Abstract
Pseudomonas aeruginosa, a Gram-negative bacterium, is one of the major pathogens implicated in human opportunistic infection and a common cause of clinically persistent infections such as cystic fibrosis, urinary tract infections, and burn infections. The main reason for the persistence of P. aeruginosa infections is due to the ability of P. aeruginosa to secrete extracellular polymeric substances such as exopolysaccharides, matrix proteins, and extracellular DNA during invasion. These substances adhere to and wrap around bacterial cells to form a biofilm. Biofilm formation leads to multiple antibiotic resistance in P. aeruginosa, posing a significant challenge to conventional single antibiotic therapeutic approaches. It has therefore become particularly important to develop anti-biofilm drugs. In recent years, a number of new alternative drugs have been developed to treat P. aeruginosa infectious biofilms, including antimicrobial peptides, quorum-sensing inhibitors, bacteriophage therapy, and antimicrobial photodynamic therapy. This article briefly introduces the process and regulation of P. aeruginosa biofilm formation and reviews several developed anti-biofilm treatment technologies to provide new directions for the treatment of P. aeruginosa biofilm infection.
Keywords: Pseudomonas aeruginosa, antibiotic resistance, biofilm, anti-biofilm molecules, alternative therapeutics
Introduction
Most microorganisms have different survival mechanisms when facing stress conditions, such as proteolytic systems and growth regulation (Kumar et al., 2017). Most bacteria live in biofilms, which provide a protective niche for the survival of microorganisms (Kumar et al., 2017; Flemming and Wuertz, 2019). Bacterial biofilms are defined as structured microbial communities encapsulated in a self-synthesizing extracellular polymeric substance (EPS) and attached to a tissue or surface (Costerton et al., 1999) that include exopolysaccharides, matrix proteins, and extracellular DNA (eDNA; Wang et al., 2020). Clinically, the long-term colonization of bacteria in humans causes chronic infections, mainly because the bacteria in biofilms are resistant to host immune responses and antibiotic therapy (Dufour et al., 2010). Research has shown that 65–80% of pathogenic infections in hospitals are associated with biofilms (Kumar et al., 2017; Shrestha et al., 2022). Although numerous antimicrobial agents are available for clinical use, these agents only inhibit infection symptoms and are unable to eradicate bacteria embedded in biofilms (Li et al., 2020).
Pseudomonas aeruginosa is a Gram-negative opportunistic pathogenic bacterium that is widely found in nature. It has been established that P. aeruginosa is involved in a diverse array of infections, both community- and hospital-acquired, including pneumonia, cystic fibrosis, urinary tract infections, and burn infections (Al-Dahmoshi et al., 2021). Antimicrobial agents approved for clinical use may be ineffective in treating P. aeruginosa infections as this bacterium has the ability to form biofilms (Azam and Khan, 2019). The formation of biofilms enables P. aeruginosa to resist external adverse environments and enhance its colonization in the host. Biofilms can also act as diffusion barriers, restricting the entry of antibiotics into bacterial cells (Drenkard, 2003; Pang et al., 2019).
As P. aeruginosa is one of the major pathogens involved in opportunistic infections in humans, the clinical treatment and control of P. aeruginosa infections have become major challenges and have been the subject of extensive research (Azam and Khan, 2019; Das et al., 2020; Shao et al., 2020). The formation of biofilm effectively aids P. aeruginosa colonization, improving bacterial resistance to antimicrobial agents and countering the host immune system (Maurice et al., 2018; Elmanama et al., 2020; Tuon et al., 2022). Therefore, conventional single antibiotic therapy is limited in the treatment of biofilm infections, and an increasing number of studies have investigated the development of new antimicrobial drugs and anti-biofilm therapeutic programs to treat P. aeruginosa infection. This review will introduce the process of biofilm formation as well as biofilm regulation and anti-biofilm therapies in P. aeruginosa.
Composition, formation, and regulation of Pseudomonas aeruginosa biofilm
The biofilm matrix components that have been identified from P. aeruginosa mainly include exopolysaccharides, eDNA, and matrix proteins, which play an important role in the structural maintenance and drug resistance of biofilms (Ghafoor et al., 2011). P. aeruginosa can synthesize at least three types of exopolysaccharides: alginate, Pel polysaccharide, and Psl polysaccharide (Mann and Wozniak, 2012). Alginate is an anionic polysaccharide of α-L-guluronic acid and β-D-mannuronic acid linked by β-1-4 glycosidic bonds (Moradali and Rehm, 2019). The overproduction of alginate is responsible for the development of excessive slimy or mucoid biofilms, while mucoid biofilms are more resistant to host immune system attack and antibiotic treatment than non-mucoid biofilms, resulting in persistent infections in the body (Moradali et al., 2017; Moradali and Rehm, 2019). The Psl polysaccharide consists of 15 co-transcribed genes (pslA to pslO) that encode proteins to synthesize Psl, enhance cell-surface and cell-to-cell adhesion in P. aeruginosa, and play an important role in the initiation and maintenance of biofilm structure (Ma et al., 2006). Pel is a positively charged exopolysaccharide composed of partially acetylated 1 → 4 glycosidic linkages of N-acetylgalactosamine and N-acetylglucosamine (Jennings et al., 2015), which is important for biofilm formation in air–liquid interfaces (Byrd et al., 2009). Pel and Psl are the major structural polysaccharides in non-mucoid and mucoid P. aeruginosa biofilms (Colvin et al., 2012; Ma et al., 2012). Cell lysis releases DNA into the environment, and this eDNA can be used as a supporting component of biofilms to provide nutrients to bacteria in biofilms during periods of nutrient deficiency (Finkel and Kolter, 2001). Aside from exopolysaccharides and eDNA, extracellular proteins are also considered to be important components of biofilm matrices, including appendages (mainly flagella and type IV fimbriae), cytoadhesions, and lectins. Studies have found that these components mainly play an auxiliary role as adhesion factors and structural support in the process of P. aeruginosa biofilm formation (Qi and Christopher, 2019).
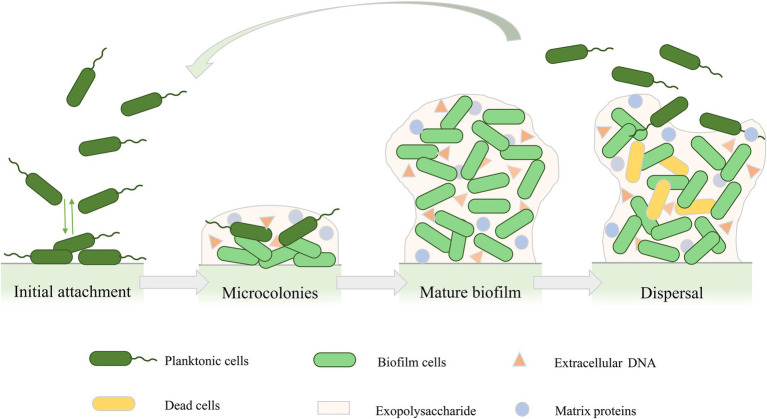
The formation of the biofilm structure of P. aeruginosa is a continuous process that includes four main stages (Figure 1; Kostakioti et al., 2013; Saxena et al., 2019). The first stage is the initial attachment. At this stage, adhesion is reversible, and bacteria can attach to the surface or revert back to planktonic cells (Olivares et al., 2019). Many early studies on the initial attachment of bacteria have suggested that bacterial cells initiate adhesion through acid–base, hydrophobic, and electrostatic interactions (Donlan, 2002). In addition, the production of polysaccharides also contributes to cell-to-cell adhesion in P. aeruginosa (Ma et al., 2006). In the second stage of biofilm development, attached bacteria gradually reproduce into a more independent structure, and bacterial cells undergo a transition from reversible to irreversible attachment (Thi et al., 2020). In this stage, bacteria grow and form microcolonies and begin to synthesize EPSs, which acts as a blockade between biofilm cells and surroundings to provide protection from various stress conditions such as antimicrobial exposure or immune cell attack (Mitchell et al., 2016; Roy et al., 2018). As the secretion of EPSs continues, the cells forming microcolonies gradually mature and undergo various physiological changes (Lee and Yoon, 2017; Roy et al., 2018), forming three-dimensional mushroom-like structures consisting of small channels that transport nutrients, water, and waste, and contribute to the distribution of nutrients and signaling molecules as well as intercellular communication (Jamal et al., 2018; Thi et al., 2020). As the biofilm matures, the bacteria produce stronger resistance to environmental stress or antibiotics (Koo et al., 2017). The final stage of biofilm development is detachment. Different detachment mechanisms have been reported, such as erosion, sloughing, and dispersal (Kim and Lee, 2016). The detachment of biofilm marks the transition of biofilm cells to the planktonic mode of growth (Rumbaugh and Sauer, 2020), which leads cells to attach to the surface of other biomolecules or start a new cycle of biofilm development (Kim and Lee, 2016).
Figure 1.
Cycle of Pseudomonas aeruginosa biofilm development. Biofilm formation includes four stages: initial attachment, microcolonies, mature biofilm, and biofilm dispersal.
Matrix components of P. aeruginosa biofilms play different regulatory roles in different formation stages (Wei and Ma, 2013; Ma et al., 2022). In the early stage of biofilm formation, Psl polysaccharides can form a fibrous matrix that is spirally anchored to the surface of bacterial cells and wraps around bacteria, thus increasing the contact between bacteria and promoting the interaction between bacterial cells, resulting in the assembly and early adhesion of biofilms (Ma et al., 2006, 2009). eDNA is also considered to be an important factor in promoting the formation of P. aeruginosa biofilm and is involved in the initial attachment of bacterial cells as well as cell–cell interconnection (Allesen-Holm et al., 2006). During the later maturation stages of biofilms, Pel can serve a structural and protective role in the biofilm matrix of P. aeruginosa and enhance resistance to aminoglycoside antibiotics (Colvin et al., 2011). The production of numerous EPSs can promote biofilm growth through providing structural scaffolds and maintaining their biofilm function (Ghafoor et al., 2011). Furthermore, quorum sensing (QS), as a cell-density-dependent bacteria-cell signaling mechanism, plays a key role in the regulation of P. aeruginosa biofilm formation (Bala et al., 2015). There are at least four QS systems in P. aeruginosa: las, rhl, pqs, and iqs (Lee and Zhang, 2015). As the two main QS systems of P. aeruginosa, both las and rhl systems use acyl homoserine lactone (AHL) as a signal molecule, which binds to the signal molecule receptor protein to play a regulatory role (Huang et al., 2019). The las system represses the pel locus, an operon encoding for the synthesis of extracellular matrix polysaccharides that induce biofilm formation and dispersion (Lin and Cheng, 2019), while the rhl system positively regulates the production of the biosurfactant rhamnolipid, which is important for late biofilm formation (Davey et al., 2003; Lequette and Greenberg, 2005; Chrzanowski et al., 2012). The P. aeruginosa pqs system uses 2-alkyl-4-quinolones (AQs) as signaling molecules. These AQs mainly include 2-heptyl-3-hydroxy-4-quinolone (PQS) and its precursor 2-heptyl-4-hydroxyquinoline (HHQ), both of which are recognized by the cognate receptor MvfR (Multiple virulence factor regulator, a P. aeruginosa quorum-sensing transcription factor, also known as PqsR; Lin et al., 2018a). Through interaction with this receptor, HHQ and PQS induce the expression of a variety of genes, including their own biosynthetic enzyme cascade and genes involved in biofilm formation (Schutz and Empting, 2018).
Anti-biofilm strategies: Current approaches and perspectives
Antibiotics have been widely used to treat biofilm infections, but clinical treatment still faces many challenges due to drug resistance issues, biofilm matrices that hinder drug penetration, and drug-microbe interactions (Kamaruzzaman et al., 2018). Therefore, many new anti-biofilm technologies have been developed (Hughes and Webber, 2017), such as combining antibiotics and using new strategies, for example, gallium, phage therapy, and antimicrobial photodynamic therapy (aPDT), to inhibit biofilm formation. Table 1 summarizes the strategies discovered in recent years for the treatment of P. aeruginosa biofilm infection and the different action mechanisms of related anti-biofilm molecules.
Table 1.
Different strategies to treat Pseudomonas aeruginosa infectious biofilms.
| Strategy | Mechanism | Molecules associated | Reference |
|---|---|---|---|
| Antibiotics | Antibiotics are used in combination with antibiotics or other substances to destroy biofilms and prevent the development of antibiotic resistance | Gentamicin/ciprofloxacin, tobramycin/clarithromycin, linolenic acid-tobramycin, gentamicin-EDTA, glutamine-ampicillin | Chauhan et al., 2012; Chanda et al., 2017; Ghorbani et al., 2017; Wang et al., 2019; Zhao et al., 2021 |
| AMPsa | Interact and penetrate with the bacterial cell membrane to cause the death of the bacteria | LL-37, P5, cationic peptide 1,037, MC1, WLBU2 | Overhage et al., 2008; de la Fuente-Nunez et al., 2012; Lin et al., 2018b Martinez et al., 2019; Yu et al., 2019 |
| QSIsb | Inhibit the QS system and interfere with signaling molecules and receptor proteins | Zingerone, trans-cinnamaldehyde and salicylic acid, chloroacetamide and maleimide analogs, halogenated furanone derivatives, M64, QSI4 | Kumar et al., 2015; Ahmed et al., 2019; O'Brien et al., 2015; Maura and Rahme, 2017; Chang et al., 2019; Schutz et al., 2021 |
| Enzymes or polysaccharides | Target extracellular polymeric substances (such as exopolysaccharides, matrix proteins, and eDNA) to disrupt biofilms | DNase I, glycoside hydrolases PelA and PslG, alginate lyase AlyP1400, A101, EPS273 | Jiang et al., 2011; Hymes et al., 2013; Zhang et al., 2018; Daboor et al., 2021; Wu et al., 2021 |
| Ga3+ | Acts as a “Trojan horse” to disrupt bacterial Fe metabolism and inhibit P. aeruginosa growth | Desferrioxamine-gallium, Ga-Cit, Ga (NO3)3 | Banin et al., 2008; Rzhepishevska et al., 2011; Kang et al., 2021 |
| NO | Creates nitrosative stress or oxidative stress in the biofilm and aids in biofilm dispersal | NO-sensing proteins | Hossain and Boon, 2017 |
| Bacteriophages | Encode enzymes to destroy the extracellular matrix | IME180, quorum quenching lactamase | Pei and Lamas-Samanamud, 2014; Mi et al., 2019 |
| aPDTc | PS binds to the biofilm matrix and generates ROS under light, initiating multi-target damage | Tetra-Py+-Me, MB + GN, ICG-APTMS@SPION/laser | Beirao et al., 2014; Bilici et al., 2020; Perez-Laguna et al., 2020 |
Antimicrobial peptides.
