Abstract
Background
Circular RNAs (circRNAs) are attractive candidates to be used as biomarkers of human cancers, including lung adenocarcinoma (LUAD). Our study aimed to investigate the functions and regulatory mechanisms of hsa_circ_0129047 in the tumorigenesis of LUAD.
Methods
Reverse transcription‐quantitative polymerase chain reaction was performed to determine the circRNA, microRNA (miRNA), and mRNA expression levels in LUAD cell lines and tissues. Tumor xenografts were established in nude mice to evaluate whether hsa_circ_0129047 affected LUAD tumor development in vivo. Cell counting kit‐8 and transwell assays were performed to assess the mechanisms by which hsa_circ_0129047 influenced the viability and migration of LUAD cells, respectively. Apoptosis was evaluated via determination of the levels of the apoptotic markers, B‐cell lymphoma‐2, and Bcl‐2‐associated X, via Western blotting. Dual‐luciferase reporter assay, RNA immunoprecipitation assay, and Pearson's correlation analysis were performed to determine the relationships among miR‐375 and hsa_circ_0129047 and activin A receptor‐like type 1 (ACVRL1).
Results
Downregulation of hsa_circ_0129047 levels was observed in LUAD cell lines and tissues. Meanwhile, the upregulation of hsa_circ_0129047 levels repressed the proliferative, migratory, and survival capacities of LUAD cells in vitro. Hsa_circ_0129047 exerted antitumor effects during in vivo tumor development. Finally, we demonstrated that hsa_circ_0129047 sponged miR‐375. This interaction facilitated the expression of the downstream target of miR‐375, ACVRL1, whose upregulation inhibited the development and malignancy of LUAD.
Conclusion
These findings demonstrate that hsa_circ_0129047 functions as a tumor inhibitor in LUAD by modulating the miR‐375/ACVRL1 axis. Hence, hsa_circ_0129047 may be a promising biomarker and gene target for LUAD treatment.
Keywords: ACVRL1, hsa_circ_0129047, lung adenocarcinoma, miR‐375
Hsa_circ_0129047 functions as a tumor inhibitor in LUAD through the modulation of the miR‐375/ACVRL1 axis. Specifically, hsa_circ_0129047 repressed the proliferative, migratory, and survival capabilities of the LUAD cells in vitro, while played an antitumor role during in vivo tumor development.
1. INTRODUCTION
Lung adenocarcinoma (LUAD) is among the most aggressive and rapidly fatal types of cancer worldwide. 1 High resistance of LUAD to conventional radiotherapies and chemotherapies is a major challenge to its treatment efficacy. 1 If LUAD invades other organs, only approximately 13% of patients can survive for 5 years. 2 Therefore, it is necessary to identify novel biomarkers and mechanisms involved in the development and malignancy of LUAD that can be used for the effective diagnosis and treatment of patients.
Although circular RNAs (circRNAs) were discovered as covalently closed non‐coding RNAs a few years ago, their coding functions in human cancers have only been recently verified. 3 There are two key methods for modulating the translation function of circRNAs, internal ribosome entry site, and N6‐methyladenosine modification. 3 CircRNAs perform key functions, such as microRNA (miRNA) sponging and regulation of gene expression. 4 CircRNAs can potentially function as either oncogenes or tumor suppressors in human cancers. 5 Recent studies have attempted to prove the involvement of circRNAs in LUAD pathogenesis. Silencing of the circRNA, has_circ_0020850, can impede LUAD development by modulating the miR‐195‐5p/insulin receptor substrate 2 axis. 6 Hsa_circ_0000326 functions as a miR‐338‐3p sponge to enhance the progression of LUAD. 7 In addition, hsa_circ_0000211 facilitates LUAD malignancy by regulating the hypoxia‐inducible factor 1‐α by sponging hsa‐miR‐622. 8 Notably, our early microarray analysis of differentially expressed circRNAs in LUAD (GSE112214) indicated significant downregulation of hsa_circ_0129047 levels in tumor tissues compared with those in normal healthy tissues. Nonetheless, the specific functions and molecular mechanisms of hsa_circ_0129047 in LUAD development and progression remain unclear.
Activin A receptor‐like type 1 (ACVRL1) encodes the activin A type I receptor, which signals a particular transcriptional response in cooperation with activin type II receptors. ACVRL1 expression is closely related to the survival and tumor response of patients with metastatic colorectal cancer who underwent first‐line bevacizumab chemotherapy. 9 This clearly indicates that ACVRL1 can be used as a novel prognostic biomarker for patients with metastatic cancer. 9 Gene expression profiling interactive analysis (GEPIA) is a database that contains the mRNA expression profiles of LUAD and corresponding normal tissue samples. Using GEPIA, we found that ACVRL1 is differentially expressed in LUAD. Based on this fact, we wondered whether ACVRL1 could function as a biomarker for the diagnosis and prognosis of LUAD. Our previous work revealed that the hsa_circ_0129047–miRNA–ACVRL1 regulatory network consists of two miRNAs, hsa‐miR‐375, and hsa‐miR‐665. miRNAs, a type of non‐coding RNAs, are ~21 nucleotides in length. miR‐375 plays an essential role in the progression of LUAD. For example, Yoda et al. reported that miR‐375 expression is closely associated with a short survival time in non‐small‐cell lung cancer. 10 Moreover, Gan et al. demonstrated that miR‐375 exhibits significantly higher expression in LUAD tissues than in normal tissues, which indicates its essential role in LUAD progression. 11 However, it remains unclear whether and how miR‐375 is involved in the regulatory mechanisms of hsa_circ_0129047 and ACVRL1 in LUAD progression and development.
In this study, we explored the specific functions and molecular mechanisms of hsa_circ_0129047 in LUAD development and progression. The findings of this study may provide novel biomarkers for the diagnosis and prognosis of patients with LUAD.
2. MATERIALS AND METHODS
2.1. Patient samples
Forty sets of tumor and adjacent healthy tissues were collected from the Wuhan Red Cross Hospital (Wuhan, China). All tissues were pathologically confirmed to have LUAD. All samples were acquired from patients who did not previously receive any form of chemoradiotherapy and provided signed informed consent for participation in this study. The Ethics Committee of Wuhan Red Cross Hospital approved this study.
2.2. Cell culture
The LUAD (PC‐9) and healthy human lung (BEAS‐2B) cell lines were acquired from the American Type Culture Collection. Other LUAD cell lines, A549, Calu‐3, and H1975, were obtained from the Chinese Academy of Sciences. All cell lines were cultivated in the Roswell Park Memorial Institute (RPMI)‐1640 medium (KeyGen, China) supplemented with 10% fetal bovine serum (FBS; Life Technologies). The culture plates were maintained in an environment with 5% CO2 at 37°C.
2.3. Cell transfection
The overexpression plasmid of circ_0129047 (oe‐circ_0129047) and its negative control (oe‐NC) were constructed based on the pCD5‐ciR vector (Geneseed, Guangzhou). miR‐375 mimic and its negative control (mimic‐NC) were purchased from RiboBio. The coding sequence of ACVRL1 was inserted into a pcDNA3.1 vector (Invitrogen) to generate an ACVRL1 overexpression vector (oe‐ACVRL1). The pcDNA3.1 vector (Invitrogen) alone was used as the negative control (oe‐NC). Following the standard protocol, Lipofectamine 3000 Reagent (Invitrogen) was used to transfect the aforementioned vectors into A549 and Calu‐3 cells. Reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) was performed after 48‐h transfection to assess the transfection efficiency.
