Abstract
As the worldwide prevalence of colorectal cancer (CRC) increases, it is vital to reduce its morbidity and mortality through early detection. Saliva‐based tests are an ideal noninvasive tool for CRC detection. Here, we explored and validated salivary biomarkers to distinguish patients with CRC from those with adenoma (AD) and healthy controls (HC). Saliva samples were collected from patients with CRC, AD, and HC. Untargeted salivary hydrophilic metabolite profiling was conducted using capillary electrophoresis–mass spectrometry and liquid chromatography–mass spectrometry. An alternative decision tree (ADTree)‐based machine learning (ML) method was used to assess the discrimination abilities of the quantified metabolites. A total of 2602 unstimulated saliva samples were collected from subjects with CRC (n = 235), AD (n = 50), and HC (n = 2317). Data were randomly divided into training (n = 1301) and validation datasets (n = 1301). The clustering analysis showed a clear consistency of aberrant metabolites between the two groups. The ADTree model was optimized through cross‐validation (CV) using the training dataset, and the developed model was validated using the validation dataset. The model discriminating CRC + AD from HC showed area under the receiver‐operating characteristic curves (AUC) of 0.860 (95% confidence interval [CI]: 0.828‐0.891) for CV and 0.870 (95% CI: 0.837‐0.903) for the validation dataset. The other model discriminating CRC from AD + HC showed an AUC of 0.879 (95% CI: 0.851‐0.907) and 0.870 (95% CI: 0.838‐0.902), respectively. Salivary metabolomics combined with ML demonstrated high accuracy and versatility in detecting CRC.
Keywords: biomarker, colorectal cancer, metabolomics, polyamine, saliva
Saliva‐based colorectal cancer tests have been developed. Machine learning and metabolomics were used.

Abbreviations
- AD
adenoma
- ADTree
alternative decision tree
- APC
adenomatous polyposis coli
- AUC
area under ROC curves
- CA19‐9
cancer antigen 19‐9
- CE
capillary electrophoresis
- CEA
carcinoembryonic antigen
- CI
confidence interval
- CRC
colorectal cancer
- CV
cross‐validation
- HC
healthy controls
- LC
liquid chromatography
- ML
machine learning
- MLR
multiple logistic regression
- MS
mass spectrometry
- QQQ
triple‐quadrupole
- ROC
receiver‐operating characteristic
- TCA
tricarboxylic acid cycle
- TOF
time‐of‐flight
- VIP
variable importance projection
1. INTRODUCTION
Despite the advances in cancer diagnosis and management in the last decade, CRC still represents a significant global health burden. Overall, CRC ranks third in cancer morbidity and second in mortality among all cancers worldwide. 1 , 2 The prevalence of CRC is strongly associated with the westernization of eating and health habits. Furthermore, it is expected to increase further in developed countries with remarkable economic growth. 1 , 3 , 4 Therefore, cancer detection is an important issue in CRC diagnosis and treatment.
Fecal occult blood tests are the most commonly used CRC screening tests in Japan. Although these tests have contributed to the reduction in the mortality rate associated with CRC, 5 , 6 their limited sensitivity for early‐stage precancerous lesions, such as AD or CRC, indicated the need for improvement. 7 In addition, a large proportion of the at‐risk population is still often detected in advanced stages. 8 Currently used blood‐based biomarkers for CRC, such as CEA and cancer antigen 19‐9 (CA19‐9), are suitable for surveillance or prognostic indicator in CRC treatment but are unsuitable for screening or diagnosing CRC due to their low sensitivity and specificity, as well as the association with other types of gastrointestinal cancers, including gastric cancer, pancreatic cancer, or gynecological cancer such as ovarian cancer. 9 Therefore, developing a convenient novel screening method with higher sensitivity and specificity is paramount.