Quorum-sensing inhibitors.
Antimicrobial photodynamic therapy.
Antibiotics
It is well known that antibiotic therapy is the most important and effective measure to control bacterial infection. However, bacterial biofilms are highly resistant to antibiotic treatment and immune response (Shrivastava et al., 2018; Srinivasan et al., 2021). Due to the low outer membrane permeability of P. aeruginosa and its own adaptive mechanisms, it is less susceptible to most antibiotics and readily achieves clinical resistance. The use of single antibiotics to treat P. aeruginosa biofilm infections therefore presents significant challenges, and various strategies have been developed to treat biofilms and prevent the development of antibiotic resistance, including increasing antibiotic concentrations or the use of antibiotics in combination (Khan et al., 2020). Here, we divide the mechanism of action of combined antibiotics into two main categories: the synergistic effect between different antibiotics and the combined use of other substances and antibiotics.
The combined use of different antibiotics against P. aeruginosa biofilms has been described in detail in a previous review (Yadav et al., 2021). The antibiotic combinations that have been found to be effective against P. aeruginosa biofilm include gentamicin/ciprofloxacin (Wang et al., 2019) and tobramycin/clarithromycin (Ghorbani et al., 2017). These combinations all increase the therapeutic efficacy of antibiotics against P. aeruginosa biofilm. In addition to the synergistic effect between antibiotics, substances such as metal chelators, fatty acids, and amino acids combined with antibiotics will also help to prevent the formation of P. aeruginosa biofilms. Linolenic acid (LNA) is an essential fatty acid that has antibacterial effects on various microorganisms. Studies have found that LNA can not only interfere with intercellular communication and reduce the production of virulence factors, but can also enhance the potency of tobramycin and synergistically inhibits biofilm formation by affecting P. aeruginosa quorum-sensing systems (Chanda et al., 2017). The cation chelator EDTA acts as a therapeutic adjuvant to destabilize biofilm matrices (Lebeaux et al., 2015). Some studies have found that the combination of EDTA and antibiotics can quickly and persistently remove biofilms formed in vivo compared with the use of antibiotics alone (Chauhan et al., 2012; Lebeaux et al., 2015). Glutamine is an amino acid that is used as a nutritional source, and the exogenous addition of glutamine can stimulate the influx of ampicillin, resulting in the accumulation of intracellular antibiotic concentrations that exceed the amount tolerated by multidrug-resistant bacteria. Glutamine-enhanced ampicillin-mediated killing has been found to be effective against P. aeruginosa biofilms in a mouse model of urinary tract infection. Moreover, glutamine also retards the development of ampicillin resistance, which may help in the future development of effective antibiotic drugs to prevent or manage difficult-to-treat bacterial biofilms (Zhao et al., 2021). In addition, the addition of extra O2 via hyperbaric oxygen therapy (HBOT) can increase the susceptibility of pathogens to several antibiotics against metabolically active bacteria by activating aerobic respiration. When combined with tobramycin or ciprofloxacin, re-oxygenation with HBOT enhanced the killing of clinical P. aeruginosa and the eradication of biofilms (Lichtenberg et al., 2022).
In vitro and in vivo experiments showed that the minimum inhibitory concentration (MIC) and minimum bactericidal concentration of biofilm bacterial cells were usually much higher than those of planktonic cells (by about 10–1,000 times; Hoiby et al., 2010, 2011). Therefore, it is difficult to achieve the eradication of biofilms in vivo with the use of conventional single antibiotics (Wu et al., 2015; Sharma et al., 2019), while the combination of antibiotics with different antibiotics or other substances to increase their effect as a new strategy for the treatment of biofilm infections has broad prospects. In addition to antibiotics, a variety of compounds have been clinically used to treat P. aeruginosa biofilm infection (Soren et al., 2020; Pang and Zhu, 2021). These treatment methods can be roughly divided into two categories: preventing biofilm formation and destroying formed biofilms. For example, antimicrobial peptides (AMPs) and quorum-sensing inhibitors (QSIs) can prevent biofilm formation by regulating the biofilm formation process, while some exopolysaccharide hydrolases and DNases can target biofilm matrix components to destroy biofilms.
AMPs
The clinical application of AMPs is accelerating with increasing antibiotic resistance worldwide (Mahlapuu et al., 2016). AMPs are tiny macromolecules composed of amino acids that have the ability to stimulate innate immune responses and exhibit potent activity against a broad range of bacterial species, fungi, protozoa, and encapsulated viruses (Mahlapuu et al., 2016; Abdi et al., 2019; Seyfi et al., 2020). It is generally acknowledged that most AMPs can directly bind to the bacterial surface, such as the lipopolysaccharide (LPS) of Gram-negative bacteria, and then depolarize and permeate the membrane (Wen et al., 2013; Chou et al., 2019). LL-37, a classic human AMP, has been identified as capable of disrupting bacterial membranes, leading to cell death and inhibiting P. aeruginosa biofilm formation (Overhage et al., 2008), but subsequent study has shown that at sub-inhibitory concentrations it can promote P. aeruginosa DNA mutations and induce its resistance to antibiotics (Limoli et al., 2014). Despite this, as potential biofilm inhibitors, AMPs still hold great promise for the targeted elimination of biofilm proliferation in multi-drug resistant and extensively drug-resistant bacteria (Pontes et al., 2022). The target of these AMPs in the cell is typically the cell membrane. Through interacting with the bacterial cell membrane and penetrating the cell membrane, AMPs cause the death of the bacteria, thereby reducing the possibility of bacterial resistance (Annunziato and Costantino, 2020). As a promising class of compounds to overcome antimicrobial resistance, AMPs have been shown to have some advantages over traditional antibiotics.
Several studies have reported the mechanism of action of AMPs in detail (Annunziato and Costantino, 2020; Talapko and Skrlec, 2020). This review divides the anti-biofilm action mechanism of AMPs into two categories. AMPs in the first category exhibit anti-biofilm activity through their membrane dissolution mechanism, which directly affects the integrity of bacterial cell membranes and cell walls. The anti-biofilm peptide P5 has been found to have the ability to target the membrane permeability of P. aeruginosa and has synergistic and bactericidal effects with the carbapenem antibiotic meropenem (Martinez et al., 2019). An outer membrane permeability assay showed that P5 could easily permeate the cell membrane at concentrations below 0.5 × MIC, which occurred because meropenem entered the cytoplasmic space to interfere with the formation of peptidoglycans in the cell wall. In addition to meropenem, other antibiotics can also act on P. aeruginosa biofilms synergistically with AMPs, such as imipenem (Feng et al., 2015) and tobramycin (Beaudoin et al., 2018).
AMPs in the second category affect the growth pattern of biofilms by inhibiting bacterial adhesion and interfering with gene expression. While AMPs are generally considered to be membrane-active molecules that disrupt biofilms by perturbing the cell wall/membrane, AMPs also possess multifunctional activities such as protein synthesis and gene expression at multiple sites within the membrane or within the cell, enabling efficient killing (Le et al., 2017). A novel synthetic cationic peptide, 1,037, can significantly reduce P. aeruginosa biofilm formation and lead to cell death in biofilms at certain concentrations. Analysis of its effect on gene expression has revealed that 1,037 directly inhibits biofilms by reducing swimming and swarming motilities, stimulating twitching motility, and inhibiting the expression of various genes involved in biofilm formation, such as PA2204 (de la Fuente-Nunez et al., 2012). Recent studies have found that the anti-biofilm peptide MC1 can inhibit biofilm formation by down-regulating the relative expression levels of pelA, algD, and pslA genes in P. aeruginosa and reducing the synthesis of exopolysaccharides (Yu et al., 2019). Another well-studied AMP, called WLBU2, and its D-amino acid enantiomer D8 have also shown gene modulating activity against P. aeruginosa in the biofilm mode of growth, as well as increased safety, stability, and antimicrobial efficacy when substituting the L-amino acids in WLBU2 with D-amino acids (Lashua et al., 2016; Lin et al., 2018b; Di et al., 2020).
As anti-biofilm peptides can inhibit the formation of biofilms or remove mature biofilms, they have been gradually recognized by an increasing number of researchers as a potential new drug for the prevention and treatment of bacterial biofilm infections. However, there are still many obstacles to clinical application. For example, anti-biofilm peptides tend to show a certain degree of hemolysis or cytotoxicity to eukaryotic cells, and they are easily hydrolyzed by protease and cannot exist stably in vivo (Aoki and Ueda, 2013). In addition, the high production cost of anti-biofilm peptides and long drug development cycle also limit their potential clinical application.
QSIs
In recent years, an increasing number of studies have developed new antibacterial drugs by targeting specific virulence factors or their regulatory mechanisms to reduce the emergence of drug-resistant strains (Muhlen and Dersch, 2016; Kamal et al., 2017). One of these strategies is directed toward interference with QS-mediated signaling. The QS system of P. aeruginosa consists of four systems that interact to form a complex intercellular communication network that regulates the expression of its virulence-related genes and biofilm formation in a cell-density-dependent manner by generating QS signaling molecules (Rutherford and Bassler, 2012; Yong et al., 2018; Lin et al., 2018a).
QSIs can be either natural or synthetic, and both types of QSIs can inhibit the formation of biofilms by targeting different sites (Kalia, 2013). This review divides QSIs into two categories: QSIs that inhibit the expression of the QS system and QSIs that interfere with the combination between signaling molecules and receptor proteins. The first type of QSI interferes with and inhibits the expression of the QS system, altering the P. aeruginosa biofilm architecture and enabling antibiotics to better penetrate and more efficiently kill bacterial cells. In a previous study, the naturally isolated plant compounds trans-cinnamaldehyde and salicylic acid effectively down-regulated both the las and rhl QS systems, significantly inhibited the expression of QS regulatory and virulence genes in P. aeruginosa PAO1, and also reduced biofilm formation concomitantly with repressed rhamnolipid gene expression (Rajkumari et al., 2018; Ahmed et al., 2019). In addition, a host of synthetic biofilm inhibitors have also been developed. Chloroacetamide and maleimide analogs, as potent, drug-like small molecule inhibitors of QS in P. aeruginosa, are anticipated to be of significant medical interest. These inhibitors exhibit potent LasR antagonist activity and inhibit the expression of the P. aeruginosa virulence factor pyocyanin, as well as biofilm formation in PAO1 and PA14 (O'Brien et al., 2015). In addition, Chang et al. identified a new series of halogenated furanone derivatives and found that they effectively inhibited lasB expression in a dose-dependent manner and showed remarkable biofilm formation in P. aeruginosa (Chang et al., 2019).
The second category of QSI molecules functions by interfering with and inhibiting the combinations between signaling molecules and receptor proteins that are required for bacterial cell-to-cell communication, the production of virulence factors, and biofilm formation (Soukarieh et al., 2018). Ginger has been widely used as a medicinal herb with strong antimicrobial properties. Zingerone is one of the main components of dry ginger root, is found in many herbal spices, and can effectively regulate the biofilm structure of P. aeruginosa (Kumar et al., 2013). In P. aeruginosa, zingerone inhibits the las, rhl, and pqs systems by binding to their respective cognate receptors (LasR, RhlR, and MvfR), which silences the cell communication network and ultimately suppresses the virulence and biofilm formation of P. aeruginosa (Kumar et al., 2015). Furthermore, MvfR, as a crucial transcriptional regulator of the PQS system of P. aeruginosa, is considered to a potential target for inhibiting the PQS-MvfR QS system (Kitao et al., 2018). The benzamide-benzimidazole compound M64 can inhibit the P. aeruginosa QS regulator MvfR, resulting in reduced biofilm formation and the increased susceptibility of P. aeruginosa to antibiotics (Maura and Rahme, 2017). Recently, a study has reported a novel class of QSIs called QSI4, which possesses excellent activity in inhibiting pyocyanin production and the MvfR reporter-gene; when combined with antibiotics, QSI4 has a significant synergistic effect on the elimination of P. aeruginosa biofilm (Schutz et al., 2021).
As an important intercellular communication system in P. aeruginosa, the QS system plays a key role in the regulation of biofilm formation. QSIs inhibit biofilm formation through anti-virulence or a pathoblocker approach, which can synergize the efficacy of antibiotics but does not affect the viability of bacteria. Clinical application of QSIs will reduce the development of antibiotic resistance as well as some toxic side effects (Wagner et al., 2016). Therefore, QSIs are currently promising drug targets for the prevention and treatment of P. aeruginosa infection.
Targeting polysaccharides
As functionally rich and dynamically changing communities, biofilms can modify matrix components to adapt to changes in various environmental conditions and pressures. In P. aeruginosa, enzymes targeting biofilm EPSs may offer a general strategy to prevent clinical biofilm infections (Zhao et al., 2018). The bacteria themselves also synthesize polysaccharides or certain endogenous matrix-degrading enzymes to induce the dispersion of the biofilm, such as glycoside hydrolases (Srinivasan et al., 2021).
