Abstract
Background
The metabolic profile of human aortic tissues is of great importance. Among the analytical platforms utilized in metabolomics, LC‐MS provides broad metabolome coverage. The non‐targeted metabolomics can comprehensively detect the entire metabolome of an organism and find the metabolic characteristics that have significant changes in the experimental group and the control group and elucidate the metabolic pathway concerning the recognized metabolites. Employing non‐targeted metabolomics is helpful to develop biomarkers for disease diagnosis and disease pathology research; for instance, Aortic aneurysm (AA) and Aortic dissection (AD).
Aim
This study sought to describe the non‐targeted analysis of 18 aortic tissue samples, comparing between AA and AD.
Material & Methods
Our experimental flow included dividing the samples into (AA, nine samples) and (AD, nine samples), SCIEX quadrupole timeofflight tandem mass spectrometer (TripleTOF) 6600+ mass spectrometer data refinement, MetDNA database analysis, and pathway analysis. We performed an initial validation by setting quality control parameters to evaluate the stability of the analysis system during the computer operation. We then used the repeatability of the control samples to examine the stability of the instrument during the entire analysis process to ensure the reliability of the results.
Results
Our study found 138 novel metabolites involved in galactose metabolism.
Discussion
138 novel metabolites found in this study will be further studied in the future.
Conclusion
Our study found 138 novel metabolites between AA and AD, which will provide viable clinical data for future studies aimed to implement galactose markers in aortic tissue analysis.
Keywords: aortic tissue, galactose pathway, LC‐MS, nontarget metabolic analysis
Differential metabolite analysis between AA and AD groups
1. INTRODUCTION
Aortic aneurysm (AA) and Aortic dissection (AD) are a major leading cause of death among cardiovascular disorders. 1 , 2 AA is a fatal asymptomatic condition due to the progressive enlargement of the aorta that may evolve toward rupture. 2 It is reported that AA is responsible for the death of more than 10,000 cases in the USA, 3 and the survival rate of less severe AA patients remains unimproved; simultaneously, there is no known treatment for AA. 4 Recently, the prognostic methods for AA such as ultrasound, computed tomography (CT), and magnetic resonance imaging (MRI) are valid and feasible; however, there is a limited benefit of the early AA diagnosis because 90% of aneurysms detected by these methods were below the currently accepted threshold for surgical intervention. 5 On the other hand, AD causes chest pain that results from tearing of the local endothelium that gradually progresses because of the blood flow and forms lesion within the aorta that manifest tear‐like pain. 6 , 7 Recently, the incident rate of AD has been increasing; it is estimated that the AD cases in the USA are around 25–30 million, 8 while the mortality rate in China has been reported to be around 17.7%. 9 The current diagnostic methods for AD are magnetic resonance imaging (MRI), computed tomographic angiography (CTA), and digital subtraction angiography (DSA). 10 , 11 However, these methods are being utilized only after the onset of the disease. Although there are abundant biomarkers for AD, the specificity and interpretation of these biomarkers in the setting of acute AD remain clinically challenging. 12 Therefore, the identification of AA and AD markers mainly focuses on early peripheral blood biomarkers for vascular endothelial injury and inflammatory reactions and repair in the early stage of the disease. 13 , 14
Metabolomics provides a comprehensive and systemic insight into the rule of metabolite profile changes of an organism. 15 It has been utilized in various fields such as disease diagnosis, drug discovery, and drug toxicity analysis. 16 , 17 Recently, there have been multiple techniques used in metabolomics analysis such as LC‐MS, NMR, GC‐MS, and CE‐MS; they are highly sensitive, precise, require low sample size, and have good reproducibility. Nevertheless, LC‐MS is one of the most potent tools in the metabolic analysis. 18 , 19
In the present study, we used the LC‐MS analysis platform to conduct metabolomics research on 18 human aortic tissue samples. The sample first undergoes pre‐processing to extract metabolites and then test and collect information in the positive and negative ion modes of LC‐MS to obtain MS, MS/MS, and RT information of metabolites. Finally, get a list of metabolites and data matrix, combined with T‐test and VIP (PLS‐DA), screen out different metabolites, and conduct advanced analysis such as pathway analysis and cluster analysis to mine the biological information of differential metabolism. We found that 138 metabolites were identified in our analysis (Table 1), and our finding would supply new biomarkers for AD treatment.
TABLE 1.