Frequently mutated genes have been identified in association with CRC, including APC, CTNNB1, KRAS, BRAF, and SMAD4. 10 The epigenetic variation in CRC changes the hyper‐ and hypomethylation, which inactivates the tumor suppressor genes and activates oncogenes, leading to epithelial cell growth to cancerous tumor formulation. 11 In addition to these genetic changes, malignant cancers, including CRC, have shown drastic metabolic shifts. For instance, regardless of oxygen availability, tumor cells activate the glycolysis pathway to produce adenosine triphosphate (Warburg effect). 12 In addition, oxidative phosphorylation upregulation has been reported in several cancers. 13 Glutamine is used as a carbon source alternative to glucose via the TCA in proliferating tumor cells to synthesize purines and pyrimidines. 14 In addition, holistic changes in the metabolic pathways have been reported, including amino acid, pentose phosphate, urea cycle, polyamine, and nucleotide pathways. 15 , 16 , 17 Therefore, the metabolites in biofluids including blood and saliva that reflect these metabolic aberrances associated with CRC have been analyzed to establish a novel set of biomarkers. 18 , 19 , 20 , 21 , 22 , 23 , 24
Saliva is an ideal biofluid that enables various disease detections. 25 However, salivary components are expected to be fragile compared with those from other biofluids. 26 , 27 , 28 , 29 Therefore, strict protocols must be established for processing saliva samples for reproducible quantification. For example, the unstimulated saliva collection, overnight fasting duration, restriction of any oral treatments before the sample collection, and frozen sample storage should follow standard protocols. 30 , 31 However, saliva tests allow noninvasive sampling, which is beneficial as cancer screening. Metabolomic biomarkers in saliva samples have been shown to represent a potential medium for cancer detection. 32 , 33 Biomarkers for oral cancer and cancers in the organs far from the oral cavity, such as breast cancer and pancreatic cancers, have been reported. 34 , 35 , 36 , 37 To enhance the discriminability of multiple biomarkers, ML is a cornerstone. 38 , 39 Using urinary polyamine profiles, we previously used an alternative decision tree (ADTree)–based prediction method to detect CRC. 22 Salivary polyamines with this ML method showed high discriminability for breast cancers. 37
In this study, we performed salivary metabolomic profiling of saliva collected from patients with CRC, patients with AD, and HC. We developed ML models to determine the combination of metabolite concentrations that could discriminate among these groups. Mainly, we drew two types of comparison: comparison between CRC + AD and HC and comparison between CRC and AD + HC. More than 2000 samples were examined, and the data were divided into two datasets. One dataset was used for the ML model development, and the other dataset was used to validate the ML model. Our approach has shown the screening potential of salivary metabolomic profiles to detect CRC.
2. MATERIALS AND METHODS
2.1. Subjects
This study was approved by the Ethics Committee of Tokyo Medical University (Nos. 2346 and 3405) and conducted in accordance with the Declaration of Helsinki. Written informed consent was obtained from all participants who agreed to serve as saliva donors. Patients with CRC who underwent chemotherapy and patients with chronic metabolic diseases, such as diabetes, were excluded. Patients histopathologically diagnosed with colorectal adenocarcinoma were included, and patients with all other types of cancer (adenosquamous cell carcinoma, endocrine carcinoma, lymphoma, etc.) were excluded.
The resected specimens were pathologically classified according to the 7th edition of the Union for International Cancer Control TNM Classification of Malignant Tumors. 40 All patients with AD were histologically diagnosed as having AD after polypectomy. In addition, samples of HC were collected at the Center for Health Surveillance and Preventive Medicine, Tokyo Medical University.
2.2. Saliva collection
Subjects were allowed only water intake after 9:00 p.m. on the day before collection. Salivary samples were collected between 9:00 and 11:00 a.m. They were required to brush their teeth without toothpaste on the day of collection and could not use lipstick, drink water, smoke, brush their teeth, or exercise intensively 1 hour before saliva collection. A polypropylene straw 1.1 cm in diameter was used to assist in the saliva collection. Approximately 400 μl of unstimulated saliva was collected and stored in 50 ml polypropylene tubes on ice to prevent the degeneration of salivary metabolites. 30 After collection, saliva samples were immediately stored at −80°C. Visibly cloudy and highly bubbly saliva was eliminated by visual inspection, and another saliva sample was collected 5 min later.