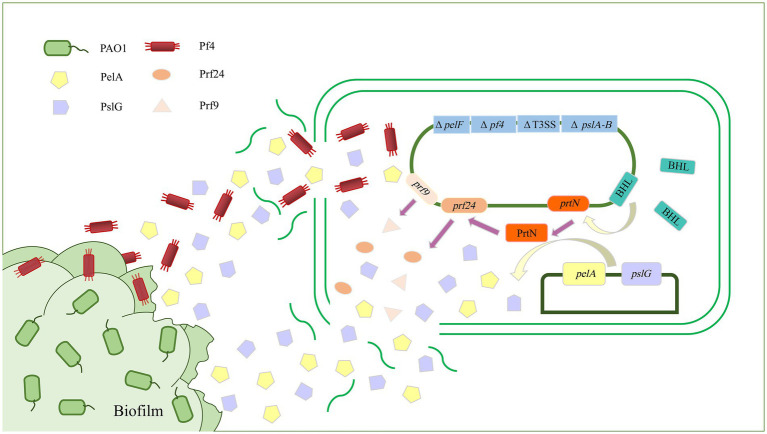
A key component of biofilm formation is the biosynthesis of the exopolysaccharides Pel, Psl, and alginate (Limoli et al., 2015). Enzymes targeting the extracellular matrix could serve as targets to disrupt biofilms. Alginate lyase can degrade alginate through the β-elimination of glycosidic bonds to disrupt the structure and integrity of biofilms and significantly increase biofilm diffusion (Kovach et al., 2020). Recently, a study has reported an alginate lyase (AlyP1400) purified from a marine bacterial Pseudoalteromonas species that can treat P. aeruginosa infections in cystic fibrosis lungs or other P. aeruginosa biofilm-related infections by combining the use of the alginate lyase with antibiotics (Daboor et al., 2021). The glycoside hydrolases alpha-amylase and cellulase can also break down complex polysaccharides, convert the bacteria into a planktonic state, effectively destroy P. aeruginosa biofilm, and increase antibiotic efficacy (Fleming et al., 2017; Redman et al., 2020). In addition, PelA and PslG, as naturally derived glycoside hydrolases, can selectively target and degrade exopolysaccharides in the biofilm matrix, thus destroying the biofilm (Baker et al., 2016; Zhang et al., 2018). In a previous study, overexpressed or exogenously supplied PslG prevented biofilm formation by degrading Psl (Yu et al., 2015). As a hydrolase, PelA can scavenge polysaccharides in the periplasm, and its deacetylase activity is related to the formation of biofilms and the morphology of wrinkled colonies (Colvin et al., 2013; Marmont et al., 2017). Recently, our team has used P. aeruginosa as the starting strain to construct an engineered bacterium for the targeted transport and delivery of functional proteins that can use two polysaccharide hydrolases, PelA and PslG, to target biofilms (Figure 2). First, the engineered bacterium was constructed through synthetic biology so that it could initiate the lysis of its own cells, and then, the recombinant vectors were introduced to overexpress the two exopolysaccharide hydrolase genes pelA and pslG. Finally, the effect of engineered bacteria on P. aeruginosa biofilm was detected by biofilm formation experiments. It was found that overexpression of pelA and pslG could accumulate polysaccharide hydrolases in the intracellular matrix and release them to the extracellular matrix through the cell lysis site to disrupt the biofilm cytoskeletal components Psl and Pel, eventually destroying the biofilm and preventing further biofilm formation (Wang et al., 2021).
Figure 2.
Schematic diagram of targeted delivery of extracellular glycoside hydrolase by engineered bacteria to destroy Pseudomonas aeruginosa biofilm. The biofilm formation and virulence-related genes pelA-B, pelF, and T3SS of P. aeruginosa were knocked out as parental strains, and exogenous recombinant vectors were introduced to overexpress the exopolysaccharide hydrolases PelA and PslG. PelA and PslG accumulated in cells and were then released into the extracellular matrix through cell lysis. There are two ways in which hydrolases are released into the extracellular matrix. The first is through regulating the prtN gene to activate the expression of cell lysis protein genes, thereby releasing PelA and PslG. The second is by deleting the Pf4 filamentous prophage-encoding gene cluster to sensitize it to the Pf4 phage in biofilms, thereby initiating the passive lysis mechanism of its own cells to release PelA and PslG. The PelA and PslG are released to the extracellular matrix to destroy the biofilm skeleton components Pel and Psl through enzymatic hydrolysis, thereby destroying the P. aeruginosa biofilm.
The extracellular biofilm matrix of P. aeruginosa is mainly composed of exopolysaccharides, which are involved in the formation and maintenance of the structural biofilm (Franklin et al., 2011). However, some bacterial exopolysaccharides can perform functions that inhibit or destabilize the biofilm (Rendueles et al., 2013). A former study showed that P. aeruginosa extracellular products, mainly polysaccharides, disrupted established biofilms (Qin et al., 2009). Recently, more exopolysaccharides have been found to show negative activity against biofilm formation in P. aeruginosa. A bacterial exopolysaccharide (A101) not only inhibits the biofilm formation of many bacteria but also disrupts established biofilms. In addition, A101 increased the ability of aminoglycoside antibiotics to eliminate P. aeruginosa biofilm, which may indicate that A101 has potential in the design of new therapeutic strategies for bacterial biofilm-associated infections and in limiting biofilm formation on medical indwelling devices (Jiang et al., 2011). A recent experiment has reported an exopolysaccharide, EPS273, that reduces biofilm formation in P. aeruginosa by reducing the expression levels of the two-component system phoP–phoQ, which then regulates the expression levels of the QS systems lasI/lasR and rhlI/rhlR. The QS system further regulates the genes involved in biofilm formation, such as the genes involved in the production of the extracellular matrix and virulence factors, genes involved in flagella and cell motility, and genes involved in iron acquisition (Wu et al., 2021).
DNase
eDNA plays a structural role in the formation of biofilms (Allesen-Holm et al., 2006). eDNA can serve as a source of nutrients for bacteria under starvation and is involved in facilitating the twitching motility-mediated expansion of biofilms (Finkel and Kolter, 2001; Wei and Ma, 2013). In addition, eDNA interacts with Psl to form eDNA-Psl to provide structural support for biofilms (Wang et al., 2015). Based on these factors, drug pathways for targeting biofilm matrices via eDNA are emerging.
Deoxyribonuclease I (DNase I) is the only enzyme used clinically to disrupt the biofilm of P. aeruginosa. This therapeutic enzyme disrupts biofilms through the hydrolysis of DNA in the extracellular matrix (Hymes et al., 2013). DNase is involved in breaking the phosphodiester linkage in the eDNA molecular backbone in biofilms. As eDNA is essential for the initial attachment and aggregation of EPSs on the surface, this makes it difficult for bacteria to form an intact biofilm (Das et al., 2010; Srinivasan et al., 2021). Immature P. aeruginosa biofilms are therefore more sensitive to DNase treatment than mature biofilms (Hymes et al., 2013). Some studies have found that L-methionine (L-Met) can destroy P. aeruginosa biofilms through up-regulating DNase genes and inducing the expression of DNase, thereby targeting eDNA in biofilms (Gnanadhas et al., 2015). In addition, some DNase-like proteins have also been found to prevent and destroy bacterial biofilms. DNase1-like 2 (DNase1L2) expressed in human stratum corneum has enzymatic activity, can degrade eDNA, and can effectively inhibit P. aeruginosa biofilm formation (Eckhart et al., 2007). In addition, DNase I coatings are used as antimicrobial coatings for modern medical equipment. DNase Ι immobilization on surfaces has shown promise in reducing bacterial adhesion to surfaces, as this enzyme targets single biofilms and can effectively cleave eDNA on bacterial cell surfaces that are essential for bacterial adhesion (Swartjes et al., 2013; Sharma and Pagedar Singh, 2018). With the ongoing rise in antibiotic resistance, DNase Ι coating may provide a timely, potent new approach to prevent the formation of biofilms on biomaterial implants and devices.
When the biofilm matures to a certain extent, DNase treatment is no longer effective (Whitchurch et al., 2002). This resistance to DNase may be due to the replacement or supplementation of eDNA by other extracellular matrix components, or the binding of eDNA to another component that protects it from enzymatic degradation (Okshevsky et al., 2015). Therefore, enzymatic hydrolysis methods that destabilize biofilms by enzymatically degrading eDNA should also target polysaccharides or proteins bound to eDNA. For example, the interactions between eDNA with Psl and Type IV pili play important roles in biofilm formation (Barken et al., 2008), and disrupting these interactions could also be a potentially interesting target for biofilm treatment. In addition to DNase, the accumulation of eDNA itself in biofilms and infection sites can acidify the local environment (Wilton et al., 2016). The acidic environment stimulated increased P. aeruginosa biofilm formation, promoted faster bacterial evolution to improve antibiotic resistance, and increased the expression of multiple biofilm/virulence-related genes. Therefore, the use of simple pH-buffering agents alongside antibiotics may be a novel treatment strategy for combating chronic infection in the acidic, DNA-enriched lungs of clinic patients (Lin et al., 2021).
Gallium (Ga)
Recently, Ga ions have shown excellent anti-Pseudomonal activity and have been used as a novel biofilm treatment approach (Smith et al., 2017; Tummler, 2019; Tovar-Garcia et al., 2020). Previous research has used Ga as a “Trojan horse” to disrupt bacterial iron metabolism and exploit the Fe stress of in vivo environments because Ga has an ionic radius nearly identical to that of Fe, and many biological systems are unable to distinguish Ga3+ from Fe3+ (Kaneko et al., 2007). While Ga3+ chemically resembles Fe3+, unlike iron, Ga cannot be reduced under physiological conditions, which also inhibits some of its basic functions (Minandri et al., 2014). Iron is not only a necessary element for growth but is also a cue in biofilm formation (Banin et al., 2005). Thus, interfering with bacterial iron homeostasis may serve as a potential therapeutic target that can block P. aeruginosa virulence in both the free-living and biofilm states.
Previous research found that low concentrations of Ga inhibited P. aeruginosa growth and prevented biofilm formation. This inhibition was mediated by the repression of the transcriptional regulator pvdS, as overriding pvdS repression partially protected bacteria from the growth-inhibitory action of Ga (Kaneko et al., 2007). Besides Ga alone, some complexes with Ga can also target P. aeruginosa iron metabolism to inhibit biofilm formation, such as desferrioxamine-Ga (DFO-Ga) and Ga citrate (Ga-Cit). Desferrioxamine (DFO) is a hydroxamate-based siderophore that induces proteins related to iron citrate and iron DFO uptake in iron-starved P. aeruginosa (Llamas et al., 2006; Marshall et al., 2009). The complex DFO-Ga, which can kill free-living bacteria and prevent biofilm formation, has been approved for clinical use (Banin et al., 2008). However, recent research indicates that Ga-Cit has an anti-biofilm effect and is more bactericidal than Ga-DFO (Rzhepishevska et al., 2011). Like other Ga complexes, Ga-Cit significantly inhibited biofilm production in P. aeruginosa at concentrations as low as 10 μM (Rzhepishevska et al., 2011). Moreover, several studies have explored the activity of Ga in combination with antibiotics in search of useful synergistic effects. Ga nitrate has been found to be an effective antimicrobial agent that inhibits P. aeruginosa growth. A recent study found that Ga(NO3)3 and tetracycline alone had a bactericidal effect, and the combined use of the two strongly inhibited the formation of P. aeruginosa biofilm (Kang et al., 2021).
Ga3+ is effective in destroying bacterial biofilms, and many drugs based on Ga have been approved for clinical treatment with remarkable results (Goss et al., 2018). For example, Ga(NO3)3, which is approved by the US Food and Drug Administration (FDA) for the treatment of infections, can be used to treat both acute and chronic pneumonia caused by P. aeruginosa infection (Bonchi et al., 2015). In addition to Ga and its compounds, combined use with antibiotics has achieved remarkable results in clinical treatment. Therefore, Ga, as a target for destroying biofilms, has potential in the development of more effective drugs for the treatment of biofilm infections.
Nitric oxide
Pseudomonas aeruginosa is a facultative anaerobe that can breathe under anaerobic conditions and denitrify in the presence of nitrate and nitrite. These abilities are associated with the virulence of bacteria (Cutruzzola and Frankenberg-Dinkel, 2016). NO is a radical diatomic gas molecule that plays important signaling roles in both eukaryotes and bacteria at low concentrations (Arora et al., 2015). NO has also been demonstrated to be an effective P. aeruginosa biofilm disruption agent that creates nitrosative or oxidative stress in the biofilm and induces the dispersal of P. aeruginosa and other bacterial biofilms by reducing c-di-GMP levels (Williams and Boon, 2019).
An early study of the effect of NO on biofilm formation showed that nanomolar NO caused biofilm dispersal in P. aeruginosa and enhanced the efficacy of antimicrobial compounds when combined with antibiotics (Barraud et al., 2006). This finding was confirmed in the clinical treatment of cystic fibrosis patients (Howlin et al., 2017). However, another study found that the exogenous addition of high concentrations of iron inhibited the diffusion of NO-induced biofilms and promoted the rapid attachment of plankton cells and resumed diffusion after the addition of iron chelator agents (Zhu et al., 2019). This is not due to the scavenging of NO by free iron but was related to an iron-induced cellular response that led to the increased production of the exopolysaccharide Psl and restored P. aeruginosa biofilm diffusion. Very recently, a novel family of heme-based NO binding proteins termed NO-sensing proteins (NosP) have been discovered in P. aeruginosa in the same operon as PA1976 (NahK). NahK has been identified as a NosP-associated histidine kinase, and it has been previously associated with biofilm regulation (Hossain and Boon, 2017). Experiments have suggested that NosP binds to NO, which controls the phosphorylation of a histidine-containing phosphotransfer domain, thus resulting in biofilm dispersal. However, the specific players involved in the signaling pathway have yet to be identified.
