Abstract
Background
COL10A1 is a secreted, short‐chain collagen found in several types of cancer. Studies have shown that COL10A1 aberrant expression is considered an oncogenic factor. However, its underlying mechanisms and regulation of gastric cancer remain undefined.
Methods
The data on the expression of COL10A1, clinicopathological characteristics, and outcome of patients with GC were obtained from The Cancer Genome Atlas. The ALGGEN‐PROMO database defined the related transcription factors. Quantitative real‐time reverse transcription‐polymerase chain reaction and western blotting analysis were used to identify the differential expression levels of COL10A1 and related transcription factors.
Results
We found that high COL10A1 expression is an independent risk factor for gastric cancer. Upregulation of LEF1 and Wnt2 was also observed in gastric cancer, suggesting a potential correlation between LEF1/COL10A1 regulation in the Wnt2 signaling pathway.
Conclusion
High COL10A1 expression may contribute to poor outcomes via upregulation of LEF1 and Wnt2 in gastric cancer.
Keywords: COL10A1, gastric cancer, LEF1, outcome, Wnt2
We performed bioinformatic analysis and experiments to identify the differential expression levels of aimed molecules. In addition, we found that high COL10A1 expression is an independent risk factor for GC. Upregulation of LEF1 and Wnt2 was also observed in GC, suggesting a potential correlation between LEF1/COL10A1 regulation in the Wnt2 signaling pathway.
![]()
1. INTRODUCTION
Gastric cancer (GC) is the fifth most common gastrointestinal malignant tumor, despite its decreasing incidence, which is predicted to account for 1.4% of total new cancer cases in 2021. 1 Patients with GC may experience high metastasis and mortality rates and low 5‐year survival rates. 2 Despite the rapid development of treatments for GC, including chemotherapy and immunotherapy, surgical resection remains the mainstay treatment. 3 However, given the absence of specific signs of early GC, a remarkable proportion of patients tend to develop advanced‐stage disease and lose the opportunity for surgical resection. 4
The collagen type X alpha 1 (COL10A1) gene is located at 6q22.1 and encodes a secreted, short‐chain collagen. 5 COL10A1 participates in cell growth, differentiation, apoptosis, migration, endochondral bone formation, and bone marrow formation. 6 The aberrant expression of COL10A1 is found in GC, colorectal cancer, breast cancer, and lung cancer. 7 , 8 , 9 , 10 Studies showed that COL10A1 was highly expressed in GC cells and promoted cell proliferation, invasion, and migration, contributing to tumor progression and poor survival. 11 Patients with breast cancer and high COL10A1 expression might benefit less from neoadjuvant chemotherapy. 12 Nevertheless, the functions and the mechanisms of COL10A1 regulation in GC remain poorly understood. In the present study, we investigated the prognostic role of COL10A1 in GC and its potential mechanisms by which COL10A1 regulates tumor progression.
2. MATERIALS AND METHODS
2.1. Clinical data
Data of expression, clinicopathological characteristics, and outcome for 301 patients with GC were obtained from The Cancer Genome Atlas (TCGA) database (https://xena.ucsc.edu); 301 GC tissue samples and 29 adjacent normal ones were analyzed. Ten paired tissues were stored in the Affiliated Lihuili Hospital, Ningbo University. The Ethics Committee of the Affiliated Lihuili Hospital, Ningbo University approved the study (KY2020PJ136). This study was performed in accordance with the tenets of the Declaration of Helsinki. Written informed consent was obtained from all participants. The clinicopathologic characteristics of 301 patients with GC are shown in Table 1, according to COL10A1 expression.
TABLE 1.
The correlation of COL10A1 expression with clinicopathologic characteristics of GC patients
| Variables | Cases (n) | COL10A1 | p value | |
|---|---|---|---|---|
| High | Low | |||
| Total | 301 | 151 | 150 | |
| Age (years) | ||||
| ≥60 | 206 | 106 | 100 | |
| <60 | 95 | 45 | 50 | 0.537 |
| Gender | ||||
| Male | 189 | 95 | 94 | |
| Female | 112 | 56 | 56 | 1.000 |
| Pathological stage | ||||
| I/II | 145 | 75 | 70 | |
| III/IV | 156 | 76 | 80 | 0.645 |
| T stage | ||||
| T1/T2 | 80 | 30 | 50 | |
| T3/T4 | 221 | 121 | 100 | 0.009 |
| N stage | ||||
| Negative | 104 | 60 | 44 | |
| Positive | 197 | 91 | 106 | 0.069 |
| M stage | ||||
| Negative | 272 | 137 | 135 | |
| Positive | 29 | 14 | 15 | 0.848 |
2.2. Identification of transcription factors associated with COL10A1
The ALGGEN‐PROMO database was used to identify the transcription factors (TFs) related to COL10A1 expression (http://alggen.lsi.upc.es). Factors were predicted within a dissimilarity margin less or equal to 5%.
