Abstract
The mechanism of action of isoniazid (INH), a first-line antituberculosis drug, is complex, as mutations in at least five different genes (katG, inhA, ahpC, kasA, and ndh) have been found to correlate with isoniazid resistance. Despite this complexity, a preponderance of evidence implicates inhA, which codes for an enoyl-acyl carrier protein reductase of the fatty acid synthase II (FASII), as the primary target of INH. However, INH treatment of Mycobacterium tuberculosis causes the accumulation of hexacosanoic acid (C26:0), a result unexpected for the blocking of an enoyl-reductase. To test whether inactivation of InhA is identical to INH treatment of mycobacteria, we isolated a temperature-sensitive mutation in the inhA gene of Mycobacterium smegmatis that rendered InhA inactive at 42°C. Thermal inactivation of InhA in M. smegmatis resulted in the inhibition of mycolic acid biosynthesis, a decrease in hexadecanoic acid (C16:0) and a concomitant increase of tetracosanoic acid (C24:0) in a manner equivalent to that seen in INH-treated cells. Similarly, INH treatment of Mycobacterium bovis BCG caused an inhibition of mycolic acid biosynthesis, a decrease in C16:0, and a concomitant accumulation of C26:0. Moreover, the InhA-inactivated cells, like INH-treated cells, underwent a drastic morphological change, leading to cell lysis. These data show that InhA inactivation, alone, is sufficient to induce the accumulation of saturated fatty acids, cell wall alterations, and cell lysis and are consistent with InhA being a primary target of INH.
Isoniazid (INH) remains one of the key drugs in global control strategies to treat tuberculosis, despite the increasing numbers of primary and secondary resistance (8). INH was first discovered to have antituberculosis activity in 1952 in random screens, though its target of action was unknown (5, 11). Early studies had shown that INH inhibited the synthesis of mycolic acids, α-alkyl β-hydroxy long-chain fatty acids (60 to 90 carbons in length) that cover the surface of mycobacteria (34, 40). Using lipid fractionation methodologies, Takayama and colleagues later demonstrated that INH treatment of Mycobacterium tuberculosis inhibited mycolic acid biosynthesis and caused the accumulation of hexacosanoic acid (C26:0), a saturated C26 fatty acid (36). They proposed three possible sites of action: (i) a desaturase, (ii) a cyclopropanase, and (iii) an enzyme involved in long-chain fatty acid elongation.
Using newly developed gene transfer systems for mycobacteria, we identified a novel gene, inhA, as a target for INH (1). Further work revealed that InhA encoded an NADH-specific enoyl-acyl carrier protein (ACP) reductase activity (10, 29) which converts Δ2-unsaturated to saturated fatty acids and is involved in the elongation of long-chain fatty acids to mycolic acids. This enzymatic activity led Mdluli et al. to argue that InhA was not the primary target of INH for M. tuberculosis (22, 23). Their major objection was that a block in an enoyl-ACP reductase should lead to the accumulation of monounsaturated intermediates and could not account for the accumulation of C26:0 observed following INH treatment of M. tuberculosis.
Unlike most organisms, mycobacteria have two fatty acid synthases, the fatty acid synthase I (FASI) and FASII systems (Fig. 1A). The discovery that mycobacteria had FASI was surprising, as it is the first prokaryote shown to have FASI, a multidomain enzyme that encodes all the activities necessary for fatty acid synthesis in one large polypeptide (7). The FASI system from Mycobacterium smegmatis produces saturated fatty acids in a bimodal pattern of palmitate (C16:0) and tetracosanoate (C24:0) (6, 27). In contrast, the ACP-requiring FASII system contains a series of independent enzymes, including InhA (21), and is responsible for the biosynthesis of mycolic acid by elongation of the FASI products.
FIG. 1.
Fatty acid biosynthesis in mycobacteria (A) and activation of INH (B). (A) Fatty acid synthetase I carries on the synthesis of C16:0 and C24:0. One or both fatty acid products act as substrates for the synthesis of mycolic acids by fatty acid synthetase II. Putative targets for the action of activated INH are underlined. (B) INH is activated by the action of a catalase-peroxidase encoded by the katG gene. Shown are two possible products of this reaction, involved in the formation of the INH-NAD adduct (31).
In this report, we sought to isolate mutants of M. smegmatis that would map to inhA and confer thermosensitivity. Using such an inhA(Ts) allele, it would be possible to test if thermal inactivation of InhA induces the same events leading to the death of M. smegmatis as INH treatment. Two independent mutants were identified that had an identical single point mutation within inhA. The analogous mutation was engineered into the M. tuberculosis inhA gene, and the expressed protein was demonstrated to be inactive at the nonpermissive temperature. Thermal inactivation of the two M. smegmatis strains harboring the inhA(Ts) allele resulted in mycolic acid biosynthesis inhibition and accumulation of long-chain saturated fatty acids, with no detectable amount of Δ2-unsaturated fatty acids. Similar fatty acid profiles were observed following INH treatment of M. smegmatis or Mycobacterium bovis BCG. Furthermore, both InhA thermal inactivation and INH treatment caused similar morphological changes to the cell wall, leading to cell lysis. In the context of these results, we present a model in which InhA is the primary target of INH in M. tuberculosis, M. bovis BCG, and M. smegmatis.
MATERIALS AND METHODS
Bacterial strains.
The mycobacterial strains used in this study are described in Table 1. The M. smegmatis mutants were obtained from the laboratory wild-type (wt) strain mc2155 (33).
TABLE 1.
Bacterial strains used in this study
| M. smegmatis strain | Genotype | Source or reference |
|---|---|---|
| mc2155 | ept-1 | 33 |
| mc2651 | ept-1 inhA1 | 1 |
| mc2699 | ept-1 inhA linked to aph | 1 |
| mc22354 | ept-1 attB::pYUB412::ndh | 25 |
| mc22359 | ept-1 attB::pYUB412::ndh inhA40 | INH-R, Ts mutant of mc22354 |
| mc22360 | ept-1 attB::pYUB412::ndh inhA41 | INH-R, Ts mutant of mc22354 |
| mc22361 | ept-1 attB::pYUB412::ndh inhA+ aph | I3 (mc2699) × mc22359 |
| mc22362 | ept-1 attB::pYUB412::ndh inhA+ aph | I3 (mc2699) × mc22360 |
Isolation of spontaneous INH-R mutants.
Spontaneous INH-resistant (INH-R) mutants were isolated from one nonmutagenized culture of mc22354, a merodiploid for the ndh gene of M. smegmatis, and grown in Middlebrook 7H9 broth (Difco) supplemented with 0.2% glycerol, 10% ADS enrichment, and 0.5% Tween 80. The culture was incubated while shaking for 5 days at 30°C. Tenfold serial dilutions were plated on minimal (Middlebrook 7H9 supplemented with 0.2% glycerol, 10% ADS, and 1.5% agar) and rich (Mueller-Hinton agar [Difco]) agar plates containing INH (25 μg/ml). The plates were incubated at 30°C for 6 days.
Revertant analysis.
