Abstract
Gender-bias in COVID-19 severity has been suggested by clinical data. Experimental data in cell and animal models have demonstrated the role of sex hormones, particularly estrogens, in viral infections such as in COVID-19. SARS-CoV-2 uses ACE2 as a receptor to recognize host cells, and the protease TMPRSS2 for priming the Spike protein, facilitating virus entry into cells. However, the involvement of estrogenic receptors in SARS-CoV-2 infection are still being explored. Thus, in order to investigate the role of estrogen and its receptors in COVID-19, the estrogen receptors ERα, ERβ and GPER1 were overexpressed in bronchial BEAS-2B cell, and then infected with SARS-CoV-2. Interestingly, the levels of ACE2 and TMPRSS2 mRNA were higher in SARS-CoV-2-infected cells, but no difference was observed in cells with estrogen receptors overexpression. GPER1 can be involved in virus infection or replication, since its higher levels reduces SARS-CoV-2 load. On the other hand, pharmacological antagonism of GPER1 enhanced viral load. Those data suggest that GPER1 has an important role in SARS-CoV-2 infection.
Keywords: Estrogen receptors, COVID-19, BEAS-2B, GPER1, Gender
1. Introduction
On March 11, 2020 the World Health Organization declared that COVID-19 became a pandemic. This devastating disease is caused by the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), a beta coronavirus which was first identified in Wuhan city (China) in late 2019, and rapidly spread around the world.
A common route of transmission is the respiratory tract, whose cells express the ACE2 (Angiotensin Converting Enzyme 2) as the main receptor that SARS-CoV-2 uses to recognize mammalian and non-mammalian cells (Li et al., 2005) (Hoffmann et al., 2020), via the Spike glycoprotein (composed by S1 and S2 subunits) (Walls et al., 2020), more specifically through the RBD (receptor binding domain) site (Gabutti, d'Anchera, Sandri et al., 2020).
The most frequent initial clinical characteristics of COVID-19 are fever, cough, and flu symptoms. Lung inflammation, cytokine storm and thromboembolic disease are often associated with COVID-19 patients’ deaths (Scialpi et al., 2020) (Poyiadji et al., 2020). The disease is characterized by lung injury, with a clinical finding by the ground-glass opacity in radiologic computed tomography (Guan et al., 2020), that can evolve to an acute respiratory distress syndrome (ARDS) and respiratory failure.
It is clear that there is a difference in gender-like severity of coronavirus disease (Channappanavar et al., 2017) (Sharma et al., 2020). Men are more susceptible to hospitalization by COVID-19 than women, with elevated mortality cases revealed in some epidemiological databases (https://globalhealth5050.org/the-sex-gender-and-covid-19-project/). A meta-analysis study has shown higher rates of infection and mortality in male patients (Li et al., 2020). The men's vulnerability to severe clinical stage has been hypothesised to be related to the lifestyle and health conditions, but recently published reviews are linking sex hormones, especially estradiol, to an elevated protection in women (Stilhano et al., 2020) (Suba, 2020) (Breithaupt-Faloppa et al., 2020) (Newson, Manyonda, R. et al., 2021). Estrogen is present in higher concentrations in premenopausal women than men, and it promotes several beneficial effects in organismal protection, such as inhibition of inflammation and regulating immune responses (Scully et al., 2020). Therefore, some studies have suggested that the Hormone Replacement Therapy (HRT) containing estrogen (more specifically estradiol) could be beneficial against COVID-19 cytokine storm and also reduce mortality from COVID-19 (Al-Kuraishy et al., 2021). Importantly, as SARS-CoV-2 main route of cell entry is via ACE2 binding, and since then some gender differences in ACE2 have been observed in human airway cells (Kalidhindi et al., 2020), it is important to investigate the actions of estrogens in SARS-CoV-2 infection. Regarding the estrogen intracellular signalling, it is mediated by classic genomic and rapid (non-genomic) actions. The genomic actions involve the activation of classic nuclear estrogen receptors, ERα and ERβ, which mediate the gene transcription by their own actions by ligand-receptor to ERE (estrogen response element) complex or via activation of other transcription factors (Couse and Korach, 1999) (O'Lone et al., 2004). The rapid actions of estrogen involve pathways such as ERK-MAPK and PI3K-AKT, mainly via membrane receptors activation. In addition, there are mobilization of second messengers such as cyclic AMP and Ca2+, via activation of GPER1, a G protein-coupled estrogen receptor, mediating the intracellular responses cascade (Filardo et al., 2002) (Revankar et al., 2005). These intracellular signaling mediated by estrogen can modulate several pathways, and despite the molecular mechanism involved in SARS-CoV-2 infection are unknown, several papers have shown that these receptors play an important role in lung function, in maturation and maintenance of alveolar homeostasis, and in immune system (Kalidhindi et al., 2019) (Massaro et al., 2007) (Massaro and Massaro, 2006) (Millas and Duarte Barros, 2021). In addition, recent studies have suggested that estrogen can be involved in ACE2 and TMPRSS2 expression, leading to reduction of the cellular invasion by SARS-Cov-2 (Kalidhindi et al., 2020) (Lemes et al., 2021) (Stelzig et al., 2020; Leach et al., 2021).
Despite the evidence that estrogen has a role in COVID-19, the involvement of estrogenic receptor in modulating virus infection or replication is still unclear. Thus, in this work we overexpressed estrogen receptors ERα, ERβ and GPER1 in a human lung cell line, and performed pharmacological treatment, in order to investigate the potential participation of those receptors in SARS-CoV-2 infectiveness. This could be relevant for evaluating hormone-related treatments in COVID-19.