2.3. Quantitative real‐time reverse transcription‐polymerase chain reaction (qRT‐PCR)
According to the manufacturer's protocol, total RNA from paired tissues stored in our hospital was isolated using TRIzol reagent (Invitrogen) at 1500 ng/μl. Next, total RNA was reverse transcribed into cDNAs, and the cDNA template was amplified using real‐time RT‐PCR with the SYBR Green PCR Master Mix (Roche), according to the manufacturer's instructions. The relative expression levels were calculated using the comparative Ct method (2−ΔΔCt). COL10A1‐F: 5'‐CCAGCACGCAGAATCCATCT‐3', COL10A1‐R: 5'‐ACTGTGTCTTGGTGTTGGGT‐3'.
2.4. Western blotting analysis
Tissues were prepared in SDS Lysis Buffer (Sigma) prepared for extraction of total protein. A BCA protein assay kit (Beyotime) was used to quantify protein concentrations according to the manufacturer's instructions. About 60 μg of total protein was mixed with loading buffer and loaded in each lane of SDS‐PAGE for electrophoresis (separated using 8%–12% gels). Next, proteins were transferred to PVDF membranes at 100 V constant voltage for 2 h. The membranes were blocked with 5% skim milk for 2 h and incubated with 1:1000 primary antibody (Abcam) overnight at 4°C. The membranes were then washed three times with TBST and incubated with 1:5000 secondary antibody (Thermo Pierce) for 1 h. Finally, the membranes were washed three times with TBST and exposed to enhanced chemiluminescence substrates to visualize the expression of the target protein using Image J (version 1.8.0).
2.5. Statistical analysis
Statistical analyses were conducted using SPSS software (version 20.0, IBM) and GraphPad Prism (version 8.0). Data were presented as mean ± standard deviation. Student's t‐test was used to calculate differences in the means of the two groups. The chi‐square test was used to analyze the differences in clinicopathologic characteristics between the groups. Overall survival (OS) was calculated from the date of diagnosis to the date of death from any cause. Recurrence‐free survival (RFS) was defined as the date of surgery to the date of disease recurrence. The Kaplan–Meier method and log‐rank test were used to create OS and RFS curves. A Cox regression model was generated for univariate and multivariate analysis. Pearson correlation analysis was used to assess the correlation between COL10A1 expression and TFs. The statistical test used in this study was two‐sided, and a p‐value <0.05 was considered statistically significant.
3. RESULTS
3.1. COL10A1 was highly expressed in human GC tissues
There was significantly more expression of COL10A1 in GC samples (n = 301) than adjacent normal ones (n = 29) according to TCGA sequencing data (p < 0.001). The pair‐matched samples demonstrated consistent results; COL10A1 expression was higher in GC tissues (n = 29, p < 0.001). We analyzed the ten paired tissues stored in our hospital and found that COL10A1 expression was increased in GC tissues, both at the mRNA (p < 0.001) and protein levels (Figure 1A). COL10A1 expression levels were significantly higher in patients with stage T3 and T4 (p = 0.009; Table 1).
FIGURE 1.

High COL10A1 expression was associated with poor outcome in patients with GC. (A) The analyses and identification of COL10A1 expression level by TCGA data, qRT‐PCR, and western blotting analysis. (B) The cutoff value, sensitivity, and specificity of COL10A1 expression and Kaplan–Meier survival analyses of the relationship of COL10A1 expression with survival and recurrence time in GC. patients.
3.2. High COL10A1 expression was associated with poor outcome of GC
According to the OS time, OS status, and COL10A1 expression level, we found the cutoff point at 8.698 of COL10A1 in GC samples (n = 301) using TCGA. We divided the patients into COL10A1 high‐expression (n = 151) and low‐expression groups (n = 150). We calculated the sensitivity and specificity of COL10A1 expression for predicting survival outcome using receiver operating characteristic curves and area under the curve; these were 58.42%, 54.50%, and 0.568%, respectively. Kaplan–Meier analysis revealed that patients with high COL10A1 expression had shorter survival time than those with low COL10A1 expression (p = 0.023); however, there was no significantly different tumor recurrence rate according to COL10A1 expression levels (Figure 1B). Univariate and multivariate analyses conducted using a Cox regression model showed that COL10A1 expression was an independent prognostic factor for survival time in patients with GC (HR: 1.551, p = 0.034), as were age and pathological stage (Table 2).