Although NO has attracted particular interest due to its role in biofilm dispersal, this approach still presents many practical issues in clinical trials. For example, the current inhaled NO therapy method is extremely expensive, largely owing to the difficulty in handling the gas and its incompatibility with oxygen, which results in the formation of toxic nitrogen dioxide (NO2; Yang et al., 2015). Therefore, while the bactericidal effect of antibacterial agents on biofilm-infected sites can be enhanced in a targeted manner, the NO-mediated toxicity should also be reduced so that it can be used in clinical anti-biofilm therapy (Barraud et al., 2012). In addition, the use of NO in combination with antibiotics can enhance the NO-mediated bactericidal effect and improve the specificity of NO delivery, so the use of NO is still very promising (Poh and Rice, 2022).
Bacteriophage therapy
Bacteriophages are natural bacterial viruses (Clokie et al., 2011). As they are unaffected by antibiotic resistance, bacteriophages have been used as therapeutic agents in early clinical practice (Kutter et al., 2010; Abedon et al., 2011). With the emergence of antibiotic-resistant strains, phage therapy has once again drawn attention, and a growing body of research has validated the use of bacteriophages in therapy and prophylaxis in the fight against drug-resistant bacteria (Kutateladze and Adamia, 2010). Phages can encode enzymes that degrade polymers and inhibit P. aeruginosa biofilm formation by disrupting the extracellular matrix and increasing the permeability, allowing antibiotics to reach the inner layer of biofilms (Harper et al., 2014). Therefore, an increasing number of studies have used bacteriophages to develop drugs to treat biofilm infections (Soothill, 2013).
The pathways by which phages destroy biofilms can be divided into two categories. First, phages can destroy biofilm structures by inducing the synthesis of enzymes such as polysaccharide depolymerases in P. aeruginosa (Pires et al., 2016; Chegini et al., 2020). Polysaccharide depolymerase is a polysaccharide hydrolase encoded by bacteriophages that can specifically degrade macromolecular carbohydrates on the host bacterial envelope (Yan et al., 2014). IME180, a P. aeruginosa phage isolated from a hospital, encodes an exopolysaccharide-degrading enzyme that is highly homologous to deglycans and can effectively degrade exopolysaccharides, inhibiting the formation of host bacterial biofilms and destroying established biofilms (Mi et al., 2019). In addition to polysaccharide hydrolases, bacteriophages can also produce endolysins, which inhibit bacterial cell wall synthesis by hydrolyzing peptidoglycan (Schmelcher et al., 2012). Another way in which bacteriophages inhibit biofilms is through the production of enzymes that inhibit biofilm production. A study has reported that phages can be genetically modified to induce the synthesis of quorum quenching lactamase, thereby inhibiting bacterial biofilm formation (Pei and Lamas-Samanamud, 2014). An engineered T7 phage incorporating the AHL lactonase aiiA gene can hydrolyze acyl AHL and inhibit QS activities in P. aeruginosa, ultimately inhibiting biofilm production (Whiteley et al., 2017). Unlike polysaccharide depolymerases, which can degrade one or several related polysaccharides, the T7aiiA phage can affect multiple bacteria in mixed-strain biofilms, rather than the host bacteria alone.
In addition to being directly used as a tool to destroy biofilms, phages can also indirectly aid in other strategies to destroy biofilms. In P. aeruginosa PAO1, Pf4 filamentous phages naturally parasitize by integrating into the genome and play critical roles in PAO1 virulence, biofilm development, and structural stability. Studies have shown that P. aeruginosa lacking the Pf4 filamentous phage-encoding gene cluster is highly susceptible to Pf4 filamentous phages. Therefore, we mutated the gene cluster encoding Pf4 filamentous phage when constructing the engineered bacteria, which made it very sensitive to Pf4 filamentous phage in biofilms. Upon contact, Pf4 filamentous phage could infect and lyse the engineered bacteria to release the intracellular accumulation of exopolysaccharide hydrolase PelA and PslG, thereby destroying the biofilm of P. aeruginosa (Figure 2; Wang et al., 2021).
As mentioned earlier, bacteriophages are considered to be potential drugs for the prevention and control of biofilms due to their infection diversity and specificity (Melo et al., 2019). However, the application of phages in biofilm control still has some limitations (Abedon et al., 2021). For example, a reduction in the metabolic activity of biofilm bacterial cells due to phage infection is dependent on host growth conditions (Chegini et al., 2020) and stimulates the rapid release of bacterial endotoxins, leading to an inflammatory response (Ferriol-Gonzalez and Domingo-Calap, 2020).
aPDT
aPDT is an emerging non-invasive treatment method that uses non-toxic photosensitizer (PS), specific wavelengths of visible or near-infrared light, and molecular oxygen around or inside pathogens to produce phototoxic reactions to kill pathogens (Rajesh et al., 2011). aPDT can also destroy microbial biofilms in a process that consists of two steps. The first step is the binding of PSs to the biofilm matrix. Although some types of PSs only bind to the cell surface, most types of PSs can pass through the cytoplasmic membrane and enter the cytoplasm. PSs bound to the biofilm matrix generate reactive oxygen species (ROS) under light, thereby initiating multi-target damage (Hu et al., 2018), which attacks various biofilm components, leading to disintegration, including the disintegration of lipids, proteins, DNA, and exopolysaccharides in the matrix (Dosselli et al., 2012; Martins Antunes de Melo et al., 2021). Studies have reported many examples of aPDT used in the treatment of biofilm infections, and some have been used in clinical trials (Tahmassebi et al., 2015; Liang et al., 2020).
Polysaccharides are the most abundant polymers in biofilm matrices In the presence of a certain concentration of Tetra-Py+-Me, the polysaccharide concentration in the P. aeruginosa biofilm was significantly reduced after irradiation, and its biofilm substrate was attacked by photodynamic force and destroyed (Beirao et al., 2014). aPDT targeting related proteins also affect P. aeruginosa biofilm formation. The PS methylene blue (MB) combined with the antibiotic gentamicin (GN) has a synergistic antibacterial effect on plankton. Adding GN at a concentration where MB alone does not have a significant antibacterial effect can exert a positive bactericidal effect against P. aeruginosa biofilms. This synergistic killing mechanism may be caused by GN acting on the level of protein synthesis, changing the permeability of the bacterial wall and thereby promoting the accumulation of MB, but its potential mechanism needs further research (Perez-Laguna et al., 2020). Recently, antimicrobial photothermal therapy (aPTT) was demonstrated to be a promising method to eliminate planktonic cells and biofilms (Al-Bakri and Mahmoud, 2019; Li et al., 2019). The combined use of aPDT and aPTT has also become an effective local replacement therapy for the treatment of antibiotic-resistant bacterial infections and biofilms (Bilici et al., 2020). While 3-aminopropylsilane-coated superparamagnetic iron oxide nanoparticles have no significant inhibition on biofilms without laser treatment, the addition of laser treatment significantly reduces P. aeruginosa biofilms. Furthermore, after combining nanoparticles with PS, the biofilm can be reduced again. This combination of nanoparticles and PS may enhance the treatment of drug-resistant bacteria and their biofilms through the dual aPDT/aPTT mechanism (Bilici et al., 2020).
Due to its multi-targeted damage to microbial cells and its inability to induce drug resistance, aPDT has received increasing attention as an alternative treatment (Cieplik et al., 2018), and it is also effective in combination with antibiotics (Feng et al., 2021). However, the application of aPDT has certain limitations. The limitation of light transmission conditions makes it more effective in the clinical treatment of local infections. In addition, the characteristics of PS and the corresponding light source affect its application. PS with low molecular weight and high penetrating power should be selected, and the cost should also be controlled (Wainwright et al., 2017).
Conclusions and prospective applications
Pseudomonas aeruginosa biofilm includes three main parts: exopolysaccharides, eDNA, and matrix protein. Different components play different roles in its adhesion, maturation, and dispersal processes and are regulated by factors such as the QS system. The most commonly used treatment of P. aeruginosa biofilm infection is mainly a single antibiotic treatment, but this clinical treatment faces many challenges due to drug resistance. With the development of drug-resistant strains, many promising therapeutic strategies have been developed to address these issues, such as combining antibiotics, targeting biofilms through enzymes or quorum-sensing systems, or using new photodynamic therapies and other compounds to prevent or inhibit P. aeruginosa biofilm formation by targeting the diffusion of biofilm formation. NO has also been shown to regulate biofilms through targeted diffusion (McDougald et al., 2011), exerting an inhibitory effect in the final stages of biofilm formation. However, when using dispersion as a therapeutic strategy, it is uncertain how efficient the dispersion reaction needs to be to become an effective therapeutic agent, and clinical treatments are mostly conducted on mixed biofilms (Lee and Yoon, 2017), which requires researchers to further explore diffusing agents and diffusion-inducing agents.
In addition to the above-mentioned therapeutic strategies, there are many other interesting methods that can be used to remove P. aeruginosa biofilms in clinical practice. P. aeruginosa itself can produce signal molecules that have inhibitory activity on its formed biofilm. For example, cis-2-decenoic acid, a short-chain fatty acid produced by P. aeruginosa, acts as a dispersal signal targeting the biofilm of some bacteria (Rahmani-Badi et al., 2014; Vuotto and Donelli, 2019; Jiang et al., 2020). In addition, some new materials such as nanoparticles, a class of emerging antibacterial agents, exhibit an antibacterial mechanism that includes the destruction of bacterial biofilms, and many innovative anti-biofilm nanomedicines and nanomaterials have been developed for clinical treatment (Xiu et al., 2021). A recent study developed a novel photocatalytic quantum dot-armed bacteriophage nanosystem that combined phage therapy and photodynamic therapy, not only specifically binding to host P. aeruginosa, but also targeting host bacteria through the inherent infectivity of phages, locally generating massive amounts of ROS under visible light irradiation, and thereby demonstrating potent anti-biofilm activity (Wang et al., 2022). However, microorganisms adhere to the surfaces of medical devices and are prone to forming biofilms, leading to inevitable and challenging issues with P. aeruginosa biofilm infection caused by the use of clinical medical devices (Wi and Patel, 2018). The surface modification of biomaterials has been the focus of extensive research to decrease microbial colonization and biofilm formation, and has been reviewed in detail (Yadav et al., 2021). An effective antimicrobial surface coating can prevent P. aeruginosa from adhering, achieving an anti-biofilm effect. One study achieved zero P. aeruginosa biofilm adhesion by adding lubricating fluids, consisting of perfluorinated liquids, to porous polytetrafluorethylene (PTFE) to fabricate liquid-infused surfaces (Epstein et al., 2012). In addition, the production of P. aeruginosa biofilms was reduced by four orders of magnitude after using a slippery omniphobic covalently attached liquid surface compared to polydimethylsiloxane (PDMS), a widely used medical implant material (Zhu et al., 2022). This biofilm-resistant liquid-like solid surface provides a novel strategy for the treatment of P. aeruginosa biofilms.
In addition to P. aeruginosa, there are multiple microorganisms that cause biofilm-related diseases, such as Staphylococcus aureus, Candida albicans, and Mycobacterium tuberculosis, which cause serious global health problems due to their resistance to antimicrobial agents. The rapid development of new antimicrobial agents to overcome resistance is urgent, and gaining insights into the specific mechanisms of biofilm occurrence and their interactions with the host is key to solving the problem. Although biofilms have been studied through genomics, proteomics, and RNA sequencing, the rapid evolution of microorganisms has exceeded the pace of therapeutic technology development. New technologies to monitor biofilm formation and the responses of biofilm to antibiotic therapy are required. Furthermore, direct eradication becomes difficult as pathogens evolve defenses against antimicrobial agents, and inhibiting bacterial virulence may be more effective than killing bacteria while also providing new possibilities for the treatment of biofilm infections.