2. Material and methods
2.1. Cell culture, estrogen receptors overexpression and treatments
A549 and BEAS-2B cells lines were cultivated in DMEN-F12 (Dulbecco's Modified Eagle Medium-Nutrient Mixture F-12) without phenol red, supplemented with 10% fetal bovine serum FBS (Gibco) and 0.02 mg/ml gentamicin, maintained in 5% CO2 atmosphere at 37 °C. Confluent 100 mm cell plates were trypsinized (trypsin-EDTA 0.05%), centrifuged and plated in for standard procedures. For BEAS-2B cultures, the cell plates were coated with type I collagen (Sigma), for better cell adherence. The following procedures were approved by the Institutional Ethics Committee (number 6864310320 CEP-UNIFESP).
In order to deliver the plasmids into the cells, we used the nucleofection method. For this, plated cells (1 × 106) were trypsinized and resuspended in 100 μL of Nucleofector® Solution, to which 5 μg of plasmids for estrogen receptors were added: ERα (Addgene plasmid # 49498; http://n2t.net/addgene:49498; RRID:Addgene_49498; pcDNA-HA-ER WT was a gift from Sarat Chandarlapaty); ERβ (‘pCDNA3.1-nv5-ER β' (Addgene plasmid # 22770; http://n2t.net/addgene:22770; RRID:Addgene_22770; pCDNA3.1-nv5-ER beta was a gift from Donald McDonnell); GPER1 (#RG203027, Origene) and Luciferase (pLenti CMV V5-LUC Blast (w567-1) was a gift from Eric Campeau (Addgene plasmid # 21474; http://n2t.net/addgene:21474; RRID:Addgene_21474). The cells were added into a specific cuvette and were placed in the nucleofector (Nucleofector 2b Device, Lonza). After the nucleofection, cells were carefully resuspended in culture medium containing DMEM-F12 with 10% FBS at a confluence of 1 × 105 cells per well and transferred to 24-well plates. The cell cultures were incubated for 48 h under standard conditions before being infected.
In order to assess the direct participation of GPER1, cells were pre-treated with 17β-estradiol (E2, 10−7 M), GPER1 agonist (G1, 10−7 M) and antagonist (G15, 10−8 M) compounds (Tocris Biosciences), for 1 h prior to SARS-CoV-2 infection. G15 was also pre-treated 30 min before E2 and G1 co-treatment. DMSO was considered as vehicle for all treatments since all drugs were diluted in 0.1% DMSO.
2.2. Virus expansion and infection protocol
For this study, SARS-CoV-2 isolated from a Brazilian patient (EPI_ISL_413016), was kindly provided by Prof Paolo Zanotto (University of São Paulo), Prof. Jose Proença Módena (State University of Campinas) and Prof Edison Durigon (University of São Paulo). All experiments for the propagation, titration and infection of SARS-CoV-2 in vitro were carried out at the BSL-3 lab at Biological Institute (Sao Paulo, Brazil). The expansion was performed until the fifth passage, and then the viruses were collected. For virus expansion, VERO CCL-81 (African green monkey kidney) cells plated at 100% of confluence in tissue culture flasks, were incubated with SARS-CoV-2 in DMEM-F12 medium (supplemented with 1% FBS). After 1 h, supplementation with FBS was adjusted to 10% and incubation extended to more 40 h. At the end, supernatant was collected, and plaque assay was performed (Cugola et al., 2016). A titter of 3,3 × 106 plaque-forming units (PFU/mL) was obtained and stored at −70 °C. For the infection experiments, 1 × 105 BEAS-2B cells were exposed to SARS-CoV-2 using the MOI 0.2 (Multiplicity of Infection) (for 2 h at 37 °C. After incubation, supernatants were removed, and cells rinsed with PBS. Then, cells cultured were kept for more 24 h in culture medium (DMEM-F12 and FBS 10%). At the end of experiment, the cells had their total RNA extracted for PCR-qPCR assays, as described in (Lemes et al., 2021).
2.3. Western and RT-qPCR
The Western blot assay was performed for quantification of estrogen receptors and ACE2/TMPRSS2. Briefly, 5 × 105 BEAS-2B and A549 cells were incubated for 48 h in cultured medium, and then were lysed in RIPA buffer (150 mM NaCl; 1% NP-40; 0.5% deoxycholic acid; 0.1% SDS; 50 mM Tris pH 8.0; 2 mM EDTA), with protease and phosphatase inhibitors. 15–30 μg of protein were electrophoresed in SDS-PAGE, transferred onto a PVDF membrane that were blocked (5% non-fat dry milk) and incubated with primary antibodies ERα (MC-20, sc-542, Santa Cruz Biotechnology, 1:1000), ERβ (MC10, Thermo Fisher Scientific, 1:2000), GPER1 (ab39742, Abcam, 1:2000), ACE2 (ab15348, Abcam 1:2000) and TMPRSS2 (sc515727, Santa Cruz Biotechnology, 1:1000) and secondary antibodies tagged with HRP (Horsearedish Peroxidase). The bands were revealed with ECL and photodocumented in a UVITEC system (Cambridge). For comparing protein quantification from different cell types (BEAS-2B x A549), which has different patterns of housekeeping proteins expression (data not shown), the values presented in histograms correspond to the optical density of bands. For comparing BEAS-2B protein quantification, GAPDH (68795, Sigma, 1:5000) housekeeping protein quantification and normalization was used. Band intensity levels were quantified by ImageJ software.
