Abstract
Sex determination in dioecious plants has been broadly and progressively studied with the blooming of genome sequencing and editing techniques. This provides us with a great opportunity to explore the evolution and genetic mechanisms underlining the sex-determining system in dioecious plants. In this study, comprehensively reviewing advances in sex-chromosomes, sex-determining genes, and floral MADS-box genes in dioecious plants, we proposed a convergent model that governs plant dioecy across divergent species using a cascade regulation pathway connecting sex-determining genes and MADS-box genes e.g., B-class genes. We believe that this convergent mechanism of sex determination in dioecious plants will shed light on our understanding of gene regulation and evolution of plant dioecy. Perspectives concerning the evolutionary pathway of plant dioecy are also suggested.
Keywords: plant dioecy, sex determination, sex-determining genes, MADS-box genes, convergent mechanism
Introduction
Dioecious plants have their male and female sexes individually separated. In the plant kingdom, just a small minority (5%−6%) of species are dioecious, distinct from the animal kingdom where gonochorism is common (Charlesworth, 2016). A large portion of these dioecious plants is commercially important including poplar (Populus spp.), kiwifruit (Actinidia spp.), garden asparagus (Asparagus officinalis L.), papaya (Carica papaya L.), spinach (Spinacia oleracea L.), hemp (Cannabis sativa L.), hardy rubber tree (Eucommia ulmoides Oliver), and pistachio (Pistacia vera L.). Notably, some widely-cultivated crops such as hexaploid persimmon (Diospyros kaki Thunb.), grape (Vitis vinifera L.), and strawberry (Fragaria × ananassa Duch.) are domesticated from their close dioecious relatives (Henry et al., 2018). Dioecy appears in various plant lineages, though less frequently. This provides us with a great opportunity to explore the potential convergent mechanisms in these plants (Leite Montalvão et al., 2021).
Two morphological types of unisexual flowers have been recognized since the Darwinian era in dioecious plants (Darwin, 1877; Jabbour et al., 2022). The first one, the Type I flower, is unisexual by abortion, which is bisexual at its very early stages but becomes unisexual following the developmental arrest of stamen or carpel, e.g., Diospyros lotus L. (Figure 1A). The second one, the Type II flower, is unisexual from inception, with only the primordium of stamen or carpel at separate floral meristem (Mitchell and Diggle, 2005; Diggle et al., 2011), e.g., Populus deltoids cv. “Zhonghuahongye” (Figure 1B). Interestingly, Type I flowers are more frequently observed in dioecious plants in comparison with Type II flowers.
Figure 1.
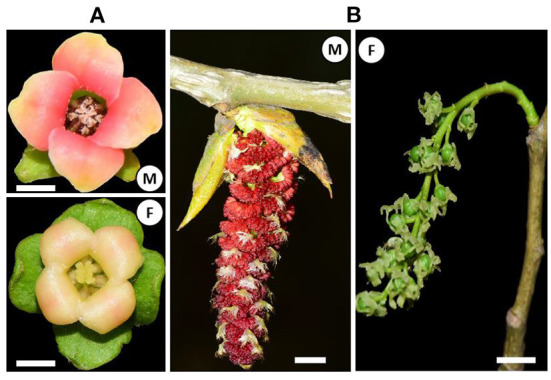
Unisexual flowers of Diospyros lotus (Type I flower) (A) and Populus deltoides (Type II flower) (B). M, male flower; F, female flower. Scale bar = 1 cm.
The rapid development of high-throughput sequencing techniques has enabled a full flourish of plant genomics. Sex-determining genes have been identified based on combined genomic and experimental approaches in certain dioecious plants with both Type I and II flowers (Carey et al., 2021; Renner and Müller, 2022). Taken together, these findings make it possible for us to elucidate the mechanism of sex determination in dioecious plants. Studies on poplar (Type II flower) (Leite Montalvão et al., 2022) and persimmon (Type I flower) (Yang et al., 2019) particularly indicate insightful clues on how the sex-determining genes are connected to the dioecious phenotype through the floral MADS-box genes. Impressively, although with distinct flowers, both studies revealed that the floral B-class genes are the downstream target of sex-determining genes (Theißen et al., 2016). This leads us to further explore the potential connection between the sex-determining genes and floral development genes and how they ultimately determine plant dioecy.
Identification of the sex-determining region (SDR) on sex chromosomes
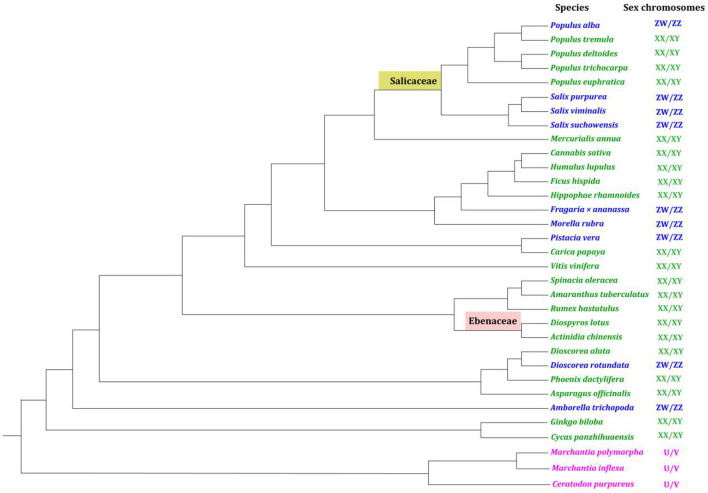
Over fifty dioecious plant species have been sequenced since 2006 when the first dioecious genome (Populus trichocarpa Hook.) was published (Tuskan et al., 2006). But in these sequenced dioecious plants, only 33 species in 22 families (Supplementary Table S1) were found to have sex chromosomes (Ming et al., 2011; Baránková et al., 2020; Leite Montalvão et al., 2021; Charlesworth, 2022) to our best knowledge. The 33 dioecious plants with available genomes and sexual systems were summarized in a phylogenetic perspective (Figure 2, Supplementary Table S1). In general, three kinds of sex systems, i.e., male-heterogametic system, female-heterogametic system, and haploid sex system have been found in dioecious plants by far, corresponding to XX/XY, ZW/ZZ, and U/V sex chromosomes respectively. The number of species with XX/XY system (21) is greater than species with ZW/ZZ system (9) and U/V system (3) (Figure 2).
Figure 2.
Dioecious plants with reference genomes and convinced sexual systems in a phylogenetic perspective. The male heterogametic system (XY/XX), female heterogametic system (ZW/ZZ), and haploid sex system (U/V) are shown in green, blue, and pink respectively. The distantly-related lineages of poplar (Salicaceae) and persimmon (Ebenaceae) are respectively highlighted in yellow and light red. The phylogenetic tree was reconstructed using chloroplast genome sequences via the online RAxML method in CIPRES (http://www.phylo.org/) (Supplementary Table S1).