Author contributions
JL conceptualized the article and critically revised the work. RY, JC, JW, and PL performed the literature search and wrote the manuscript. RY and JC prepared the figures and tables. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (32070103, 31860012, and 31700031), the Natural Science Basic Research Plan in Shaanxi Province of China (2021JM-415), the Regional Development Talent Project of the “Special Support Plan” of Shaanxi Province, a grant from the Outstanding Young Talent Support Plan of the Higher Education Institutions of Shaanxi Province, the Youth Innovation Team of Shaanxi Universities (2022), and by the Startup Foundation for Doctors of Yan’an University (YDBK2016-01).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Abdi M., Mirkalantari S., Amirmozafari N. (2019). Bacterial resistance to antimicrobial peptides. J. Pept. Sci. 25:e3210. doi: 10.1002/psc.3210 [DOI] [PubMed] [Google Scholar]
- Abedon S. T., Danis-Wlodarczyk K. M., Wozniak D. J., Sullivan M. B. (2021). Improving phage-biofilm in vitro experimentation. Viruses 13, 1175. doi: 10.3390/v13061175, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abedon S. T., Kuhl S. J., Blasdel B. G., Kutter E. M. (2011). Phage treatment of human infections. Bacteriophage 1, 66–85. doi: 10.4161/bact.1.2.15845, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed S., Rudden M., Smyth T. J., Dooley J. S. G., Marchant R., Banat I. M. (2019). Natural quorum sensing inhibitors effectively downregulate gene expression of Pseudomonas aeruginosa virulence factors. Appl. Microbiol. Biotechnol. 103, 3521–3535. doi: 10.1007/s00253-019-09618-0, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bakri A. G., Mahmoud N. N. (2019). Photothermal-induced antibacterial activity of gold nanorods loaded into polymeric hydrogel against Pseudomonas aeruginosa biofilm. Molecules 24, 2661. doi: 10.3390/molecules24142661, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Dahmoshi H., Al-Obaidi R., Al-Khafaji N. (2021). “Pseudomonas aeruginosa: diseases, biofilm and antibiotic resistance.” In: Pseudomonas aeruginosa — Biofilm Formation, Infections and Treatments. eds. Das T. (London: InTechOpen Limited; ). [Google Scholar]
- Allesen-Holm M., Barken K. B., Yang L., Klausen M., Webb J. S., Kjelleberg S., et al. (2006). A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 59, 1114–1128. doi: 10.1111/j.1365-2958.2005.05008.x, PMID: [DOI] [PubMed] [Google Scholar]
- Annunziato G., Costantino G. (2020). Antimicrobial peptides (AMPs): A patent review (2015-2020). Expert Opin. Ther. Pat. 30, 931–947. doi: 10.1080/13543776.2020.1851679, PMID: [DOI] [PubMed] [Google Scholar]
- Martins Antunes de Melo W. C., Celiesiute-Germaniene R., Simonis P., Stirke A. (2021). Antimicrobial photodynamic therapy (aPDT) for biofilm treatments. Possible synergy between aPDT and pulsed electric fields. Virulence 12, 2247–2272. doi: 10.1080/21505594.2021.1960105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki W., Ueda M. (2013). Characterization of antimicrobial peptides toward the development of novel antibiotics. Pharmaceuticals (Basel) 6, 1055–1081. doi: 10.3390/ph6081055, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora D. P., Hossain S., Xu Y., Boon E. M. (2015). Nitric oxide regulation of bacterial biofilms. Biochemistry 54, 3717–3728. doi: 10.1021/bi501476n [DOI] [PubMed] [Google Scholar]
- Azam M. W., Khan A. U. (2019). Updates on the pathogenicity status of Pseudomonas aeruginosa. Drug Discov. Today 24, 350–359. doi: 10.1016/j.drudis.2018.07.003, PMID: [DOI] [PubMed] [Google Scholar]
- Baker P., Hill P. J., Snarr B. D., Alnabelseya N., Pestrak M. J., Lee M. J., et al. (2016). Exopolysaccharide biosynthetic glycoside hydrolases can be utilized to disrupt and prevent Pseudomonas aeruginosa biofilms. Sci. Adv. 2:e1501632. doi: 10.1126/sciadv.1501632, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bala A., Kumar L., Chhibber S., Harjai K. (2015). Augmentation of virulence related traits of pqs mutants by pseudomonas quinolone signal through membrane vesicles. J. Basic Microbiol. 55, 566–578. doi: 10.1002/jobm.201400377, PMID: [DOI] [PubMed] [Google Scholar]
- Banin E., Lozinski A., Brady K. M., Berenshtein E., Butterfield P. W., Moshe M., et al. (2008). The potential of desferrioxamine-gallium as an anti-pseudomonas therapeutic agent. Proc. Natl. Acad. Sci. U. S. A. 105, 16761–16766. doi: 10.1073/pnas.0808608105, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banin E., Vasil M. L., Greenberg E. P. (2005). Iron and Pseudomonas aeruginosa biofilm formation. Proc. Natl. Acad. Sci. U. S. A. 102, 11076–11081. doi: 10.1073/pnas.0504266102, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barken K. B., Pamp S. J., Yang L., Gjermansen M., Bertrand J. J., Klausen M., et al. (2008). Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 10, 2331–2343. doi: 10.1111/j.1462-2920.2008.01658.x, PMID: [DOI] [PubMed] [Google Scholar]
- Barraud N., Hassett D. J., Hwang S. H., Rice S. A., Kjelleberg S., Webb J. S. (2006). Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J. Bacteriol. 188, 7344–7353. doi: 10.1128/JB.00779-06, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraud N., Kardak B. G., Yepuri N. R., Howlin R. P., Webb J. S., Faust S. N., et al. (2012). Cephalosporin-3'-diazeniumdiolates: targeted NO-donor prodrugs for dispersing bacterial biofilms. Angew. Chem. Int. Ed. Engl. 51, 9057–9060. doi: 10.1002/anie.201202414, PMID: [DOI] [PubMed] [Google Scholar]
- Beaudoin T., Stone T. A., Glibowicka M., Adams C., Yau Y., Ahmadi S., et al. (2018). Activity of a novel antimicrobial peptide against Pseudomonas aeruginosa biofilms. Sci. Rep. 8, 14728. doi: 10.1038/s41598-018-33016-7, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beirao S., Fernandes S., Coelho J., Faustino M. A., Tome J. P., Neves M. G., et al. (2014). Photodynamic inactivation of bacterial and yeast biofilms with a cationic porphyrin. Photochem. Photobiol. 90, 1387–1396. doi: 10.1111/php.12331, PMID: [DOI] [PubMed] [Google Scholar]
- Bilici K., Atac N., Muti A., Baylam I., Dogan O., Sennaroglu A., et al. (2020). Broad spectrum antibacterial photodynamic and photothermal therapy achieved with indocyanine green loaded SPIONs under near infrared irradiation. Biomater. Sci. 8, 4616–4625. doi: 10.1039/d0bm00821d, PMID: [DOI] [PubMed] [Google Scholar]
- Bonchi C., Frangipani E., Imperi F., Visca P. (2015). Pyoverdine and proteases affect the response of Pseudomonas aeruginosa to gallium in human serum. Antimicrob. Agents Chemother. 59, 5641–5646. doi: 10.1128/AAC.01097-15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd M. S., Sadovskaya I., Vinogradov E., Lu H., Sprinkle A. B., Richardson S. H., et al. (2009). Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol. Microbiol. 73, 622–638. doi: 10.1111/j.1365-2958.2009.06795.x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda W., Joseph T. P., Padhiar A. A., Guo X., Min L., Wang W., et al. (2017). Combined effect of linolenic acid and tobramycin on Pseudomonas aeruginosa biofilm formation and quorum sensing. Exp. Ther. Med. 14, 4328–4338. doi: 10.3892/etm.2017.5110, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y., Wang P. C., Ma H. M., Chen S. Y., Fu Y. H., Liu Y. Y., et al. (2019). Design, synthesis and evaluation of halogenated furanone derivatives as quorum sensing inhibitors in Pseudomonas aeruginosa. Eur. J. Pharm. Sci. 140:105058. doi: 10.1016/j.ejps.2019.105058, PMID: [DOI] [PubMed] [Google Scholar]
- Chauhan A., Lebeaux D., Ghigo J. M., Beloin C. (2012). Full and broad-spectrum in vivo eradication of catheter-associated biofilms using gentamicin-EDTA antibiotic lock therapy. Antimicrob. Agents Chemother. 56, 6310–6318. doi: 10.1128/AAC.01606-12, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chegini Z., Khoshbayan A., Taati Moghadam M., Farahani I., Jazireian P., Shariati A. (2020). Bacteriophage therapy against Pseudomonas aeruginosa biofilms: A review. Ann. Clin. Microbiol. Antimicrob. 19, 45. doi: 10.1186/s12941-020-00389-5, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S., Wang J., Shang L., Akhtar M. U., Wang Z., Shi B., et al. (2019). Short, symmetric-helical peptides have narrow-spectrum activity with low resistance potential and high selectivity. Biomater. Sci. 7, 2394–2409. doi: 10.1039/c9bm00044e, PMID: [DOI] [PubMed] [Google Scholar]
- Chrzanowski L., Lawniczak L., Czaczyk K. (2012). Why do microorganisms produce rhamnolipids? World J. Microbiol. Biotechnol. 28, 401–419. doi: 10.1007/s11274-011-0854-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieplik F., Deng D., Crielaard W., Buchalla W., Hellwig E., Al-Ahmad A., et al. (2018). Antimicrobial photodynamic therapy – what we know and what we don't. Crit. Rev. Microbiol. 44, 571–589. doi: 10.1080/1040841X.2018.1467876, PMID: [DOI] [PubMed] [Google Scholar]
- Clokie M. R., Millard A. D., Letarov A. V., Heaphy S. (2011). Phages in nature. Bacteriophage 1, 31–45. doi: 10.4161/bact.1.1.14942, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin K. M., Alnabelseya N., Baker P., Whitney J. C., Howell P. L., Parsek M. R. (2013). PelA deacetylase activity is required for Pel polysaccharide synthesis in Pseudomonas aeruginosa. J. Bacteriol. 195, 2329–2339. doi: 10.1128/JB.02150-12, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin K. M., Gordon V. D., Murakami K., Borlee B. R., Wozniak D. J., Wong G. C., et al. (2011). The pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog. 7:e1001264. doi: 10.1371/journal.ppat.1001264, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin K. M., Irie Y., Tart C. S., Urbano R., Whitney J. C., Ryder C., et al. (2012). The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ. Microbiol. 14, 1913–1928. doi: 10.1111/j.1462-2920.2011.02657.x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton J. W., Stewart P. S., Greenberg E. P. (1999). Bacterial biofilms: A common cause of persistent infections. Science 284, 1318–1322. doi: 10.1126/science.284.5418.1318 [DOI] [PubMed] [Google Scholar]
- Cutruzzola F., Frankenberg-Dinkel N. (2016). Origin and impact of nitric oxide in Pseudomonas aeruginosa biofilms. J. Bacteriol. 198, 55–65. doi: 10.1128/JB.00371-15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daboor S. M., Rohde J. R., Cheng Z. (2021). Disruption of the extracellular polymeric network of Pseudomonas aeruginosa biofilms by alginate lyase enhances pathogen eradication by antibiotics. J. Cyst. Fibros. 20, 264–270. doi: 10.1016/j.jcf.2020.04.006, PMID: [DOI] [PubMed] [Google Scholar]
- Das T., Manoharan A., Whiteley G., Glasbey T., Manos J. (2020). “Pseudomonas aeruginosa biofilms and infections: roles of extracellular molecules,” in New and Future Developments in Microbial Biotechnology and Bioengineering: Microbial Biofilms. eds. Yadav M. K., Singh B. P. (Amsterdam: Elsevier; ), 29–46. [Google Scholar]
- Das T., Sharma P. K., Busscher H. J., van der Mei H. C., Krom B. P. (2010). Role of extracellular DNA in initial bacterial adhesion and surface aggregation. Appl. Environ. Microbiol. 76, 3405–3408. doi: 10.1128/AEM.03119-09, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey M. E., Caiazza N. C., O'Toole G. A. (2003). Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J. Bacteriol. 185, 1027–1036. doi: 10.1128/JB.185.3.1027-1036.2003, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente-Nunez C., Korolik V., Bains M., Nguyen U., Breidenstein E. B., Horsman S., et al. (2012). Inhibition of bacterial biofilm formation and swarming motility by a small synthetic cationic peptide. Antimicrob. Agents Chemother. 56, 2696–2704. doi: 10.1128/AAC.00064-12, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Y. P., Lin Q., Chen C., Montelaro R. C., Doi Y., Deslouches B. (2020). Enhanced therapeutic index of an antimicrobial peptide in mice by increasing safety and activity against multidrug-resistant bacteria. Sci. Adv. 6, eaay6817. doi: 10.1126/sciadv.aay6817, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlan R. M. (2002). Biofilms: microbial life on surfaces. Emerg. Infect. Dis. 8, 881–890. doi: 10.3201/eid0809.020063, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosselli R., Millioni R., Puricelli L., Tessari P., Arrigoni G., Franchin C., et al. (2012). Molecular targets of antimicrobial photodynamic therapy identified by a proteomic approach. J. Proteome 77, 329–343. doi: 10.1016/j.jprot.2012.09.007, PMID: [DOI] [PubMed] [Google Scholar]
- Drenkard E. (2003). Antimicrobial resistance of Pseudomonas aeruginosa biofilms. Microbes Infect. 5, 1213–1219. doi: 10.1016/j.micinf.2003.08.009 [DOI] [PubMed] [Google Scholar]
- Dufour D., Leung V., Lévesque C. (2010). Bacterial biofilm: structure, function, and antimicrobial resistance. Endod. Top. 22, 2–16. doi: 10.1111/j.1601-1546.2012.00277.x [DOI] [Google Scholar]
- Eckhart L., Fischer H., Barken K. B., Tolker-Nielsen T., Tschachler E. (2007). DNase1L2 suppresses biofilm formation by Pseudomonas aeruginosa and Staphylococcus aureus. Br. J. Dermatol. 156, 1342–1345. doi: 10.1111/j.1365-2133.2007.07886.x, PMID: [DOI] [PubMed] [Google Scholar]