Quantitative real-time PCR to detect SARS-CoV-2 were performed as described in detail by Corman et al. (2020) (Corman et al., 2020). The plated cells were collected and had their RNA extracted by the RNeasy Mini Kit, according to (Lemes et al., 2021). For ACE2, TMPRSS2, ESR1, ESR2 and GPER1 gene expression analyses, RT-qPCR was performed. For this mRNA extracted was quantified in Nanodrop (Thermo Fisher Scientific) and used as template to cDNA synthesis by High-Capacity Kit (Thermo Fisher Scientific). For RT-qPCR assay, cDNA samples were added with the SYBR-Green reagent (Qiagen) and primer pairs for ACE2 (Fw: 5′-CATTGGAGCAAGTGTTGGATCTT-3'; Rv: 5′-GAGCTAATGCATGCCATTCTCA-3′); TMPRSS2 (Fw 5′-CCTCTAACTGGTGTGATGGCGT-3'; Rv 5′-TGCCAGGACTTCCTCTGAGATG-3′); ESR1 (Fw: 5′ CAGCGTGGCCCTGGTTGGAA-3'; Rv: 5′ACAGCAACGGTCCCCACCCT); ESR2 (Fw: 5′GGTCGGCAGACCACAAGCCC-3'; Rv 5′CTGGCGCAACGGTTCCCACT) GPER1 (Fw: 5′CTCCCTGCAAGCAGTCTTTC-3'; Rv: 5′CGGCAAATTTGTTTTCTGCT-3′); and RPL35 (Fw: 5′CGAGTCGTCCGGAAATCCAT3’; Rv: 5′ GGCTTGTACTTCTTGCCCTTG3′). The quantification method used was the relative comparison 2–ΔΔCT to the endogenous gene RPL35 (Livak and Schmittgen, 2001). For virus quantification, as described in (Lemes et al., 2021), the following primers and probes were used: SARS-CoV-2 nucleocapsid (N) primer Fw 5′ CACATTGGCACCCGCAATC 3’; primer Rv 5′ GAGGAACGAGAAGAGGCTTG 3’; Probe 5′ FAM-ACTTCCTCAAGGAACAACATTGCCA-BBQ 3’. RNase P primer Fw 5′ AGATTTGGACCTGCGAGC 3’; primer Rv 5′ GAGCGGCTGTCTCCACAAGT 3’; Probe 5′ FAMTTCTGACCTGAAGGCTCTGCGCG-BHQ 3’.
2.4. Statistical analysis
The results were presented as mean ± standard error (SEM). All data were analysed using the GraphPad Prism version 8.0 (GraphPad Software) by using One-way ANOVA and Student's unpaired t-test. It was accepted as statistically significant p < 0.05.
3. Results
3.1. Characterization of lung-derived cellular types based on estrogen receptors, ACE2 and TMPRSS2 expression
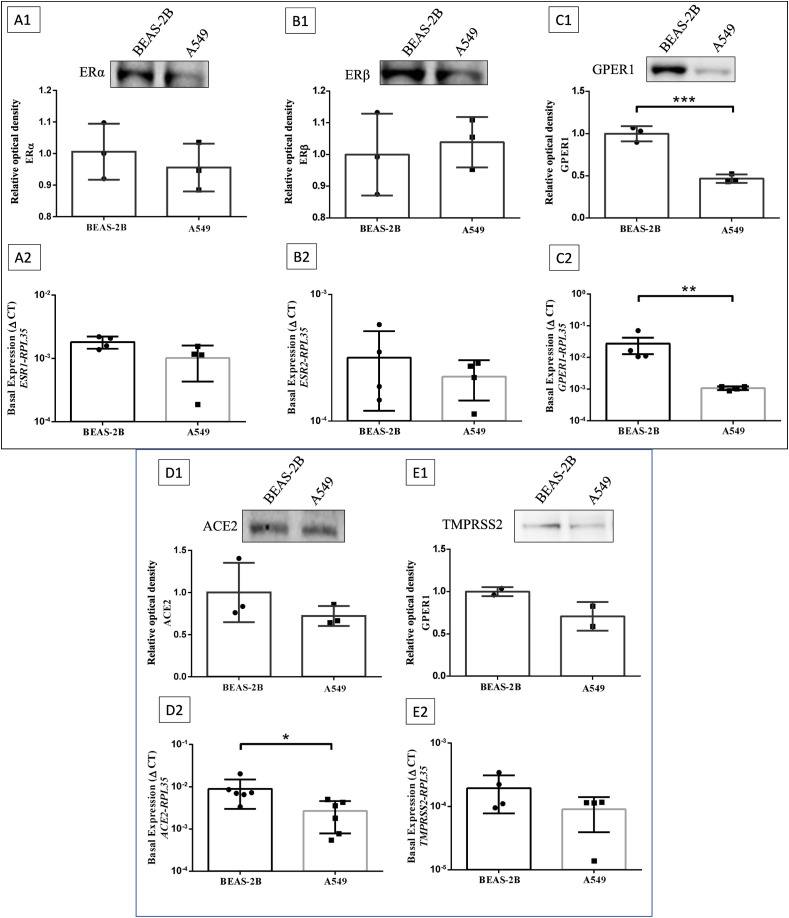
We first evaluated the basal protein and gene expression levels of ERα (Fig. 1A1; ESR1 - Fig. 1A2), ERβ (Fig. 1B1; ESR2 - Fig. 1B2), and GPER1 (Fig. 1C1; GPER1 - Fig. 1C2) in lung-derived cell lines. The data presented in Fig. 1 suggest that BEAS-2B and A549 have similar protein and gene expressions of ERα and ERβ. Regarding the expression of GPER1, the data suggest that BEAS-2B cells express significantly higher levels of GPER1 than A549 cells, both proteins and gene expression. Despite ACE2 gene expression levels are higher in BEAS-2B cells than in A549 cells (Fig. 1D2), the ACE2 protein levels are similar (Fig. 1D1). TMPRSS2 expression are similar between the cell lines (Fig. 1E1 and 1E2). Interestingly, data from our group indicate that BEAS-2B cells have less infectiveness of SARS-CoV-2 kinetics than A549 (data not shown, manuscript under revision). Thus, due to the evident differences on the basal expression of estrogen receptors (mainly GPER1) the next experiments were performed with the BEAS-2B cell line to verify the potential participation of estrogen receptors in SARS-CoV-2 infection.
Fig. 1.