In the present genomic era, deep sequencing and high-quality assembly of sex chromosomes are undoubtedly critical to reliably identifying the non-recombining sex-determining region (SDR). There are three stages in deciphering SDR of dioecious plants that have been undertaken: (i) Traditional molecular techniques stage: genetic linkage mapping and bacterial artificial chromosome (BAC) were employed to overcome the encountered difficulties, for example, when assembling the Y chromosome of papaya in 2004 (Liu et al., 2004). However, these methodologies are either labor-intensive or hard to obtain the linkage map, particularly for species with long juvenile periods e.g., E. ulmoides (Wang et al., 2020). Moreover, extra small- or large-sized SDR, such as ~59 kb in Morella rubra Lour. (Jia et al., 2019) and ~50 Mb in Ginkgo biloba L. (Gong and Filatov, 2022) were also difficult to be reliably identified or orderly aligned based on genetic linkage mapping and BAC approaches. (ii) Short-read sequencing stage: The advent of high throughput sequencing techniques that generate an enormous amount of short reads (100–300 bp) boosted the rapid development of dioecious plant genomics. Although the output of the short sequencing reads is extremely high, for instance, Illumina HiSeq 4000 platform can generate up to >1 Tb per run, it remains problematic. On the one hand, the overall sequencing quality of short reads needs to be improved, more importantly, on the other, short reads fail to span certain genomic territories such as repetitive regions (e.g., transposable elements and tandem repeats) which would normally accumulate on SDR of the sex chromosomes (Li et al., 2016; Muyle et al., 2017). It is thus understandable that both X and Y (or Z and W) sex chromosomes were either largely absent or partially assembled as a chimera in some of the published dioecious genomes (Supplementary Table S1 and Figure 2). (iii) Long-read sequencing stage: Long-read sequencing techniques including Nanopore, PacBio, and Hi-C chromatin conformation techniques that produce long or ultra-long reads (20–100 kb) largely remedied the deficiency given by short-read sequencing, which can bridge up to 15–30 kb genomic territories (Carey et al., 2021). Contigs of X and Y chromosomes of Ficus hispida L. were primarily assembled using Pacbio long reads (~20 kb) before the fineX (21.9 Mb) and Y (22.6 Mb) chromosomes were fully constructed with Hi-C reads (500–700 bp) (Zhang et al., 2020). The SDR of Cycas panzhihuaensis L. Zhou and S. Y. Yang were configured based on Nanopore long-reads and Hi-C data, giving rise to a 45.5 Mb-sized Y haplotype (Liu et al., 2022).
Many of the genome-sequenced dioecious plants have only one sex, i.e., either male or female, selected for sequencing due to technique or budget issues leaving their sex chromosomes not covered by genomic data (Baránková et al., 2020) though experimental data may be available. Nevertheless, we need to be open to the varied sequencing techniques and/or assembly methodologies to tackle practical problems in genomics studies of dioecious plants. A T2T gapless human X chromosome (Miga et al., 2020) and highly continuous human Y chromosome (Kuderna et al., 2019) have been newly constructed by long-read sequencing data. This may be also the bright direction to decipher the intractable sex chromosomes of dioecious plant species in the near future. In particular, deciphering the haplotype-phased genome of a heterogametic individual, such as the male with XY sex chromosomes in the male-heterogametic system and/or the female with ZW sex chromosomes in the female-heterogametic system, will accelerate the identification of sex-determining genes in dioecious plants.
Divergent sex-determining genes
Two-gene model
The “two-gene model” (Westergaard, 1958; Charlesworth and Charlesworth, 1978) was first proposed to explain the evolution of dioecy from a hermaphrodite ancestor. In this model, a recessive male-sterility mutation in the male-promoting factor (M) initially results in a gynodioecious population i.e., co-existence of females and hermaphrodites. Then, a dominant female sterility mutation occurs in the suppressor of femaleness (SuF). Both M and SuF locate at the SDR, leading to dioecy. This model was confirmed by practical evidence found in kiwifruit (Actinidia spp.) and garden asparagus (Actinidia officinalis) (Table 1). Most species of Actinidia are dioecious, among which Actinidia chinensis Planch is a worldwide well-known fruit crop with XX/XY chromosomes (Wang et al., 2016). It has a small-sized (~0.8 Mb) SDR housing two male-specific expressed genes, i.e., SyGI and FrBy (Akagi et al., 2019). Gene SyGI (“Shy Girl”) is a type-C cytokinin response regulator. Transgenic expression of SyGI in Arabidopsis thaliana (L.) Heynh. and Nicotiana tabacum L. resulted in suppression of carpel development but no effect on stamen (Akagi et al., 2018). Recent work using the CRISPR/Cas9 gene editing technique to mutagenize SyGl in A. chinensis male plants gave rise to hermaphrodites with functional gynoecium and androecium, validating its identity of SuF (Varkonyi-Gasic et al., 2021). The second gene FrBy (Friendly Boy) is critical for viable pollens formation, functioning as an M factor. Transgenic expression of FrBy converted A. chinensis females into hermaphroditic individuals (Akagi et al., 2019).
Table 1.
Sex-determining genes and/or strong candidates identified in dioecious plants.
| Type of unisexual flowers | Species (family) | Sex system | Sex-determining genes and function | References |
|---|---|---|---|---|
| Type I flowers: Unisexual by abortion | Actinidia spp. *(Actinidiaceae) | XX/XY | SyGI: Female suppressor FrBy: Male activator | Akagi et al. (2018, 2019) |
| Asparagus officinalis *(Asparagaceae) | XX/XY | SOFF: Female suppressor aspTDF1: Male activator | Harkess et al. (2017, 2020) | |
| Carica papaya (Caricaceae) | XX/XY | CpSVP: MADS-box gene, unknown function CpSERK and CpCAF1AL, unknown function | Urasaki et al. (2012), Lee et al. (2018) |
|
| Diospyros lotus *(Ebenaceae) | XX/XY | OGI: Male activator | Akagi et al. (2014, 2020) | |
| Ficus hispida (Moraceae) | XX/XY | FhAG2: MADS-box gene, putative male activator | Zhang et al. (2020) | |
| Fragaria virginiana F. chiloensis (Rosaceae) | ZW/ZZ | GMEW: unknown function RPP0W: unknown function | Tennessen et al. (2018) | |
| Nepenthes gracilis N. rafflesiana N. pervillei (Nepenthaceae) | XX/XY | DYT1: Putative male activator SEP1: MADS-box gene, unknown function | Scharmann et al. (2019) | |
| Phoenix spp. (Arecaceae) | XX/XY | CYP703 and GPAT3: Putative male activator LOG-like: Putative female suppressor | Torres et al. (2018) | |
| Silene latifolia (Caryophyllaceae) | XX/XY | SlAP3: MADS-box gene, unknown function SlSTM: unknown function SlCUC: unknown function | Zluvova et al. (2006); Cegan et al. (2010) | |
| Vitis vinifera * (Vitaceae) | XX/XY | VviINP1: Putative male activator VviPLATZ1: Female activator | Massonnet et al. (2020), Iocco-Corena et al. (2021) | |
| Type II flowers: Unisexual from inception |
Cycas panzhihuaensis (Cycadaceae) | XX/XY | CYCAS_034085: MADS-box gene, unknown function | Liu et al. (2022) |
| Ginkgo biloba (Ginkgoaceae) | XX/XY | GbMADS18: MADS-box gene, unknown function Gb_15883: unknown function Gb_15884: unknown function Gb_15885: unknown function Gb_15886: unknown function Gb_28587: MADS-box gene, unknown function | Zhang et al. (2019); Liao et al. (2020) | |
| Morella rubra (Myricaceae) | ZW/ZZ | MrCPS2: unknown function MrASP2: unknown function | Jia et al. (2019) | |
| Populus alba* (Salicaceae) | ZW/ZZ | ARR17: Female activator | Müller et al. (2020) | |
| Populus tremula*P. trichocarpa* (Salicaceae) | XX/XY | ΨARR17-IR : Female suppressor | Müller et al. (2020) | |
| Populus deltoids* (Salicaceae) | XX/XY | FERR-R: Female suppressor MSL: Male activator | Xue et al. (2020) | |
| Populus euphratica (Salicaceae) | XX/XY | ΨRR-IR: Putative female suppressor | Yang et al. (2021) | |
| Salix purpurea (Salicaceae) | ZW/ZZ | SpRR9: Putative female activator | Zhou et al. (2020a) | |
| Spinacia oleracea (Amaranthaceae) | XX/XY | NRT1/PTR6.4: Putative male activator | Ma et al. (2022) |
Indicates the sex-determining genes of these species have been functionally studied.