Different LC‐MS screening results
| Acquisition mode | Features | Metabolites | Total |
|---|---|---|---|
| Positive ion mode | 400 | 51 | 138 |
| Negative ion mode | 336 | 87 |
2. MATERIALS AND METHODS
2.1. Reagents and instrument
The aortic tissue samples were obtained from patients with AD and AA at Beijing Anzhen Hospital. The employed instrument in the present study was as follows: Ultra‐High‐Performance Liquid Chromatography (1290 UHPLC, Agilent), High‐resolution mass spectrometry (Triple TOF 6600plus, AB Sciex, USA), Centrifuge (5424R, Eppendorf, USA), Balance (ME204E, Mettler, ***), Vortex (Maxi Mix 2, Thermo Fisher Scientific, USA), Ultrasound system (FB15055, Thermo Fisher Scientific, USA), Column (ACQUITY UPLC BEH Amide 1.7 μm 2.1*100 mm, Waters, USA). The reagents used in this study were as follows: Methanol (CAS# 67561, LCMS grade, Honeywell, USA), Acetonitrile (CAS# 75058, LCMS grade, Merck, USA), Water (CAS# 7732185, LCMS grade, Honeywell, USA).
2.2. Samples preparation
The aortic tissues samples were cut on dry ice, and the tissue weight was recorded, making sure <10% error across different samples. Two‐hundred μl water was added to 20 mg tissue and homogenized with cooled N2 gas flow from liquid N2 for three cycles; each cycle was set to 5500 rpm for 20 s and repeated three times. Then, 800 μl MeOH: ACN (v:v, 1:1) was added to 200 μl of the homogenized sample, vortexed for 30 s, and sonicated for 10 min. Proteins were prone to precipitate by being incubated for 1 h at −20°C. Then the sample was centrifuged at 13,000 rpm for 15 min at 4°C, and the supernatant was collected and evaporated to dryness in a vacuum concentrator. The dry extracts were then resuspended in 100 μl of 1:1 ACN: water and sonicated 10 min. The mixture was centrifuged at 13,000 rpm for 15 min at 4°C, and the supernatant was collected and stored at −80°C.
2.3. LC‐MS/MS setup/analysis
The supernatant was analyzed by HPLC–MS/MS on TripleTOF™6600plus mass spectrometer (AB SCIEX, USA) coupled to an Agilent 1290 liquid chromatography system (Agilent, USA). For LC separation, the ACQUITY UPLC BEH C18 column (100 mm × 2.1 mm id, 1.7 μm; Waters) was used. Five μl sample was injected and separated with a 12 min gradient. The column flow rate was maintained at 500 μl/min with the column temperature of 40°C. The chromatographic gradient is shown in Table 2. The electrospray ionization mass spectra were acquired in positive ion mode and negative ion mode, respectively. Information‐dependent acquisition (IDA) was used to collect full scan MS and MSMS information simultaneously. Mass data were collected between m/z 60 and 1200 Da. The ion spray voltage was set to 5000 V for positive mode and 4000 V for negative mode, and the heatedcapillary temperature was maintained at 600°C. The curtain gas flow, nebulizer, and heater gas were set to 35, 60, and 60 arbitrary units, respectively. The collision energy was set to 30 V. The chromatographic column for positive and negative ion detection mode is Waters Acquity UPLC, BEH Amide (1.7 μm, 2.1*100 mm; Waters); mobile phase A is water (including 25 mM ammonium acetate and 25 mM ammonia), mobile phase B is pure acetonitrile. The flow rate is 0.50 ml/min, the injection volume is 2 μl, and the column temperature is 25°C. The mass spectrum signal acquisition adopts positive ion (ESI+) and negative ion (ESI−) scanning modes. The primary parameter electrospray capillary voltage is 5000 V (ESI+) And − 4000 V (ESI−), TOF MS scanning range: 60–1200 m/z, ion source temperature: 600°C, atomizing gas, auxiliary heating gas, and curtain airflow are set separately at 60 psi, 60 psi, and 35 psi. The specific mass spectrometer parameters in positive and negative ion modes are shown in Tables 2 and 3.
TABLE 2.