Characterization of protein and gene expressions of estrogen receptors, ACE2 and TMPRSS2 in the BEAS-2B and A549 cell lines. Representative Western blot image for ERα (A1), ERβ (B1) and GPER1 (C1) of BEAS-2B and A549 cells cultured during 48 h under standard conditions (37 °C and 5% CO2), along with basal gene expression (by RT-qPCR) of ESR1 (A2), ESR2 (B2) and GPER1 (C2). D and E are relative values of the gene (by RT-qPCR) and protein (Western blot) expression of ACE2 (D) and TMPRSS2 (E) of BEAS-2B and A549 cells cultured for 48 h under standard conditions, according to the expression of the endogenous gene (RPL53) by RT-qPCR, or relative optical density for Western blot. Values correspond to the mean ± SEM. Student's unpaired t-test, parametric between BEAS-2B and A549 groups was applied. p*≤0.05, p**≤0.001, p***≤0.0001. N = 2 (E1), N = 3 (A1, B1, C1, D1), N = 4 (A2, B2, C2, E2), N = 6 (D2).
3.2. Overexpression of ERα, ERβ and GPER1 in BEAS-2B – effects on ACE2 and TMPRSS2 and viral loads
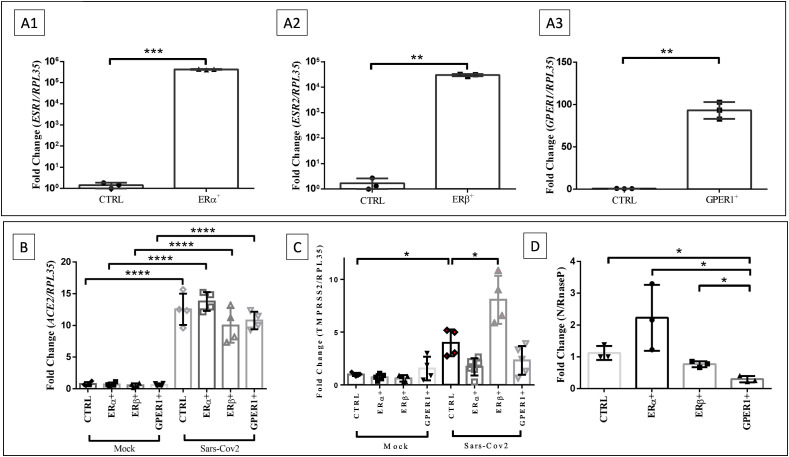
In order to evaluate the potential activity of each estrogen receptor in SARS-CoV-2 infection, BEAS-2B cell line were nucleofected with plasmids containing the genes that encodes ERα, ERβ and GPER1 (ESR1, ESR2 and GPER1), using the Luciferase plasmid as nucleofection plasmid control. Gene expression data indicate that plasmid overexpression was highly efficient (Fig. 2 A) in BEAS-2B cells.
Fig. 2.
Overexpression of ERα, ERβ and GPER1 does not significantly module ACE2 and TMPRSS2 in BEAS-2B cells. However, estrogen receptors overexpression revealed the participation of GPER1 in the modulation of viral load in BEAS-2B cells. A) Histogram of gene expression by RT-qPCR of ESR1 (A1), ESR2 (A2) and GPER1 (A3) between plasmid control cells group (plasmid containing Luciferase) and nucleofected cells group with plasmids containing ESR1, ESR2 and GPER1 genes, respectively, after 48 h of incubation. Data were normalized to the RPL35 gene. B, C) Histogram of gene expression by RT-qPCR of ACE2 (B) and TMPRSS2 (C) in cells that overexpress ERα, ERβ, GPER1 or Luciferase (control plasmid) after 24 h of infection with SARS-CoV-2 or not (mock). D) Histogram of gene expression by RT-qPCR (N gene) of cell extract from groups overexpressing Luciferase (plasmid control), ERα, ERβ or GPER1, after 24 h of infection with SARS-CoV2 (MOI 0.2). N gene expression data were normalized in relation to the RNAse P gene. Values correspond to mean ± SEM. Significant difference was considered, with *p ≤ 0.05 - one-way ANOVA, with Tukey's post-test or Student's unpaired t-test. N = 3 (A1, A2, A3, D), N = 4 (B), N = 6 (C).
The next step was infecting the cell models with SARS-CoV-2. After 48 h of nucleofection with plasmids containing the genes of ERα, ERβ and GPER1 and Luciferase (plasmid control), cells were infected or not with SARS-CoV-2 for 24 h at MOI 0.2. As shown in Fig. 2B, despite the infection per se enhanced the expression of ACE2, it was observed similar levels of ACE2 expression among cells overexpressing estrogen receptors, compared to the control plasmid. Regarding TMPRSS2 expression, SARS-CoV-2 infection elevated this gene expression, and only the group of cells overexpressing ERβ presented higher TMPRSS2 levels, compared to control plasmid (Fig. 2C). Finally, the analysis of viral infection indicated a modulation of viral load in cells that overexpressed GPER1 compared to the control group. The reduction of viral load in cells overexpressing GPER1 may indicate the participation of this receptor during SARS-CoV-2 infection or replication (Fig. 2D). Nevertheless, these data do not allow to assume a direct correlation between the decrease in viral load observed in the overexpression of GPER1 and the expression of ACE2 or TMPRSS2 (Fig. 2B and C).