The thermosensitive strains were grown at 30°C to an optical density (OD) of 1.5 to 2.0. Tenfold serial dilutions were plated on rich and minimal media. The plates were incubated at 30°C (for titration of the cultures) and 42°C for 5 days. The frequency of reversion was determined by the number of CFU at 42°C divided by the titer.
Transformation experiments.
Cultures were grown at 30°C (OD of 1.0), spun down, washed twice with cold 10% glycerol, and resuspended in 10% glycerol (0.5 ml). To the cold cell suspension (100 μl) was added pYUB1019 (a 3-kb fragment containing orf1-inhA-orf3 subcloned into pMD31, a kanamycin-resistant [Kanr] Escherichia coli mycobacterial shuttle plasmid; 1 μl). The cold suspension was electroporated (2.5 V, 25 μFd, 1,000 Ω), plated on kanamycin (20 μg/ml)-containing plates, and incubated at 30°C for 7 days.
Transduction experiments.
Cultures were grown in 20 ml of minimal medium containing INH (25 μg/ml) at 30°C (OD of 1.0), spun down, and resuspended in 2 ml of SM buffer (0.1 M NaCl, 8 mM MgSO4 · 7H2O, 50 mM Tris hydrochloride, pH 7.5) containing 0.5% Tween 80 and 10 mM CaCl2. The cell suspension (200 μl) was mixed with 200-μl aliquots of high-titer I3 transducing lysate dilutions (108 and 109 phage-forming units/ml) and incubated without shaking at 30°C for 30 min. After addition of Luria-Bertani broth (Difco) (400 μl), the cell suspension was incubated with shaking at 30°C for 90 min and plated on Luria-Bertani medium containing 10 mM sodium citrate, and the plates were incubated at 30°C for 6 days. Isolated transductants were freed of phage by streaking on Luria-Bertani medium containing 10 mM sodium citrate.
PCR amplification.
The inhA gene was amplified from chromosomal DNA, using the primers TW175 (5′-CCAAATGACAGGACTACTCG-3′) and TW176 (5′-TCACAACAGATGCGTGCTGG-3′) along with amplification of this gene from the wt mc2155 chromosome. These products were directly sequenced bidirectionally and aligned against each other.
Construction of a V238F mutant M. tuberculosis InhA protein and enzymatic characterization.
The V238F mutation was introduced into the pET M. tuberculosis inhA expression vector (10) using an inverse PCR methodology (2) and verified by DNA sequence analysis. The InhA protein was purified as previously described (29). All enzyme assays were done in 30 mM piperazine-N,N′-bis(2-ethanesulfonic) acid (PIPES buffer), pH 6.8, using 2-trans-octenoyl-coenzyme A (CoA) (0.6 mM) as a substrate. The extinction coefficients of NADH at 340 and 380 nm were 6,220 and 1,128 M−1 cm−1, respectively. The enzyme activity was defined by the number of micromoles of NADH turned over per minute per milligram of protein. Enzyme activities of the InhA(Ts) (V238F) mutant were measured at various temperatures and compared with those of wt InhA. Small aliquots of enzyme solution were injected to assay solution in a quartz cell. After mixing vigorously, a decrease of absorbency at 380 nm for the Ts mutant and 340 nm for the wt enzyme was recorded for 5 min at 20, 37, and 42°C. Final concentrations of InhA and NADH were 1.08 μM (per active site) and 800 μM for the Ts mutant and 0.034 μM (per active site) and 5 μM for wt InhA, respectively. In this experiment, NADH concentrations were set around Km values for NADH because enzyme activity was significantly inhibited at higher NADH concentration (>1 mM) for the Ts mutant as described below. Enzyme activity was measured at various NADH concentrations from 0.1 to 1.6 mM at 20°C. The decrease of absorbance was recorded at 380 nm. Kinetic parameters, Km and Vmax, were calculated from initial rates at each NADH concentrations by using KaleidaGraph software (Synergy Software Inc.).
Time course assays.
Time course assays were determined spectrophotometrically by monitoring NADH oxidation at 340 nm using a Shimadzu UV-1201 spectrophotometer with a time course program pack. Shimadzu PC 1201 personal spectroscopy software was used to automate the monitoring of the reactions. All reactions were run in 30 mM PIPES buffer, pH 6.8. Reactions were conducted at fixed concentrations of NADH (180 μM) and 2-trans-dodecenoyl-CoA (50 μM). Cell lysate from mc22359 was diluted, substrates were added, and the spectrophotometer was zeroed upon addition of NADH. Reactions were monitored for 5 min at 0.5-s intervals and were conducted at 25, 37, and 42°C.
Radiolabeling of lipids with [1-14C]acetate.
Cultures from M. smegmatis or BCG were grown in Middlebrook 7H9 broth supplemented with 0.2% glycerol, 10% ADS enrichment, and Tween 80 (0.5% for M. smegmatis and 0.1% for BCG Pasteur) to an OD at 600 nm of ∼0.8 and then incubated at 42°C for 1 h with or without INH (25 μg/ml for M. smegmatis and 1 μg/ml for BCG). [1-14C]Acetate (0.3 μCi/ml) was added, and the incubation was continued for another hour. Cells were collected by centrifugation and lyophilized.
Analysis of fatty acids by TLC.
Fatty acid methyl esters (FAMEs) and mycolic acid methyl esters (MAMEs) were obtained by alkaline treatment of the pellet, esterification with methyl iodide, and extraction with dichloromethane. After concentration to dryness, the residue was resuspended in diethyl ether. Radiolabeled fatty acid extracts were spotted (10 μl of lipid extract; ∼20,000 cpm) by silica gel thin-layer chromatography (TLC) and eluted (double elution with hexane-ethyl acetate [9:1, vol/vol]). Detection of radiolabeled species was done by autoradiography. The autoradiograms were obtained after exposure at −80°C for 2 days on X-ray film.
Analysis of fatty acids by HPLC.
Samples were prepared by saponification of the cell pellets, acidification, extraction with chloroform, evaporation, and derivatization to their UV-absorbing p-bromophenacyl esters using the p-bromophenacyl ester kit (part no. 18036) from Alltech Associates, Inc. High-performance liquid chromatography (HPLC) analysis of the p-bromophenacyl fatty acid esters was conducted on a Hewlett-Packard model HP1100 gradient chromatograph equipped with an HP1100 series thermostated column compartment, an HP1100 series diode array detector, and an IN/US β-RAM model 2B flowthrough radioisotope beta-gamma radiation detector. The data were collected and processed with HP Chemstation software (version A.04.01). Separation of the p-bromophenacyl fatty acid esters was achieved using an Alltech all-guard column (part no. 77082 and 96080) coupled to a reverse-phase C18 column (4.6 by 150 mm; 3-μm column diameter; Alltima C18 [Alltech]). The column temperature was set at 45°C, the flow rate was set at 2 ml/min, the sample injection volume was set at 95 μl, and the wavelength was set at 260 nm. The mobile phase was acetonitrile-water employed as an isocratic elution (83:17, vol/vol) for 20 min (24), followed by a linear increase to 100% acetonitrile in 2 min and held at 100% acetonitrile for 18 min. The 14C-labeled fatty acid esters were detected using In-FlowTM ES (IN/US Systems, Inc., Tampa, Fla.) at a 3:1 (vol/vol) ratio with the column eluant. The chromatograms' peaks were identified by comparison with chromatograms of p-bromophenacyl fatty acid ester standards. Saturated fatty acid standards were purchased from Sigma. 2-Alkenoic acid standards were obtained as described below.
