–652. doi: 10.3233/JAD-200972. [DOI] [PubMed] [Google Scholar]
- 26.Sun J, Wen S, Zhou J, Ding S. Association between malnutrition and hyperhomocysteine in Alzheimer's disease patients and diet intervention of betaine. J. Clin. Lab. Anal. 2017;31:e22090. doi: 10.1002/jcla.22090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jaaskelainen O, Hall A, Tiainen M, van Gils M, Lotjonen J, Kangas AJ, Helisalmi S, Pikkarainen M, Hallikainen M, Koivisto A, Hartikainen P, Hiltunen M, Ala-Korpela M, Soininen P, Soininen H, Herukka SK. Metabolic profiles help discriminate mild cognitive impairment from dementia stage in Alzheimer's disease. J. Alzheimers Dis. 2020;74:277–286. doi: 10.3233/JAD-191226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yilmaz A, Ustun I, Ugur Z, Akyol S, Hu WT, Fiandaca MS, Mapstone M, Federoff H, Maddens M, Graham SF. A Community-based study identifying metabolic biomarkers of mild cognitive impairment and Alzheimer's disease using artificial intelligence and machine learning. J. Alzheimers Dis. 2020;78:1381–1392. doi: 10.3233/JAD-200305. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.