Garden asparagus (A. officinalis, Asparagaceae) also has XX/XY sex chromosomes and a similar small-sized SDR (~1 Mb) as seen in kiwifruit (Harkess et al., 2017). Two SDR-encoded genes, i.e., SOFF (Suppressor of Female Function) and aspTDF1 (Defective in Tapetum Development and Function 1) expressed specifically in males (Harkess et al., 2020). Knockout of SOFF by gamma irradiation converted males into hermaphrodites. Additionally, mutagenesis of aspTDF1 in males by Ethyl Methanesulfonate (EMS) resulted in asexual neuters where neither carpels nor stamens developed normally. When both SOFF and aspTDF1 genes underwent knockout, males directly changed into females. All these findings provided robust evidence that the two Y-encoded genes SOFF (SuF factor) and aspTDF1 (M factor) were physically linked at SDR to co-determine the sex of the garden asparagus. Most notably, the male-promoting genes in kiwifruit (FrBy) and asparagus (aspTDF1) both function in tapetum for male flower fertility (Andreuzza, 2020), suggesting a convergent mechanism.
No direct experimental data were available for the identified sex-determining candidates in grape (V. vinifera) and date palm (Phoenix dactylifera L.), but the “two-gene model” can most likely be applied to them (Table 1). In grape, two SDR-encoded genes might be potential sex-determining genes: VviINP1 likely participated in pollen fertility and acted as an M factor (Massonnet et al., 2020; Zou et al., 2021), and VviPLATZ1 could suppress carpel development and most likely functioned as SuF factor (Iocco-Corena et al., 2021). In 13 Phoenix species, three genes (CYP703, GPAT3, and LOG-like) that were specifically expressed in male flowers were determined as strong sex-determining candidates (Torres et al., 2018). CYP703 and GPAT3 might affect pollen development while LOG-like genes likely suppressed carpel development (Torres et al., 2018).
One-gene model
In contrast, sex determination of several other dioecious plants (Table 1) is known to follow the “one-gene model” that was firstly reported in persimmon (Diospyros, Ebenaceae) (Akagi et al., 2014; Charlesworth, 2016; Renner and Müller, 2022). In this model, one single gene located at SDR is critical to determine the sex via an epistatic gene interaction, rather than physical linkage with downstream genes. The SDR (~1 Mb) of Caucasian persimmon (D. lotus) contained one pseudogene OGI (Oppressor of MeGI) which exhibited male-specific expression. The OGI had an autosome-located duplicate gene, MeGI (Male Growth Inhibitor) which showed female-biased expression in floral buds. Importantly, OGI housed two identical inverted repeat sequences (IRs) which generated hairpin RNAs and 21-nt small RNAs (sRNAs) to return silence MeGI via RNA interference (RNAi) (Akagi et al., 2014, 2016, 2020; Akagi and Charlesworth, 2019). MeGI could repress androecium development, causing femaleness in transgenic A. thaliana, Nicotiana benthamiana Domin, and N. tabacum. When OGI was introduced in the MeGI transgenic plants, male sterility was restored. These results demonstrated that OGI functioned alone as the sex-determining gene in persimmon (Akagi et al., 2014; Akagi and Charlesworth, 2019).
Nearly all the Populus species in Salicaceae are dioecious (Yang et al., 2021) possessing both XX/XY and ZW/ZZ sexual systems, which makes it a model group for the study of plant sex determination (Cronk and Müller, 2020). Geraldes and colleagues (2015) (Geraldes et al., 2015) firstly performed whole-genome re-sequencing of P. trichocarpa and conducted a genome-wide association study (GWAS). They consequently revealed an XX/XY system presence in species belonging to both section Tacamahaca (P. trichocarpa and Populus balsamifera L.) and Aigeiros (Populus deltoids Marshall and Populus nigra L.). A small-sized (~100 Kb) SDR was identified in the Y chromosome (Chr. 19) of P. trichocarpa. It contained 12 protein-coding genes, including A. thaliana orthologs ARR17 (Arabidopsis Response Regulator 17) and MET1 (Methyltransferase 1). Subsequent studies on P. balsamifera further revealed that ARR17 probably played a direct role in poplar sex determination (Bräutigam et al., 2017; Sanderson et al., 2019; Cronk et al., 2020; Zhou et al., 2020b). These aforementioned studies however lacked experimental data to evidence the biological function of ARR17 in Populus. Only recently, Müller et al. (2020) demonstrated the feminizing function of ARR17 by CRISPR/Cas9-induced mutations in Populus tremula L. (Müller et al., 2020). ARR17 in Populus is also a cytokinin-related gene, similar to the above-mentioned MeGI gene of Diospyros (Akagi et al., 2020). CRISPR/Cas9 knockout of ARR17 converted female P. tremula into males but made the males unchanged (Müller et al., 2020). Another group (Xue et al., 2020) independently performed the transgenic analyses of ARR17 from P. deltoids (here named FERR) (Xue et al., 2020). They revealed stigma exaggeration and carpel-like sepal phenotypes in transgenic plants of A. thaliana, confirming the feminizing function of ARR17 (Xue et al., 2020). Interestingly, the ARR17 gene itself was found in the pseudo autosomal region of P. deltoids and P. tremula, but not in the SDR (Müller et al., 2020; Xue et al., 2020). Instead, a Y-specific pseudogene of ARR17 (called ΨARR17-IR) was seated at the SDR of P. deltoids and P. tremula (Müller et al., 2020; Xue et al., 2020). Significantly, as a partial duplicate of ARR17, ΨARR17-IR could produce 24-nt sRNAs to silence ARR17 reversely through RNA-directed DNA methylation (RdDM) (Müller et al., 2020; Xue et al., 2020).
The “one gene model” has been applied to several other dioecious plants beyond Diospyros and Populus (Table 1). For example, Salix purpurea L. (Zhou et al., 2020a) and Salix viminalis L. (Almeida et al., 2020) carried homologs of ARR17 (SpRR9) at the SDR of the W chromosome (Zhou et al., 2020a). F. hispida had an SDR-located FhAG2 gene which was identified as the putative sex-determining gene (Zhang et al., 2020). Spinacia oleracea contained a Y-specific gene NRT1/PTR6.4 that was considered to be a strong candidate for sex determination (Ma et al., 2022). More recently the gymnosperm species C. panzhihuaensis was reported to have one SDR-located sex-determining candidate CYCAS_034085 (Liu et al., 2022). It has been proposed that the evolution of dioecy from monoecy likely preferred the flexible “one-gene model,” whereas that from hermaphrodite might be accomplished by the “two-gene model” (Cronk, 2022). Regardless of the number of genes, both one- and two-gene models include the genetic connection of two independent genes. The difference rests on that whether the two sex-determining genes are physically linked together at the SDR of the Y (or W) chromosome (two-gene model), as FrBy/SyGl in kiwifruit (Akagi et al., 2019) and SOFF/aspTDF1 in garden asparagus (Harkess et al., 2020), or the SDR-located sex-determining gene epistatically regulates its (pseudo)autosome-located homologous counterpart (one-gene model), as OGI/MeGI in persimmon (Akagi et al., 2014, 2020; Akagi and Charlesworth, 2019) and ΨARR17-IR/ARR17 in poplar (Müller et al., 2020; Xue et al., 2020; Yang et al., 2021).