Synthesis of 2-trans-alkenoic acids.
Oxidation of the n-alcohols (C14 to C22) with pyridinium chlorochromate in dichloromethane for 4 h at room temperature gave the corresponding aldehydes in 90 to 97% yield (9). The Wittig reaction of the aldehydes with ethyl (triphenylphosphoranylidene)acetate in dry acetonitrile for 72 h at room temperature afforded ethyl 2-trans-alkenoates in 50 to 80% yield (26). The esters were saponified with a 10% methanolic solution of potassium hydroxide for 2 h at reflux. The resulting 2-trans-alkenoic acids were purified by flash chromatography and recrystallized in hexane (80 to 90%). The structures of the final products and intermediates were confirmed by proton and carbon nuclear magnetic resonance spectroscopy.
Scanning electron microscopy (SEM).
Strains were grown in Mueller-Hinton or 7H9 broth at 30°C to an OD of ∼0.2, INH (25 μg/ml) was added to the M. smegmatis mc2155 culture, and then the cultures were shifted to 42°C. Samples (50 to 100 μl) were taken at 0, 3, 6, and 9 h after the temperature shift. The samples were fixed in 2.5% glutaraldehyde in 0.1 M sodium cacodylate, pH 7.4, plated out on poly-l-lysine-coated coverslips, dehydrated through a graded series of ethanol, critical point dried using liquid carbon dioxide in a Tousimis Samdri (Rockville, Md.) 790 critical point drier, and then sputter coated with gold-palladium in a Denton (Cherry Hill, N.J.) vacuum desk-1 sputter coater. The samples were examined in a JEOL (Peabody, Mass.) JSM6400 scanning electron microscope using an accelerating voltage of 15 kV.
RESULTS
Isolation of a Ts allele of inhA in M. smegmatis.
A Ts allele of inhA, the inhA(Ts) allele, was identified by screening a large set of spontaneous INH-R mutants of M. smegmatis using a strain of M. smegmatis containing a second copy of ndh. This strategy was based on our previous work in which INH-R and Ts mutants were isolated, but surprisingly, their mutations mapped to ndh, the gene encoding an NADH dehydrogenase (25). These mutations had pleiotropic phenotypes including coresistance to INH and ethionamide (ETH), temperature sensitivity for growth, and auxotrophies as a result of increased NADH/NAD ratios in the cell (25). We hypothesized that INH-R, Ts mutations in inhA would be present in a population of spontaneous mutants, but much less frequent than mutations in ndh. Therefore, mutations in inhA could be enriched by using a strain that was a merodiploid for ndh and by plating on minimal medium with INH, which would select against the auxotrophic phenotype associated with many ndh mutants.
The M. smegmatis ndh gene was introduced into mc2155 at the chromosomal attB site for the mycobacterial phage L5 using an integration-proficient vector to make mc22354. Spontaneous INH-R mutants were isolated from a nonmutagenized culture of mc22354 at a frequency of 4 × 10−7 on 7H9 minimal agar medium and 4 × 10−6 on a rich Mueller-Hinton agar medium. The majority of colonies from both the rich (47 of 52) and the minimal (50 of 52) media were coresistant to both INH and ETH, with a higher frequency of Ts phenotypes found from INH-R mutants isolated on the minimal medium (11 of 52) compared to the rich medium (3 of 53). Three of the mutants failed to generate cultures in liquid media and were not analyzed further. Eleven of the remaining mutants were INH-R, ETH-R, and Ts and were subjected to further analysis.
Each of the 11 INH-R, ETH-R, and Ts mutants was transformed with the M. smegmatis inhA gene on multicopy plasmid. Two of the 11 mutants, one isolated from the minimal medium, mc22359, and one isolated from the complex medium, mc22360, were able to grow at 42°C when transformed with the multicopy inhA. To further test if the mutations conferring all three phenotypes mapped to inhA, an allelic exchange I3 transductional analysis was performed using a donor strain, mc2699, which had the wild-type inhA allele closely linked to an aph gene of a mini-Tn5 (1). The frequency of transduction of Kanr was between 10−7 and 10−8 per mc22359 or mc22360 input cell in five different experiments (Table 2). The majority of the Kanr transductants (66 to 100%) had acquired the ability to grow at 42°C, demonstrating a tight linkage of the Kanr marker with inhA that was previously reported (1). Moreover, 100% of the transductants that acquired the ability to grow at 42°C were sensitive to both INH and ETH. The cosegregation of the three phenotypes strongly suggests that a mutation within the inhA gene was responsible for all three phenotypes.
TABLE 2.
I3 transductional analysis with inhA linked to aph in mc22359 and mc22360
| Cross and expt no. | INH-S, Tra colonies/ Kanr colonies (%) |
|---|---|
| I3 (mc2699) × mc22359 [inhA(Ts)] | |
| 1 | 12/18 (66) |
| 2 | 46/50 (92) |
| I3 (mc2699) × mc22360 [inhA(Ts)] | |
| 1 | 15/15 (100) |
| 2 | 47/50 (94) |
| 3 | 7/8 (88) |
Tr, temperature resistant.
Both inhA(Ts) mutants showed high resistance to INH (the MIC was greater than 100 μg/ml, determined by plating onto medium containing different concentrations of INH [25] [data not shown]). Sequence analysis revealed a single point mutation, a G-to-T conversion (G712T), that caused a Val-to-Phe amino acid substitution (V238F) in InhA in both mc22359 and mc22360. Notably, the amino acid sequence of the InhA protein was found to be highly homologous to the amino acid sequence of the fabI-encoded enoyl-ACP reductase from E. coli and that similar substitution (Ser-to-Phe amino acid change) at a comparable position (position 241) in FabI from E. coli and Salmonella enterica serovar Typhimurium caused a Ts phenotype in these organisms (4). This V238F mutation encodes an amino acid substitution that is located in the NADH binding site of the InhA protein. All previously defined mutations that cause an alteration in the NADH binding site of InhA have been found to correlate with INH-R and ETH-R (1, 2). If the G712T mutation is solely responsible for the INH-R, ETH-R, and, Ts phenotypes, then the Kanr transductants found to have regained the wt phenotypes as described above should have regained the G at position 712 in the inhA gene, regenerating the wt inhA sequence. This change could be readily detected, as the G712T mutation alters a PshAI restriction site (5′ … GACNNNNGTC … 3′ [mutation underlined]) within the inhA gene. Three independent transductants of mc22359 and mc22360 were analyzed by PCR amplification of a segment of inhA and digestion with PshAI. All transductants regained the PshAI site like the mc22354 parent, while both Ts mutants were left uncut by PshAI. Thus, all transductants that regained the three parental phenotypes—(i) ability to grow at 42°C, (ii) INH-S, and (iii) ETH-S—regained the wt DNA sequence at position 712. This result is consistent with the premise that the G712T mutation in inhA is solely responsible for the INH-R, ETH-R, and Ts phenotypes.