TABLE 2.
Cox regression analysis of COL10A1 expression as survival predictor
| Variables | Univariate Cox regression analysis | Multivariate Cox regression analysis | ||
|---|---|---|---|---|
| RR (95% CI) | p value | RR (95% CI) | p value | |
| Age (years) | ||||
| <60 vs. ≥60 | 1.743 (1.104–2.751) | 0.017 | 1.850 (1.169‐2.928) | 0.009 |
| Gender | ||||
| Male vs. Female | 1.251 (0.824–1.899) | 0.293 | NA | NA |
| Pathological stage | ||||
| III/IV vs.I/II | 1.931 (1.272–2.932) | 0.002 | 2.081 (1.367‐3.168) | 0.001 |
| T stage | ||||
| T3 + T4 vs. T1 + T2 | 1.584 (0.967‐2.593) | 0.068 | NA | NA |
| N staging | ||||
| Positive vs. Negative | 1.522 (0.978–2.367) | 0.063 | NA | NA |
| M stage | ||||
| Positive vs. Negative | 1.641 (0.915–2.943) | 0.096 | NA | NA |
| COL10A1 expression | ||||
| High vs. Low | 1.595 (1.062–2.395) | 0.024 | 1.551 (1.033–2.329) | 0.034 |
Abbreviation: NA, not analyzed.
3.3. Transcription factor‐LEF1 was positively associated with COL10A1 expression
Based on the ALGGEN‐PROMO database, we selected the top 16 TFs for further investigation. The expression data were downloaded from TCGA database, and the results of TFs expression levels between GC samples and adjacent normal ones were shown in Figure 2A, as well were pair‐matched ones. We found nine TFs showing aberrant expression in GC samples (GATA1, GATA2, FOXP3, YY1, IRF2, VDR, LEF1, USF2, and HNF1A). Five levels were increased (FOXP3, YY1, VDR, LEF1, and HNF1A), and four were decreased (GATA1, GATA2, IRF2, and USF2). The Pearson correlation analysis between COL10A1 expression and these TFs are shown in Figure 2B, suggesting that LEF1, FOXP3, GATA2, and USF2 had significantly positive relationships with COL10A1 expression. Because of the highest correlation found between LEF1 and COL10A1 expression (r = 0.3685, p < 0.0001), we chose LEF1 for further investigation.
FIGURE 2.
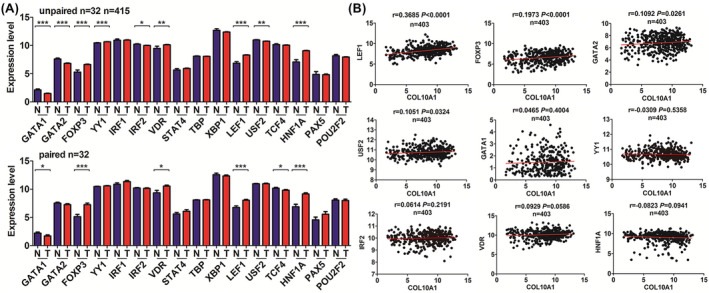
Expression pattern of transcription factors (TFs) and their correlations with COL10A1 expression in GC. (A) The expression levels of TFs in the GC samples compared with adjacent normal ones. (B) Pearson's correlation between the expression of COL10A1 and differential expressed TFs in GC.
3.4. Wnt2 signaling pathway acted downstream of LEF1/COL10A1
We further measured LEF1 and Wnt2/4 protein expression levels using the same batch of samples and found similar trends in expression level changes with COL10A1. LEF1 and Wnt2 were both upregulated in GC tissues; however, there was no significant expression level change in Wnt4 (Figure 3). The corresponding upregulation of LEF1 and Wnt2 indicated the possibility of LEF1/COL10A1 regulation in the Wnt2 signaling pathway.
FIGURE 3.