2.4. Real‐time quantitative reverse transcription‐polymerase chain reaction (RT‐qPCR)
Total RNA was extracted from the cells and tissues using TRIzol reagent (Invitrogen). Using the ReverTra Ace qPCR RT Kit (Toyobo), cDNA was synthesized via reverse transcription of total RNA. Oligo (dT) and random primers were used for the quantification of mRNA and circRNA, whereas stem‐loop RT primers were used for the quantification of miRNA. RT‐PCR was performed using the SYBR Green PCR Kit (Takara). Glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH) was used to normalize the circRNA and mRNA levels, while uracil 6 (U6) was used to normalize the miRNA levels. Finally, the 2−ΔΔCt approach 12 was used for calculations. Primer sequences used in this study are listed in Table 1.
TABLE 1.
Primers for qRT‐PCR
| Gene | Forward | Reverse |
|---|---|---|
| circ_0129047 | 5’‐ATTCCAGCTCTTTCACATGG‐3′ | 5’‐TATCCAGGATGGTCGTTTCAA‐3′ |
| miR‐375 | 5’‐CGGAATTCAGGGTGGCTGGGAAAGG‐3′ | 5’‐CCGCTCGAGCCGTATTACGACGCAGAATG‐3′ |
| ACVRL1 | 5’‐AGGGCAAACCAGCCATTG‐3′ | 5’‐GGTTGCTCTTGACCAGCACAT‐3′ |
| U6 | 5’‐GCTTCGGCAGCACATATACTAAAAT‐3′ | 5’‐CGCTTCACGAATTTGCGTGTCAT‐3’ |
| GAPDH | 5’‐GTCAAGGCTGAGAACGGGAA‐3’ | 5’‐AAATGAGCCCCAGCCTTCTC‐3’ |
2.5. Subcellular fractionation
The fractions of cytoplasm and nucleus from A549 and Calu‐3 were isolated using Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific, CA, USA). RT‐qPCR was performed to determine the expression levels of has_circ_0129047, GAPDH (cytoplasm), and U6 (nucleus).
2.6. RNase R enrichment
One unit of RNase R (Sigma‐Aldrich) was used to digest 1 μg of RNA. Hsa_circ_0129047 and circ_0129047 mRNA were individually mixed with RNase R and placed in a water bath at 37°C. RNA was then precipitated with phenol/chloroform and ethanol before RT‐qPCR quantification analysis.
2.7. Cell counting kit (CCK)‐8 assay
The wells of a 96‐well culture plate were seeded with 5 × 103 stably transfected A549 and Calu‐3 cells. After the indicated durations of 0, 24, 48, and 72 h, 10 μl of CCK‐8 solution (Sigma) was pipetted into each well for another 4 h of culture. Cell viability was assessed by measuring the absorbance using a microplate spectrophotometer (BioTek) with a 450 nm filter.
2.8. Transwell assay
Transwell chambers (Corning Costar, New York, USA) were utilized in this assay for the evaluation of LUAD cell migration. First, transfected cells (1 × 104) suspended in 200 μl serum‐free medium were pipetted into the upper transwell chambers of a 24‐well plate. The lower chambers were then filled with 200 μl RPMI‐1640 medium supplemented with 10% FBS. The plates were maintained at 37°C with 5% CO2 for 48 h. Cells at the bottom were fixed with 95% ethanol and stained with 0.1% crystal violet. Finally, the number of cells was counted using an optical microscope (Thermo Fisher Scientific, CA, USA).
2.9. Western blotting
Total proteins were isolated using the radioimmunoprecipitation assay buffer (Sangon Biotech). A BCA kit (Beyotime) was used to quantify the total protein levels. Equal amounts of protein (30 μg) were subjected to 10% sodium dodecyl sulfate‐polyacrylamide gel electrophoresis, transferred onto polyvinylidene fluoride membranes, and blocked with 5% non‐fat milk for an hour. The proteins were then incubated overnight at 4°C with the following primary antibodies acquired from Abcam: B‐cell lymphoma‐2 (Bcl‐2; 1:1000; ab32123), Bcl‐2‐associated X (Bax; 1:1000; ab182733), ACVRL1 (1:1500; ab263902), and GAPDH (1:2000; ab8245) antibodies. The following day, the membranes were incubated for an hour in the dark with the corresponding secondary antibodies (1:5000; catalog: 7076, Cell Signaling). A chemiluminescence detection kit (Millipore) was employed to visualize the protein bands. Finally, ImageJ software (NIH, Bethesda, MD, USA) was used for protein quantification.
2.10. In vivo assay
Four‐ to five‐week‐old BALB/c nude mice were sourced from the Guangdong Medical Laboratory Animal Center, China. A549 (4 × 106) cells stably transfected with either oe‐circ or oe‐NC vector were injected into BALB/c nude mice (five mice per group). The mice were kept well, and the width (W) and length (L) of their tumors were measured and recorded weekly. Five weeks later, the mice were sacrificed and their tumors were extracted and weighed. Tumor volumes (V) were calculated using the following formula: V = W2 × L/2. The Institutional Animal Care and Use Committee of Wuhan Red Cross Hospital approved all animal experiments performed in this study.
2.11. Dual‐luciferase reporter assay
Hsa_circ_0129047 and ACVRL1 3′‐untranslated region (UTR) sequences that contained the wild‐type (circ_0129047‐WT and ACVRL1‐WT, respectively) and corresponding predicted mutated binding sites (circ_0129047‐MUT and ACVRL1‐MUT, respectively) for miR‐375 were cloned into Promega pmirGLO vectors. The above‐mentioned vectors were transfected along with a miR‐375 mimic or miR‐NC into A549 and Calu‐3 cells. Transfection was accomplished using Lipofectamine 3000. After 48 h, the luciferase reporter activity was assessed using a dual‐luciferase reporter assay system (Promega).
2.12. RNA immunoprecipitation (RIP) assay
The Magnetic RIP RNA‐Binding Protein Immunoprecipitation Kit, IgG control, and anti‐argonaute 2 (Ago2) antibody (Millipore) were used for the RIP assay. Following the manufacturer's instructions, immunoprecipitated RNAs were isolated and purified prior to RT‐qPCR analysis.
2.13. Statistical analysis
All data are presented as the mean ± standard deviation. Statistical significance was set at p < 0.05. GraphPad Prism software (GraphPad, USA) was used for all statistical analyses. One‐way analysis of variance and Student's t test were used to evaluate the differences among multiple groups or between two groups, respectively. Pearson's correlation coefficient was used to assess the association between hsa_circ_0129047 and miR‐375 as well as that between miR‐375 and ACVRL1.
3. RESULTS
3.1. LUAD cell lines and tissues exhibit downregulation of hsa_circ_0129047 levels
To investigate the function of hsa_circ_0129047 in LUAD, we measured its expression levels in LUAD tissues and cells using RT‐qPCR. The results revealed a decrease in hsa_circ_0129047 expression levels in tumor tissues and cell lines (Calu‐3, H1975, A549, and PC9) of LUAD compared with those in the normal tissues and cell line (BEAS‐2B). Among LUAD cell lines, Calu‐3 and A549 showed the lowest expression levels of hsa_circ_0129047; hence, they were chosen for subsequent experiments (Figure 1A,B). Upon determining the levels of hsa_circ_0129047 in the nuclear and cytoplasmic fractions of Calu‐3 and A549 cells, we found that hsa_circ_0129047 was predominantly expressed in the cytoplasm (Figure 1C). Furthermore, RNase R treatment was performed to enrich the circRNAs in A549 and Calu‐3 cells. This was followed by the analysis of hsa_circ_0129047 expression, with circ_0129047 mRNA expression serving as a negative control. As shown in Figure 1D, RNase R treatment significantly decreased the circ_0129047 mRNA levels, but did not influence the hsa_circ_0129047 levels. These findings suggest that hsa_circ_0129047 expression is reduced in LUAD cells and its upregulation may repress LUAD progression.