To further confirm that this mutation within inhA was responsible for temperature sensitivity and antibiotic resistance, we sought revertants that map to the inhA gene. Revertants could be consistently obtained by plating cells for growth at 42°C, but all at very low frequencies of less than 10−8. All 19 revertants analyzed acquired single point mutations within the mutated inhA gene (Table 3). The majority of mutants had acquired mutations that caused amino acid substitutions at amino acid 238. One class became INH-S (F238V, F238L, and F238C), and another remained INH-R (F238I). One additional class of revertants encodes an intramolecular suppressor mutation causing a glycine-to-cysteine amino acid substitution at amino acid 102. While this suppressing mutation allowed the revertants to grow at 42°C, both of these revertants retained their INH-R and ETH-R phenotypes.
TABLE 3.
Sequence analysis of inhA alleles from temperature-resistant (Tr) revertants
| Strain | Phenotype | DNA change(s) | Amino acid change | No. with phenotype/no. of mutants analyzed |
|---|---|---|---|---|
| Revertants of mc22359 [inhA(Ts)] | ||||
| mc22363, mc22364 | Tr INH-Sa | 712TTA, 712TTG | F238L | 4/9 |
| mc22365 | Tr INH-S | 712TGC | F238C | 1/9 |
| mc22366 | Tr INH-R | 304TGT | G102C | 4/9 |
| Revertants of mc22360 [inhA(Ts)] | ||||
| mc22367 | Tr INH-S | 712GTC | F238V | 1/10 |
| mc22368, mc22369, mc22370 | Tr INH-S | 712TTA, 712TTG, 712CTC | F238L | 4/10 |
| mc22371 | Tr INH-S | 712TGC | F238C | 2/10 |
| mc22372 | Tr INH-R | 712ATC | F238I | 1/10 |
| mc22373 | Tr INH-R | 304TGT | G102C | 2/10 |
INH-S, INH susceptible.
The revertants having a F238V or F238L amino acid change had the same growth rate at 42°C as the wt mc2155 or the parental strain mc22354, while other revertants (F238C, F238I, and G102C) had a growth defect as shown by a longer doubling time (data not shown).
The V238F-altered InhA enoyl-ACP reductase loses activity at 42°C.
E. coli-expressed V238F M. tuberculosis InhA was purified to homogeneity, and its ability to catalyze the reduction of 2-trans-octenoyl-CoA was tested at various temperatures (Table 4). The V238F mutant showed activity similar to the wt enzyme at the permissive temperature of 20°C under saturating conditions of NADH (29). The Km and Vmax values of the InhA(Ts) mutant were determined to be 0.88 ± 0.15 mM and 1.22 ± 0.12 U/mg of protein, respectively. The Km of the InhA(Ts) is about 200 times larger than that of the wt enzyme (4 μM), and the Vmax is less than one-half that of the wt enzyme (ca. 3 U/mg protein). In addition, InhA(Ts) mutant activity was significantly inhibited at higher NADH concentrations (>1 mM). The V238F mutant M. tuberculosis InhA enzyme also showed a significant reduction in its catalytic activity at 37°C and a complete inactivation at 42°C. Similar results were obtained for the V238F mutant of M. smegmatis, with a reduction of InhA activity at 37°C (by 33%) and a total loss of InhA activity at 42°C.
TABLE 4.
Enzymatic activities of wt InhA and InhA(Ts) (V238F)
| Temperature (°C)a | Enzymatic activity (U/mg of protein) (% residual activityb) of:
|
|
|---|---|---|
| wt InhA | InhA(Ts) (V238F) | |
| 20 | 1.51 (100) | 0.70 (100) |
| 37 | 0.98 (65) | 0.051 (7) |
| 42 | 0.51 (34) | NDc (0) |
Two experiments were done for each temperature.
Numbers in parentheses are percentages of residual activity at each temperature, assuming enzyme activity at 20°C to be 100%.
ND, not detected.
Thermal inactivation of InhA inhibits mycolic acid biosynthesis and causes an accumulation of FASI end products.
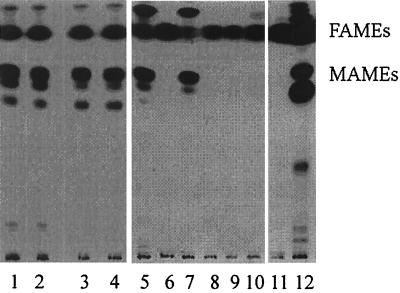
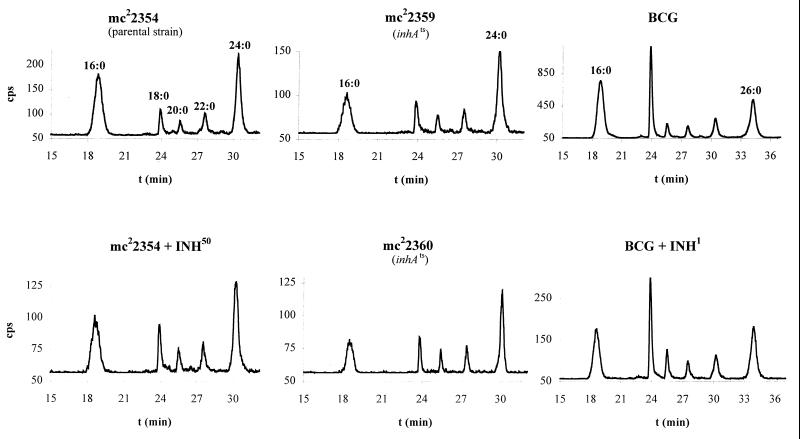
Takayama and colleagues had shown that M. tuberculosis accumulates C26:0 following 1 h of INH treatment (36). We hypothesize that the accumulated C26:0 is an end product generated by FASI following the inhibition of the FASII pathway through inhibition of InhA. The in vitro activities of FASI from M. smegmatis and M. bovis BCG were characterized by Bloch (27) and Kikuchi et al. (16), respectively. A bimodal fatty acid pattern of C16:0 and C24:0 was reported for the fast grower M. smegmatis (27), while the slow grower BCG displayed a bimodal fatty acid pattern of C16:0 and C26:0 (16). To test our hypothesis, we first examined INH treatment of BCG. One hour of INH treatment inhibited the synthesis of mycolic acids (MAMEs) and resulted in an accumulation of nonhydroxylated fatty acids (FAMEs) by TLC (Fig. 2, lane 12). To identify the fatty acids that accumulated following INH treatment, the radiolabeled materials were analyzed by HPLC. The nonhydroxylated fatty acids, present in the radiolabeled samples, were found to be saturated fatty acids with no detectable levels of Δ2-unsaturated products (Fig. 3). These fatty acids had chain lengths ranging from 16 to 26 carbons, similar to the fatty acid pattern described for BCG FASI in vitro (16). There was a dramatic shift in the INH-treated cells in the amounts of C26:0 compared to C16:0 (Fig. 4). While the percentage of other FASI intermediates, namely, C18:0, C20:0, C22:0, and C24:0, remained mostly unchanged (22, 5, 5, and 12%, respectively), we observed a 25% decrease in the level of C16:0 and a 36% increase in the level of C26:0 when the cells were treated with INH.