- Elmanama A. A., Al-Sheboul S., Abu-Dan R. I. (2020). Antimicrobial resistance and biofilm formation of Pseudomonas aeruginosa. Int. Arabic J. Antimicrob. Agents 10, 3. doi: 10.3823/846 [DOI] [Google Scholar]
- Epstein A. K., Wong T. S., Belisle R. A., Boggs E. M., Aizenberg J. (2012). Liquid-infused structured surfaces with exceptional anti-biofouling performance. Proc. Natl. Acad. Sci. U. S. A. 109, 13182–13187. doi: 10.1073/pnas.1201973109, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Coradi Tonon C., Ashraf S., Hasan T. (2021). Photodynamic and antibiotic therapy in combination against bacterial infections: efficacy, determinants, mechanisms, and future perspectives. Adv. Drug Deliv. Rev. 177:113941. doi: 10.1016/j.addr.2021.113941, PMID: [DOI] [PubMed] [Google Scholar]
- Feng Q., Huang Y., Chen M., Li G., Chen Y. (2015). Functional synergy of alpha-helical antimicrobial peptides and traditional antibiotics against gram-negative and gram-positive bacteria in vitro and in vivo. Eur. J. Clin. Microbiol. Infect. Dis. 34, 197–204. doi: 10.1007/s10096-014-2219-3, PMID: [DOI] [PubMed] [Google Scholar]
- Ferriol-Gonzalez C., Domingo-Calap P. (2020). Phages for biofilm removal. Antibiotics (Basel) 9, 268. doi: 10.3390/antibiotics9050268, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel S. E., Kolter R. (2001). DNA as a nutrient: novel role for bacterial competence gene homologs. J. Bacteriol. 183, 6288–6293. doi: 10.1128/JB.183.21.6288-6293.2001, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming D., Chahin L., Rumbaugh K. (2017). Glycoside hydrolases degrade polymicrobial bacterial biofilms in wounds. Antimicrob. Agents Chemother. 61, e01998-16. doi: 10.1128/AAC.01998-16, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming H. C., Wuertz S. (2019). Bacteria and archaea on earth and their abundance in biofilms. Nat. Rev. Microbiol. 17, 247–260. doi: 10.1038/s41579-019-0158-9, PMID: [DOI] [PubMed] [Google Scholar]
- Franklin M. J., Nivens D. E., Weadge J. T., Howell P. L. (2011). Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, alginate, Pel, and Psl. Front. Microbiol. 2, 167. doi: 10.3389/fmicb.2011.00167, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghafoor A., Hay I. D., Rehm B. H. (2011). Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture. Appl. Environ. Microbiol. 77, 5238–5246. doi: 10.1128/AEM.00637-11, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghorbani H., Memar M. Y., Sefidan F. Y., Yekani M., Ghotaslou R. (2017). In vitro synergy of antibiotic combinations against planktonic and biofilm Pseudomonas aeruginosa. GMS Hyg. Infect. Control. 12, Doc17. doi: 10.3205/dgkh000302, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnanadhas D. P., Elango M., Datey A., Chakravortty D. (2015). Chronic lung infection by Pseudomonas aeruginosa biofilm is cured by L-methionine in combination with antibiotic therapy. Sci. Rep. 5, 16043. doi: 10.1038/srep16043, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss C. H., Kaneko Y., Khuu L., Anderson G. D., Ravishankar S., Aitken M. L., et al. (2018). Gallium disrupts bacterial iron metabolism and has therapeutic effects in mice and humans with lung infections. Sci. Transl. Med. 10, eaat7520. doi: 10.1126/scitranslmed.aat7520, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper D., Parracho H., Walker J., Sharp R., Hughes G., Werthén M., et al. (2014). Bacteriophages and Biofilms. Antibiotics 3, 270–284. doi: 10.3390/antibiotics3030270 [DOI] [Google Scholar]
- Hoiby N., Bjarnsholt T., Givskov M., Molin S., Ciofu O. (2010). Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 35, 322–332. doi: 10.1016/j.ijantimicag.2009.12.011 [DOI] [PubMed] [Google Scholar]
- Hoiby N., Ciofu O., Johansen H. K., Song Z. J., Moser C., Jensen P. O., et al. (2011). The clinical impact of bacterial biofilms. Int. J. Oral Sci. 3, 55–65. doi: 10.4248/IJOS11026, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain S., Boon E. M. (2017). Discovery of a novel nitric oxide binding protein and nitric-oxide-responsive signaling pathway in Pseudomonas aeruginosa. ACS Infect. Dis. 3, 454–461. doi: 10.1021/acsinfecdis.7b00027, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlin R. P., Cathie K., Hall-Stoodley L., Cornelius V., Duignan C., Allan R. N., et al. (2017). Low-dose nitric oxide as targeted anti-biofilm adjunctive therapy to treat chronic Pseudomonas aeruginosa infection in cystic fibrosis. Mol. Ther. 25, 2104–2116. doi: 10.1016/j.ymthe.2017.06.021, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Huang Y. Y., Wang Y., Wang X., Hamblin M. R. (2018). Antimicrobial photodynamic therapy to control clinically relevant biofilm infections. Front. Microbiol. 9, 1299. doi: 10.3389/fmicb.2018.01299, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Shao X., Xie Y., Wang T., Zhang Y., Wang X., et al. (2019). An integrated genomic regulatory network of virulence-related transcriptional factors in Pseudomonas aeruginosa. Nat. Commun. 10, 2931. doi: 10.1038/s41467-019-10778-w, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes G., Webber M. A. (2017). Novel approaches to the treatment of bacterial biofilm infections. Br. J. Pharmacol. 174, 2237–2246. doi: 10.1111/bph.13706, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hymes S. R., Randis T. M., Sun T. Y., Ratner A. J. (2013). DNase inhibits Gardnerella vaginalis biofilms in vitro and in vivo. J. Infect. Dis. 207, 1491–1497. doi: 10.1093/infdis/jit047, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamal M., Ahmad W., Andleeb S., Jalil F., Imran M., Nawaz M. A., et al. (2018). Bacterial biofilm and associated infections. J. Chin. Med. Assoc. 81, 7–11. doi: 10.1016/j.jcma.2017.07.012 [DOI] [PubMed] [Google Scholar]
- Jennings L. K., Storek K. M., Ledvina H. E., Coulon C., Marmont L. S., Sadovskaya I., et al. (2015). Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc. Natl. Acad. Sci. U. S. A. 112, 11353–11358. doi: 10.1073/pnas.1503058112, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Geng M., Bai L. (2020). Targeting biofilms therapy: current research strategies and development hurdles. Microorganisms 8, 1222. doi: 10.3390/microorganisms8081222, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P., Li J., Han F., Duan G., Lu X., Gu Y., et al. (2011). Antibiofilm activity of an exopolysaccharide from marine bacterium vibrio sp. QY101. PLoS One 6:e18514. doi: 10.1371/journal.pone.0018514, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia V. C. (2013). Quorum sensing inhibitors: An overview. Biotechnol. Adv. 31, 224–245. doi: 10.1016/j.biotechadv.2012.10.004 [DOI] [PubMed] [Google Scholar]
- Kamal A. A. M., Maurer C. K., Allegretta G., Haupenthal J., Empting M., Hartmann R. W. (2017). “Quorum sensing inhibitors as pathoblockers for Pseudomonas aeruginosa infections: A new concept in anti-infective drug discovery,” in Antibacterials, Topics in Medicinal Chemistry. eds. Fisher J., Mobashery S., Miller M. (Cham: Springer; ), 185–210. [Google Scholar]
- Kamaruzzaman N. F., Tan L. P., Mat Yazid K. A., Saeed S. I., Hamdan R. H., Choong S. S., et al. (2018). Targeting the bacterial protective armour; challenges and novel strategies in the treatment of microbial biofilm. Materials (Basel) 11, 1705. doi: 10.3390/ma11091705, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko Y., Thoendel M., Olakanmi O., Britigan B. E., Singh P. K. (2007). The transition metal gallium disrupts Pseudomonas aeruginosa iron metabolism and has antimicrobial and antibiofilm activity. J. Clin. Invest. 117, 877–888. doi: 10.1172/JCI30783, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D., Revtovich A. V., Deyanov A. E., Kirienko N. V. (2021). Pyoverdine inhibitors and gallium nitrate synergistically affect Pseudomonas aeruginosa. mSphere 6:e0040121. doi: 10.1128/mSphere.00401-21, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan F., Pham D. T. N., Kim Y. M. (2020). Alternative strategies for the application of aminoglycoside antibiotics against the biofilm-forming human pathogenic bacteria. Appl. Microbiol. Biotechnol. 104, 1955–1976. doi: 10.1007/s00253-020-10360-1, PMID: [DOI] [PubMed] [Google Scholar]
- Kim S. K., Lee J. H. (2016). Biofilm dispersion in Pseudomonas aeruginosa. J. Microbiol. 54, 71–85. doi: 10.1007/s12275-016-5528-7 [DOI] [PubMed] [Google Scholar]
- Kitao T., Lepine F., Babloudi S., Walte F., Steinbacher S., Maskos K., et al. (2018). Molecular insights into function and competitive inhibition of Pseudomonas aeruginosa multiple virulence factor regulator. mBio 9, e02158-17. doi: 10.1128/mBio.02158-17, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H., Allan R. N., Howlin R. P., Stoodley P., Hall-Stoodley L. (2017). Targeting microbial biofilms: current and prospective therapeutic strategies. Nat. Rev. Microbiol. 15, 740–755. doi: 10.1038/nrmicro.2017.99, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostakioti M., Hadjifrangiskou M., Hultgren S. J. (2013). Bacterial biofilms: development, dispersal, and therapeutic strategies in the dawn of the postantibiotic era. Cold Spring Harb. Perspect. Med. 3:a010306. doi: 10.1101/cshperspect.a010306, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach K. N., Fleming D., Wells M. J., Rumbaugh K. P., Gordon V. D. (2020). Specific disruption of established Pseudomonas aeruginosa biofilms using polymer-attacking enzymes. Langmuir 36, 1585–1595. doi: 10.1021/acs.langmuir.9b02188, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Alam A., Rani M., Ehtesham N. Z., Hasnain S. E. (2017). Biofilms: survival and defense strategy for pathogens. Int. J. Med. Microbiol. 307, 481–489. doi: 10.1016/j.ijmm.2017.09.016, PMID: [DOI] [PubMed] [Google Scholar]
- Kumar L., Chhibber S., Harjai K. (2013). Zingerone inhibit biofilm formation and improve antibiofilm efficacy of ciprofloxacin against Pseudomonas aeruginosa PAO1. Fitoterapia 90, 73–78. doi: 10.1016/j.fitote.2013.06.017, PMID: [DOI] [PubMed] [Google Scholar]
- Kumar L., Chhibber S., Kumar R., Kumar M., Harjai K. (2015). Zingerone silences quorum sensing and attenuates virulence of Pseudomonas aeruginosa. Fitoterapia 102, 84–95. doi: 10.1016/j.fitote.2015.02.002, PMID: [DOI] [PubMed] [Google Scholar]
- Kutateladze M., Adamia R. (2010). Bacteriophages as potential new therapeutics to replace or supplement antibiotics. Trends Biotechnol. 28, 591–595. doi: 10.1016/j.tibtech.2010.08.001 [DOI] [PubMed] [Google Scholar]
- Kutter E., De Vos D., Gvasalia G., Alavidze Z., Gogokhia L., Kuhl S., et al. (2010). Phage therapy in clinical practice: treatment of human infections. Curr. Pharm. Biotechnol. 11, 69–86. doi: 10.2174/138920110790725401 [DOI] [PubMed] [Google Scholar]
- Lashua L. P., Melvin J. A., Deslouches B., Pilewski J. M., Montelaro R. C., Bomberger J. M. (2016). Engineered cationic antimicrobial peptide (eCAP) prevents Pseudomonas aeruginosa biofilm growth on airway epithelial cells. J. Antimicrob. Chemother. 71, 2200–2207. doi: 10.1093/jac/dkw143, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le C. F., Fang C. M., Sekaran S. D. (2017). Intracellular targeting mechanisms by antimicrobial peptides. Antimicrob. Agents Chemother. 61, e02340-16. doi: 10.1128/AAC.02340-16, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebeaux D., Leflon-Guibout V., Ghigo J. M., Beloin C. (2015). In vitro activity of gentamicin, vancomycin or amikacin combined with EDTA or l-arginine as lock therapy against a wide spectrum of biofilm-forming clinical strains isolated from catheter-related infections. J. Antimicrob. Chemother. 70, 1704–1712. doi: 10.1093/jac/dkv044, PMID: [DOI] [PubMed] [Google Scholar]
- Lee K., Yoon S. S. (2017). Pseudomonas aeruginosa biofilm, a programmed bacterial life for fitness. J. Microbiol. Biotechnol. 27, 1053–1064. doi: 10.4014/jmb.1611.11056, PMID: [DOI] [PubMed] [Google Scholar]
- Lee J., Zhang L. (2015). The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 6, 26–41. doi: 10.1007/s13238-014-0100-x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lequette Y., Greenberg E. P. (2005). Timing and localization of rhamnolipid synthesis gene expression in Pseudomonas aeruginosa biofilms. J. Bacteriol. 187, 37–44. doi: 10.1128/JB.187.1.37-44.2005, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Geng X., Liu D., Li Z. (2019). Near-infrared light-enhanced protease-conjugated gold nanorods as a photothermal antimicrobial agent for elimination of exotoxin and biofilms. Int. J. Nanomedicine 14, 8047–8058. doi: 10.2147/IJN.S212750, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Xiao P., Wang Y., Hao Y. (2020). Mechanisms and control measures of mature biofilm resistance to antimicrobial agents in the clinical context. ACS Omega 5, 22684–22690. doi: 10.1021/acsomega.0c02294, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X., Zou Z., Zou Z., Li C., Dong X., Yin H., et al. (2020). Effect of antibacterial photodynamic therapy on Streptococcus mutans plaque biofilm in vitro. J. Innov. Opt. Health Sci. 13, 2050022. doi: 10.1142/S1793545820500224 [DOI] [Google Scholar]