FIGURE 1.
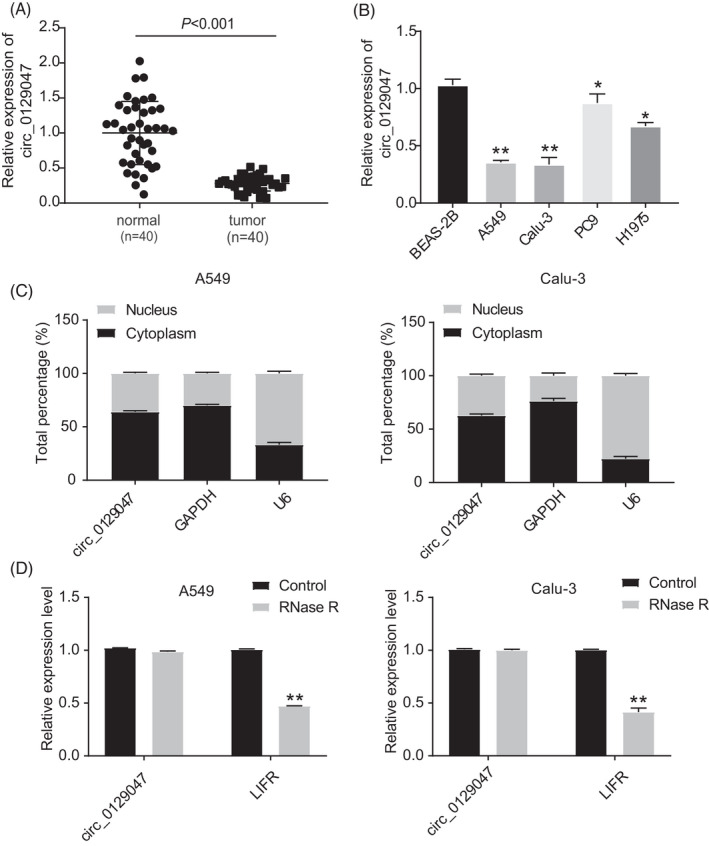
Hsa_circ_0129047 was downregulated in LUAD tissues and cell lines. (A) Real‐time quantitative reverse transcription‐polymerase chain reaction (RT‐qPCR) was used to measure hsa_circ_0129047 expression in tissues from 40 LUAD patients and normal lung tissues. (B) hsa_circ_0129047 expression level in LUAD cell lines (A549, Calu‐3, PC9 and H1975) and normal lung cell line (BEAS‐2B) was detected by RT‐qPCR. *p < 0.05, **p < 0.001 vs. BEAS‐2B. (C) Subcellular fractionation analysis was conducted to determine the subcellular localization of hsa_circ_0129047 in LUAD. (D) Hsa_circ_0129047 and circ_0129047 mRNA expression was detected by RT‐qPCR after RNase R treatment. **p < 0.001 vs. Control
3.2. Hsa_circ_0129047 overexpression attenuates the cell proliferation and migration, while enhancing the apoptosis of LUAD cells in vitro and inhibits LUAD tumor formation in vivo
To investigate the potential regulatory functions of hsa_circ_0129047 in LUAD cells, we transfected oe‐circ_0129047 and its negative control (NC) into A549 and Calu‐3 cells. As shown in Figure 2A, considerably higher levels of hsa_circ_0129047 were detected among cells transfected with oe‐circ_0129047, unlike those transfected with oe‐NC. This indicates that stable Calu‐3 and A549 cell lines overexpressing hsa_circ_0129047 were successfully constructed. CCK‐8 assay revealed that hsa_circ_0129047 overexpression decreased the viability of Calu‐3 and A549 cells (Figure 2B). To scrutinize the influence of hsa_circ_0129047 on the apoptosis of LUAD cells, related protein marker levels were measured. Western blotting demonstrated that hsa_circ_0129047 overexpression elevated the expression levels of the pro‐apoptotic protein, Bax, while decreasing the levels of the anti‐apoptotic protein, Bcl‐2 (Figure 2C). This indicates that hsa_circ_0129047 facilitates the apoptosis of Calu‐3 and A549 cells. Furthermore, transwell assays were performed to assess whether hsa_circ_0129047 repressed the migration of LUAD cells. The results revealed that hsa_circ_0129047 overexpression considerably hampered the migration of Calu‐3 and A549 cells (Figure 2D). Nude mice were injected with stably transfected A549 cells carrying either oe‐circ_0129047 or oe‐NC. The volumes of the xenograft tumors over 5 weeks were monitored, and at the end of the fifth week, tumor weights were recorded after excision (Figure 3A–C). These results reveal that overexpression of hsa_circ_0129047 suppresses the formation of LUAD tumors in vivo. Thus, hsa_circ_0129047 overexpression suppresses the growth, migration, and survival of LUAD cells both in vivo and in vitro.
FIGURE 2.