FIG. 2.
Autoradiographic TLC of 14C-labeled fatty acids of M. smegmatis following INH treatment or InhA inactivation. In lanes 1 to 4, the labelings were done at 30°C: lane 1, wt mc2155; lane 2, ndh merodiploid mc22354; lane 3, mc22359; lane 4, mc22360. In lanes 5 to 12, the labelings were done at 42°C: lane 5, wt mc2155; lane 6, wt mc2155 plus INH (50 μg/ml); lane 7, mc22354; lane 8, mc22354 plus INH (50 μg/ml); lane 9, mc22359; lane 10, mc22360; lane 11, M. bovis BCG Pasteur plus INH (1 μg/ml); lane 12, M. bovis BCG Pasteur.
FIG. 3.
HPLC analysis of 14C-labeled fatty acids from INH-treated or InhA-inactivated samples. The retention times for the p-bromophenacyl 2-alkanoyl esters using the elution conditions described in Materials and Methods are as follows: C16:0, 18.6 min; C18:0, 23.9 min; C20:0, 25.5 min; C22:0, 27.5 min; C24:0, 30.2 min. The retention times for p-bromophenacyl 2-alkenoyl esters are the following: Δ2-C16, 15.4 min; Δ2-C18, 23.0 min; Δ2-C20, 24.6 min; and Δ2-C24, 28.8 min.
FIG. 4.
INH treatment or InhA inactivation causes accumulation of C24:0. The bars represent the percentage of C16:0 and C24:0 for the M. smegmatis strains and C16:0 and C26:0 for the BCG strain, obtained from the corresponding signals in the HPLC radiochromatograms. INH50 and INH1, INH at 50 and 1 μg/ml, respectively.
The same experiment was conducted with the fast-growing M. smegmatis. The fatty acids produced by M. smegmatis during INH treatment at 42°C also consisted of saturated fatty acids with no detectable levels of Δ2-unsaturated products (Fig. 3). The only notable difference between the INH treatment of BCG and M. smegmatis was that the longest fatty acid isolated was C24:0 for M. smegmatis and C26:0 for BCG. This is consistent with the previous in vitro characterizations of FASI activities of M. smegmatis and BCG (16, 27). Following INH treatment of M. smegmatis cells, the fatty acid distribution became similar to that of BCG (Fig. 4). While the percentages of C18:0, C20:0, and C22:0 remained constant (10, 5, and 9%, respectively), we observed an 18% decrease in the level of C16:0 and up to 29% increase in the level of C24:0 when the cells were treated with INH.
To test if FASI end products accumulate following InhA inactivation, we performed the same analyses on the inhA(Ts) mutants. Both mc22359 and mc22360 mutants have the same fatty acid profile as the wt M. smegmatis and their parent strain mc22354 at the permissive temperature (Fig. 2, lanes 1 to 4). Their fatty acid profiles were then examined following a 1-h incubation at 42°C. Analysis by TLC revealed that both inhA(Ts) mutants failed to make mycolates and accumulated nonhydroxylated fatty acids (Fig. 2, lanes 9 and 10), identical to what was observed for INH-treated M. smegmatis cultures (Fig. 2, lanes 6 and 8). The accumulated products were analyzed by HPLC, revealing the presence of saturated fatty acids with no Δ2-unsaturated fatty acids (Fig. 3). As for INH-treated M. smegmatis, we observed a 26% decrease in C16:0 and up to 50% increase in C24:0 (Fig. 4), while the other intermediates stayed unchanged. This analysis clearly establishes that a block in the inhA-encoded enoyl-ACP reductase induces an accumulation of a long-chain saturated fatty acid, comparable to INH treatment of M. smegmatis cells, with no detectable levels of Δ2-unsaturated products.
Lysis of M. smegmatis is induced by thermal inactivation of InhA.
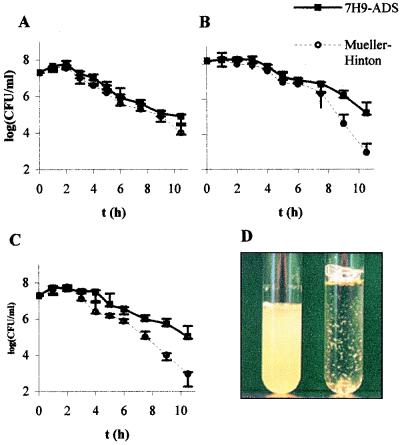
Takayama and colleagues had previously demonstrated that the viability of M. tuberculosis cells was unaffected after the first hour of INH treatment and gradually declined to 18% viability after 3 h of treatment (34). To test if InhA inactivation followed similar death kinetics as that seen in INH-treated cells, we compared INH treatment of M. smegmatis to InhA thermal inactivation in minimal medium. In both cases, we observed an increase in the number of CFU during the first hour, followed by up to 66% decrease after 3 h of treatment. The viability continued to decrease up to 3 orders of magnitude after 7 h for wt M. smegmatis mc2155 treated with INH and for each of the mutants after thermal inactivation (Fig. 5A to C). Faster death kinetics were observed when the cells were incubated in Mueller-Hinton medium compared to 7H9 minimal medium (Fig. 5B and C). Notably, the cultures reproducibly displayed significant lysis 9 h postinduction in both media (Fig. 5D).
FIG. 5.
Viability of M. smegmatis following INH treatment or InhA inactivation. The strains were grown at 30°C up to an early log phase (OD ≈ 0.2), INH (25 μg/ml) was added to wt mc2155, and then all the cultures were shifted to 42°C. Samples were taken every 60 to 90 min, diluted, and plated in duplicate on Mueller-Hinton plates. The plates were incubated at 30°C for 6 days, and the CFU were then counted. The mean CFU per milliliter from four independent experiments for InhA inactivation are shown: wt M. smegmatis following incubation with 25 μg/ml INH (A), mc22359 (Ts mutant) (B), and mc22360 (Ts mutant) (C). (D) Cell lysis of M. smegmatis in Mueller-Hinton broth following thermal inactivation of InhA. Left, wt M. smegmatis incubated at 42°C for 9 h; right, mc22359 incubated at 42°C for 9 h.
To visualize the events leading to lysis, scanning electron microscopy was performed on representative cells at various times after thermal inactivation or INH treatment (Fig. 6). At initial times, cells appeared normal with a smooth and long shape, with dimensions of 0.5 μm in diameter and 3 to 5 μm in length. At 3 h post-InhA inactivation or -INH treatment, the cells began to form blebs at sporadic sites across the cell surface. With increasing time, the blebs appeared to turn into filaments and eventually shred from the cell wall surface. Concomitant with the massive shredding, the cells appeared to shorten in length, widen in cell diameter, and undergo lysis.
FIG. 6.
SEM of inhA(Ts) mutant of M. smegmatis mc22359. Shown are mc22359 at 0 h (A), 3 h (C), 6 h (E), and 9 h (F) and INH-treated wt M. smegmatis at 0 h (B), 3 h (D), 6 h (F), and 9 h (H) after the temperature shift. Several fields were examined at each time point, and representative samples are shown in each panel. The bar represents 1.0 μm.