3.3. Infection of SARS-CoV-2 in cells is modulated by GPER1 ligands
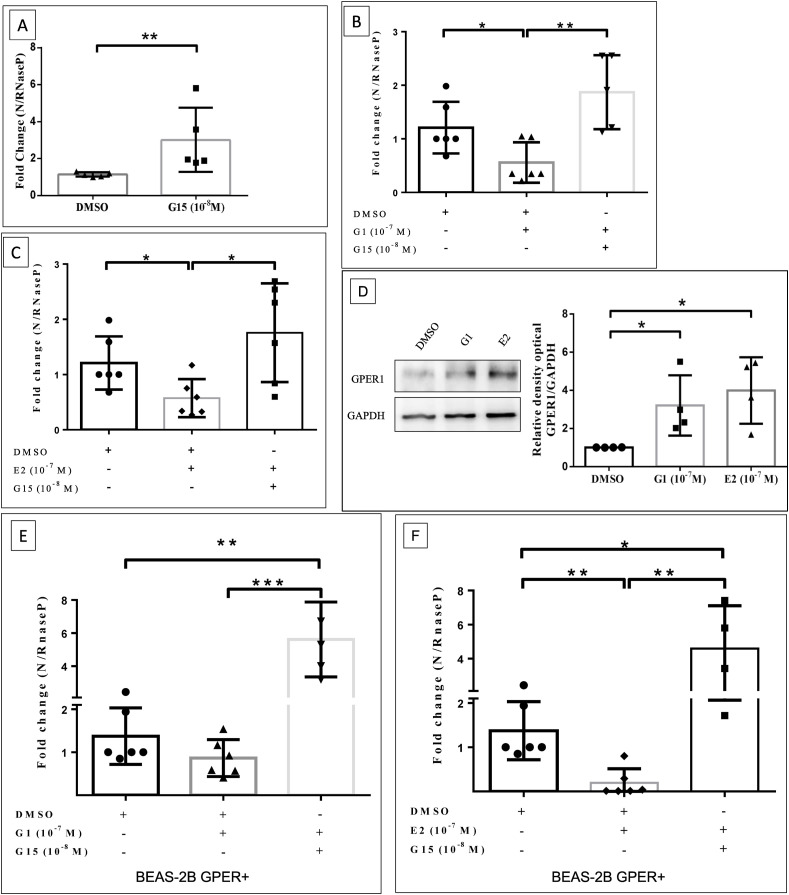
The participation of the GPER1 receptor was also evaluated using pharmacological tools, as the overexpression of this receptor showed a significant difference in the reduction of SARS-CoV-2 viral load (Fig. 2D). Initially wild type BEAS-2B cells were exposed to GPER1 antagonist, G15 at 10−8 M during 1 h prior to SARS CoV-2 infection. This concentration was chosen based on Pisolato et al. (2016) (Pisolato et al., 2016) and previous standard experiments (data not shown). The antagonism of GPER1 (with G15 treatment) promoted an increase in viral load in BEAS-2B cells, compared to the control vehicle (DMSO) (Fig. 3 A). On the other hand, the treatment of cells with G1 (GPER1 selective agonist) at 10−7 M (chosen based on previous standard experiments) during 1 h prior to SARS CoV-2 infection has significantly reduced SARS-CoV-2 viral load (Fig. 3B), indicating that GPER1 function is also negatively correlated to coronavirus infection or replication. In order to reinforce the contribution of GPER1 to the estrogenic signaling and its participation in SARS-CoV-2 infection, we treated cells with 17β-estradiol (E2 -10−7 M), based on previous study published by our group (Lemes et al., 2021), showing a reduction in viral load, compared to control vehicle (Fig. 3C). In addition, it was noted that both G1 and E2 treatment for 1 h significantly increased GPER1 expression (Fig. 3D), indicating that upon those treatments there are an upregulation of GPER1, which can contribute to reduce viral loads. Furthermore, the pre-treatment with G15 antagonist (for 30 min before agonists) blocked the effects of both G1 (Fig. 3B) and E2 (Fig. 3C) in reducing the viral load, which strengthen the hypothesis that GPER1 function is involved in inhibition of coronavirus infection or replication.
Fig. 3.
GPER1 receptor overexpression and its agonist treatment promotes reduction in SARS-CoV-2 viral load in BEAS-2B cells, and it is reversed by treatment with GPER1 antagonist. A-C) Histogram of gene expression by RT-qPCR (N gene) of cell extract in BEAS-2B cells, after 1 h of G15 (A), G1+G15 (B), E2+G15 (C) pre-treatment followed by 24 h of infection with SARS-CoV-2 (MOI 0.2). D) Representative Western blot image for GPER1 in BEAS-2B treated for 1 h with G1 or E2. E-F) Histogram of gene expression by RT-qPCR (N gene) of cell extract in BEAS-2B cells overexpressing GPER1, after 1 h of G1+G15 (E), E2+G15 (F) pre-treatment followed by 24 h of infection with SARS-CoV-2 (MOI 0.2). N gene expression data were normalized in relation to the RNAse P gene and GPER1 were normalized by GAPDH housekeeping protein. Values correspond to mean ± SEM. Significant difference was considered, with *p ≤ 0.05 - one-way ANOVA, with Tukey's post-test or Student's unpaired t-test. N = 4 (D), N = 5 (A), N = 6 (B, C, E, F).
Finally, BEAS-2B cells overexpressing GPER1 were treated with the E2 and G1 agonists. It is possible to observe that E2 treatment and GPER1 overexpression have a synergistic effect in reducing the viral load (Fig. 3F), suggesting that not only the expression of the receptor but also its function can be combined to protect cells from SARS-CoV-2. In addition, pre-treatment with G15 in the presence of agonists increased the viral load compared to the control vehicle, evidencing that the synergistic effects of pharmacological treatment and genetic modulation were blocked by the GPER1 antagonist (Fig. 3E and F).
4. Discussion
Gender differences in COVID-19 have been recently explored due their severity discrepancies. Studies suggests that sex hormones, especially estrogen, may have a protective role on this variability, and it is important to investigate the participation of estrogen receptors in SARS-CoV-2 pathology. In this study, we aimed to evaluate the receptors involved in estrogenic signaling in a pulmonary cell line, by overexpressing estrogen receptors and evaluating SARS-CoV-2 infection. We observed that no alterations in ACE2 and TMPRSS2 expression was found in cells that overexpressed estrogen receptors, despite higher levels of ACE2 due to the SARS-CoV-2 infection per se were found. Interestingly, GPER1 overexpression promoted a reduction in virus load and the same was observed with GPER1 agonism, which highlights the importance of investigating the cellular signaling related to estrogen in COVID-19.