2.3. Saliva preparation and metabolomics analyses
Saliva samples were analyzed via two methods. CE‐TOF‐MS (TOF‐MS) was used for nontargeted analyses of hydrophilic metabolites, and LC–triple quadrupole MS (QQQ‐MS) was used for accurate quantification of polyamines as described previously with slight modifications. 26 , 30 Frozen saliva was thawed at 4°C for approximately 1.5 hours and subsequently dissolved using a Vortex mixer at 25°C. Ten microliters of each sample were used in the LC‐MS analysis, and the rest in the CE‐MS analysis.
For LC‐MS analysis, saliva was mixed with methanol (90 μl) containing 149.6 mM ammonium hydroxide [1% (v/v) ammonia solution] and 0.9 μM internal standards (d8‐spermine, d8‐spermidine, d6‐N 1‐acetylspermidine, d3‐N 1‐acetylspermine, d6‐N 1,N 8‐diacetylspermidine, d6‐N 1,N 12‐diacetylspermine, hypoxanthine‐13C,15N, 1,6‐diaminohexane, 13C,15N‐Arg, 13C,15N‐Lys, 13C,15N‐Met, 13C,15N‐Pro, 13C,15N‐Trp, d3‐Leu, and d5‐Phe). After centrifugation at 15,780 × g for 10 minutes at 4°C, the supernatant was transferred to a fresh tube and vacuum dried. The sample was reconstituted with 90% methanol (10 μl) and water (30 μl) and then vortexed and centrifuged at 15,780 × g for 10 minutes at 4°C. One microliter of supernatant was then injected into the LC‐MS.
For CE‐MS, saliva was centrifuged through a 5‐kDa‐cutoff filter (EMD Millipore) at 9100 × g for at least 2.5 hours at 4°C. The filtrate (45 μl) was transferred to a 1.5‐ml Eppendorf tube with 2 mM of internal standards (methionine sulfone, 2‐[N‐morpholino]‐ethanesulfonic acid [MES], D‐camphol‐10‐sulfonic acid, sodium salt, 3‐aminopyrrolidine, and trimesate). The instrumentation and measurement conditions used for LC‐QQQ‐MS and CE‐TOF‐MS were as described previously. 26 , 27 , 30
Raw data processing was conducted by following the typical data processing flow. 41 LC‐MS data were processed using Agilent MassHunter Qualitative Analysis and Quantitative Analysis software, including the MassHunter Optimizer and the Dynamic Multiple Reaction Monitoring Mode (DMRM) software (version B.08.00; Agilent Technologies). Polyamine concentrations were calculated based on the peak area of corresponding internal standards. CE‐MS data were analyzed using MasterHands (Keio University) 33 , 41 with noise filtering, subtraction of baselines, peak integration for each sliced electropherogram, estimation of accurate m/z in MS, alignment of multiple datasets to generate peak matrices, and identification of each peak by matching m/z values and corrected migration times to corresponding entries in a standard library. Metabolite concentrations in CE‐MS were calculated based on the ratio of peak area divided by the area of the internal standards in the samples and standard compound mixtures. Polyamine LC‐MS data were used for subsequent analyses because both methods redundantly detected their peaks.
2.4. Data analysis
The collected data were randomly split into training and validation datasets (Figure 1A ). The metabolites detected in more than 95% of samples with P values < 0.05 (Mann‐Whitney test) between CRC and HC were selected. Fold changes (FC) of averaged concentrations between HC and CRC were calculated. Only the metabolites showing higher FC than the average FC were used for clustering analyses. To evaluate the overall difference in the metabolite profile between HC and CRC, we conducted partial least squares discriminant analysis (PLS‐DA) 42 and pathway analyses. PLS‐DA is a classification method frequently used in the metabolomics field, which aims to maximize the covariance between the independent variables (metabolites) and the corresponding dependent variables (groups) by finding the subspace of the explanatory variables, for example, independent components. 43 PLS‐DA can generate score plots and VIP score plots. 44
FIGURE 1.