The mobile phase elution gradient
| Time (min) | Flow rate (ml/min) | A (%) | B (%) |
|---|---|---|---|
| 0 | 0.5 | 5 | 95 |
| 0.5 | 0.5 | 5 | 95 |
| 7 | 0.5 | 35 | 65 |
| 8 | 0.5 | 60 | 40 |
| 9 | 0.5 | 60 | 40 |
| 9.1 | 0.5 | 5 | 95 |
| 12 | 0.5 | 5 | 95 |
TABLE 3.
Mass spectrometry condition parameters (ESI+) positive ion mode and mass spectrometry condition parameters (ESI−) negative ion mode
| Scan mode | ||
|---|---|---|
| Full scan (TOF MS) | IDA (Product Ion) | |
| (ESI+) | ||
| Quality range | m/z 601,200 Da | m/z 251,200 Da |
| Atomizing gas (GS1) | 60 psi | 60 psi |
| Auxiliary heating gas (GS2) | 60 psi | 60 psi |
| Curtain air (CUR) | 35 psi | 35 psi |
| Temperature (TEM) | 600°C | 600°C |
| Capillary voltage (IS) | 5000 V | 5000 V |
| Declustering voltage (DP) | 60 V | 60 V |
| Collision energy (CE) | 10 V | 30 V |
| (ESI−) | ||
| Quality range | m/z 601,200 Da | m/z 251,200 Da |
| Atomizing gas (GS1) | 60 psi | 60 psi |
| Auxiliary heating gas (GS2) | 60 psi | 60 psi |
| Curtain air (CUR) | 35 psi | 35 psi |
| Temperature (TEM) | 600°C | 600°C |
| Capillary voltage (IS) | 4000 V | 5000 V |
| Declustering voltage (DP) | 60 V | 60 V |
2.4. Quality control sample preparation
The tissues were lysed and dissolved in boiled water. Then, samples were incubated at −20°C for 1 h and then centrifuged (13,000 r/min) for 15 min. Finally, the supernatants were collected and stored at −80°C.
2.5. Quality control analysis
In order to evaluate the stability of the analysis system during the computer operation, a quality control sample (Quality Control, QC) will be prepared during the experiment. The test samples are mixed with equal volumes. In the process of instrument analysis, one QC sample is inserted for every six analysis samples. During data analysis, the QC sample can be passed, and repeatability is used to examine the stability of the instrument during the entire analysis process to ensure the reliability of the results. The quality control analysis has analyzed using Chromatogram analysis of sample base peak (BPC) and Sample QC RSD analysis. By superimposing the BPC spectra of the QC samples, it can be seen that the chromatographic peak intensity and retention time of each QC sample are basically the same, indicating that the stability of the system and the experiment and the repeatability are very good (Figure S1A). Through the relative standard deviation (RSD) distribution of the features in the QC sample, the repeatability of the QC sample can be evaluated, reflecting the stability and stability of data collection. We use the ratio of features with an RSD of less than 30% to all feature data as the main criterion for evaluation. As shown in Figure S1B, it can be seen that the detection among the 72,228 features, the number of features whose RSD is less than 30% is higher than 97%, indicating that the system stability and the repeatability of the experiment are very good.
2.6. Statistical analysis
(O)PLS‐DA was performed in statistical analysis. Student's T‐test was used for comparisons between the groups. p < 0.05 and FC > 1.5 suggested a significant change.
3. RESULTS
3.1. LC‐MS analysis showed a significant difference between the AA and AD groups in their metabolome profile
We analyzed 18 aortic tissue samples, nine samples with aortic aneurysm (AA), and nine samples with aortic dissection (AD) using LC‐MS (SCIEX quadrupole timeofflight tandem mass spectrometer (TripleTOF) 6600+ mass spectrometer). The output data from the LC‐MS has been processed using ProteoWizard (version 3.0.6150). The peak identification and retention time alignment were made using XCMS (version 1.46.0). We identified the metabolites using MetDNA (http://metdna.zhulab.cn/) and statistically analyzed them using MetaboAnalyst (https://www.metaboanalyst.ca). Principal Component Analysis (PCA) and Partial Least Squares Discrimination Analysis (PLS‐DA) were performed using normalized peak table by total intensity to investigate the possible separation of metabolite profiles between the two groups and fold changes, and pvalues (assessed by the Student's ttest) were computed. The discovery data set contained 72,228 features, and 736 features were significantly differentiated with fold changes greater than 1.5, p‐value less than 0.05, and VIP > 1. Hundred and thirty eight of these differential features were identified successfully (Figure 1A–C).
FIGURE 1.