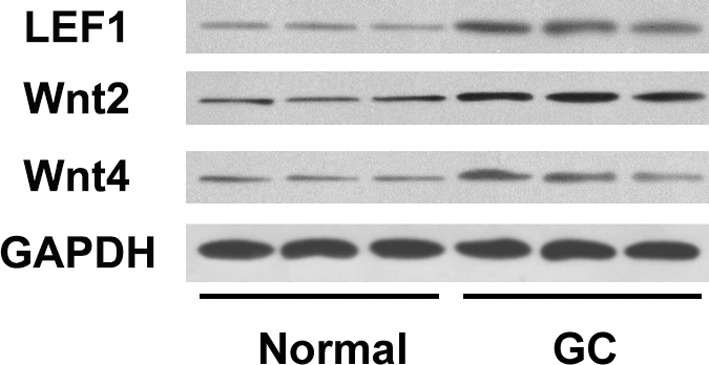
Western blotting analysis of LEF1, Wnt2, and Wnt4 in GC tissues
4. DISCUSSION
We investigated the expression modes and prognostic value of COL10A1 in GC. There was markedly higher COL10A1 expression in GC samples than in adjacent normal tissues, according to TCGA database. We also performed qRT‐PCR and western blotting analyses with GC tissues stored in our hospital; these findings were consistent with the previous results: COL10A1 was upregulated at the mRNA and protein levels. However, there were no significant alterations in genetic or epigenetic dysregulation of COL10A1 in GC, which may account for the aberrant COL10A1 expression (Figure S1). The predictive analysis indicated that COL10A1 acted as an independent risk factor for survival time in patients with GC. Recently, studies showed that aberrant expression of COL10A1 in several types of cancer might result in tumor progression. Some groups reported that high COL10A1 expression promoted cell proliferation, invasion, and migration in GC. 7 , 11 High plasma levels of COL10A1 indicated poor survival and acted as a potential biomarker for early detection of GC. 13 Huang et al. 8 found that COL10A1 expression was higher in colorectal cancer tissues. It induced tumor progression, considered an independent risk factor of OS in patients with colorectal cancer. In lung adenocarcinoma, COL10A1 upregulation positively correlated with lymph node metastasis, and COL10A1 became a novel target for lung cancer. 10 Patients with breast cancer may benefit less from neoadjuvant chemotherapy than those with high COL10A1 expression. 12
To account for the aberrant expression of COL10A1, we identified several related TFs. Based on the differential expression levels and Pearson correlation analysis, we found associations between COL10A1 expression and TFs expression. Lymphoid enhancer‐binding factor 1 (LEF1) ranked first and was positively correlated with COL10A1 expression. Western blot analysis confirmed the upregulation of LEF1 in GC tissues. Recent studies showed that LEF1 is involved in the highly‐conserved Wnt/β‐catenin classical pathway, which mediates cell proliferation, differentiation, and apoptosis, resulting in tumor development. 14 LEF1 was upregulated in lung adenocarcinoma, colon cancer, and breast cancer. 15 , 16 , 17 High LEF1 expression may relate to cell proliferation, invasion, and migration of GC. Gastrin 17 promoted cell proliferation and angiogenesis by increasing the expression of LEF1. 18 OSR1 inhibited cell proliferation by regulating the cell cycle, and overexpression of OSR1 decreased the expression of β‐catenin, TCF1, and LEF1, reversibly. 19 Activation of TCF1 and LEF1 may promote cell invasion and metastasis. 20 There is some evidence that COL10A1 functions with LEF1 in bone formation and development. Yan et al. reported that bony ankylosis was characterized by higher mRNA expression trends in Wnt2b, Wnt5a, β‐catenin, Lef1, Runx2 Col10a1 than in fibrous ankylosis. 21 In osteoarthritis, decreased LEF1 expression reduced COL10A1 expression by the observed strong binding of LEF1 to the COL10A1 promoter. 22
Given the potential association between LEF1 expression and COL10A1 expression in GC, we measured Wnt2/4 expression levels and LEF1 protein levels using the same tissues stored at our hospital. We found a possible correlation between LEF1/COL10A1 and the Wnt2 signaling pathway. Recent studies reported increased Wnt2 expression in GC; the latter may contribute to proliferation, migration, invasion, and metastasis in vitro or in vivo. 23 , 24 Wnt2 upregulation may lead to tumorigenesis through the activation of the Wnt/β‐catenin pathway, 25 , 26 participating in several biological processes. The Wnt2‐specific antagonist was a novel target for the chemoprevention or treatment of GC. 25 Tian et al. 27 reported Wnt2 expression level changes with dysregulated SERPINH1 expression and suggested that SERPINH1 regulated epithelial‐to‐mesenchymal transition and GC progression via the Wnt/β‐catenin pathway. Cheng et al. 28 illustrated the relationship between Wnt2 expression and β‐catenin trans‐localization in GC. Cao et al. 24 reported that CircLMO7/miR‐30a‐3p interaction affected the Wnt2/β‐catenin pathway in GC, and CircLMO7 may function in the glutamine metabolism through the Wnt2/β‐catenin pathway. Our analysis was based on the large sample size from TCGA, and our study and the previous studies supported the results. LEF1 and Wnt2 were both components of canonical Wnt/β‐catenin signaling; 29 therefore, we suggest that LEF1/COL10A1 may regulate the Wnt2 signaling pathway in GC.