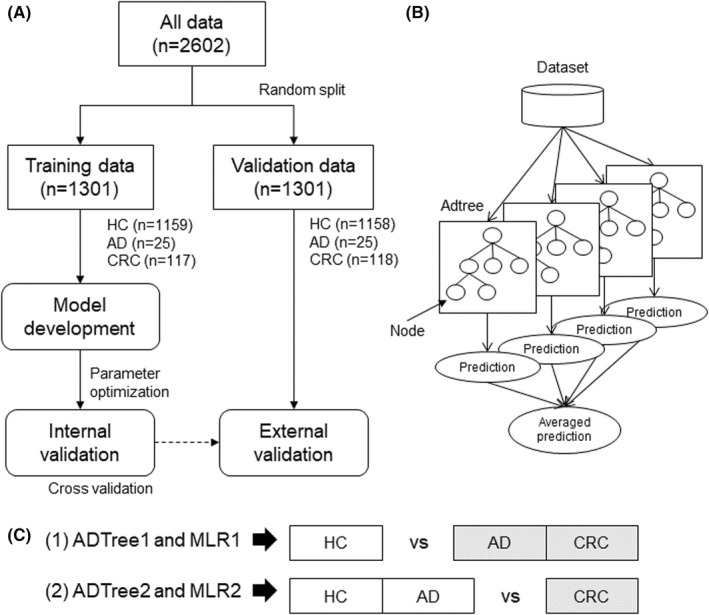
Data analysis design. (A), Data used in this study. Data were randomly split into training and validation datasets. Machine learning (ML) models were developed using the cross‐validation (CV) of the training dataset and validated using the validation dataset. (B), The ensemble alternative decision tree (ADTree) models. Each model has several nodes. The averaged predictions of multiple ADTree are used as the final prediction. (C), Depiction of the comparisons drawn in the study. The gray and white boxes indicate positive and negative groups, respectively. HC and CRC are negative and positive groups, respectively. AD is considered as a positive group in comparison (1) and as a negative group in comparison (2). ADTree1 and MLR1 are developed for comparison (1), and ADTree2 and MLR2 are developed for comparison (2). AD, adenoma; CRC, colorectal cancer; HC, healthy controls
We evaluated the discrimination ability of multiple combinations of metabolites. As one of the multivariate analyses, MLR was used. Stepwise feature forward selection was used to eliminate colinearity of the independent variables. This method limits the number of variables and deals with only linear relationships between independent (metabolites) and dependent variables (groups). Therefore, we also used the ADTree algorithm, an ML method, which is an improved form of the conventional “if‐then” decision tree–based method. 45 In addition, multiple datasets were generated by random sampling allowing redundant selection, and ADTree models corresponding to each of the generated datasets were developed. Predictive ability of each ADTree model was averaged to enhance prediction accuracy (ensemble method; Figure 1B). 46 The number of nodes and ADtrees were optimized via k‐fold CV using the training dataset. The developed model was validated using the validation dataset (Figure 1A).
In this study, although data were obtained from three groups (HC, AD, and CRC), we used MLR and ADTree as a two‐group classification method. Therefore, we drew two types of comparison for discriminating CRC + AD from HC (1) and CRC from AD + HC (2). ADTree models were developed for (1) and (2), named ADTree1 and ADTree2, respectively. In addition, MLR models were developed for (1) and (2), named MLR1 and MLR2, respectively (Figure 1C).
The discrimination ability was evaluated using the area under the ROC curves (AUC). The quantified values, such as metabolite concentration and predicted probabilities, were evaluated using the Mann‐Whitney test for two‐group comparisons and the Kruskal‐Wallis test with Dunn's post‐tests for ≥ 3 group comparisons.
JMP Pro (ver. 14.1.0; SAS Institute Inc.), GraphPad Prism (ver. 7.0.3; GraphPad Software, Inc.), MeV TM4 (ver. 4.9.0; http://mev.tm4.org), Weka (ver. 3.6.15; the University of Waikato), and Metaboanalysis (v.5.0, https://www.metaboanalyst.ca) 47 were used for the analyses.