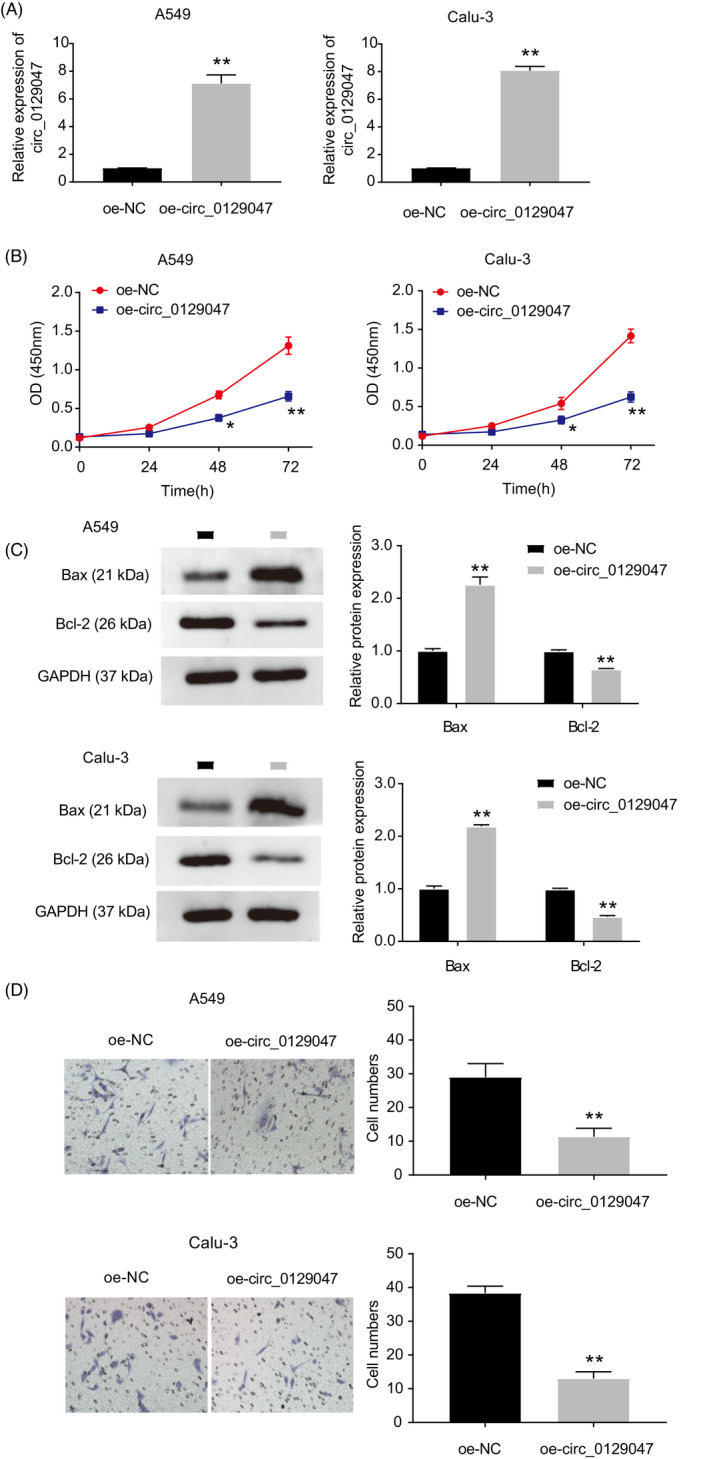
Hsa_circ_0129047 overexpression inhibited LUAD cell viability, migration and promoted cell apoptosis. (A) Hsa_circ_0129047 overexpression efficiency verified by RT‐qPCR after transfection with oe‐circ_0129047 vector. **p < 0.001 vs. Oe‐NC. (B) CCK‐8 assay was adapted to evaluate the cell viability in A549 and Calu‐3 cells transfected with oe‐NC and oe‐circ_0129047. *p < 0.05, **p < 0.001 vs. Oe‐NC. (C) Western blot experiment showed the expression of anti‐apoptotic marker Bcl‐2 and pro‐apoptotic marker Bax in A549 and Calu‐3 cells transfected with oe‐NC and oe‐circ_0129047. **p < 0.001 vs. Oe‐NC. (D) Cell migration ability was determined by Transwell assay in A549 and Calu‐3 cells transfected with oe‐NC and oe‐circ_0129047. **p < 0.001 vs. oe‐NC
FIGURE 3.
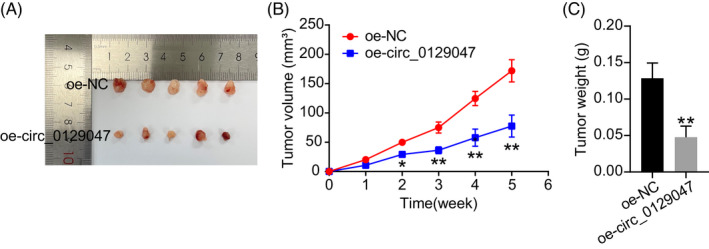
Hsa_circ_0129047 overexpression suppressed LUAD cell growth in vivo. (A) Tumors derived from nude mice injected with A549 cells stably transfected with either oe‐NC or oe‐circ 0129047 are shown. (B) Tumor volume (mm3) was recorded every week. *p < 0.05, **p < 0.001 vs. oe‐NC. (C) On the 35th day after injection, the nude mice were euthanized and the tumors were excised and weighed. **p < 0.001 vs. oe‐NC
3.3. Hsa_circ_0129047 sponges miR‐375
To explore the molecular mechanism of hsa_circ_0129047, we utilized CircInteractome to identify potential miRNA‐binding sites on hsa_circ_0129047. Figure 4A shows the putative binding sites for miR‐375 on hsa_circ_0129047. The dual‐luciferase reporter experiment demonstrated that the miR‐375 mimic significantly diminished the luciferase activity of hsa_circ_0129047‐WT, but did not influence the activity of hsa_circ_0129047‐MUT (Figure 4B). These findings confirmed the interaction between miR‐375 and hsa_circ_0129047. Additionally, an RIP assay was performed using an Ago2‐specific antibody to further verify their interactions. As shown in Figure 4C, compared with the anti‐IgG control, both miR‐375 and hsa_circ_0129047 were remarkably enriched in the anti‐Ago2 group. This verified the close association between miR‐375 and hsa_circ_0129047. miR‐375 expression levels were higher in the LUAD tissues (Figure 4D) and cell lines (Figure 4E) than in the corresponding normal tissues and cells. Furthermore, Pearson's correlation coefficient indicated a negative association between miR‐375 and hsa_circ_0129047 expression (Figure 4F). Additionally, we discovered a significant decline in miR‐375 expression levels in A549 and Calu‐3 cells transfected with the hsa_circ_0129047 mimic (Figure 4G). These changes were reversed after the introduction of the miR‐375 mimic. Our findings indicate that hsa_circ_0129047 can reduce miR‐375 expression in LUAD. In summary, hsa_circ_0129047 binds to miR‐375 to repress miR‐375 expression in LUAD.
FIGURE 4.
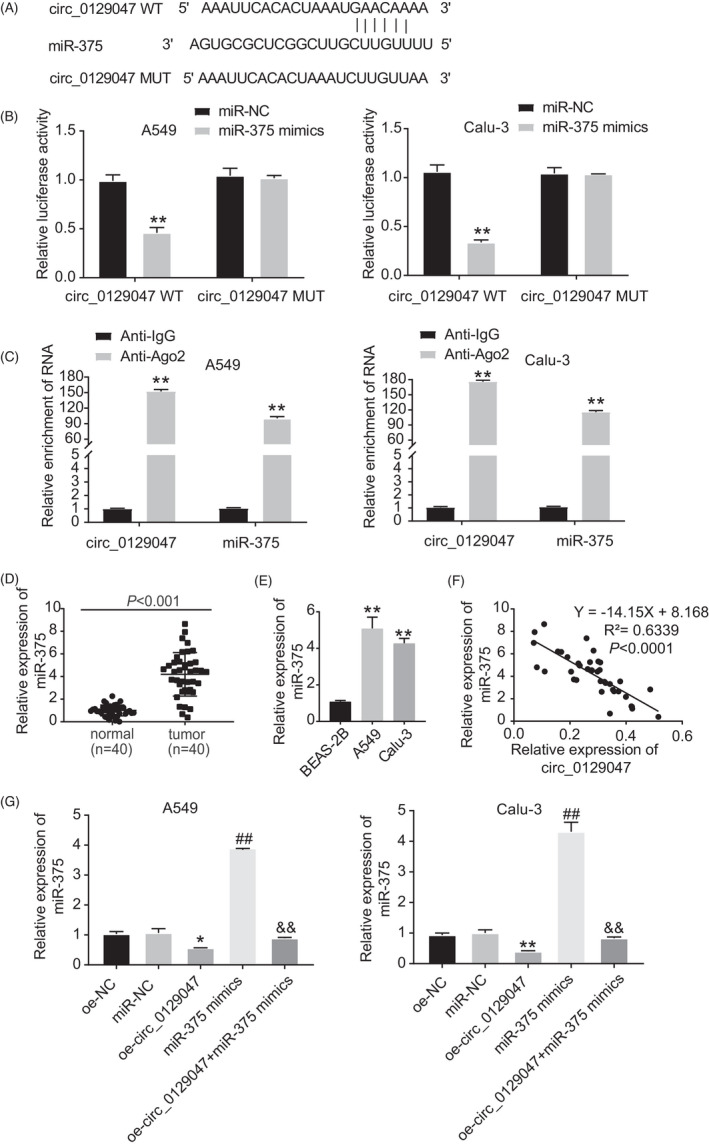
Hsa_circ_0129047 acted as a sponge for miR‐375. (A) The binding site between hsa_circ_0129047 and miR‐375 was predicted by circInteractome (https://ngdc.cncb.ac.cn/databasecommons/database/id/4778). (B) Luciferase reporter experiment was performed to validate the interaction of miR‐375 and hsa_circ_0129047 in the A549 and Calu‐3 cells. **p < 0.001 vs. miR‐NC. (C) RIP assay was used to validate the interaction between hsa_circ_0129047 and miR‐375. **p < 0.001 vs. anti‐IgG. (D) MiR‐375 expression level in LUAD tissues and normal adjacent tissues were detected by RT‐qPCR. (E) RT‐qPCR was conducted to measure miR‐375 expression level in LUAD cell lines (A549 and Calu‐3) and normal lung cell line (BEAS‐2B). **p < 0.001 vs. BEAS‐2B. (F) Correlation between hsa_circ_0129047 and miR‐375 was determined via Pearson's correlation coefficient. (G) The expression of miR‐375 in A549 and Calu‐3 cells transfected with Oe‐NC, mimic‐NC, Oe‐circ, mimic, and Oe + mimic was detected by RT‐qPCR. *p < 0.05 and **p < 0.001 vs. Oe‐NC; # p < 0.05 and ## p < 0.001 vs. miR‐NC; & p < 0.05 and && p < 0.001 vs. miR‐375 mimic