- Lichtenberg M., Jakobsen T. H., Kuhl M., Kolpen M., Jensen P. O., Bjarnsholt T. (2022). The structure-function relationship of Pseudomonas aeruginosa in infections and its influence on the microenvironment. FEMS Microbiol. Rev. doi: 10.1093/femsre/fuac018 [Epub ahead of print], PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limoli D. H., Jones C. J., Wozniak D. J. (2015). Bacterial extracellular polysaccharides in biofilm formation and function. Microbiol. Spectr. 3, MB-0011–2014. doi: 10.1128/microbiolspec.MB-0011-2014, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limoli D. H., Rockel A. B., Host K. M., Jha A., Kopp B. T., Hollis T., et al. (2014). Cationic antimicrobial peptides promote microbial mutagenesis and pathoadaptation in chronic infections. PLoS Pathog. 10:e1004083. doi: 10.1371/journal.ppat.1004083, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Cheng J. (2019). “Quorum sensing in Pseudomonas aeruginosa and its relationship to biofilm development,” in Introduction to Biofilm Engineering. eds. Rathinam N. K., Sani R. K. (Washington, DC: ACS Publications; ), 1–16. [Google Scholar]
- Lin J., Cheng J., Wang Y., Shen X. (2018a). The pseudomonas quinolone signal (PQS): not just for quorum sensing anymore. Front. Cell. Infect. Microbiol. 8, 230. doi: 10.3389/fcimb.2018.00230, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q., Deslouches B., Montelaro R. C., Di Y. P. (2018b). Prevention of ESKAPE pathogen biofilm formation by antimicrobial peptides WLBU2 and LL37. Int. J. Antimicrob. Agents 52, 667–672. doi: 10.1016/j.ijantimicag.2018.04.019, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q., Pilewski J. M., Di Y. P. (2021). Acidic microenvironment determines antibiotic susceptibility and biofilm formation of Pseudomonas aeruginosa. Front. Microbiol. 12:747834. doi: 10.3389/fmicb.2021.747834, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llamas M. A., Sparrius M., Kloet R., Jimenez C. R., Vandenbroucke-Grauls C., Bitter W. (2006). The heterologous siderophores ferrioxamine B and ferrichrome activate signaling pathways in Pseudomonas aeruginosa. J. Bacteriol. 188, 1882–1891. doi: 10.1128/JB.188.5.1882-1891.2006, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Conover M., Lu H., Parsek M. R., Bayles K., Wozniak D. J. (2009). Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog. 5:e1000354. doi: 10.1371/journal.ppat.1000354, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Jackson K. D., Landry R. M., Parsek M. R., Wozniak D. J. (2006). Analysis of Pseudomonas aeruginosa conditional Psl variants reveals roles for the Psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J. Bacteriol. 188, 8213–8221. doi: 10.1128/JB.01202-06, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L. Z., Wang D., Liu Y., Zhang Z., Wozniak D. J. (2022). Regulation of biofilm exopolysaccharide biosynthesis and degradation in Pseudomonas aeruginosa. Annu. Rev. Microbiol. 76, 413–433. doi: 10.1146/annurev-micro-041320-111355 [DOI] [PubMed] [Google Scholar]
- Ma L., Wang S., Wang D., Parsek M. R., Wozniak D. J. (2012). The roles of biofilm matrix polysaccharide Psl in mucoid Pseudomonas aeruginosa biofilms. FEMS Immunol. Med. Microbiol. 65, 377–380. doi: 10.1111/j.1574-695X.2012.00934.x, PMID: [DOI] [PubMed] [Google Scholar]
- Mahlapuu M., Hakansson J., Ringstad L., Bjorn C. (2016). Antimicrobial peptides: An emerging category of therapeutic agents. Front. Cell. Infect. Microbiol. 6, 194. doi: 10.3389/fcimb.2016.00194, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann E. E., Wozniak D. J. (2012). Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol. Rev. 36, 893–916. doi: 10.1111/j.1574-6976.2011.00322.x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmont L. S., Whitfield G. B., Rich J. D., Yip P., Giesbrecht L. B., Stremick C. A., et al. (2017). PelA and PelB proteins form a modification and secretion complex essential for Pel polysaccharide-dependent biofilm formation in Pseudomonas aeruginosa. J. Biol. Chem. 292, 19411–19422. doi: 10.1074/jbc.M117.812842, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall B., Stintzi A., Gilmour C., Meyer J. M., Poole K. (2009). Citrate-mediated iron uptake in Pseudomonas aeruginosa: involvement of the citrate-inducible FecA receptor and the FeoB ferrous iron transporter. Microbiology (Reading) 155, 305–315. doi: 10.1099/mic.0.023531-0, PMID: [DOI] [PubMed] [Google Scholar]
- Martinez M., Goncalves S., Felicio M. R., Maturana P., Santos N. C., Semorile L., et al. (2019). Synergistic and antibiofilm activity of the antimicrobial peptide P5 against carbapenem-resistant Pseudomonas aeruginosa. Biochim. Biophys. Acta Biomembr. 1861, 1329–1337. doi: 10.1016/j.bbamem.2019.05.008, PMID: [DOI] [PubMed] [Google Scholar]
- Maura D., Rahme L. G. (2017). Pharmacological inhibition of the Pseudomonas aeruginosa MvfR quorum-sensing system interferes with biofilm formation and potentiates antibiotic-mediated biofilm disruption. Antimicrob. Agents Chemother. 61, e01362-17. doi: 10.1128/AAC.01362-17, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurice N. M., Bedi B., Sadikot R. T. (2018). Pseudomonas aeruginosa biofilms: Host response and clinical implications in lung infections. Am. J. Respir. Cell Mol. Biol. 58, 428–439. doi: 10.1165/rcmb.2017-0321TR, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDougald D., Rice S. A., Barraud N., Steinberg P. D., Kjelleberg S. (2011). Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat. Rev. Microbiol. 10, 39–50. doi: 10.1038/nrmicro2695 [DOI] [PubMed] [Google Scholar]
- Melo L. D. R., Pires D. P., Monteiro R., Azeredo J. (2019). “Phage therapy of infectious biofilms: challenges and strategies,” in Phage Therapy: A Practical Approach. eds. Górski A., Międzybrodzki R., Borysowski J. (Cham: Springer; ), 295–313. [Google Scholar]
- Mi L., Liu Y., Wang C., He T., Gao S., Xing S., et al. (2019). Identification of a lytic Pseudomonas aeruginosa phage depolymerase and its anti-biofilm effect and bactericidal contribution to serum. Virus Genes 55, 394–405. doi: 10.1007/s11262-019-01660-4, PMID: [DOI] [PubMed] [Google Scholar]
- Minandri F., Bonchi C., Frangipani E., Imperi F., Visca P. (2014). Promises and failures of gallium as an antibacterial agent. Future Microbiol. 9, 379–397. doi: 10.2217/fmb.14.3, PMID: [DOI] [PubMed] [Google Scholar]
- Mitchell K. F., Zarnowski R., Andes D. R. (2016). Fungal super glue: the biofilm matrix and its composition, assembly, and functions. PLoS Pathog. 12:e1005828. doi: 10.1371/journal.ppat.1005828, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradali M. F., Ghods S., Rehm B. H. (2017). Pseudomonas aeruginosa lifestyle: A paradigm for adaptation, survival, and persistence. Front. Cell. Infect. Microbiol. 7, 39. doi: 10.3389/fcimb.2017.00039, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradali M. F., Rehm B. H. (2019). “The role of alginate in bacterial biofilm formation,” in Extracellular Sugar-Based Biopolymers Matrices, Biologically-Inspired Systems. eds. Cohen E., Merzendorfer H. (Cham: Springer; ), 517–537. [Google Scholar]
- Muhlen S., Dersch P. (2016). Anti-virulence strategies to target bacterial infections. Curr. Top. Microbiol. Immunol. 398, 147–183. doi: 10.1007/82_2015_490 [DOI] [PubMed] [Google Scholar]
- O'Brien K. T., Noto J. G., Nichols-O'Neill L., Perez L. J. (2015). Potent irreversible inhibitors of LasR quorum sensing in Pseudomonas aeruginosa. ACS Med. Chem. Lett. 6, 162–167. doi: 10.1021/ml500459f, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okshevsky M., Regina V. R., Meyer R. L. (2015). Extracellular DNA as a target for biofilm control. Curr. Opin. Biotechnol. 33, 73–80. doi: 10.1016/j.copbio.2014.12.002, PMID: [DOI] [PubMed] [Google Scholar]
- Olivares E., Badel-Berchoux S., Provot C., Prevost G., Bernardi T., Jehl F. (2019). Clinical impact of antibiotics for the treatment of Pseudomonas aeruginosa biofilm infections. Front. Microbiol. 10, 2894. doi: 10.3389/fmicb.2019.02894, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overhage J., Campisano A., Bains M., Torfs E. C., Rehm B. H., Hancock R. E. (2008). Human host defense peptide LL-37 prevents bacterial biofilm formation. Infect. Immun. 76, 4176–4182. doi: 10.1128/IAI.00318-08, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Z., Raudonis R., Glick B. R., Lin T. J., Cheng Z. (2019). Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 37, 177–192. doi: 10.1016/j.biotechadv.2018.11.013, PMID: [DOI] [PubMed] [Google Scholar]
- Pang Z., Zhu Q. (2021). Traditional Chinese medicine is an alternative therapeutic option for treatment of Pseudomonas aeruginosa infections. Front. Pharmacol. 12:737252. doi: 10.3389/fphar.2021.737252, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei R., Lamas-Samanamud G. R. (2014). Inhibition of biofilm formation by T7 bacteriophages producing quorum-quenching enzymes. Appl. Environ. Microbiol. 80, 5340–5348. doi: 10.1128/AEM.01434-14, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Laguna V., Garcia-Luque I., Ballesta S., Perez-Artiaga L., Lampaya-Perez V., Rezusta A., et al. (2020). Photodynamic therapy using methylene blue, combined or not with gentamicin, against Staphylococcus aureus and Pseudomonas aeruginosa. Photodiagn. Photodyn. Ther. 31:101810. doi: 10.1016/j.pdpdt.2020.101810 [DOI] [PubMed] [Google Scholar]
- Pires D. P., Oliveira H., Melo L. D., Sillankorva S., Azeredo J. (2016). Bacteriophage-encoded depolymerases: their diversity and biotechnological applications. Appl. Microbiol. Biotechnol. 100, 2141–2151. doi: 10.1007/s00253-015-7247-0, PMID: [DOI] [PubMed] [Google Scholar]
- Poh W. H., Rice S. A. (2022). Recent developments in nitric oxide donors and delivery for antimicrobial and anti-biofilm applications. Molecules 27, 674. doi: 10.3390/molecules27030674, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes J. T. C., Toledo Borges A. B., Roque-Borda C. A., Pavan F. R. (2022). Antimicrobial peptides as an alternative for the eradication of bacterial biofilms of multi-drug resistant bacteria. Pharmaceutics 14, 642. doi: 10.3390/pharmaceutics14030642, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L., Christopher G. F. (2019). Role of flagella, type IV Pili, biosurfactants, and extracellular polymeric substance polysaccharides on the formation of pellicles by Pseudomonas aeruginosa. Langmuir 35, 5294–5304. doi: 10.1021/acs.langmuir.9b00271, PMID: [DOI] [PubMed] [Google Scholar]
- Qin Z., Yang L., Qu D., Molin S., Tolker-Nielsen T. (2009). Pseudomonas aeruginosa extracellular products inhibit staphylococcal growth, and disrupt established biofilms produced by Staphylococcus epidermidis. Microbiology (Reading) 155, 2148–2156. doi: 10.1099/mic.0.028001-0, PMID: [DOI] [PubMed] [Google Scholar]
- Rahmani-Badi A., Sepehr S., Mohammadi P., Soudi M. R., Babaie-Naiej H., Fallahi H. (2014). A combination of cis-2-decenoic acid and antibiotics eradicates pre-established catheter-associated biofilms. J. Med. Microbiol. 63, 1509–1516. doi: 10.1099/jmm.0.075374-0, PMID: [DOI] [PubMed] [Google Scholar]
- Rajesh S., Koshi E., Philip K., Mohan A. (2011). Antimicrobial photodynamic therapy: An overview. J. Indian Soc. Periodontol. 15, 323–327. doi: 10.4103/0972-124X.92563, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkumari J., Borkotoky S., Murali A., Suchiang K., Mohanty S. K., Busi S. (2018). Cinnamic acid attenuates quorum sensing associated virulence factors and biofilm formation in Pseudomonas aeruginosa PAO1. Biotechnol. Lett. 40, 1087–1100. doi: 10.1007/s10529-018-2557-9, PMID: [DOI] [PubMed] [Google Scholar]