Floral MADS-box genes regulated by sex-determining genes
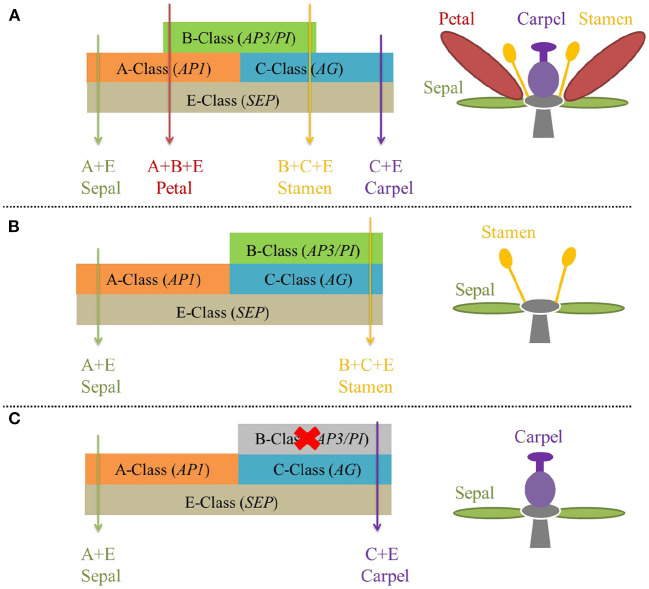
Floral MADS-box genes are well known for their significant role in flower development according to the extended ABCDE model of perfect flower development (Theißen, 2001). The four whorls of perfect flowers from outside to inside i.e., sepal, petal, stamen, and carpel are specified by class A + E, A + B + E, B + C + E, and C + E genes respectively in the ABCDE model (Figure 3A) (Theißen, 2001). The frequently seen A-class gene AP1 (APETALA1), B-class genes AP3 (APETALA3) and PI (PISTILLATA), and C-class gene AG (AGAMOUS) were first discovered in A. thaliana, the model plant (Theißen et al., 2016). Notably, the spatial expression domains of the floral MADS-box genes are flexible (Soltis et al., 2007). For example, the expression of B-class genes has been expanded to whorl-1 i.e., covering the A-class domain in basal eudicot Aquilegia (Kramer et al., 2007) and monocot Liliaceae (Kanno et al., 2003), leading to petal-like sepals. The B-class gene QsPI was exclusively expressed in male flowers of Quercus suber L. and QsPI transgene in pi-1 mutant A. thaliana successfully rescued the stamen function (Sobral and Costa, 2017). Therefore, when the expression domain of B- and C-class genes are overlapped, unisexual flowers e.g., male (Figure 3B) or female (Figure 3C) develop via switching on or off B-class genes (AP3 and PI) (Soltis et al., 2007; Shan et al., 2019; Jabbour et al., 2022).
Figure 3.
The “ABCDE model” of flower development, with D genes that specify ovule unshown. (A) B-class gene's normal expression makes perfect flowers. (B) Shifted and expanded expression domain of B-class genes permits male flowers. (C) Repression of B-class genes gives rise to female flowers.
Floral MADS-box genes played key roles in the unisexual flower development of dioecious plants (Jabbour et al., 2022). For instance, B-class genes SpAP3 and SpPI were found to express specifically in male floral primordial of S. oleracea, but absent in female flowers (Pfent et al., 2005). Subsequent functional analysis of B- (SpAP3/SpPI) and C-class (SpAG) genes documented that suppression of B-class expression converted male flowers into a female, indicating that regulation of B-class genes was associated with dioecy establishment in spinach (Sather et al., 2010). Similarly, B-class genes (ThdAP3-2a/b) of Thalictrum dioicum L. (Ranunculaceae) were known to be only expressed in male flowers but not in any stage of female flowers (Di Stilio et al., 2005). Besides, our group initially found one B-class gene, EuAP3, with male-biased expression in E. ulmoides (Wang and Zhang, 2017), which was confirmed by a different group working on the floral development of E. ulmoides (Zhu, 2019). This indicates that B-class genes were probable key sex regulatory factors of E. ulmoides (Wang et al., 2018, 2020). Moreover, an intact floral B-class gene SlAP3Y in Silene latifolia Poir. was found to be Y-specific and had an autosomal paralog SlAP3A (Matsunaga et al., 2003). Expression analysis revealed that SlAP3Y was highly expressed in stamens, but SlAP3A was specifically expressed in petals. This suggested that the B-class genes themselves might reside at the SDR of S. latifolia and act as masculinizing factors.
Recently, the regulatory pathway that bridges sex-determining genes and floral MADS-box genes were uncovered in Populus (Salicaceae) and Diospyros (Ebenaceae). In P. tremula, B-class gene PI was highly expressed in the arr17 CRISPR mutants (males), in contrast to its low expression level in the isogenic females (ARR17) (Leite Montalvão et al., 2022). Notably, the meristem identity gene UFO (Unusual Floral Organs) was simultaneously and highly derepressed in arr17 mutants. On the other hand, UFO encoded an F-box protein as a transcriptional cofactor with LFY (LEAFY) to activate the expression of B-class genes (Ng and Yanofsky, 2001; Theißen et al., 2016). Accordingly, it is probable that the genetic cascade of (ΨARR17-IR)-ARR17-UFO-PI controlled the sex of P. tremula (Leite Montalvão et al., 2022). In females, the normally expressed ARR17 gene repressed downstream UFO-PI module, giving rise to female flowers; whereas in males, Y-specific ΨARR17-IR generated a large number of sRNAs to silence ARR17, restoring the expression of UFO-PI to produce male flowers (Leite Montalvão et al., 2022). Interestingly, the female-biased expression of ARR17 was consistently correlated with the male-biased expression of AP3 and PI in P. balsamifera (Cronk et al., 2020), suggesting a conserved sex-determination network in poplar.
Likewise, in persimmon, the feminizing gene MeGI also negatively regulated the B-class gene PI (Yang et al., 2019). MeGI directly controlled the expression of SVP (Short Vegetative Phase) which is an upstream repressor of floral B- and C-class genes (Theißen et al., 2016). In diploid dioecious D. lotus, the stable expression of OGI caused accumulation of sRNAs which in return silenced MeGI, so the B-class genes could normally express to produce male flowers. On the contrary, when B-class genes were repressed by the upstream MeGI-SVP module, only female flowers developed (Yang et al., 2019). Additionally, the OGI-MeGI-SVP-PI regulation cascade was applicable in monoecious hexaploid persimmon (D. kaki). In D. kaki, a 268-bp SINE-like element inserted into the promoter region of the OGI gene repressed its expression and subsequently produced female flowers (Akagi et al., 2016). In short, the aforementioned studies give a broad hint that the B-class genes play significant roles in determining the sex of dioecious plants.
A convergent model underlying sex determination
Although a total of 21 and 16 sex-determining genes/candidates were identified in species with Type I and II flowers respectively (Table 1 and Figure 1), consensus gene sequences were lacking between phylogenetically divergent species that either share the same or have different flower types. Remarkably it seems that those divergent genes are functionally convergent, for example, a group of male-promoting e.g., FrBy in kiwifruit (Varkonyi-Gasic et al., 2021), aspTDF1 in garden asparagus (Harkess et al., 2020), CYP703 and GPAT3 in date palm (Torres et al., 2018), VviINP1 in grape (Massonnet et al., 2020), and DYT1 in nepenthes (Scharmann et al., 2019) are all involved in the pollen development. This is indicative of a potential convergent mechanism governing the sex determination of dioecious plants.