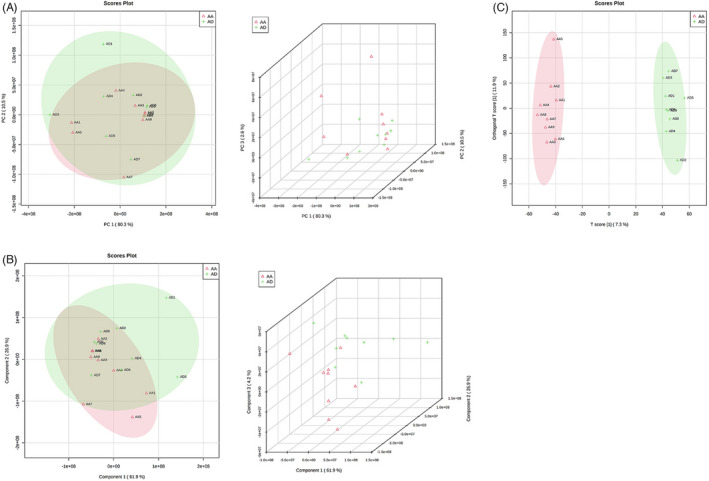
Differential metabolite analysis between AA and AD groups. (A) The PCA analysis comparison result between the AA and AD groups. (B) PLS‐DA analysis of the comparison between the AA and AD groups. (C) OPLS‐DA analysis of the comparison between the AA and AD groups
3.2. Galactose pathway‐related biomarkers were identified among the AA and AD group
We analyzed the significant metabolic differences between the AA and AD groups. As seen in Figure 2A, the difference between the two groups was significant and indicated the presence of distinct biomarkers in each group of samples. Further confirmation on the differential metabolite findings has been done using a correlation heat map. We found a significant difference between the expressed metabolites in each group (Figure 2B,C). Pathway annotation was performed using MetaboAnalyst for KEGG annotation to identify the related pathway of the found metabolites. As shown in Figure 2D, the galactose pathway has been identified as a potential signaling pathway for the found metabolites. Enrichment analysis has been done to determine the upregulated and downregulated metabolites as seen in Figure 2E; the bubble chart presents the yellow dots as the metabolites that are upregulated, and the blue dots are the metabolites that are downregulated, while the abscissa is the pathway influence value of pathway topology analysis.
FIGURE 2.
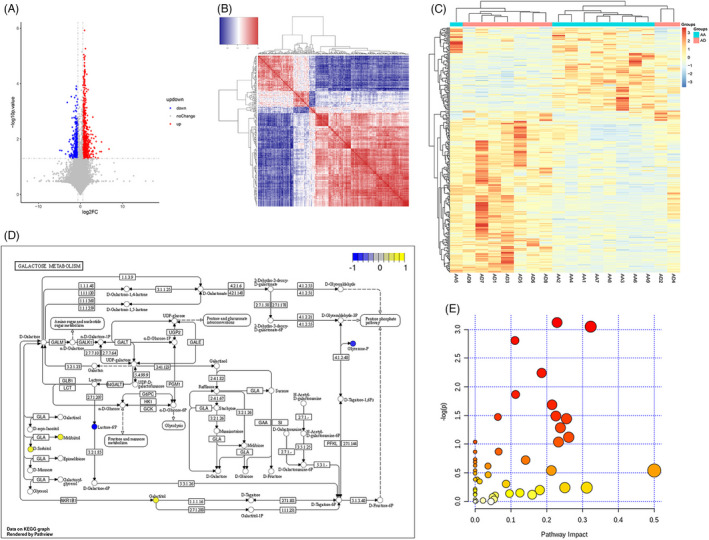
Different metabolites correlations between the AA and AD groups. (A) Volcano map illustrating the different metabolites between the AA and AD groups. (B) Heat map chart of the correlation of different metabolites between the AA and AD groups. (C) Heat map chart of the comparison group. (D) Pathway analysis of the differential metabolites. (E) Bubble chart of the enrichment of metabolic pathways of different metabolites
4. DISCUSSION
Aortic aneurysm (AA) and aortic dissection (AD) are acute diseases that are complicated by the blood streamflow. However, their pathogenesis remains unclear, and the alteration in the body metabolism concerning the AA and AD is poorly discussed. It is also challenging to diagnose them at an early stage, leaving a big chance for these diseases to progress beyond treatment. Previously, it has been reported that distinct metabolites are found in the plasma of AA patients. 20 , 21 Also, the biomarker studies of AD have focused on peripheral blood markers but yet to be fully disseminated in clinical applications due to cost, diagnosis interval, and testing conditions. 22 , 23 In this regard, establishing standard settings to identify novel biomarkers for AA and AD remains a challenge.