In conclusion, our bioinformatics analysis suggests that high COL10A1 expression is an independent risk factor in GC, and it may be regulated by transcription factor‐LEF1 through the Wnt2 signaling pathway.
AUTHOR CONTRIBUTIONS
MZ and WZ conceived and designed the study. MZ and MJ wrote the manuscript. MZ, MJ, QZ, and WY acquired and analyzed the data. MZ and WZ revised the manuscript for important intellectual content. All authors read and approved the final manuscript.
FUNDING INFORMATION
This work was supported by a grant from the Natural Science Foundation of Ningbo (Grant No. 202003N4201) and Natural Science Foundation of Zhejiang Province (Grant LQ19E030011).
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
Supporting information
Figure S1
Data S1
ACKNOWLEDGEMENT
We would like to acknowledge the open‐access bioinformatics database for providing platforms, as well as our colleagues for collecting data and perform the experiments.
Zhang M, Jin M, Gao Z, Yu W, Zhang W . High COL10A1 expression potentially contributes to poor outcomes in gastric cancer with the help of LEF1 and Wnt2. J Clin Lab Anal. 2022;36:e24612. doi: 10.1002/jcla.24612
Miaozun Zhang and Ming Jin contributed equally to the study.
DATA AVAILABILITY STATEMENT
The datasets analyzed during the current study are available on The Cancer Genome Atlas (TCGA) program website (https://xena.ucsc.edu) and ALGGEN‐PROMO database (http://alggen.lsi.upc.es/). The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Siegel RL, Miller KD, Fuchs HE, et al. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71(1):7‐33. [DOI] [PubMed] [Google Scholar]
- 2. Song Z, Wu Y, Yang J, et al. Progress in the treatment of advanced gastric cancer. Tumour Biol. 2017;39(7):1010428317714626. [DOI] [PubMed] [Google Scholar]
- 3. Das M. Neoadjuvant chemotherapy: survival benefit in gastric cancer. Lancet Oncol. 2017;18(6):e307. [DOI] [PubMed] [Google Scholar]
- 4. Tan Z. Recent advances in the surgical treatment of advanced gastric cancer: a review. Med Sci Monit. 2019;25:3537‐3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kielty CM, Kwan AP, Holmes DF, et al. Type X collagen, a product of hypertrophic chondrocytes. Biochem J. 1985;227(2):545‐554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chapman KB, Prendes MJ, Sternberg H, et al. COL10A1 expression is elevated in diverse solid tumor types and is associated with tumor vasculature. Future Oncol. 2012;8(8):1031‐1040. [DOI] [PubMed] [Google Scholar]
- 7. Li T, Huang H, Shi G, et al. TGF‐β1‐SOX9 axis‐inducible COL10A1 promotes invasion and metastasis in gastric cancer via epithelial‐to‐mesenchymal transition. Cell Death Dis. 2018;9(9):849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huang H, Li T, Ye G, et al. High expression of COL10A1 is associated with poor prognosis in colorectal cancer. Onco Targets Ther. 2018;11:1571‐1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang M, Chen H, Wang M, et al. Bioinformatics analysis of prognostic significance of COL10A1 in breast cancer. Biosci Rep. 2020;40(2):BSR20193286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liang Y, Xia W, Zhang T, et al. Upregulated collagen COL10A1 remodels the extracellular matrix and promotes malignant progression in lung adenocarcinoma. Front Oncol. 2020;10:573534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li HH, Wang JD, Wang W, et al. Effect of miR‐26a‐5p on gastric cancer cell proliferation, migration and invasion by targeting COL10A1. Eur Rev Med Pharmacol Sci. 2020;24(3):1186‐1194. [DOI] [PubMed] [Google Scholar]
- 12. Brodsky AS, Xiong J, Yang D, et al. Identification of stromal ColXα1 and tumor‐infiltrating lymphocytes as putative predictive markers of neoadjuvant therapy in estrogen receptor‐positive/HER2‐positive breast cancer. BMC Cancer. 2016;16:274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Necula L, Matei L, Dragu D, et al. High plasma levels of COL10A1 are associated with advanced tumor stage in gastric cancer patients. World J Gastroenterol. 2020;26(22):3024‐3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Santiago L, Daniels G, Wang D, et al. Wnt signaling pathway protein LEF1 in cancer, as a biomarker for prognosis and a target for treatment. Am J Cancer Res. 2017;7(6):1389‐1406. [PMC free article] [PubMed] [Google Scholar]