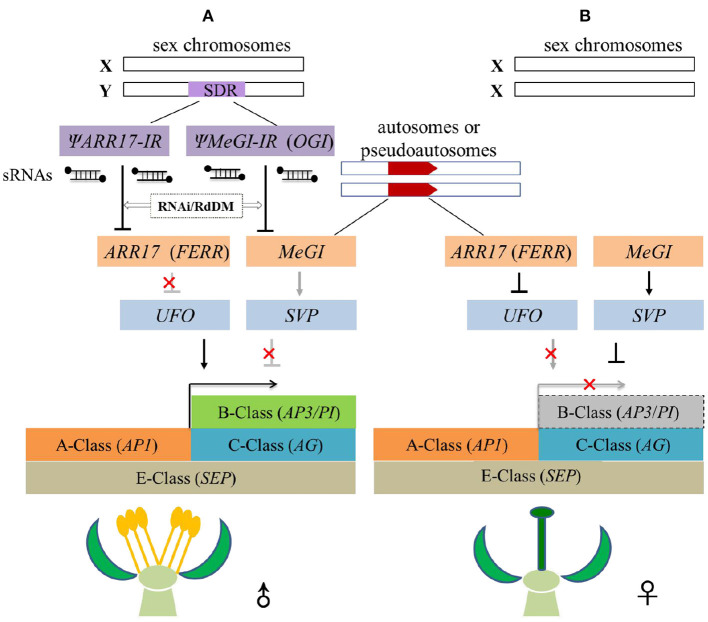
Persimmon (Ebenaceae) and poplar (Salicaceae) diverged ~150 million years ago and possess distinct unisexual flowers as described previously (Figure 1) (Diggle et al., 2011; Li et al., 2019), but they share similar sex-regulation mechanisms. On the one hand, sex determination systems in Populus and Diospyros both support the “one-gene model,” both involving a non-SDR-located sex-switch regulator (ARR17 in Populus, MeGI in Diospyros) that is toggled on off by its SDR-located pseudogene (ΨARR17-IR in Populus, OGI in Diospyros) on the Y chromosome. On the other hand, in both lineages, sex-determining genes ultimately function on B-class genes forming a regulatory cascade, i.e., (ΨARR17-IR)-ARR17-UFO-PI in Populus and OGI-MeGI-SVP-PI in Diospyros. Moreover, earlier work on S. latifolia (Matsunaga et al., 2003), S. oleracea (Pfent et al., 2005; Sather et al., 2010), T. dioicum (Di Stilio et al., 2005), and our current work on E. ulmoides (Wang and Zhang, 2017; Wang et al., 2018, 2020) all suggest the possibility of a universal convergent model that connects upstream sex-determining genes with downstream dioecious phenotypes (Figure 4). In this model, the spatial expression domain of B-class genes (AP3/PI) overlaps with the C-class gene (AG) and ultimately gets fixed at the population level. Only male flowers develop when B-class genes are normally expressed (Figure 4A), and only female flowers appear if B-class genes are repressed (Figure 4B).
Figure 4.
Convergent model of sex-regulation cascades from upstream sex-determining genes to downstream B-class genes in dioecious plants. (A) Duplicated pseudogenes (ΨARR17-IR or ΨMeGI-IR) in the SDR of the Y chromosome generate sRNAs that dominantly repress their counterpart feminizing factors (ARR17 or MeGI) on autosomes/pseudo autosomes via epigenetic RNAi/RdDM pathway. Downstream B-class genes are activated either by the expression of activator (UFO) or the suppression of repressor (SVP), producing only male flowers. (B) In XX individual, no pseudogenes of feminizing factors are available, repressor (SVP) of B-class genes are thus expressed, or activator (UFO) is repressed, and the expression of B-class genes are suppressed forming only female flowers.
Two steps are indispensable for the evolution of dioecy in this model (Figure 4). First, a feminizing mutation happens and gets fixed in the ancestral co-sexual (hermaphrodite or monoecy) populations. This feminizing mutation most likely occurs in the upstream negative regulators of B-class genes (AP3/PI), such as the ARR17 in poplar (Müller et al., 2020) or MeGI in persimmon (Akagi et al., 2014). The interaction between feminized factors and B-class genes may be indirect via intermediate processes, for example through floral meristem genes UFO (Leite Montalvão et al., 2022) and SVP (Yang et al., 2019). Second, an SDR-located masculinizing mutation arises to repress the feminized genes and co-segregates with sex. This masculinizing mutation probably occurs in paralogs of the feminized factors, such as the ΨARR17-IR in P. tremula and P. deltoids (Müller et al., 2020; Xue et al., 2020), and OGI (here in this model referred to as ΨMeGI-IR) in D. lotus (Akagi et al., 2020). The masculinized gene thus could silence its paralogous feminized gene in trans via epigenetic regulations e.g., RNAi or RdDM (Akagi et al., 2014, 2020; Müller et al., 2020; Xue et al., 2020). These two mutations eventually achieve to determine the floral sexuality by toggling on/off the expression of B class genes, resulting in dioecy (Figure 4).
Speculatively, this model could be further extended to dioecious species that follow the “one-gene model.” Recent genomics studies on spinach (Ma et al., 2022), cycad (Liu et al., 2022), and Banyan tree (Zhang et al., 2020) all identified only one SDR-located gene that showed a male-specific expression pattern (Table 1). Therefore, all these species probably follow the convergent model we proposed here. Indeed, the sex determination mechanism via a single SDR-located gene is relatively more flexible and efficient compared to the classical “two-gene model” (Cronk and Müller, 2020; Renner and Müller, 2021). For instance, the “one-gene model” explained well the phenomena of sexual system transition between XY/XX (P. tremula) and ZZ/ZW (P. alba) in a simple way (Müller et al., 2020). The plant “one-gene model” was usually thought of as reminiscent of the human sex-determining system that has SRY (Sex-Determining Region Y) gene acting alone as a sex determinant (Sinclair et al., 1990; Graves, 2016). This convergent model thus resembles the bottom-up regulatory model of animal sex determination (Feng et al., 2020). Both models suggested that the sex-determining genes might be divergent across different species, but the regulatory cascades eventually converged to a conserved downstream hub that directly affects the development of reproductive organs. The floral B-class genes are most likely the conserved hub to control the unisexual flower development and sex of the individual (Figure 4).
Additionally, this convergent model itself can be varied. First, the feminization gene per se may locate at SDR and co-segregate with sex. This is the case of P. alba which elided the top module of epigenetic regulation (ΨARR17-IR/ARR17) (Müller et al., 2020). In the next place, B-class genes (AP3/PI) may be directly regulated by the feminized genes, omitting the middle module of activators (e.g., UFO in poplar) or repressors (e.g., SVP in persimmon). Yet this possibility remains to be further verified. And third, plant sex determination may also affect other important traits in addition to floral characteristics (Sanderson et al., 2019). For example, the flower odor-compounds of S. latifolia (Zluvova et al., 2010) and the leaf rubber content of E. ulmoides (Wang and Zhang, 2017) differed significantly between the sexes. To our knowledge, the MADS-box gene family contains several gene members, e.g., c.100 in E. ulmoides (Wang and Zhang, 2017), that function fundamentally and broadly in plant growth and development beyond flower development (Theißen, 2001; Soltis et al., 2007). Therefore, it is logical to speculate that different MADS-box gene members that influence important traits or phenotypes could be regulated by sex-determining genes as well.
Conclusion and outlook
Dioecy has independently evolved more than hundreds of times across various plant lineages (Henry et al., 2018). Multiple-omics studies on dioecious plants accelerate the discovery of sex-determining genes, supporting both the “two-gene model” and “one-gene model” (Charlesworth, 2016; Renner, 2016). Recent findings from poplar (Leite Montalvão et al., 2022) and persimmon (Yang et al., 2019), as well as studies in other dioecious species (Wang and Zhang, 2017; Zhang et al., 2020; Liu et al., 2022; Ma et al., 2022), led us to propose a convergent model of plant sex determination. According to this model, the sex-determining genes may vary across divergent species, but they eventually converged into floral MADS-box genes, B-class genes, in particular, to determine sexuality.
Despite rapid advances in sex determination in dioecious plants, more issues need to be addressed. On the one hand, to determine SDR on sex chromosomes and identify the SDR-located sex-determining genes should be performed in a broad range, e.g., via involving distantly-related dioecious species or incorporating certain dioecious plants and their close non-dioecious relatives. This would assist us in better understanding the evolution of the sex determination mechanism. On the other hand, genome-wide identification and functional analysis of the MADS-box genes in dioecious species are encouraged to be conducted. Novel molecular techniques e.g., long-read sequencing in combination with genome-editing approaches will extensively facilitate sex chromosome assembly and sex-determining genes identification (Masuda et al., 2020, 2022). In conclusion, a comprehensive understanding of the sex determination mechanism will not only shed light on the repeated evolution of dioecy in the plant kingdom but also will promote the breeding application of sex regulatory genes in economically important dioecious plant species.