3.4. Hsa_circ_0129047 overexpression reverses the stimulating influence of miR‐375 on the development and malignancy of LUAD
To further investigate the underlying mechanisms of miR‐375 and hsa_circ_0129047 in LUAD development, we performed rescue assays by transfecting miR‐375 mimic and/or oe‐circ_0129047 into A549 and Calu‐3 cells. First, we evaluated the viability of LUAD cells among the five groups: oe‐NC, mimic‐NC, miR‐375 mimic, oe‐circ_0129047, and oe‐circ_0129047 + miR‐375 mimic. CCK‐8 assay revealed that cells transfected with miR‐375 exhibited high viability, and this effect was reversed via the co‐transfection with circ_0129047 (Figure 5A). In addition, Western blotting analysis revealed that the Bax levels declined, whereas Bcl‐2 levels were significantly upregulated in the miR‐375 mimic group. This suppressive effect of the miR‐375 mimic on the apoptosis of LUAD cells was reversed by hsa_circ_0129047 overexpression (Figure 5B). Transwell assay results showed that hsa_circ_0129047 overexpression reduced the boosting effects of the miR‐375 mimic on the migration of the two LUAD cell lines (Figure 5C). These findings indicate that hsa_circ_0129047 reduces the proliferation and malignancy of LUAD cells by sponging miR‐375.
FIGURE 5.
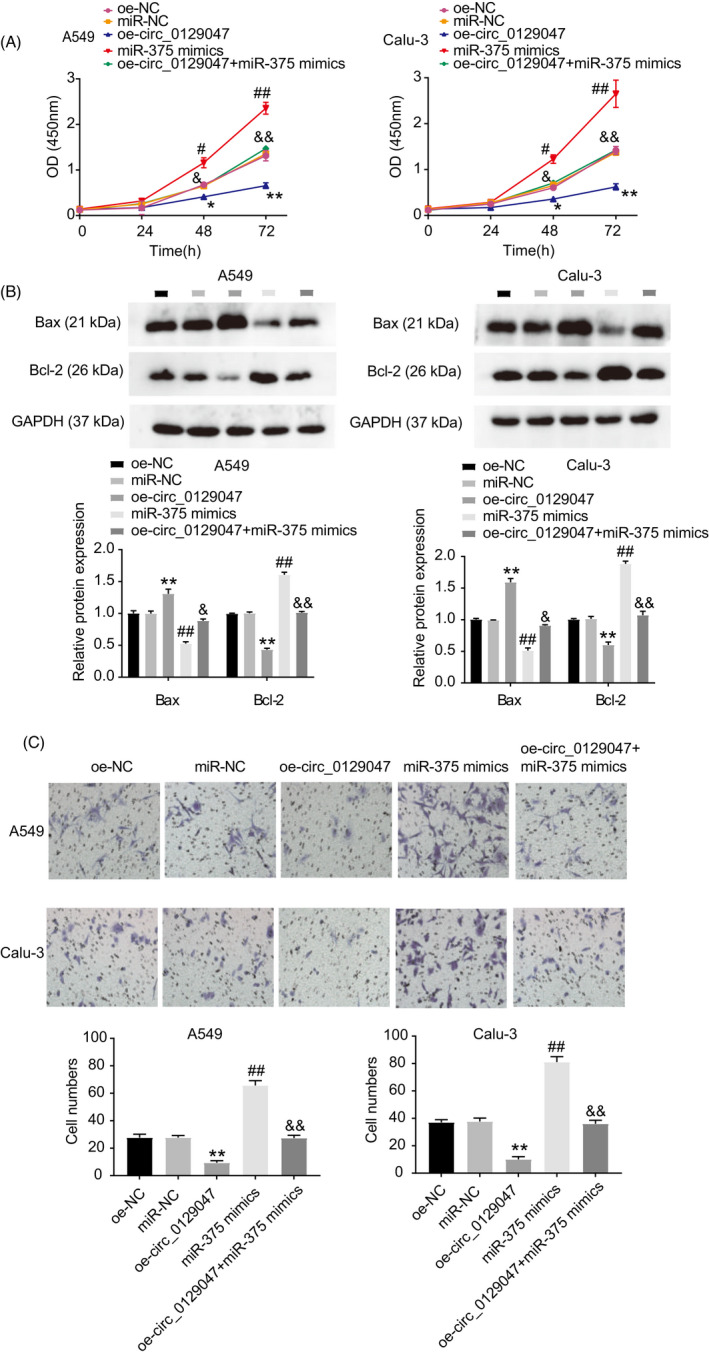
Overexpression of hsa_circ_0129047 inhibited LUAD progression by sponging miR‐375. Five groups were established: oe‐NC, miR‐NC, miR‐375 mimic, oe‐ circ_0129047, and oe‐circ_0129047 + miR‐375 mimic were transfected into the A549 and Calu‐3 cells in five groups, respectively. (A) CCK‐8 experiment was carried out to assess the cell viability in different groups. (B) Bax and Bcl‐2 protein levels in transfected A549 and Calu‐3 cells were detected in different groups. (C) Transwell assay showed the cell ability of migration in different groups. *p < 0.05 and **p < 0.001 vs. OE‐NC; # p < 0.05 and ## p < 0.001 vs. miR‐NC; & p < 0.05 and && p < 0.001 vs. miR‐375 mimic
3.5. ACVRL1 is a target gene for miR‐375
To investigate the downstream regulatory mechanism of the hsa_circ_0129047/miR‐375 axis in LUAD development, we conducted a bioinformatics analysis. Figure 6A shows the predicted functional miR‐375 binding site on ACVRL1 3′‐UTR. This binding site was further verified using dual‐luciferase experiments. The results revealed reduced luciferase reporter activity of the ACVRL1‐WT vectors after transfection with the miR‐375 mimic. In contrast, the luciferase reporter activity of the ACVRL1‐MUT vectors was not affected (Figure 6B), indicating that miR‐375 can specifically bind to ACVRL1. Moreover, RT‐qPCR revealed that LUAD tissues (Figure 6C) and cells (Figure 6D) had significantly lower levels of ACVRL1 than the corresponding controls. Pearson's correlation coefficient analysis revealed an inverse association between miR‐375 and ACVRL1 expression (Figure 6E). Calu‐3 and A549 cells transfected with the miR‐375 and oe‐ACVRL1 + miR‐375 mimics showed reduced ACVRL1 expression levels compared with those transfected with mimic‐NC and oe‐ACVRL1 (Figure 6F). This suggests that miR‐375 decreases the expression levels of ACVRL1 in LUAD cells. These results demonstrate that miR‐375 downregulates ACVRL1 expression in LUAD cells by binding to ACVRL1 3′‐UTR.