DISCUSSION
Direct selection for thermosensitive mutations in genes encoding drug targets.
Ts mutants represent powerful tools to identify genes encoding essential functions and to study metabolic pathways. In our case, we wished to isolate a Ts allele specifically in inhA, a gene encoding the enoyl-ACP reductase of the FASII system of mycobacteria. Typically, libraries of mutagenized cells are screened for mutants that fail to grow at nonpermissive temperatures and then the resulting mutants are genetically characterized to identify the mutated gene. This approach has been successfully used for M. smegmatis to identify a number of essential genes, but the screening of 200,000 mutagenized population of mutants thus far has failed to yield a Ts allele in inhA (3, 17). Since INH-R could be mediated by mutations in the inhA gene (1), we reasoned that Ts alleles of inhA could be identified from a large set of spontaneous INH-R mutants isolated at 30°C, as an amino acid substitution conferring INH-R might also confer thermosensitivity to the InhA enzyme. Skold had successfully used a similar strategy to isolate Ts alleles in the gene encoding dihydropteroate synthase, the target of sulfonamides (32). Since we had previously shown that the mutations conferring resistance to INH that are Ts map to ndh, a gene encoding an NADH dehydrogenase (25), we reasoned ndh(Ts) alleles could be eliminated from our screen by starting with a strain that was merodiploid for ndh. Using this approach, spontaneous INH-R mutants were isolated at frequencies of 4 × 10−7 to 4 × 10−6. Approximately 14% of the INH-R mutants were Ts for growth, and 2% were found to map to inhA. Based on our success and that of Skold, we would suggest that this approach could be generally applied to isolate Ts alleles of any gene encoding a drug target.
Inactivation of the FASII enoyl-reductase inhibits mycolic acid biosynthesis and induces cell lysis.
The two independent inhA(Ts) mutants isolated are identical mutants, both having a single point mutation in inhA (V238F) which confers resistance to INH and ETH and renders the InhA enzyme inactive at 42°C. This V238F substitution occurs in the NADH binding pocket of the InhA protein, like other alleles conferring INH-R and ETH-R, and likely reduces the binding of the INH-NAD adduct to mediate the resistance. Our inability to isolate extragenic suppressors establishes that InhA has no redundant function in M. smegmatis. The selective inhibition of InhA in M. smegmatis upon thermal inactivation results in an inhibition of mycolic acid biosynthesis and accumulation of the long-chain saturated fatty acid C24:0. The inhibition of mycolic acid biosynthesis confirms that the FASII pathway, which InhA is part of, is primarily responsible for the elongation of fatty acids to mycolic acids in vivo. The saturated fatty acids that accumulate following InhA inactivation and FASII inactivation leads us to conclude that these fatty acids emanate from FASI since these are the same products observed for FASI activity in vitro (27). Most importantly, InhA inactivation induces the lysis of M. smegmatis cells. The death kinetics of M. smegmatis cells following InhA inactivation are very similar to those seen in M. smegmatis cells treated with INH. In both cases, a common series of cell surface blebbing followed by shredding preceded the cell lysis. These morphological changes are strikingly similar to those reported for M. tuberculosis treated with INH (35).
Is InhA the primary target of INH in M. tuberculosis?
M. tuberculosis is 100-fold more sensitive to INH than M. smegmatis is. To explain this difference, Mdluli et al. have postulated that M. tuberculosis has a different target for INH than M. smegmatis (22). They argue that InhA can not be the target as the inhibition of inhA-encoded enoyl reductase could not account for the accumulation of C26:0 observed following treatment of M. tuberculosis with INH (22). However, since this present work demonstrates that InhA inactivation leads to the accumulation of FASI end products and in vitro studies have established that FASI of M. bovis BCG synthesizes C26:0 (16), we believe it is reasonable to extrapolate that InhA inactivation in M. tuberculosis would also lead to an accumulation of C26:0. Mdluli et al. have also provided evidence for another target by demonstrating that radioactive INH covalently attaches to AcpM, the ACP of FASII, in a complex with the kasA-encoded β-keto acyl synthase in M. tuberculosis (23). While this establishes binding, this is but one criterion for a target. A drug target for a bactericidal drug is an enzyme of the bacterium (i) that binds the drug, (ii) that is inhibited by the drug, and (iii) whose inhibition induces the death of the bacterium. All three criteria have been demonstrated for InhA. INH is a prodrug which, when activated by the catalase-peroxidase katG (Fig. 1B), attacks NAD, and the resulting INH-NAD adduct is the actual drug which binds the InhA enzyme (20, 31, 38) and inhibits InhA function (2, 15, 41). In this study, we demonstrate the third criterion by showing that the inactivation of InhA is alone sufficient to induce the lysis of M. smegmatis in a manner similar to INH action. While a similar study needs to be performed for M. tuberculosis, analysis of mutations in clinical isolates of INH-R strains suggests that this will hold true. First, mutations have been identified within the structural inhA gene from M. tuberculosis and their resulting altered enzymes have been shown to be resistant to KatG-activated INH (2, 10, 30). Moreover, two different mutations that map to the expression region of the mabA-inhA operon of M. tuberculosis have been found in as many as 30% of INH-R isolates and consistently correlate with resistance to INH and ETH (14, 37). Such expression-enhancing mutations would increase InhA levels inside a cell, and overexpressed InhA genes from M. tuberculosis, M. bovis, and M. smegmatis all confer INH-R and ETH-R to M. smegmatis cells and INH-R to inhibition of cell extracts (1). In contrast, no evidence has yet been reported that M. tuberculosis KasA activity is inhibited by activated INH nor that mutant KasA proteins are resistant to inhibition. Genetic evidence has not consistently supported the argument that kasA is a target. Initially four mutations in the kasA gene were found to correlate with INH-R, but two independent reports have demonstrated that two of these mutations are found in INH-S strains of M. tuberculosis (19, 28). Moreover, while kasA and kasB have been shown to confer resistance to thiolactomycin when cloned on multicopy expression vectors in BCG, no resistance to INH was observed (18). While our results suggest that inhibition of any of the enzymes of the FASII pathway might lead to lysis, it will be necessary to demonstrate that KasA activity is not duplicated by the highly homologous KasB protein. Even if binding and inhibition are established for KasA, like InhA, the primary target would likely represent the rate-limiting step for the FASII pathway. For E. coli, this rate-limiting step has been shown to be the enoyl-reductase (13). It appears to be the same for the FASII system of M. tuberculosis as kasA has been shown to be induced following INH treatment, while inhA expression is not (23, 39). Thus, if KasA is not the rate-limiting step, an alternative explanation for INH-R, if it is mediated by the KasA-AcpM binding of INH, might be that the KasA-ACP complex titrates the activated INH species to prevent inhibition of the rate-limiting InhA activity. Such a role would be secondary to InhA.