3. RESULTS
3.1. Overview of profiled metabolites
Table 1 summarizes training and validation data of the subjects enrolled in this study. A total of 2602 unstimulated saliva samples were collected from 235 subjects with CRC, 50 subjects with ADs, and 2317 HC. All data were randomly assigned to training (n = 1301) and validation datasets (n = 1301; Figure 1A). Among the 122 quantified metabolites, 63 metabolites showed a p < 0.05 (Mann‐Whitney test) between HC and CRC. Among them, 23 metabolites showing an FC > 1.71 (average FC) were selected for clustering analyses (Figure 2). The CRC metabolites showed higher concentrations than those of HC. Several metabolites associated with AD also showed higher concentrations than those of HC. The FC between HC and CRC is visualized in Figure S1. The acetylated polyamines, such as N‐acetylputrescine, N 1‐acetylspermine, N 1,N 8‐diacetylspermidine, N 8‐acetylspermidine, N 1‐acetylspermidine, and N 1,N 12‐diacetylspermine, were included, and the first two polyamines showed relatively high FCs in both datasets. Two glycolysis metabolites, pyruvate and lactate, two citrate cycle metabolites, succinate and malate, and four amino acids were also included.
TABLE 1.
Subject information
| Training data (n = 1301) | Validation data (n = 1301) | |||||
|---|---|---|---|---|---|---|
| HC | AD | CRC | HC | AD | CRC | |
| n | 1159 | 25 | 117 | 1158 | 25 | 118 |
| Age | ||||||
| Mean | 45.65 | 66.30 | 67.42 | 45.19 | 61.81 | 69.63 |
| ±SD | 10.15 | 11.07 | 11.24 | 10.10 | 10.40 | 12.14 |
| Gender | ||||||
| Male | 318 | 21 | 64 | 338 | 20 | 66 |
| Female | 841 | 4 | 53 | 820 | 5 | 52 |
| Stage | ||||||
| 0/I/II(N1)/II(N2)/Iva | 2/30/36/25/14/10 | 2/31/36/25/14/10 | ||||
Abbreviations: AD, adenoma; CRC, colorectal cancer; HC, healthy controls.
FIGURE 2.
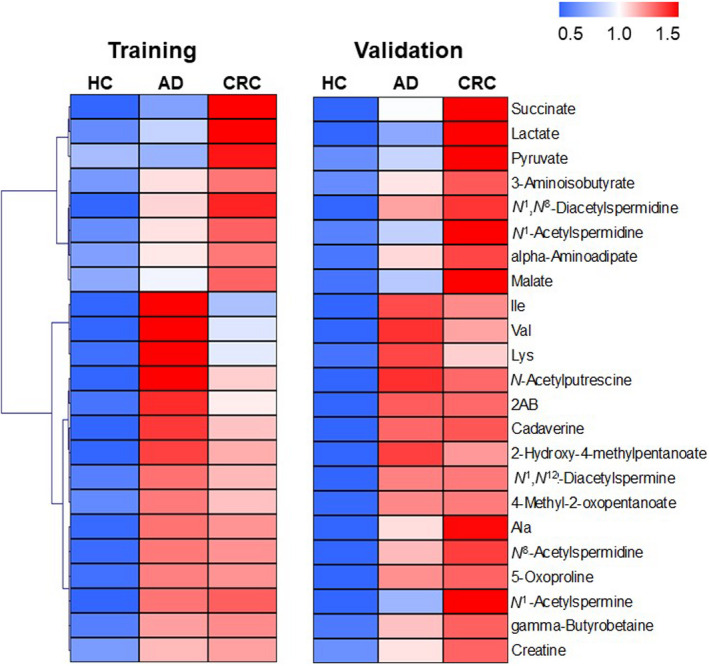
Heatmap illustrating salivary metabolite concentrations. Each metabolite concentration was divided by its average for the training and validation dataset. These data were averaged again for each group
3.2. Partial least squares discriminant analysis and pathway analysis
The overall differences in metabolomic profiles between HC and CRC were evaluated using PLS‐DA (Figure 3A,B) and pathway analysis (Figure 3C). The score plots showed separated HC and CRC (Figure 3A). N‐acetylputrescine and N 1‐acetylspermine showed high VIP scores, thus highly contributing to this discrimination (Figure 3B). Pathway analysis detected three significantly enriched pathways, including (1) amino sugar and nucleotide sugar metabolism, (2) alanine, aspartate, and glutamate metabolism, and (3) arginine as well as proline metabolism. However, the pathway impact of pathway (1) was small, while those of (2) and (3) were relatively high.