Author contributions
XZ and WW: drafted the manuscript. XZ: original draft. LP, WG, YL, and WW: inputs and revision. All authors read and approved the submitted version.
Funding
We acknowledge funding from the Rural Revitalization Project of Guangdong Province (KTP20210181) and the Innovation Project for Forestry Science and Technology in Guangdong (2021KJCX015).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Xinxin Zhu from Xinyang Normal University for kindly sharing the flower photos of persimmon and poplar.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.953445/full#supplementary-material
References
- Akagi T., Charlesworth D. (2019). Pleiotropic effects of sex-determining genes in the evolution of dioecy in two plant species. Proc. R. Soc. B. 286, 20191805. 10.1098/rspb.2019.1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akagi T., Henry I. M., Kawai T., Comai L., Tao R. (2016). Epigenetic regulation of the sex determination gene MeGI in polyploid persimmon. Plant Cell 28, 2905–2915. 10.1105/tpc.16.00532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akagi T., Henry I. M., Ohtani H., Morimoto T., Beppu K., Kataoka I., et al. (2018). Y-encoded suppressor of feminization arose via lineage-specific duplication of a cytokinin response regulator in kiwifruit. Plant Cell 30, 780–795. 10.1105/tpc.17.00787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akagi T., Henry I. M., Tao R., Comai L. A. (2014). Y-chromosome–encoded small RNA acts as a sex determinant in persimmons. Science 346, 646–650. 10.1126/science.1257225 [DOI] [PubMed] [Google Scholar]
- Akagi T., Pilkington S. M., Varkonyi-Gasic E., Henry I. M., Sugano S. S., Sonoda M., et al. (2019). Two Y-chromosome-encoded genes determine sex in kiwifruit. Nat. Plants 5, 801–809. 10.1038/s41477-019-0489-6 [DOI] [PubMed] [Google Scholar]
- Akagi T., Shirasawa K., Nagasaki H., Hirakawa H., Tao R., Comai L., et al. (2020). The persimmon genome reveals clues to the evolution of a lineage-specific sex determination system in plants. PLoS Genet. 16, e1008566. 10.1371/journal.pgen.1008566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida P., Proux-Wera E., Churcher A., Soler L., Dainat J., Pucholt P., et al. (2020). Genome assembly of the basket willow, Salix viminalis, reveals earliest stages of sex chromosome expansion. BMC Biol. 18, 1–18. 10.1186/s12915-020-00808-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreuzza S. Y. (2020). Y keep your X? Insights into the genetic basis of plant sex chromosome evolution. Plant Cell 32, 1769–1770. 10.1105/tpc.20.00265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baránková S., Pascual-Díaz J. P., Sultana N., Alonso-Lifante M. P., Balant M., Barros K., et al. (2020). Sex-chrom, a database on plant sex chromosomes. New Phytol. 227, 1594–1604. 10.1111/nph.16635 [DOI] [PubMed] [Google Scholar]
- Bräutigam K., Soolanayakanahally R., Champigny M., Mansfield S., Douglas C., Campbell M. M., et al. (2017). Sexual epigenetics: gender-specific methylation of a gene in the sex determining region of Populus balsamifera. Sci. Rep. 7, 1–8. 10.1038/srep45388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey S., Yu Q., Harkess A. (2021). The diversity of plant sex chromosomes highlighted through advances in genome sequencing. Genes 12, 381. 10.3390/genes12030381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cegan R., Marais G. A., Kubekova H., Blavet N., Widmer A., Vyskot B., et al. (2010). Structure and evolution of Apetala3, a sex-linked gene in Silene latifolia. BMC Plant Biol. 10:180. 10.1186/1471-2229-10-180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Charlesworth D. A. (1978). A model for the evolution of dioecy and gynodioecy. Am. Nat. 112, 975–997. 10.1086/283342 [DOI] [Google Scholar]
- Charlesworth D. (2016). Plant sex chromosomes. Ann. Rev. Plant Biol. 67, 397–420. 10.1146/annurev-arplant-043015-111911 [DOI] [PubMed] [Google Scholar]
- Charlesworth D. (2022). The mysterious sex chromosomes of haploid plants. Heredity 129, 17–21. 10.1038/s41437-022-00524-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronk Q. (2022). The distribution of sexual function in the flowering plant: from monoecy to dioecy. Philos. Trans. R. Soc. B. 377, 20210486. 10.1098/rstb.2021.0486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronk Q., Müller N. A. (2020). Default sex and single gene sex determination in dioecious plants. Front. Plant Sci. 11:1162. 10.3389/fpls.2020.01162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronk Q., Soolanayakanahally R., Bräutigam K. (2020). Gene expression trajectories during male and female reproductive development in balsam poplar (Populus balsamifera L.). Sci. Rep. 10, 1–14. 10.1038/s41598-020-64938-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C. R. (1877). The Different Forms of Flowers on Plants of the Same Species. New York, NY: D. Appleton and Company. 10.5962/bhl.title.110054 [DOI] [Google Scholar]
- Di Stilio V. S., Kramer E. M., Baum D. A. (2005). Floral MADS box genes and homeotic gender dimorphism in Thalictrum dioicum (Ranunculaceae) - a new model for the study of dioecy. Plant J. 41, 755–766. 10.1111/j.1365-313X.2005.02336.x [DOI] [PubMed] [Google Scholar]
- Diggle P. K., Stilio D. i, Gschwend V. S., Golenberg A. R., Moore E. M., et al. (2011). Multiple developmental processes underlie sex differentiation in angiosperms. Trends Genet. 27, 368–376. 10.1016/j.tig.2011.05.003 [DOI] [PubMed] [Google Scholar]
- Feng G., Sanderson B. J., Keefover-Ring K., Liu J., Ma T., Yin T., et al. (2020). Pathways to sex determination in plants: how many roads lead to Rome? Curr. Opin. Plant Biol. 54, 61–68. 10.1016/j.pbi.2020.01.004 [DOI] [PubMed] [Google Scholar]
- Geraldes A., Hefer C. A., Capron A., Kolosova N., Martinez-Nuñez F., et al. (2015). Recent Y chromosome divergence despite ancient origin of dioecy in poplars (Populus). Mol. Ecol. 24, 3243–3256. 10.1111/mec.13126 [DOI] [PubMed] [Google Scholar]
- Gong W., Filatov D. A. (2022). Evolution of the sex-determining region in Ginkgo biloba. Philos. Trans. R. Soc. B. 377, 20210229. 10.1098/rstb.2021.0229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves J. A. M. (2016). How Australian mammals contributed to our understanding of sex determination and sex chromosomes. Aust. J. Zool. 64, 267–276. 10.1071/ZO16054 [DOI] [Google Scholar]
- Harkess A., Huang K., van der Hulst R., Tissen B., Caplan J. L., Koppula A., et al. (2020). Sex determination by two Y-linked genes in garden asparagus. Plant Cell 32, 1790–1796. 10.1105/tpc.19.00859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkess A., Zhou J., Xu C., Bowers J. E., Van der Hulst R., Ayyampalayam S., et al. (2017). The asparagus genome sheds light on the origin and evolution of a young Y chromosome. Nat. Commun. 8, 1279. 10.1038/s41467-017-01064-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry I. M., Akagi T., Tao R., Comai L. (2018). One hundred ways to invent the sexes: theoretical and observed paths to dioecy in plants. Annu. Rev. Plant Biol. 69, 553–575. 10.1146/annurev-arplant-042817-040615 [DOI] [PubMed] [Google Scholar]