- Redman W. K., Welch G. S., Rumbaugh K. P. (2020). Differential efficacy of glycoside hydrolases to disperse biofilms. Front. Cell. Infect. Microbiol. 10, 379. doi: 10.3389/fcimb.2020.00379, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendueles O., Kaplan J. B., Ghigo J. M. (2013). Antibiofilm polysaccharides. Environ. Microbiol. 15, 334–346. doi: 10.1111/j.1462-2920.2012.02810.x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R., Tiwari M., Donelli G., Tiwari V. (2018). Strategies for combating bacterial biofilms: A focus on anti-biofilm agents and their mechanisms of action. Virulence 9, 522–554. doi: 10.1080/21505594.2017.1313372, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumbaugh K. P., Sauer K. (2020). Biofilm dispersion. Nat. Rev. Microbiol. 18, 571–586. doi: 10.1038/s41579-020-0385-0, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford S. T., Bassler B. L. (2012). Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2:a012427. doi: 10.1101/cshperspect.a012427, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzhepishevska O., Ekstrand-Hammarstrom B., Popp M., Bjorn E., Bucht A., Sjostedt A., et al. (2011). The antibacterial activity of Ga3+ is influenced by ligand complexation as well as the bacterial carbon source. Antimicrob. Agents Chemother. 55, 5568–5580. doi: 10.1128/AAC.00386-11, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena P., Joshi Y., Rawat K., Bisht R. (2019). Biofilms: Architecture, resistance, quorum sensing and control mechanisms. Indian J. Microbiol. 59, 3–12. doi: 10.1007/s12088-018-0757-6, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelcher M., Donovan D. M., Loessner M. J. (2012). Bacteriophage endolysins as novel antimicrobials. Future Microbiol. 7, 1147–1171. doi: 10.2217/fmb.12.97, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutz C., Empting M. (2018). Targeting the pseudomonas quinolone signal quorum sensing system for the discovery of novel anti-infective pathoblockers. Beilstein J. Org. Chem. 14, 2627–2645. doi: 10.3762/bjoc.14.241, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutz C., Ho D. K., Hamed M. M., Abdelsamie A. S., Rohrig T., Herr C., et al. (2021). A new PqsR inverse agonist potentiates tobramycin efficacy to eradicate Pseudomonas aeruginosa biofilms. Adv. Sci. (Weinh) 8:e2004369. doi: 10.1002/advs.202004369, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyfi R., Kahaki F. A., Ebrahimi T., Montazersaheb S., Eyvazi S., Babaeipour V., et al. (2020). Antimicrobial peptides (AMPs): roles, functions and mechanism of action. Int. J. Pept. Res. Ther. 26, 1451–1463. doi: 10.1007/s10989-019-09946-9 [DOI] [Google Scholar]
- Shao X., Xie Y., Zhang Y., Liu J., Ding Y., Wu M., et al. (2020). Novel therapeutic strategies for treating Pseudomonas aeruginosa infection. Expert Opin. Drug Discov. 15, 1403–1423. doi: 10.1080/17460441.2020.1803274, PMID: [DOI] [PubMed] [Google Scholar]
- Sharma D., Misba L., Khan A. U. (2019). Antibiotics versus biofilm: An emerging battleground in microbial communities. Antimicrob. Resist. Infect. Control 8, 76. doi: 10.1186/s13756-019-0533-3, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K., Pagedar Singh A. (2018). Antibiofilm effect of DNase against single and mixed species biofilm. Foods 7, 42. doi: 10.3390/foods7030042, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha L., Fan H. M., Tao H. R., Huang J. D. (2022). Recent strategies to combat biofilms using antimicrobial agents and therapeutic approaches. Pathogens 11, 292. doi: 10.3390/pathogens11030292, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastava S., Shrivastava P. S., Ramasamy J. (2018). World health organization releases global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. J. Med. Soc. 32, 76. doi: 10.4103/jms.jms_25_17 [DOI] [Google Scholar]
- Smith W. D., Bardin E., Cameron L., Edmondson C. L., Farrant K. V., Martin I., et al. (2017). Current and future therapies for Pseudomonas aeruginosa infection in patients with cystic fibrosis. FEMS Microbiol. Lett. 364, fnx121. doi: 10.1093/femsle/fnx121, PMID: [DOI] [PubMed] [Google Scholar]
- Soothill J. (2013). Use of bacteriophages in the treatment of Pseudomonas aeruginosa infections. Expert Rev. Anti Infect. Ther. 11, 909–915. doi: 10.1586/14787210.2013.826990, PMID: [DOI] [PubMed] [Google Scholar]
- Soren O., Rineh A., Silva D. G., Cai Y., Howlin R. P., Allan R. N., et al. (2020). Cephalosporin nitric oxide-donor prodrug DEA-C3D disperses biofilms formed by clinical cystic fibrosis isolates of Pseudomonas aeruginosa. J. Antimicrob. Chemother. 75, 117–125. doi: 10.1093/jac/dkz378, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukarieh F., Williams P., Stocks M. J., Camara M. (2018). Pseudomonas aeruginosa quorum sensing systems as drug discovery targets: current position and future perspectives. J. Med. Chem. 61, 10385–10402. doi: 10.1021/acs.jmedchem.8b00540, PMID: [DOI] [PubMed] [Google Scholar]
- Srinivasan R., Santhakumari S., Poonguzhali P., Geetha M., Dyavaiah M., Xiangmin L. (2021). Bacterial biofilm inhibition: A focused review on recent therapeutic strategies for combating the biofilm mediated infections. Front. Microbiol. 12:676458. doi: 10.3389/fmicb.2021.676458, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartjes J. J., Das T., Sharifi S., Subbiahdoss G., Sharma P. K., Krom B. P., et al. (2013). A functional DNase I coating to prevent adhesion of bacteria and the formation of biofilm. Adv. Funct. Mater. 23, 2843–2849. doi: 10.1002/adfm.201202927 [DOI] [Google Scholar]
- Tahmassebi J. F., Drogkari E., Wood S. R. (2015). A study of the control of oral plaque biofilms via antibacterial photodynamic therapy. Eur. Arch. Paediatr. Dent. 16, 433–440. doi: 10.1007/s40368-014-0165-5, PMID: [DOI] [PubMed] [Google Scholar]
- Talapko J., Skrlec I. (2020). The principles, mechanisms, and benefits of unconventional agents in the treatment of biofilm infection. Pharmaceuticals (Basel) 13, 299. doi: 10.3390/ph13100299, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thi M. T. T., Wibowo D., Rehm B. H. A. (2020). Pseudomonas aeruginosa biofilms. Int. J. Mol. Sci. 21, 8671. doi: 10.3390/ijms21228671, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovar-Garcia A., Angarita-Zapata V., Cazares A., Jasso-Chavez R., Belmont-Diaz J., Sanchez-Torres V., et al. (2020). Characterization of gallium resistance induced in a Pseudomonas aeruginosa cystic fibrosis isolate. Arch. Microbiol. 202, 617–622. doi: 10.1007/s00203-019-01777-y, PMID: [DOI] [PubMed] [Google Scholar]
- Tummler B. (2019). Emerging therapies against infections with Pseudomonas aeruginosa. F1000Res 8:1371. doi: 10.12688/f1000research.19509.1, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuon F. F., Dantas L. R., Suss P. H., Tasca Ribeiro V. S. (2022). Pathogenesis of the Pseudomonas aeruginosa biofilm: A review. Pathogens 11, 300. doi: 10.3390/pathogens11030300, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuotto C., Donelli G. (2019). Novel treatment strategies for biofilm-based infections. Drugs 79, 1635–1655. doi: 10.1007/s40265-019-01184-z, PMID: [DOI] [PubMed] [Google Scholar]
- Wagner S., Sommer R., Hinsberger S., Lu C., Hartmann R. W., Empting M., et al. (2016). Novel strategies for the treatment of Pseudomonas aeruginosa infections. J. Med. Chem. 59, 5929–5969. doi: 10.1021/acs.jmedchem.5b01698, PMID: [DOI] [PubMed] [Google Scholar]
- Wainwright M., Maisch T., Nonell S., Plaetzer K., Almeida A., Tegos G. P., et al. (2017). Photoantimicrobials-are we afraid of the light? Lancet Infect. Dis. 17, e49–e55. doi: 10.1016/s1473-3099(16)30268-7, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Di Luca M., Tkhilaishvili T., Trampuz A., Gonzalez Moreno M. (2019). Synergistic activity of Fosfomycin, ciprofloxacin, and gentamicin Against Escherichia coli and Pseudomonas aeruginosa biofilms. Front. Microbiol. 10, 2522. doi: 10.3389/fmicb.2019.02522, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Fan X., Gonzalez Moreno M., Tkhilaishvili T., Du W., Zhang X., et al. (2022). Photocatalytic quantum dot-armed bacteriophage for combating drug-resistant bacterial infection. Adv. Sci. (Weinh) 9:e2105668. doi: 10.1002/advs.202105668, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Gao Q., Cheng J., Lin J. (2020). Regulation of Pseudomonas aeruginosa biofilms by quorum sensing systems and c-di-GMP. Acta Microbiol. Sin. 61, 1106–1122. doi: 10.13343/j.cnki.wsxb.20200367 [DOI] [Google Scholar]
- Wang S., Liu X., Liu H., Zhang L., Guo Y., Yu S., et al. (2015). The exopolysaccharide Psl-eDNA interaction enables the formation of a biofilm skeleton in Pseudomonas aeruginosa. Environ. Microbiol. Rep. 7, 330–340. doi: 10.1111/1758-2229.12252, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Niu Y., Zhang H., Li P., Zhang N., Cheng J., et al. (2021). An engineered bacterium for the targeted delivery of proteins to destroy Pseudomonas aeruginosa biofilms. Acta Microbiol. Sin. 61, 2726–2748. doi: 10.13343/j.cnki.wsxb.20200680 [DOI] [Google Scholar]
- Wei Q., Ma L. Z. (2013). Biofilm matrix and its regulation in Pseudomonas aeruginosa. Int. J. Mol. Sci. 14, 20983–21005. doi: 10.3390/ijms141020983, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y. L., Wu B. J., Kao P. H., Fu Y. S., Chang L. S. (2013). Antibacterial and membrane-damaging activities of beta-bungarotoxin B chain. J. Pept. Sci. 19, 1–8. doi: 10.1002/psc.2463, PMID: [DOI] [PubMed] [Google Scholar]
- Whitchurch C. B., Tolker-Nielsen T., Ragas P. C., Mattick J. S. (2002). Extracellular DNA required for bacterial biofilm formation. Science 295, 1487. doi: 10.1126/science.295.5559.1487 [DOI] [PubMed] [Google Scholar]
- Whiteley M., Diggle S. P., Greenberg E. P. (2017). Progress in and promise of bacterial quorum sensing research. Nature 551, 313–320. doi: 10.1038/nature24624, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wi Y. M., Patel R. (2018). Understanding biofilms and novel approaches to the diagnosis, prevention, and treatment of medical device-associated infections. Infect. Dis. Clin. N. Am. 32, 915–929. doi: 10.1016/j.idc.2018.06.009, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams D. E., Boon E. M. (2019). Towards understanding the molecular basis of nitric oxide-regulated group behaviors in pathogenic bacteria. J. Innate Immun. 11, 205–215. doi: 10.1159/000494740, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilton M., Charron-Mazenod L., Moore R., Lewenza S. (2016). Extracellular DNA acidifies biofilms and induces aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 60, 544–553. doi: 10.1128/AAC.01650-15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Moser C., Wang H. Z., Hoiby N., Song Z. J. (2015). Strategies for combating bacterial biofilm infections. Int. J. Oral Sci. 7, 1–7. doi: 10.1038/ijos.2014.65, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Zheng R., Zhang J., Wu S. (2021). Transcriptional profiling of Pseudomonas aeruginosa PAO1 in response to anti-biofilm and anti-infection agent exopolysaccharide EPS273. J. Appl. Microbiol. 130, 265–277. doi: 10.1111/jam.14764, PMID: [DOI] [PubMed] [Google Scholar]
- Xiu W., Shan J., Yang K., Xiao H., Yu W., Li H., et al. (2021). Recent development of nanomedicine for the treatment of bacterial biofilm infections. Viewpoints 2, 20200065. doi: 10.1002/VIW.20200065 [DOI] [Google Scholar]
- Yadav J., Kumari R. M., Verma V., Nimesh S. (2021). Recent development in therapeutic strategies targeting Pseudomonas aeruginosa biofilms – a review. Mater. Today Proc. 46, 2359–2373. doi: 10.1016/j.matpr.2021.05.245 [DOI] [Google Scholar]
- Yan J., Mao J., Xie J. (2014). Bacteriophage polysaccharide depolymerases and biomedical applications. BioDrugs 28, 265–274. doi: 10.1007/s40259-013-0081-y, PMID: [DOI] [PubMed] [Google Scholar]
- Yang Y., Qi P. K., Yang Z. L., Huang N. (2015). Nitric oxide based strategies for applications of biomedical devices. Biosurf. Biotribol. 1, 177–201. doi: 10.1016/j.bsbt.2015.08.003 [DOI] [Google Scholar]
- Yong V. F. L., Soh M. M., Jaggi T. K., Mac Aogain M., Chotirmall S. H. (2018). The microbial endocrinology of Pseudomonas aeruginosa: inflammatory and immune perspectives. Arch. Immunol. Ther. Exp. 66, 329–339. doi: 10.1007/s00005-018-0510-1, PMID: [DOI] [PubMed] [Google Scholar]
- Yu Z., Kong Y., Luo Z., Liu T., Lin J. (2019). Anti-bacterial activity of mutant chensinin-1 peptide against multidrug-resistant Pseudomonas aeruginosa and its effects on biofilm-associated gene expression. Exp. Ther. Med. 17, 2031–2038. doi: 10.3892/etm.2019.7182, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S., Su T., Wu H., Liu S., Wang D., Zhao T., et al. (2015). PslG, a self-produced glycosyl hydrolase, triggers biofilm disassembly by disrupting exopolysaccharide matrix. Cell Res. 25, 1352–1367. doi: 10.1038/cr.2015.129, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., He J., Zhai C., Ma L. Z., Gu L., Zhao K. (2018). Effects of PslG on the surface movement of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 84, e00219-18. doi: 10.1128/AEM.00219-18, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X. L., Chen Z. G., Yang T. C., Jiang M., Wang J., Cheng Z. X., et al. (2021). Glutamine promotes antibiotic uptake to kill multidrug-resistant uropathogenic bacteria. Sci. Transl. Med. 13, eabj0716. doi: 10.1126/scitranslmed.abj0716, PMID: [DOI] [PubMed] [Google Scholar]
- Zhao T., Zhang Y., Wu H., Wang D., Chen Y., Zhu M. J., et al. (2018). Extracellular aminopeptidase modulates biofilm development of Pseudomonas aeruginosa by affecting matrix exopolysaccharide and bacterial cell death. Environ. Microbiol. Rep. 10, 583–593. doi: 10.1111/1758-2229.12682, PMID: [DOI] [PubMed] [Google Scholar]
- Zhu Y., McHale G., Dawson J., Armstrong S., Wells G., Han R., et al. (2022). Slippery liquid-Like solid surfaces with promising antibiofilm performance under both static and flow conditions. ACS Appl. Mater. Interfaces 14, 6307–6319. doi: 10.1021/acsami.1c14533, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Rice S. A., Barraud N. (2019). Nitric oxide and iron signaling cues have opposing effects on biofilm development in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 85, e02175-18. doi: 10.1128/AEM.02175-18, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]