FIGURE 6.
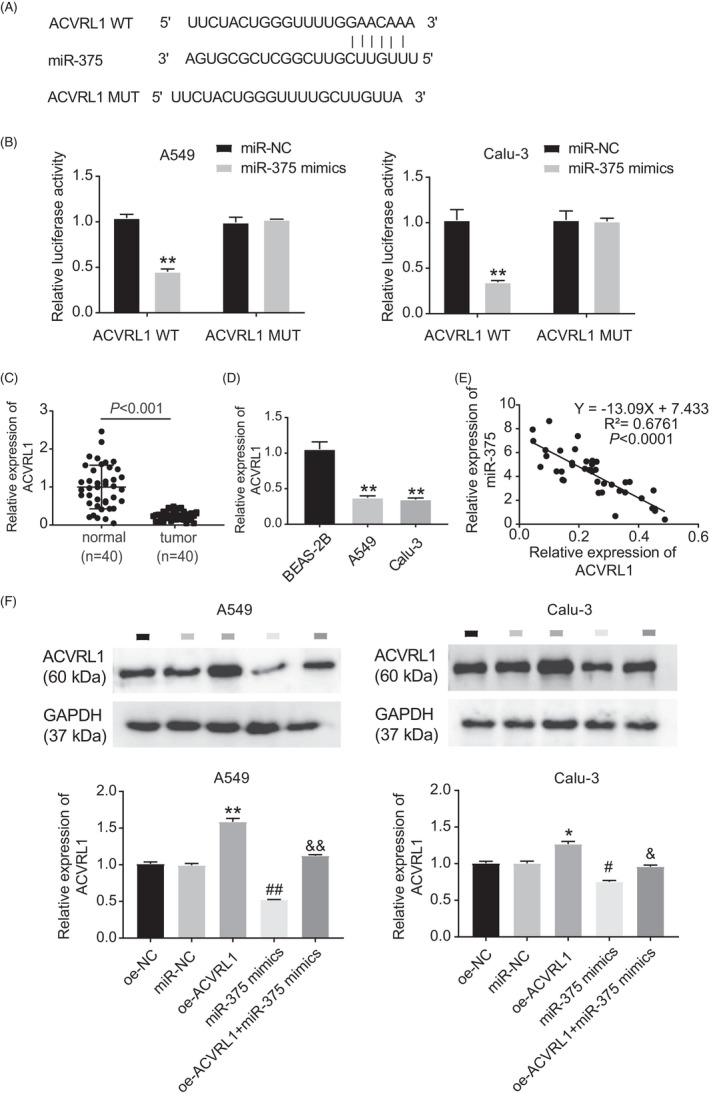
ACVRL1 was a target gene of miR‐375. (A) MiR‐375 targeted ACVRL1 3’UTR with a potential binding site by starBase. (B) Dual‐luciferase reporter assay was used to measure the relationship between miR‐375 and ACVRL1. **p < 0.001 vs. miR‐NC. (C) RT‐qPCR was performed to measure ACVRL1 expression in LUAD tissues and adjacent normal tissues. (D) RT‐qPCR was used to measure ACVRL1 expression in LUAD cell lines (A549 and Calu‐3) and normal cell line (BEAS‐2B). **p < 0.01 vs. BEAS‐2B. (E) Pearson's correlation analysis was carried out to evaluate the relationship between miR‐375 and ACVRL1 expression. (F) Western blotting was used to measure ACVRL1 expression in A549 and Calu‐3 cells in the presence of oe‐NC, mimic‐NC, mimic, oe‐ACVRL1, and oe‐ACVRL1 + mimic. *p < 0.05, **p < 0.001 vs. oe‐NC; # p < 0.05 and ## p < 0.001 vs. miR‐NC; & p < 0.05 and && p < 0.001 vs. miR‐375 mimic
3.6. miR‐375 facilitates the proliferation and migration of LUAD cells, and inhibits their apoptosis by reducing ACVRL1 expression
To further investigate the functions of ACVRL1 in LUAD cells, we first overexpressed ACVRL1 in Calu‐3 and A549 cells. This was accomplished by transfecting the cells with the oe‐ACVRL1 vector and co‐transfecting them with the miR‐375 mimic to uncover the underlying regulatory mechanism of the miR‐375/ACVRL1 axis in LUAD cells. Experiments were performed to analyze the viability (CCK‐8 assay), apoptosis (western blotting), and migration (transwell assay) of LUAD cells in different groups. As shown in Figure 7A, ACVRL1 overexpression significantly reduced the cell viability, which was recovered via co‐transfection with an miR‐375 mimic. Additionally, cells overexpressing ACVRL1 showed enhanced Bax and reduced Bcl‐2 protein levels. This indicates that ACVRL1 promotes cell apoptosis, whereas the co‐transfection with the miR‐375 mimic attenuates this effect (Figure 7B). Transwell assay showed that miR‐375 restored the migratory capacities of cells that were suppressed by the overexpression of ACVRL1 (Figure 7C). These results demonstrate that ACVRL1 overexpression exerts antitumor effects in LUAD cells, which can be counteracted by miR‐375 overexpression.
FIGURE 7.