The primary site of SARS-CoV-2 infection is the respiratory tract, and the cytokine storm that occurs rapidly in lungs contributes to the death of these patients (Mehta et al., 2020). In regard to the beneficial effects of sex hormones in the respiratory tract, it is important to know that the protective effect of estrogen on pulmonary inflammation has already been studied in different diseases that lead to an exacerbated inflammatory response (Fuentes et al., 2019). In COVID-19 research, for instance, a large world database study conducted by Seeland et al. (2020) have found that perimenopause and menopause women taking 17β-estradiol has more than 50% of reduced risk to develop severe COVID-19, and for young and middle-aged women the risk of fatality were not correlated to hormone replacement probably due to the high estrogen levels at this age (Seeland et al., 2020). Therefore, the role of estrogen and its receptors must be further investigated in COVID-19 (Stilhano et al., 2020).
Estrogen treatment can modulate the expression of important genes and proteins related to SARS-CoV-2 entry in cells. For example, Stelzig et al. (2020) have reported that 17β-estradiol reduced ACE2 gene expression in human bronchial epithelial cells from a female donor (Stelzig et al., 2020), and our group have shown a reduction in TMPRSS2 gene expression in VERO E6 cells after 17β-estradiol treatment (Lemes et al., 2021). Kalidhindi et al. (2020) have shown that, in airway smooth muscle cells exposed to testosterone, ACE2 expression was increased in both men and women, while treatment with estrogen resulted in decreased ACE2 levels (Kalidhindi et al., 2020). In fact, those studies revealed an important role of nuclear receptors in modulating the expression of ACE2 and TMPRSS2. Wang et al. (2021) have shown a positive correlation of ESR1 and ACE2 gene expression in the human atrium (Wang, Sun, J et al., 2021). Our data showed that SARS-CoV-2 infection in BEAS-2B cells per se (without pharmacological treatment) increased ACE2 and TMPRSS2 mRNA (Fig. 2B and C), corroborating with our previous data in VERO E6 cells (Lemes et al., 2021). Moreover, we showed that virus infection further increased TMPRSS2 gene expression levels in cells overexpressing Erβ (compared to the control) (Fig. 2C), although it did not result in modulation of SARS-CoV-2 viral load (Fig. 2D). As the TMPRSS2 expression was not directly modulated by ERβ overexpression (mock group, Fig. 2C), it is plausible to conceive that other mechanisms mediated by estrogenic signaling can be involved in SARS-CoV-2 viral load alterations, and further investigations are necessary to understand this complex signaling in COVID-19. For instance, no further modulation of ACE2 and TMPRSS2 gene expression was seen in cells that overexpressed GPER1 (Fig. 2B and C), although a sharp reduction in viral load was observed in this group (Fig. 2D). Altogether those findings reinforce the existence of multiple factors other than TMPRSS2 and ACE2 gene expression modulation to determine the virus entry into the cells, such as seen in the presence of GPER1 overexpression (Fig. 2D).
There are very few reports associating the role of GPER1 in virus infections. Accordingly, a very recent work has shown the importance of GPER1 for Influenza A infection. Harding et al. (2021) have found that GPER1 is found in foetal tissues, which in turn protect the foetus against organism response by suppressing interferon I, counteracting immune overstimulation that could be harmful for the foetus. With G15 treatment, an antagonist of GPER1, the viral infection worsened and eventually led to death of new-borns, reinforcing the role of GPER1 in organism protection (Harding et al., 2021). In addition, according to the Consensus database (The Protein Atlas - https://www.proteinatlas.org/ENSG00000164850-GPER1/tissue) there is a high expression of GPER1 gene in lungs, reinforcing the importance of deeper investigation of this receptor in coronavirus infection. The data presented here show that treatment with G15 increased SARS-CoV-2 viral load in bronchial cell line (Fig. 3A), while in the presence of the G1 (GPER1 agonist) the SARS-CoV-2 was reduced in this cell line (Fig. 3B). Importantly, estrogen (17β-estradiol) treatment also promoted a reduction in viral load (corroborating our previous data in Lemes et al. (2021) (Lemes et al., 2021). Additionally, when both treatments were associated in the presence of G15 (GPER1 antagonist), this protective effect (reduction in viral load) was reversed (Fig. 3B and C), thus supporting the hypothesis that GPER1 can be adjusting the cellular response against coronavirus. Our results showed that 17β-estradiol and G1 treatment led to the upregulation of GPER1 (Fig. 3D), but it is not known how GPER1 overexpression would be acting. A possible explanation is that this receptor may be translocated to the cell membrane, which could prevent the virus entry or replicate through the activation of the rapid intracellular signalling pathways, such as via EGFR (epidermal growth factor receptor) signaling or the PI3K-Akt pathway (Prossnitz and Barton, 2009). Finally, here we showed that in cells overexpressing GPER1, 17β-estradiol treatment further reduced SARS-CoV-2 viral load compared to control group (and this effect was blocked by antagonist G15) (Fig. 3F), suggesting that both the presence of GPER1 receptors and its functionality are important to reduce virus infection or replication in BEAS-2B cells.
5. Conclusion
Overall, the collective data suggest that GPER1 has a crucial role in SARS-CoV-2 infection in vitro, and additional studies in other cell lines, animal models and translational approaches should be further investigated to unravel the potentially beneficial role of estrogen in COVID-19.
Funding
This work was supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo - FAPESP: 2016/20796–2 (RPU), 2020/04709–8 (RPU), 2018/06088–0 (CMP), 2020/13480–4 (CMP), 2020/06153–7 (RMRL), 2006/60402–1 (RMB), 2019/10922–9 (RSS), 2018/02762–9 (RBO); Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - CAPES: code 001 (AJC, CSB, TAN); Conselho Nacional de Desenvolvimento Científico e Tecnológico - CNPq: 303035/2018–8 (CMP); FAP-FCMSCSP 2019/2021.