- 15. Wang ZX, Zhao Y, Yu Y, et al. Effects of lncRNA SNHG20 on proliferation and apoptosis of non‐small cell lung cancer cells through Wnt/β‐catenin signaling pathway. Eur Rev Med Pharmacol Sci. 2020;24(1):230‐237. [DOI] [PubMed] [Google Scholar]
- 16. Hao YH, Lafita‐Navarro MC, Zacharias L, et al. Induction of LEF1 by MYC activates the WNT pathway and maintains cell proliferation. Cell Commun Signal. 2019;17(1):129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Blazquez R, Rietkötter E, Wenske B, et al. LEF1 supports metastatic brain colonization by regulating glutathione metabolism and increasing ROS resistance in breast cancer. Int J Cancer. 2020;146(11):3170‐3183. [DOI] [PubMed] [Google Scholar]
- 18. Tang E, Wang Y, Liu T, et al. Gastrin promotes angiogenesis by activating HIF‐1α/β‐catenin/VEGF signaling in gastric cancer. Gene. 2019;704:42‐48. [DOI] [PubMed] [Google Scholar]
- 19. Otani K, Dong Y, Li X, et al. Odd‐skipped related 1 is a novel tumour suppressor gene and a potential prognostic biomarker in gastric cancer. J Pathol. 2014;234(3):302‐315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li P, Lin X, Zhang JR, et al. The expression of presenilin 1 enhances carcinogenesis and metastasis in gastric cancer. Oncotarget. 2016;7(9):10650‐10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yan YB, Li JM, Xiao E, et al. A pilot trial on the molecular pathophysiology of traumatic temporomandibular joint bony ankylosis in a sheep model. Part II: The differential gene expression among fibrous ankylosis, bony ankylosis and condylar fracture. J Craniomaxillofac Surg. 2014;42(2):e23‐e28. [DOI] [PubMed] [Google Scholar]
- 22. Papathanasiou I, Malizos KN, Tsezou A. Bone morphogenetic protein‐2‐induced Wnt/β‐catenin signaling pathway activation through enhanced low‐density‐lipoprotein receptor‐related protein 5 catabolic activity contributes to hypertrophy in osteoarthritic chondrocytes. Arthritis Res Ther. 2012;14(2):R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang Z, Wang J, Dong X. Wnt2 contributes to the progression of gastric cancer by promoting cell migration and invasion. Oncol Lett. 2018;16(3):2857‐2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cao J, Zhang X, Xu P, et al. Circular RNA circLMO7 acts as a microRNA‐30a‐3p sponge to promote gastric cancer progression via the WNT2/β‐catenin pathway. J Exp Clin Cancer Res. 2021;40(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Katoh M. WNT2 and human gastrointestinal cancer (review). Int J Mol Med. 2003;12(5):811‐816. [PubMed] [Google Scholar]
- 26. Katoh M. Frequent up‐regulation of WNT2 in primary gastric cancer and colorectal cancer. Int J Oncol. 2001;19(5):1003‐1007. [DOI] [PubMed] [Google Scholar]
- 27. Tian S, Peng P, Li J, et al. SERPINH1 regulates EMT and gastric cancer metastasis via the Wnt/β‐catenin signaling pathway. Aging (Albany NY). 2020;12(4):3574‐3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cheng XX, Wang ZC, Chen XY, et al. Correlation of Wnt‐2 expression and beta‐catenin intracellular accumulation in Chinese gastric cancers: relevance with tumour dissemination. Cancer Lett. 2005;223(2):339‐347. [DOI] [PubMed] [Google Scholar]
- 29. Zhang M, Shi J, Huang Y, et al. Expression of canonical WNT/β‐CATENIN signaling components in the developing human lung. BMC Dev Biol. 2012;12:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Data S1
Data Availability Statement
The datasets analyzed during the current study are available on The Cancer Genome Atlas (TCGA) program website (https://xena.ucsc.edu) and ALGGEN‐PROMO database (http://alggen.lsi.upc.es/). The data that support the findings of this study are available from the corresponding author upon reasonable request.