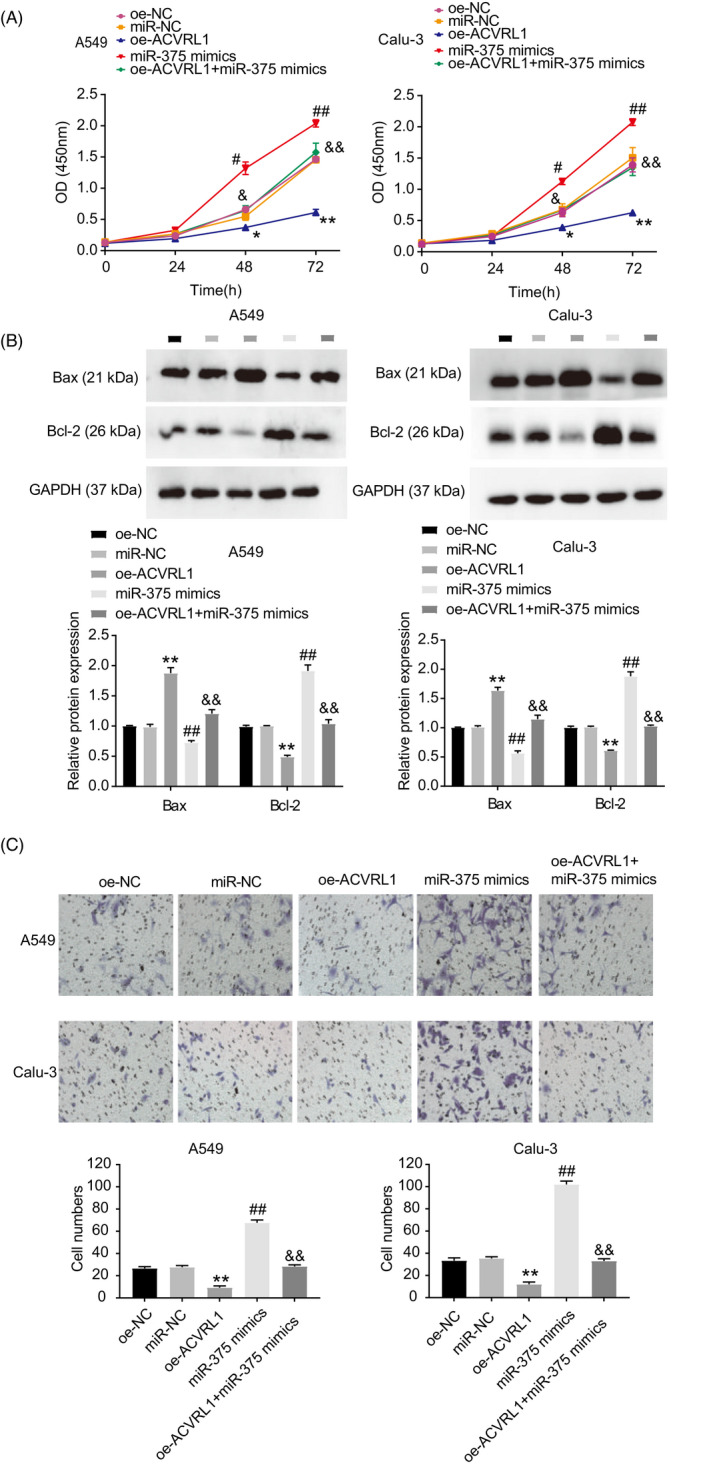
MiR‐375 facilitated the proliferation and migration of LUAD cells and inhibited their apoptosis by reducing ACVRL1 expression. Five groups (oe‐NC, miR‐NC, miR‐375 mimic, oe‐ACVRL1, and oe‐ACVRL1 + miR‐375 mimic) were established in the following experiments. (A) CCK‐8 experiment was conducted to evaluate the viability of cells from the different groups. (B) Cell apoptosis was detected by analyzing the expression of Bax and Bcl‐2 protein in different groups. (C) Transwell assay was adapted to determine the cell migration in different groups. *p < 0.05 and **p < 0.001 vs. Oe‐NC; # p < 0.05 and ## p < 0.001 vs. miR‐NC; & p < 0.05 and && p < 0.001 vs. miR‐375 mimic
4. DISCUSSION
CircRNAs have covalently closed single‐stranded structures that confer better resistance to RNase R than linear RNAs. 13 Hence, most circRNAs are used as potential diagnostic and prognostic biomarkers for various diseases, including human cancer. Extensive research has recognized the critical role of circRNAs in the carcinogenesis and malignancy of human cancers, such as gastric, 14 lung, 15 pancreatic, 16 and cervical 17 cancers. In this study, we identified a circRNA, hsa_circ_0129047, and investigated its function and molecular regulatory mechanisms in the pathogenesis of LUAD. Our experiments demonstrated that hsa_circ_0129047 overexpression suppressed the proliferation and migration, while enhancing the apoptosis of LUAD cells. More importantly, hsa_circ_0129047 exerted these antitumor effects in LUAD by regulating the miR‐375/ACVRL1 axis.
We found considerable downregulation of hsa_circ_0129047 levels in LUAD cells and tissues. Hsa_circ_0129047 functions as a tumor suppressor, and its overexpression inhibits the proliferation and migration of LUAD cells and facilitates their apoptosis. Consistent with our results, Wang et al. documented a significant decrease in hsa_circ_0008274 levels in LUAD. 18 Moreover, overexpression of hsa_circ_0008274 reduces the growth and invasion of LUAD cells. 18 Similarly, circ‐000881 shows poor expression in LUAD, whereas its overexpression diminishes the proliferative and invasive capacities of LUAD cells. 19 Another study by Zhou et al. demonstrated that the upregulation of circ_cMras levels hampered LUAD tumorigenesis and progression in vitro and in vivo 20 . Interestingly, in this study, subcellular fractionation analysis showed that hsa_circ_0129047 was predominantly located within the cytoplasm of LUAD cells. This implies that hsa_circ_0129047 may serve as an miRNA sponge for LUAD. circRNAs can act as miRNA sponges, bind to miRNAs, and release the mRNAs of their target genes. For instance, Chen et al. showed that hsa_circ_100395 sponges miR‐1228, which targets the transcription factor 21 in lung cancer. 21 Similarly, circ_0008193 sponges miR‐1180‐3p to regulate the expression of tripartite motif containing 62. 22 However, no study has focused on the effects of hsa_circ_0129047 in human cancers. In this study, bioinformatics analysis identified the putative binding site of miR‐375 and hsa_circ_0129047, which was validated via a dual‐luciferase reporter assay. Furthermore, Pearson's correlation coefficient analysis revealed an inverse relationship between hsa_circ_0129047 and miR‐375 expression. RT‐qPCR results suggest that miR‐375 is highly expressed in LUAD cells and tissues.
To determine the biological functions and regulatory mechanisms of miR‐375 in LUAD cells, we performed a series of rescue experiments. We discovered that miR‐375 serves as an oncogene for LUAD as it counteracts the inhibitory influence of hsa_circ_0129047 on the development and malignancy of LUAD. Consistent with our findings, Zhao et al. reported a significant elevation in miR‐375 expression in small‐cell lung cancer, suggesting that it could be a novel biomarker for the diagnosis of patients with small‐cell lung cancer. 23 A similar study revealed that miR‐375 expression is upregulated and plays a crucial role in the tumorigenesis and development of breast cancer. 24 Another study found that miR‐375 has clinical value and may be a possible signaling pathway for LUAD by utilizing the Cancer Genome Atlas and Gene Expression Omnibus databases. 11 Notably, the present study is the first to confirm the role of miR‐375 in the regulation of hsa_circ_0129047 in LUAD.
Finally, we found that ACVRL1 levels were significantly downregulated in LUAD cells and tissues. In line with our findings, a study based on microarray data reported that ACVRL1 is expressed differentially in LUAD tissues, revealing its diagnostic value for patients with LUAD. 25 Similarly, Hanna et al. showed that ACVRL1 may regulate tumor angiogenesis and function as a prognostic biomarker for patients with metastatic colorectal cancer. 9 miR‐375 has a binding site on ACVRL1 3′‐UTR, which was verified via the dual‐luciferase assay. Furthermore, ACVRL1 functions as a tumor suppressor by inhibiting the ability of LUAD cells to grow, migrate, and survive, which can be recovered by miR‐375 expression. Altogether, our findings support the hypothesis that hsa_circ_0129047 inhibits the progression of LUAD by releasing ACVRL1 via sponging of miR‐375.