CRediT authorship contribution statement
Angelica Jardim Costa: Conceptualization, Methodology, Data curation, Investigation, Validation, Writing – review & editing. Robertha Mariana Rodrigues Lemes: Investigation, Visualization, Formal analysis, Data curation, Validation, Writing – original draft. Cynthia Silva Bartolomeo: Investigation, Writing – review & editing. Tamires Alves Nunes: Investigation, Writing – review & editing. Gabriela Cruz Pereira: Investigation, Writing – review & editing. Rafaela Brito Oliveira: Investigation, Validation, Writing – review & editing. Alexandre Lopes Gomes: Investigation, Writing – review & editing. Soraya Soubhi Smaili: Resources, Visualization, Writing – review & editing. Rui Monteiro de Barros Maciel: Resources, Visualization, Writing – review & editing. Louise Newson: Visualization, Writing – review & editing. Ana Lopez Ramirez: Visualization, Writing – review & editing. Liria Hiromi Okuda: Resources, Visualization, Writing – review & editing. Carla Máximo Prado: Conceptualization, Methodology, Validation, Formal analysis, Writing – review & editing, Visualization, Supervision. Roberta Sessa Stilhano: Conceptualization, Methodology, Validation, Formal analysis, Writing – review & editing, Visualization, Supervision. Rodrigo Portes Ureshino: Conceptualization, Methodology, Validation, Formal analysis, Data curation, Writing – original draft, Visualization, Supervision, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare that there are no conflicts of interest in this article.
Acknowledgements
The authors thank to Ilda Kunii and Angela Faria for technical assistance.
Data availability
Data will be made available on request.
References
- Al-Kuraishy H.M., Al-Gareeb A.I., Faidah H., Al-Maiahy T.J., Cruz-Martins N., Batiha G.E. The looming effects of estrogen in Covid-19: a rocky rollout. Front. Nutr. 2021;8 doi: 10.3389/fnut.2021.649128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breithaupt-Faloppa A.C., Correia C.J., Prado C.M., Stilhano R.S., Ureshino R.P., Moreira L.F.P. 17beta-Estradiol, a potential ally to alleviate SARS-CoV-2 infection. Clinics. 2020;75 doi: 10.6061/clinics/2020/e1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Fett C., Mack M., Ten Eyck P.P., Meyerholz D.K., Perlman S. Sex-based differences in susceptibility to severe acute respiratory syndrome coronavirus infection. J. Immunol. 2017;198:4046–4053. doi: 10.4049/jimmunol.1601896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brunink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couse J.F., Korach K.S. Estrogen receptor null mice: what have we learned and where will they lead us? Endocr. Rev. 1999;20:358–417. doi: 10.1210/edrv.20.3.0370. [DOI] [PubMed] [Google Scholar]
- Cugola F.R., Fernandes I.R., Russo F.B., Freitas B.C., Dias J.L., Guimaraes K.P., Benazzato C., Almeida N., Pignatari G.C., Romero S., Polonio C.M., Cunha I., Freitas C.L., Brandao W.N., Rossato C., Andrade D.G., Faria Dde P., Garcez A.T., Buchpigel C.A., Braconi C.T., Mendes E., Sall A.A., Zanotto P.M., Peron J.P., Muotri A.R., Beltrao-Braga P.C. The Brazilian Zika virus strain causes birth defects in experimental models. Nature. 2016;534:267–271. doi: 10.1038/nature18296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filardo E.J., Quinn J.A., Frackelton A.R., Jr., Bland K.I. Estrogen action via the G protein-coupled receptor, GPR30: stimulation of adenylyl cyclase and cAMP-mediated attenuation of the epidermal growth factor receptor-to-MAPK signaling axis. Mol. Endocrinol. 2002;16:70–84. doi: 10.1210/mend.16.1.0758. [DOI] [PubMed] [Google Scholar]
- Fuentes N., Nicoleau M., Cabello N., Montes D., Zomorodi N., Chroneos Z.C., Silveyra P. 17beta-Estradiol affects lung function and inflammation following ozone exposure in a sex-specific manner. Am. J. Physiol. Lung Cell Mol. Physiol. 2019;317:L702–L716. doi: 10.1152/ajplung.00176.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabutti G., d'Anchera E., Sandri F., Savio M., Stefanati A. Coronavirus: update related to the Current outbreak of COVID-19. Infect. Dis. Ther. 2020:1–13. doi: 10.1007/s40121-020-00295-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan C.S., Wei L.G., Xie R.M., Lv Z.B., Yan S., Zhang Z.X., Chen B.D. CT findings of COVID-19 in follow-up: comparison between progression and recovery. Diagn Interv Radiol. 2020;26:301–307. doi: 10.5152/dir.2019.20176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding A.T., Goff M.A., Froggatt H.M., Lim J.K., Heaton N.S. GPER1 is required to protect fetal health from maternal inflammation. Science. 2021;371:271–276. doi: 10.1126/science.aba9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280 e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalidhindi R.S.R., Ambhore N.S., Bhallamudi S., Loganathan J., Sathish V. Role of estrogen receptors alpha and beta in a murine model of asthma: exacerbated airway hyperresponsiveness and remodeling in ERbeta knockout mice. Front. Pharmacol. 2019;10:1499. doi: 10.3389/fphar.2019.01499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalidhindi R.S.R., Borkar N.A., Ambhore N.S., Pabelick C.M., Prakash Y.S., Sathish V. Sex steroids skew ACE2 expression in human airway: a contributing factor to sex differences in COVID-19? Am. J. Physiol. Lung Cell Mol. Physiol. 2020;319:L843–L847. doi: 10.1152/ajplung.00391.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach D.A., Brooke G.N., Bevan C.L. Roles of steroid receptors in the lung and COVID-19. Essays Biochem. 