- Iocco-Corena P., Chaïb J., Torregrosa L., Mackenzie D., Thomas M. R., Smith H. M., et al. (2021). VviPLATZ1 is a major factor that controls female flower morphology determination in grapevine. Nat. Commun. 12, 1–10. 10.1038/s41467-021-27259-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabbour F., Espinosa F., Dejonghe Q., Le Péchon T. (2022). Development and evolution of unisexual flowers: a review. Plants 11, 155. 10.3390/plants11020155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H. M., Jia H. J., Cai Q. L., Wang Y., Zhao H. B., Yang W. F., et al. (2019). The red bayberry genome and genetic basis of sex determination. Plant Biotechnol. J. 17, 397–409. 10.1111/pbi.12985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno A., Saeki H., Kameya T., Saedler H., Theissen G. (2003). Heterotopic expression of class B floral homeotic genes supports a modified ABC model for tulip (Tulipa gesneriana). Plant Mol. Biol. 52, 831–841. 10.1023/A:1025070827979 [DOI] [PubMed] [Google Scholar]
- Kramer E. M., Holappa L., Gould B., Jaramillo M. A., Santiago S. P. M. (2007). Elaboration of B gene function to include the identity of novel floral organs in the lower eudicot Aquilegia. Plant Cell 19, 750–766. 10.1105/tpc.107.050385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuderna L. F., Lizano E., Juli,à E., Gomez-Garrido J., Serres-Armero A., Kuhlwilm M., et al. (2019). Selective single molecule sequencing and assembly of a human Y chromosome of African origin. Nat. Commun. 10, 1–8. 10.1038/s41467-018-07885-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. -Y., Lin H. -J., Viswanath K. K., Lin C. -P., Chang B. C. -H., Chiu P. -H., et al. (2018). The development of functional mapping by three sex related loci on the third whorl of different sex types of Carica papaya L. PLoS ONE 13:e0194605. 10.1371/journal.pone.0194605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leite Montalvão A. P., Kersten B., Fladung M., Müller N. A. (2021). The diversity and dynamics of sex determination in dioecious plants. Front. Plant Sci. 11:580488. 10.3389/fpls.2020.580488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leite Montalvão A. P., Kersten B., Kim G., Fladung M., Müller N. A. (2022). ARR17 controls dioecy in Populus by repressing B-class MADS-box gene expression. Philos. Trans. R. Soc. B. 377, 20210217. 10.1098/rstb.2021.0217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.-T., Yi T.-S., Gao L.-M., Ma P.-F., Zhang T., Yang J. B., et al. (2019). Origin of angiosperms and the puzzle of the Jurassic gap. Nat. Plants 5, 461–470. 10.1038/s41477-019-0421-0 [DOI] [PubMed] [Google Scholar]
- Li S.-F., Zhang G.-J., Yuan J.-H., Deng C.-L., Gao W. J. (2016). Repetitive sequences and epigenetic modification: inseparable partners play important roles in the evolution of plant sex chromosomes. Planta 243, 1083–1095. 10.1007/s00425-016-2485-7 [DOI] [PubMed] [Google Scholar]
- Liao Q., Du R., Gou J., Guo L., Shen H., Liu H., et al. (2020). The genomic architecture of the sex determining region and sex-related metabolic variation in Ginkgo biloba. Plant J. 104, 1399–1409. 10.1111/tpj.15009 [DOI] [PubMed] [Google Scholar]
- Liu Y., Wang S., Li L., Yang T., Dong S., Wei T., et al. (2022). The Cycas genome and the early evolution of seed plants. Nat. Plants. 8:389–401. 10.1038/s41477-022-01129-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Moore P. H., Ma H., Ackerman C. M., Ragiba M., Yu Q., et al. (2004). A primitive Y chromosome in papaya marks incipient sex chromosome evolution. Nature 427, 348. 10.1038/nature02228 [DOI] [PubMed] [Google Scholar]
- Ma X., Yu L. A., Fatima M., Wadlington W. H., Hulse-Kemp A. M., Zhang X., et al. (2022). The spinach YY genome reveals sex chromosome evolution, domestication, and introgression history of the species. Genome Biol. 23, 1–30. 10.1186/s13059-022-02633-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massonnet M., Cochetel N., Minio A., Vondras A. M., Lin J., Muyle A., et al. (2020). The genetic basis of sex determination in grapes. Nat. Commun. 11, 1–12. 10.1038/s41467-020-16700-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda K., Fujita N., Yang H.-. W, Ushijima K., Kubo Y, et al. Molecular mechanism underlying derepressed male production in hexaploid persimmon. Front. Plant Sci. (2020) 11:567249. 10.3389/fpls.2020.567249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda K., Ikeda Y., Matsuura T., Kawakatsu T., Tao R., Kubo Y., et al. (2022). Reinvention of hermaphroditism via activation of a RADIALIS-like gene in hexaploid persimmon. Nat. Plants 8, 217–224. 10.1038/s41477-022-01107-z [DOI] [PubMed] [Google Scholar]
- Matsunaga S., Isono E., Kejnovsky E., Vyskot B., Dolezel J., Kawano S., et al. (2003). Duplicative transfer of a MADS box gene to a plant Y chromosome. Mol. Biol. Evol. 20, 1062–1069. 10.1093/molbev/msg114 [DOI] [PubMed] [Google Scholar]
- Miga K. H., Koren S., Rhie A., Vollger M. R., Gershman A., Bzikadze A., et al. (2020). Telomere-to-telomere assembly of a complete human X chromosome. Nature 585, 79–84. 10.1038/s41586-020-2547-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming R., Bendahmane A., Renner S. S. (2011). Sex chromosomes in land plants. Annu. Rev. Plant Biol. 62, 485–514. 10.1146/annurev-arplant-042110-103914 [DOI] [PubMed] [Google Scholar]
- Mitchell C. H., Diggle P. K. (2005). The evolution of unisexual flowers: morphological and functional convergence results from diverse developmental transitions. Am. J. Bot. 92, 1068–1076. 10.3732/ajb.92.7.1068 [DOI] [PubMed] [Google Scholar]
- Müller N. A., Kersten B., Leite Montalvão A. P., Mähler N., Bernhardsson C., Bräutigam K., et al. (2020). A single gene underlies the dynamic evolution of poplar sex determination. Nat. Plants 6, 630–637. 10.1038/s41477-020-0672-9 [DOI] [PubMed] [Google Scholar]
- Muyle A., Shearn R., Marais G. A. (2017). The evolution of sex chromosomes and dosage compensation in plants. Genome Biol. Evol. 9, 627–645. 10.1093/gbe/evw282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng M., Yanofsky M. F. (2001). Activation of the Arabidopsis B class homeotic genes by APETALA1. Plant Cell. 13, 739–753. 10.2307/3871337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfent C., Pobursky K. J., Sather D. N., Golenberg E. M. (2005). Characterization of SpAPETALA3 and SpPISTILLATA, B class floral identity genes in Spinacia oleracea, and their relationship to sexual dimorphism. Dev. Genes Evol. 215, 132–142. 10.1007/s00427-004-0459-4 [DOI] [PubMed] [Google Scholar]
- Renner S. S. (2016). Pathways for making unisexual flowers and unisexual plants: moving beyond the “two mutations linked on one chromosome” model. Am. J. Bot. 103, 587–589. 10.3732/ajb.1600029 [DOI] [PubMed] [Google Scholar]
- Renner S. S., Müller N. A. (2021). Plant sex chromosomes defy evolutionary models of expanding recombination suppression and genetic degeneration. Nat. Plants 7, 392–402. 10.1038/s41477-021-00884-3 [DOI] [PubMed] [Google Scholar]
- Renner S. S., Müller N. A. (2022). Sex determination and sex chromosome evolution in land plants. Philos. Trans. R. Soc. B. 377, 20210210. 10.1098/rstb.2021.0210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson B. J., Wang L., Tiffin P., Wu Z., Olson M. S. (2019). Sex-biased gene expression in flowers, but not leaves, reveals secondary sexual dimorphism in Populus balsamifera. New Phytol. 221, 527–539. 10.1111/nph.15421 [DOI] [PubMed] [Google Scholar]