Our previous work revealed that miR‐665 is involved in the hsa_circ_0129047–miRNA–ACVRL1 regulatory network. Notably, a recent study reported the upregulation of miR‐665 levels in lung cancer and metastatic disease samples. 26 Based on these findings, the regulatory mechanism of hsa_circ_0129047/miR‐665/ACVRL1 in the progression of LUAD should be investigated further in future studies. Our findings demonstrate that hsa_circ_0129047 serves as a tumor suppressor in LUAD as it strongly inhibits the tumorigenesis and malignancy of LUAD by modulating the miR‐375/ACVRL1 axis. Our findings highlight the novel mechanism and denote the promising value of hsa_circ_0129047 for the treatment of patients with LUAD.
AUTHOR CONTRIBUTION
JXF and XFX performed the experiments and analysis of data. JXF and ZJF conceived and designed the study. ZJF acquired the data. JXF conducted the analysis and interpretation of data. All authors have read and approved this article.
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest.
PATIENT CONSENT
All patients signed written informed consent.
CONSENT FOR PUBLICATION
Consent for publication was obtained from all participants.
ACKNOWLEDGEMENTS
None.
Fan J, Xia X, Fan Z. Hsa_circ_0129047 regulates the miR‐375/ACVRL1 axis to attenuate the progression of lung adenocarcinoma. J Clin Lab Anal. 2022;36:e24591. doi: 10.1002/jcla.24591
Jinxiu Fan and Xiaofan Xia contribute equally to this work.
DATA AVAILABILITY STATEMENT
All data that have been generated or analyzed during this study are included in this article.
REFERENCES
- 1. Denisenko TV, Budkevich IN, Zhivotovsky B. Cell death‐based treatment of lung adenocarcinoma. Cell Death Dis. 2018;9(2):117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Khaltaev N, Axelrod S. Global lung cancer mortality trends and lifestyle modifications: preliminary analysis. Chin Med J (Engl). 2020;133(13):1526‐1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lu Y, Li Z, Lin C, Zhang J, Shen Z. Translation role of circRNAs in cancers. J Clin Lab Anal. 2021;35(7):e23866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675‐691. [DOI] [PubMed] [Google Scholar]
- 5. Chen B, Huang S. Circular RNA: an emerging non‐coding RNA as a regulator and biomarker in cancer. Cancer Lett. 2018;418:41‐50. [DOI] [PubMed] [Google Scholar]
- 6. Xin T, Li S, Zhang Y, Kamali X, Liu H, Jia T. circRNA Hsa_circ_0020850 silence represses the development of lung adenocarcinoma via regulating miR‐195‐5p/IRS2 axis. Cancer Manag Res. 2020;12:10679‐10692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xu Y, Yu J, Huang Z, et al. Circular RNA hsa_circ_0000326 acts as a miR‐338‐3p sponge to facilitate lung adenocarcinoma progression. J Exp Clin Cancer Res. 2020;39(1):57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Feng D, Xu Y, Hu J, Zhang S, Li M, Xu L. A novel circular RNA, hsa‐circ‐0000211, promotes lung adenocarcinoma migration and invasion through sponging of hsa‐miR‐622 and modulating HIF1‐α expression. Biochem Biophys Res Commun. 2020;521(2):395‐401. [DOI] [PubMed] [Google Scholar]
- 9. Hanna DL, Loupakis F, Yang D, et al. Prognostic value of ACVRL1 expression in metastatic colorectal cancer patients receiving first‐line chemotherapy with bevacizumab: results from the triplet plus bevacizumab (TRIBE) study. Clin Colorectal Cancer. 2018;17(3):e471‐e488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yoda S, Soejima K, Hamamoto J, et al. Claudin‐1 is a novel target of miR‐375 in non‐small‐cell lung cancer. Lung Cancer. 2014;85(3):366‐372. [DOI] [PubMed] [Google Scholar]
- 11. Gan TQ, Chen WJ, Qin H, et al. Clinical value and prospective pathway signaling of MicroRNA‐375 in lung adenocarcinoma: a study based on the cancer genome atlas (TCGA), gene expression omnibus (GEO) and bioinformatics analysis. Med Sci Monit. 2017;23:2453‐2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C[T]) method. Methods. 2001;25(4):402‐408. [DOI] [PubMed] [Google Scholar]
- 13. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453‐461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li XW, Yang WH, Xu J. Circular RNA in gastric cancer. Chin Med J (Engl). 2020;133(15):1868‐1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang C, Ma L, Niu Y, et al. Circular RNA in lung cancer research: biogenesis, functions, and roles. Int J Biol Sci. 2020;16(5):803‐814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rong Z, Xu J, Shi S, et al. Circular RNA in pancreatic cancer: a novel avenue for the roles of diagnosis and treatment. Theranostics. 2021;11(6):2755‐2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chaichian S, Shafabakhsh R, Mirhashemi SM, Moazzami B, Asemi Z. Circular RNAs: a novel biomarker for cervical cancer. J Cell Physiol. 2020;235(2):718‐724. [DOI] [PubMed] [Google Scholar]
- 18. Wang M, Ma M, Yang Y, et al. Overexpression of hsa_circ_0008274 inhibited the progression of lung adenocarcinoma by regulating HMGA2 via sponging miR‐578. Thoracic Cancer. 2021;12(16):2258‐2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang C, Yue W, Li L, et al. Circular RNA hsa‐circ‐000881 suppresses the progression of lung adenocarcinoma in vitro via a miR‐665/PRICKLE2 axis. Ann Transl Med. 2021;9(6):498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhou Q, Sun Y. Circular RNA cMras suppresses the progression of lung adenocarcinoma through ABHD5/ATGL Axis using NF‐κB signaling pathway. Cancer Biother Radiopharm. 2020. doi: 10.1089/cbr.2020.3709 [DOI] [PubMed] [Google Scholar]
- 21. Chen D, Ma W, Ke Z, Xie F. CircRNA hsa_circ_100395 regulates miR‐1228/TCF21 pathway to inhibit lung cancer progression. Cell Cycle. 2018;17(16):2080‐2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen M, Huang X, Li L, Huang M, Cai R, Liao X. A regulatory axis of circ_0008193/miR‐1180‐3p/TRIM62 suppresses proliferation, migration, invasion, and Warburg effect in lung adenocarcinoma cells under hypoxia. Med Sci Monit. 2020;26:e922900. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23. Zhao H, Zhu L, Jin Y, Ji H, Yan X, Zhu X. miR‐375 is highly expressed and possibly transactivated by achaete‐scute complex homolog 1 in small‐cell lung cancer cells. Acta Biochim Biophys Sin. 2012;44(2):177‐182. [DOI] [PubMed] [Google Scholar]
- 24. Tang W, Li GS, Li JD, et al. The role of upregulated miR‐375 expression in breast cancer: an in vitro and in silico study. Pathol Res Pract. 2020;216(1):152754. [DOI] [PubMed] [Google Scholar]
- 25. Xu Z, Chen C. Abnormal expression and prognostic significance of bone morphogenetic proteins and their receptors in lung adenocarcinoma. Biomed Res Int. 2021;2021:6663990‐6663923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang Z, Lin M, He L, Qi H, Shen J, Ying K. Exosomal lncRNA SCIRT/miR‐665 transferring promotes lung cancer cell metastasis through the inhibition of HEYL. J Oncol. 2021;2021:9813773‐9813713. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data that have been generated or analyzed during this study are included in this article.