Metabolomics is a new approach in systematic biology, and it can identify the changes in the metabolic profile of an entire organism. 24 , 25 The untargeted metabolic analysis identifies a broad spectrum of metabolites and compares their features among different groups/species. 25
In the present study, a non‐targeted LC‐MS‐based metabolomics approach has been employed to analyze the metabolic profile variation between aortic aneurysm (AA) and aortic dissection (AD) tissue samples. We used tissue samples to directly encounter the metabolic alteration in each sample regardless of the testing condition; this shall help overcome the limitation regarding the difference among each disease progression stage. The LC‐MS results have been analyzed using different statistical approaches after being controlled to guarantee their accuracy. We found that there are significant metabolomic changes between the AA and AD group (Figure 1). Also, our investigation has concluded that 138 metabolites were engaged in the galactose pathway (Figure 2). These metabolites present a step toward a better understanding the difference between AA and AD and their potential biomarkers for diagnosis and treatment. Different statistical analyses and quality control setups provided rigorous evidence upon the results from LC‐MS; this can help overcome the false‐positive results during analysis.
Our study has several limitations. First, the metabolic comparison has been made between two diseased groups (AA and AD) without proper control. The second is the limited small sample number. Our goal from this study was to prove that using LC‐MS accompanied with rigor statistical analysis and pre‐set quality control measures will result in identifying novel metabolites that will help in the diagnosis and treatment of AA and AD. Sequential sampling will permit patients to serve as their own controls in longitudinal studies that correlate metabolites with aneurysmal or dissection progression rates. A further detailed investigation will be planned to overcome the present limitation of our study and to present more details about our found metabolites and their role in AA and AD.
5. CONCLUSION
The utilization of LC‐MS analysis of aortic tissue samples (AA and AD) has led to the identification of 138 novel metabolites. Our findings have concluded that the metabolites are engaged in the galactose pathway, which can be used as biomarkers for the diagnosis and treatment of AA and AD. Also, we illustrated that the use of the LC‐MS combined with one‐dimensional and multi‐dimensional analysis is more feasible to predict different significant metabolites between different groups. Our data shall provide an insight into novel biomarkers between AA and AD that could be used as a potential diagnostic and treatment for AA and AD.
CONFLICT OF INTEREST
These authors declared no competing interests in this study.
Supporting information
Figure S1
Zhang K, Pan X, Zheng J, Liu Y, Sun L. The metabolic analysis in human aortic tissues of aortic dissection. J Clin Lab Anal. 2022;36:e24623. doi: 10.1002/jcla.24623
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Regeer MV, Martina B, Versteegh MI, et al. Prognostic implications of descending thoracic aorta dilation after surgery for aortic dissection. J Cardiovasc Comput Tomogr. 2017;11:1‐7. [DOI] [PubMed] [Google Scholar]
- 2. Touat Z, Ollivier V, Dai J, et al. Renewal of mural thrombus releases plasma markers and is involved in aortic abdominal aneurysm evolution. Am J Pathol. 2006;168:1022‐1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Klink A, Hyafil F, Rudd J, et al. Diagnostic and therapeutic strategies for small abdominal aortic aneurysms. Nat Rev Cardiol. 2011;8:338‐347. [DOI] [PubMed] [Google Scholar]
- 4. Miyake T, Morishita R. Pharmacological treatment of abdominal aortic aneurysm. Cardiovasc Res. 2009;83:436‐443. [DOI] [PubMed] [Google Scholar]