In contrast to M. smegmatis having a different target for INH compared to M. tuberculosis, we propose that the 100-fold-increased sensitivity for INH in M. tuberculosis compared to M. smegmatis results from an increased ability to generate the InhA inhibitor, the INH-NAD adduct. Two different mechanisms could be proposed to prevent formation of the adduct leading to INH-R: (i) the loss of the activator KatG (42) or (ii) increased NADH/NAD ratios (25, 38). Definitive elucidation of the roles that inhA and kasA play in INH-R will require the construction of isogenic strains of M. tuberculosis differing by single point mutations and subsequent product analysis. Although this has been unachievable to date for technical reasons, we have developed a novel specialized transduction system which should enable allelic exchanges involving the transfer of specific point mutations linked to a selectable marker gene (12; S. Bardarov et al., unpublished data).
FASII enzymes represent attractive drug targets for mycobacteria.
Ultimately, the knowledge that InhA inactivation induces the lysis of mycobacteria underscores the importance of the InhA enzyme for further drug development to kill pathogenic mycobacteria, including INH-R variants of M. tuberculosis. For example, it should be possible to design compounds that require no activation step to inhibit InhA activity. Moreover, the availability of the inhA(Ts) mutant, the first fully characterized Ts mutant in the FASII system of mycobacteria, provides a system that should lead to a better understanding of the events leading to the death of a mycobacterial cell. A better understanding of the mechanism(s) underlying cell death of INH-treated mycobacteria should allow the design of not only improved enoyl-ACP reductase inhibitors but also drugs that synergize to augment the mycobacterial killing process. Indeed, all the FASII enzymes might represent attractive targets for development of novel antimycobacterial agents.
ACKNOWLEDGMENTS
C. Vilchèze and H. R. Morbidoni contributed equally to this work.
We thank Lynn Miesel for providing I3 transducing lysates and I3-transduction protocol, Stoyan Bardarov for providing PCR products, and the Analytical Imaging Facility of AECOM for assistance with SEM. We thank David Alland, Del Besra, John Blanchard, Michael Glickman, and Annaik Quémard for their critical reading of the manuscript and helpful comments.
This work was supported by NIH grant AI43268.
REFERENCES
- 1.Banerjee A, Dubnau E, Quemard A, Balasubramanian V, Um K S, Wilson T, Collins D, de Lisle G, Jacobs W R., Jr inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science. 1994;263:227–230. doi: 10.1126/science.8284673. [DOI] [PubMed] [Google Scholar]
- 2.Basso L A, Zheng R, Musser J M, Jacobs W R, Jr, Blanchard J S. Mechanisms of isoniazid resistance in Mycobacterium tuberculosis: enzymatic characterization of enoyl reductase mutants identified in isoniazid-resistant clinical isolates. J Infect Dis. 1998;178:769–775. doi: 10.1086/515362. [DOI] [PubMed] [Google Scholar]
- 3.Belanger A E, Hatfull G F. Exponential-phase glycogen recycling is essential for growth of Mycobacterium smegmatis. J Bacteriol. 1999;181:6670–6678. doi: 10.1128/jb.181.21.6670-6678.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergler H, Hogenauer G, Turnowsky F. Sequences of the envM gene and of two mutated alleles in Escherichia coli. J Gen Microbiol. 1992;138:2093–2100. doi: 10.1099/00221287-138-10-2093. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein J, Lott W A, Steinberg B A, Yale H L. Chemotherapy of experimental tuberculosis. Am Rev Tuberc. 1952;65:357–374. doi: 10.1164/art.1952.65.4.357. [DOI] [PubMed] [Google Scholar]
- 6.Bloch K. Control mechanisms for fatty acid synthesis in Mycobacterium smegmatis. Adv Enzymol Relat Areas Mol Biol. 1977;45:1–84. doi: 10.1002/9780470122907.ch1. [DOI] [PubMed] [Google Scholar]
- 7.Brindley D N, Matsumura S, Bloch K. Mycobacterium phlei fatty acid synthetase-A bacterial multienzyme complex. Nature. 1969;224:666–669. [Google Scholar]
- 8.Cohn D L, Bustreo F, Raviglione M C. Drug-resistant tuberculosis: review of the worldwide situation and the WHO/IUATLD global surveillance project. Clin Infect Dis. 1997;24:S121–S130. doi: 10.1093/clinids/24.supplement_1.s121. [DOI] [PubMed] [Google Scholar]
- 9.Corey E J, Suggs J W. Pyridinium chlorochromate. An efficient reagent for oxidation of primary and secondary alcohols to carbonyl compounds. Tetrahedron Lett. 1975;31:2647–2650. [Google Scholar]
- 10.Dessen A, Quemard A, Blanchard J S, Jacobs W R, Jr, Sacchettini J C. Crystal structure and function of the isoniazid target of Mycobacterium tuberculosis. Science. 1995;267:1638–41. doi: 10.1126/science.7886450. [DOI] [PubMed] [Google Scholar]
- 11.Fox H H. The chemical approach to the control of tuberculosis. Science. 1952;116:129–134. doi: 10.1126/science.116.3006.129. [DOI] [PubMed] [Google Scholar]
- 12.Glickman M S, Cox J S, Jacobs W R., Jr A novel mycolic acid cyclopropane synthetase is required for cording, persistence, and virulence of Mycobacterium tuberculosis. Mol Cell. 2000;5:717–727. doi: 10.1016/s1097-2765(00)80250-6. [DOI] [PubMed] [Google Scholar]
- 13.Heath R J, Rock C O. Enoyl-acyl carrier protein reductase (fabI) plays a determinant role in completing cycles of fatty acid elongation in Escherichia coli. J Biol Chem. 1995;270:26538–26542. doi: 10.1074/jbc.270.44.26538. [DOI] [PubMed] [Google Scholar]
- 14.Heym B, Honore N, Truffot-Pernot C, Banerjee A, Schurra C, Jacobs W R, Jr, van Embden J D, Grosset J H, Cole S T. Implications of multidrug resistance for the future of short-course chemotherapy of tuberculosis: a molecular study. Lancet. 1994;344:293–298. doi: 10.1016/s0140-6736(94)91338-2. [DOI] [PubMed] [Google Scholar]
- 15.Johnsson K, King D S, Schultz P G. Studies on the mechanisms of action of isoniazid and ethionamide in the chemotherapy of tuberculosis. J Am Chem Soc. 1995;117:5009–5010. [Google Scholar]
- 16.Kikuchi S, Rainwater D L, Kolattukudy P E. Purification and characterization of an unusually large fatty acid synthase from Mycobacterium tuberculosis var. bovis BCG. Arch Biochem Biophys. 1992;295:318–326. doi: 10.1016/0003-9861(92)90524-z. [DOI] [PubMed] [Google Scholar]