- Sather D. N., Jovanovic M., Golenberg E. M. (2010). Functional analysis of B and C class floral organ genes in spinach demonstrates their role in sexual dimorphism. BMC Plant Biol. 10, 1–14. 10.1186/1471-2229-10-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharmann M., Grafe T. U., Metali F., Widmer A. (2019). Sex is determined by XY chromosomes across the radiation of dioecious Nepenthes pitcher plants. Evol. Lett. 3, 586–597. 10.1002/evl3.142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan H., Cheng J., Zhang R., Yao X., Kong H. (2019). Developmental mechanisms involved in the diversification of flowers. Nat. Plants. 5, 917–923. 10.1038/s41477-019-0498-5 [DOI] [PubMed] [Google Scholar]
- Sinclair A. H., Berta P., Palmer M. S., Hawkins J. R., Griffiths B. L., Smith M. J., et al. (1990). A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature 346, 240–244. 10.1038/346240a0 [DOI] [PubMed] [Google Scholar]
- Sobral R., Costa M. M. R. (2017). Role of floral organ identity genes in the development of unisexual flowers of Quercus suber L. Sci. Rep. 7, 1–15. 10.1038/s41598-017-10732-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis D. E., Hong M., Frohlich M. W., Soltis P. S., Albert V. A., Oppenheimer D. G., et al. (2007). The floral genome: an evolutionary history of gene duplication and shifting patterns of gene expression. Trends Plant. Sci. 12, 358–367. 10.1016/j.tplants.2007.06.012 [DOI] [PubMed] [Google Scholar]
- Tennessen J. A., Wei N., Straub S. C. K., Govindarajulu R., Liston A., Ashman T.-L. (2018). Repeated translocation of a gene cassette drives sex chromosome turnover in strawberries. PLoS Biol. 16:e2006062. 10.1371/journal.pbio.2006062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theißen G. (2001). Development of floral organ identity: stories from the MADS house. Curr. Opin. Plant Biol. 4, 75–85. 10.1016/S1369-5266(00)00139-4 [DOI] [PubMed] [Google Scholar]
- Theißen G., Melzer R., Rümpler F. (2016). MADS-domain transcription factors and the floral quartet model of flower development: linking plant development and evolution. Development 143, 3259–3271. 10.1242/dev.134080 [DOI] [PubMed] [Google Scholar]
- Torres M. F., Mathew L. S., Ahmed I., Al-Azwani I. K., Krueger R., Rivera-Nuñez D., et al. (2018). Genus-wide sequencing supports a two-locus model for sex-determination in Phoenix. Nat. Commun. 9, 1–9. 10.1038/s41467-018-06375-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan G. A., DiFazio S., Jansson S., Bohlmann J., Grigoriev I., Hellsten U., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604. 10.1126/science.1128691 [DOI] [PubMed] [Google Scholar]
- Urasaki N., Tarora K., Shudo A., Ueno H., Tamaki M., Miyagi N., et al. (2012). Digital transcriptome analysis of putative sex-determination genes in papaya (Carica papaya). PLoS ONE. 7, e40904. 10.1371/journal.pone.0040904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkonyi-Gasic E., Wang T., Cooney J., Jeon S., Voogd C., Douglas M. J., et al. (2021). Shy girl, a kiwifruit suppressor of feminization, restricts gynoecium development via regulation of cytokinin metabolism and signalling. New Phytol. 230, 1461–1475. 10.1111/nph.17234 [DOI] [PubMed] [Google Scholar]
- Wang W., Chen S., Zhang X. (2018). Whole-genome comparison reveals heterogeneous divergence and mutation hotspots in chloroplast genome of Eucommia ulmoides Oliver. Int. J. Mol. Sci. 19, 1037. 10.3390/ijms19041037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Yang G., Deng X., Shao F., Li Y., Guo W., et al. (2020). Molecular sex identification in the hardy rubber tree (Eucommia ulmoides Oliver) via ddRAD markers. Int. J. Genomics. 2020, 2420976. 10.1155/2020/2420976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Zhang X. (2017). Identification of the sex-biased gene expression and putative sex-associated genes in Eucommia ulmoides Oliver using comparative transcriptome analyses. Molecules 22, 2255. 10.3390/molecules22122255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. C., Chen S. Y., Zhang X. Z. (2016). Chloroplast genome evolution in Actinidiaceae: clpP loss, heterogenous divergence and phylogenomic practice. PLoS ONE 11, e0162324. 10.1371/journal.pone.0162324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westergaard M. (1958). The mechanism of sex determination in dioecious flowering plants. Adv. Genet. 9, 217–281. 10.1016/S0065-2660(08)60163-7 [DOI] [PubMed] [Google Scholar]
- Xue L., Wu H., Chen Y., Li X., Hou J., Lu J., et al. (2020). Evidences for a role of two Y-specific genes in sex determination in Populus deltoides. Nat. Commun. 11, 1–12. 10.1038/s41467-020-19559-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H. W., Akagi T., Kawakatsu T., Tao R. (2019). Gene networks orchestrated by MeGI: a single-factor mechanism underlying sex determination in persimmon. Plant J. 98, 97–111. 10.1111/tpj.14202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Wang D., Li Y., Zhang Z., Tong S., Li M., et al. (2021). General model to explain repeated turnovers of sex determination in the Salicaceae. Mol. Biol. Evol. 38, 968–980. 10.1093/molbev/msaa261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Zhang R., Yang X., Gu K. -J., Chen W., Chang Y., et al. (2019). Recent origin of an XX/XY sex determination system in the ancient plant lineage Ginkgo biloba. bioRxiv [Preprint]. 10.1101/517946 [DOI] [Google Scholar]
- Zhang X., Wang G., Zhang S., Chen S., Wang Y., Wen P., et al. (2020). Genomes of the banyan tree and pollinator wasp provide insights into fig-wasp coevolution. Cell 183, 875–889. e817. 10.1016/j.cell.2020.09.043 [DOI] [PubMed] [Google Scholar]
- Zhou R., Macaya-Sanz D., Carlson C. H., Schmutz J., Jenkins J. W., Kudrna D., et al. (2020a). A willow sex chromosome reveals convergent evolution of complex palindromic repeats. Genome Biol. 21, 1–19. 10.1186/s13059-020-1952-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R., Macaya-Sanz D., Schmutz J., Jenkins J. W., Tuskan G. A., DiFazio S. P., et al. (2020b). Sequencing and analysis of the sex determination region of Populus trichocarpa. Genes 11, 843. 10.3390/genes11080843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L. (2019). Analysis of mRNAs and non-coding RNAs expression involved in the female and male floral develoment of Eucommia ulmoides. (PhD Thesis). Central South University of Forestry and Technology. [Google Scholar]
- Zluvova J., Nicolas M., Berger A., Negrutiu I., Monéger F. (2006). Premature arrest of the male flower meristem precedes sexual dimorphism in the dioecious plant Silene latifolia. Proc. Natl. Acad. Sci. USA. 103, 18854–18859. 10.1073/pnas.0606622103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zluvova J., Zak J., Janousek B., Vyskot B. (2010). Dioecious Silene latifolia plants show sexual dimorphism in the vegetative stage. BMC Plant Biol. 10, 1–5. 10.1186/1471-2229-10-208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou C., Massonnet M., Minio A., Patel S., Llaca V., Karn A., et al. (2021). Multiple independent recombinations led to hermaphroditism in grapevine. Proc. Natl. Acad. Sci. 118, e2023548118. 10.1073/pnas.2023548118 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.