- 5. Ciborowski M, Teul J, Martin‐Ventura JL, Egido J, Barbas C. Metabolomics with LC‐QTOF‐MS permits the prediction of disease stage in aortic abdominal aneurysm based on plasma metabolic fingerprint. PLoS One. 2012;7:e31982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chiu P, Miller DC. Evolution of surgical therapy for Stanford acute type a aortic dissection. Ann Cardiothorac Surg. 2016;5:275‐295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ziganshin BA, Dumfarth J, Elefteriades JA. Natural history of type B aortic dissection: ten tips. Ann Cardiothorac Surg. 2014;3:247‐254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Roger VL, Go AS, Lloyd‐Jones DM, et al. Heart disease and stroke statistics‐‐2011 update: a report from the American Heart Association. Circulation. 2011;123:e18‐e209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li Y, Yang N, Duan W, Liu S, Yu S, Yi D. Acute aortic dissection in China. Am J Cardiol. 2012;110:1056‐1061. [DOI] [PubMed] [Google Scholar]
- 10. De Leon Ayala IA, Chen YF. Acute aortic dissection: an update. Kaohsiung J Med Sci. 2012;28:299‐305. [DOI] [PubMed] [Google Scholar]
- 11. Garcia A, Ferreiros J, Santamaria M, Bustos A, Abades JL, Santamaria N. MR angiographic evaluation of complications in surgically treated type a aortic dissection. Radiographics. 2006;26:981‐992. [DOI] [PubMed] [Google Scholar]
- 12. Ren Y, Tang Q, Liu W, Tang Y, Zhu R, Li B. Serum biomarker identification by mass spectrometry in acute aortic dissection. Cell Physiol Biochem. 2017;44:2147‐2157. [DOI] [PubMed] [Google Scholar]
- 13. Qu F, Zheng SJ, Liu S, Wu CS, Duan ZP, Zhang JL. Serum sphingolipids reflect the severity of chronic HBV infection and predict the mortality of HBV‐acute‐on‐chronic liver failure. PLoS One. 2014;9:e104988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shulaev V. Metabolomics technology and bioinformatics. Brief Bioinform. 2006;7:128‐139. [DOI] [PubMed] [Google Scholar]
- 15. Nicholson JK, Lindon JC, Holmes E. ‘Metabonomics’: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29:1181‐1189. [DOI] [PubMed] [Google Scholar]
- 16. Keun HC. Metabonomic modeling of drug toxicity. Pharmacol Ther. 2006;109:92‐106. [DOI] [PubMed] [Google Scholar]
- 17. Lindon JC, Holmes E, Nicholson JK. Metabonomics and its role in drug development and disease diagnosis. Expert Rev Mol Diagn. 2004;4:189‐199. [DOI] [PubMed] [Google Scholar]
- 18. Lindon JC, Nicholson JK. Spectroscopic and statistical techniques for information recovery in metabonomics and metabolomics. Annu Rev Anal Chem (Palo Alto Calif). 2008;1:45‐69. [DOI] [PubMed] [Google Scholar]
- 19. Liu JL, Wang HL, Zhang LF, et al. Metabonomics study of brain‐specific human S100B transgenic mice by using high‐performance liquid chromatography coupled with quadrupole time of flight mass spectrometry. Biol Pharm Bull. 2011;34:871‐876. [DOI] [PubMed] [Google Scholar]
- 20. Aoyagi K, Shahrzad S, Iida S, et al. Role of nitric oxide in the synthesis of guanidinosuccinic acid, an activator of the N‐methyl‐D‐aspartate receptor. Kidney Int Suppl. 2001;78:S93‐S96. [DOI] [PubMed] [Google Scholar]
- 21. Zhang F, Jia Z, Gao P, et al. Metabonomics study of atherosclerosis rats by ultra fast liquid chromatography coupled with ion trap‐time of flight mass spectrometry. Talanta. 2009;79:836‐844. [DOI] [PubMed] [Google Scholar]
- 22. Wen JJ, Zago MP, Nunez S, Gupta S, Burgos FN, Garg NJ. Serum proteomic signature of human chagasic patients for the identification of novel potential protein biomarkers of disease. Mol Cell Proteomics. 2012;11:435‐452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. White L, Ma J, Liang S, Sanchez‐Espiridion B, Liang D. LC–MS/MS determination of d‐mannose in human serum as a potential cancer biomarker. J Pharm Biomed Anal. 2017;137:54‐59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fiehn O, Kopka J, Dormann P, Altmann T, Trethewey RN, Willmitzer L. Metabolite profiling for plant functional genomics. Nat Biotechnol. 2000;18:1157‐1161. [DOI] [PubMed] [Google Scholar]
- 25. Zhao L, Zhang H, Wang J, et al. C60 Fullerols enhance copper toxicity and Alter the leaf metabolite and protein profile in cucumber. Environ Sci Technol. 2019;53:2171‐2180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