- 17.Klann A G, Belanger A E, Abanes-De Mello A, Lee J Y, Hatfull G F. Characterization of the dnaG locus in Mycobacterium smegmatis reveals linkage of DNA replication and cell division. J Bacteriol. 1998;180:65–72. doi: 10.1128/jb.180.1.65-72.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kremer L, Douglas J D, Baulard A R, Morehouse C, Guy M R, Alland D, Dover L G, Lakey J H, Jacobs W R, Jr, Brennan P J, Minnikin D E, Besra G S. Thiolactomycin and related-analogues as novel anti-mycobacterial agents targeting KasA and KasB condensing enzymes in Mycobacterium tuberculosis. J Biol Chem. 2000;275:16857–16864. doi: 10.1074/jbc.M000569200. [DOI] [PubMed] [Google Scholar]
- 19.Lee A S, Lim I H, Tang L L, Telenti A, Wong S Y. Contribution of kasA analysis to detection of isoniazid-resistant Mycobacterium tuberculosis in Singapore. Antimicrob Agents Chemother. 1999;43:2087–2089. doi: 10.1128/aac.43.8.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lei B, Wei C-J, Tu S-C. Action mechanism of antitubercular isoniazid. J Biol Chem. 2000;275:2520–2526. doi: 10.1074/jbc.275.4.2520. [DOI] [PubMed] [Google Scholar]
- 21.Marrakchi H, Lanéelle G, Quémard A. InhA, a target of the antituberculous drug isoniazid, is involved in a mycobacterial fatty acid elongation system, FAS-II. Microbiology. 2000;146:289–296. doi: 10.1099/00221287-146-2-289. [DOI] [PubMed] [Google Scholar]
- 22.Mdluli K, Sherman D R, Hickey M J, Kreiswirth B N, Morris S, Stover C K, Barry C E., 3rd Biochemical and genetic data suggest that InhA is not the primary target for activated isoniazid in Mycobacterium tuberculosis. J Infect Dis. 1996;174:1085–1090. doi: 10.1093/infdis/174.5.1085. [DOI] [PubMed] [Google Scholar]
- 23.Mdluli K, Slayden R A, Zhu Y, Ramaswamy S, Pan X, Mead D, Crane D D, Musser S V, Barry C E., 3rd Inhibition of a Mycobacterium tuberculosis β-ketoacyl ACP synthase by isoniazid. Science. 1998;280:1607–1610. doi: 10.1126/science.280.5369.1607. [DOI] [PubMed] [Google Scholar]
- 24.Mehta A, Oeser A M, Carlson M G. Rapid quantitation of free fatty acids in human plasma by high-performance liquid chromatography. J Chromatogr B. 1998;719:9–23. doi: 10.1016/s0378-4347(98)00403-4. [DOI] [PubMed] [Google Scholar]
- 25.Miesel L, Weisbrod T R, Marcinkeviciene J A, Bittman R, Jacobs W R., Jr NADH dehydrogenase defects confer isoniazid resistance and conditional lethality in Mycobacterium smegmatis. J Bacteriol. 1998;180:2459–2467. doi: 10.1128/jb.180.9.2459-2467.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monteagudo E S, Burton G, Gros E G. Synthesis of 24-methylene [24-C-14]-cholesterol and 24-methylidene [7-H-13] cholesterol. Helv Chim Acta. 1990;73:2097–2100. [Google Scholar]
- 27.Peterson D O, Bloch K. Mycobacterium smegmatis fatty acid synthetase. Long chain transacylase chain length specificity. J Biol Chem. 1977;252:5735–5739. [PubMed] [Google Scholar]
- 28.Piatek A S, Telenti A, Murray M R, El-Hajj H, Jacobs W R, Jr, Kramer F R, Alland D. Genotypic analysis of Mycobacterium tuberculosis in two distinct populations using molecular beacons: implications for rapid susceptibility testing. Antimicrob Agents Chemother. 2000;44:103–110. doi: 10.1128/aac.44.1.103-110.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quemard A, Sacchettini J C, Dessen A, Vilcheze C, Bittman R, Jacobs W R, Jr, Blanchard J S. Enzymatic characterization of the target for isoniazid in Mycobacterium tuberculosis. Biochemistry. 1995;34:8235–8241. doi: 10.1021/bi00026a004. [DOI] [PubMed] [Google Scholar]
- 30.Ristow M, Mohlig M, Rifai M, Schatz H, Feldmann K, Pfeiffer A. New isoniazid/ethionamide resistance gene mutation and screening for multidrug-resistant Mycobacterium tuberculosis strains. Lancet. 1995;346:502–503. doi: 10.1016/s0140-6736(95)91351-3. [DOI] [PubMed] [Google Scholar]
- 31.Rozwarski D A, Grant G A, Barton D H R, Jacobs W R, Jr, Sacchettini J C. Modification of the NADH of the isoniazid target (InhA) from Mycobacterium tuberculosis. Science. 1998;279:98–102. doi: 10.1126/science.279.5347.98. [DOI] [PubMed] [Google Scholar]
- 32.Skold O. R-factor-mediated resistance to sulfonamides by a plasmid-borne, drug-resistant dihydropteroate synthase. Antimicrob Agents Chemother. 1976;9:49–54. doi: 10.1128/aac.9.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snapper S B, Melton R E, Mustafa S, Kieser T, Jacobs W R., Jr Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 34.Takayama K, Wang L, David H L. Effect of isoniazid on the in vivo mycolic acid synthesis, cell growth, and viability of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1972;2:29–35. doi: 10.1128/aac.2.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takayama K, Wang L, Merkal R S. Scanning electron microscopy of the H37Ra strain of Mycobacterium tuberculosis exposed to isoniazid. Antimicrob Agents Chemother. 1973;4:62–65. doi: 10.1128/aac.4.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takayama K, Schnoes H K, Armstrong E L, Boyle R W. Site of inhibitory action of isoniazid in the synthesis of mycolic acids in Mycobacterium tuberculosis. J Lipid Res. 1975;16:308–317. [PubMed] [Google Scholar]
- 37.Telenti A, Honore N, Bernasconi C, March J, Ortega A, Heym B, Takiff H E, Cole S T. Genotypic assessment of isoniazid and rifampin resistance in Mycobacterium tuberculosis: a blind study at reference laboratory level. J Clin Microbiol. 1997;35:719–723. doi: 10.1128/jcm.35.3.719-723.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilming M, Johnsson K. Spontaneous formation of the bioactive form of the tuberculosis drug isoniazid. Angew Chem Int Ed. 1999;38:2588–2590. doi: 10.1002/(sici)1521-3773(19990903)38:17<2588::aid-anie2588>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 39.Wilson M, DeRisi J, Kristensen H H, Imboden P, Rane S, Brown P O, Schoolnik G K. Exploring drug-induced alterations in gene expression in Mycobacterium tuberculosis by microarray hybridization. Proc Natl Acad Sci USA. 1999;96:12833–12838. doi: 10.1073/pnas.96.22.12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winder F G, Collins P B. Inhibition by isoniazid of synthesis of mycolic acids in Mycobacterium tuberculosis. J Gen Microbiol. 1970;63:41–48. doi: 10.1099/00221287-63-1-41. [DOI] [PubMed] [Google Scholar]
- 41.Zabinski R F, Blanchard J S. The requirement for manganese and oxygen in the isoniazid-dependent inactivation of Mycobacterium tuberculosis enoyl reductase. J Am Chem Soc. 1997;119:2331–2332. [Google Scholar]
- 42.Zhang Y, Heym B, Allen B, Young D, Cole S. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature. 1992;358:591–593. doi: 10.1038/358591a0. [DOI] [PubMed] [Google Scholar]