2021;65:1025–1038. doi: 10.1042/EBC20210005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemes R.M.R., Costa A.J., Bartolomeo C.S., Bassani T.B., Nishino M.S., Pereira G., Smaili S.S., Maciel R.M.B., Braconi C.T., da Cruz E.F., Ramirez A.L., Maricatto J.T., Janini L.M.R., Prado C.M., Stilhano R.S., Ureshino R.P. 17beta-estradiol reduces SARS-CoV-2 infection in vitro. Phys. Rep. 2021;9 doi: 10.14814/phy2.14707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Zhang C., Sui J., Kuhn J.H., Moore M.J., Luo S., Wong S.K., Huang I.C., Xu K., Vasilieva N., Murakami A., He Y., Marasco W.A., Guan Y., Choe H., Farzan M. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 2005;24:1634–1643. doi: 10.1038/sj.emboj.7600640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L.Q., Huang T., Wang Y.Q., Wang Z.P., Liang Y., Huang T.B., Zhang H.Y., Sun W., Wang Y. COVID-19 patients' clinical characteristics, discharge rate, and fatality rate of meta-analysis. J. Med. Virol. 2020;92:577–583. doi: 10.1002/jmv.25757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Massaro D., Massaro G.D. Estrogen receptor regulation of pulmonary alveolar dimensions: alveolar sexual dimorphism in mice. Am. J. Physiol. Lung Cell Mol. Physiol. 2006;290:L866–L870. doi: 10.1152/ajplung.00396.2005. [DOI] [PubMed] [Google Scholar]
- Massaro D., Clerch L.B., Massaro G.D. Estrogen receptor-alpha regulates pulmonary alveolar loss and regeneration in female mice: morphometric and gene expression studies. Am. J. Physiol. Lung Cell Mol. Physiol. 2007;293:L222–L228. doi: 10.1152/ajplung.00384.2006. [DOI] [PubMed] [Google Scholar]
- Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J., Hlh Across Speciality Collaboration U.K. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millas I., Duarte Barros M. Estrogen receptors and their roles in the immune and respiratory systems. Anat. Rec. 2021;304:1185–1193. doi: 10.1002/ar.24612. [DOI] [PubMed] [Google Scholar]
- Newson L., Manyonda I., L R., P R., P S., Seeland U. Sensitive to infection but strong in defense—female sex and the power of oestradiol in the COVID-19 pandemic. Frontiers in Global Women’s Health. 2021;2:651752. doi: 10.3389/fgwh.2021.651752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Lone R., Frith M.C., Karlsson E.K., Hansen U. Genomic targets of nuclear estrogen receptors. Mol. Endocrinol. 2004;18:1859–1875. doi: 10.1210/me.2003-0044. [DOI] [PubMed] [Google Scholar]
- Pisolato R., Lombardi A.P., Vicente C.M., Lucas T.F., Lazari M.F., Porto C.S. Expression and regulation of the estrogen receptors in PC-3 human prostate cancer cells. Steroids. 2016;107:74–86. doi: 10.1016/j.steroids.2015.12.021. [DOI] [PubMed] [Google Scholar]
- Poyiadji N., Cormier P., Patel P.Y., Hadied M.O., Bhargava P., Khanna K., Nadig J., Keimig T., Spizarny D., Reeser N., Klochko C., Peterson E.L., Song T. Acute pulmonary embolism and COVID-19. Radiology. 2020;297:E335–E338. doi: 10.1148/radiol.2020201955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prossnitz E.R., Barton M. Signaling, physiological functions and clinical relevance of the G protein-coupled estrogen receptor GPER. Prostag. Other Lipid Mediat. 2009;89:89–97. doi: 10.1016/j.prostaglandins.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revankar C.M., Cimino D.F., Sklar L.A., Arterburn J.B., Prossnitz E.R. A transmembrane intracellular estrogen receptor mediates rapid cell signaling. Science. 2005;307:1625–1630. doi: 10.1126/science.1106943. [DOI] [PubMed] [Google Scholar]
- Scialpi M., Scialpi S., Piscioli I., Battista Scalera G., Longo F. Pulmonary thromboembolism in critical ill COVID-19 patients. Int. J. Infect. Dis. 2020;95:361–362. doi: 10.1016/j.ijid.2020.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully E.P., Haverfield J., Ursin R.L., Tannenbaum C., Klein S.L. Considering how biological sex impacts immune responses and COVID-19 outcomes. Nat. Rev. Immunol. 2020;20:442–447. doi: 10.1038/s41577-020-0348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeland U., Coluzzi F., Simmaco M., Mura C., Bourne P.E., Heiland M., Preissner R., Preissner S. Evidence for treatment with estradiol for women with SARS-CoV-2 infection. BMC Med. 2020;18:369. doi: 10.1186/s12916-020-01851-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma G., Volgman A.S., Michos E.D. Sex differences in mortality from COVID-19 pandemic: are men vulnerable and women protected? JACC Case Rep. 2020;2:1407–1410. doi: 10.1016/j.jaccas.2020.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzig K.E., Canepa-Escaro F., Schiliro M., Berdnikovs S., Prakash Y.S., Chiarella S.E. Estrogen regulates the expression of SARS-CoV-2 receptor ACE2 in differentiated airway epithelial cells. Am. J. Physiol. Lung Cell Mol. Physiol. 2020;318:L1280–L1281. doi: 10.1152/ajplung.00153.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stilhano R.S., Costa A.J., Nishino M.S., Shams S., Bartolomeo C.S., Breithaupt-Faloppa A.C., Silva E.A., Ramirez A.L., Prado C.M., Ureshino R.P. SARS-CoV-2 and the possible connection to ERs, ACE2, and RAGE: focus on susceptibility factors. Faseb. J. 2020;34:14103–14119. doi: 10.1096/fj.202001394RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suba Z. Prevention and therapy of COVID-19 via exogenous estrogen treatment for both male and female patients. J. Pharm. Pharmaceut. Sci. 2020;23:75–85. doi: 10.18433/jpps31069. [DOI] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 Spike glycoprotein. Cell. 2020;181:281–292 e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Sun X., J L.V., Kon N.D., Ferrario C.M., Groban L. Estrogen receptors are linked to angiotensin-converting enzyme 2 (ACE2), ADAM metallopeptidase domain 17 (ADAM-17), and transmembrane protease serine 2 (TMPRSS2) expression in the human atrium: insights into COVID-19. Hypertens. Res. 2021 doi: 10.1038/s41440-021-00626-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.