FIGURE 3.
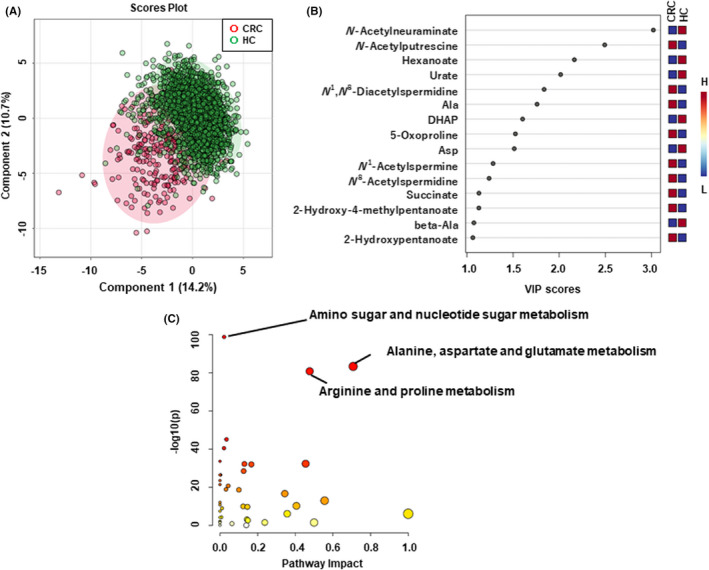
The difference in salivary metabolites between healthy controls (HC) and colorectal cancer (CRC). (A), Score plots of partial least squares discriminant analysis (PLS‐DA). 42 x‐ and y‐axes indicate the first and the second PLS component. Each plot corresponds to one sample. The plots with shorter distances indicate high similarity of the metabolomics profile of these samples. (B), Variable importance projection (VIP) score of PLS‐DA. (C), Pathway analysis. The metabolite concentration of each sample was divided by its median value. Subsequently, the data were log2‐transformed and translated into Z‐scores. For PLS‐DA, the 10‐fold cross‐validation with five components showed the highest generalization value (R 2 = 0.552 and Q 2 = 0.524)
3.3. Alternative decision tree models
The discrimination abilities of multiple combinations of metabolites were analyzed. Two ADTree models were developed for comparison (1) to discriminate AD + CRC from HC (ADTree1) and for comparison (2) to discriminate CRC from AD + HC (ADTree2; Figure 1C). CV using training data resulted in 16 trees and 7 nodes for ADTree1 and 12 trees and 8 nodes for ADTree2. The ADTree1 discriminability was 0.933 (95% CI: 0.916‐0.951) for all data (Figure 4A) and 0.860 (95% CI: 0.828‐0.891) for CV (Figure 4B) in the training dataset. The generalization ability of the developed ADTree1 was 0.870 (95% CI: 0.870‐0.870) in the validation dataset (Figure 4C). This value was similar to that observed for CV in the training datasets. The stage‐specific differences in probabilities of AD + CRC predicted by ADTree1 of training data and validation data are depicted in Figure 4D,E. The significant differences at all stages except for stage 0 (n = 2) between HC were observed (Dunn's post‐tests of Kruskal‐Wallis test). The ADTree2 proved similar discrimination abilities of 0.951 (95% CI: 0.936‐0.965) for all data (Figure 4F) and 0.879 (95% CI: 0.851‐0.907) for CV (Figure 4G) in the training datasets, and 0.870 (95% CI: 0.838‐0.902) in the validation dataset (Figure 4H). The probabilities of CRC predicted by ADTree1 also showed similar patterns (Figure 4I,J). The used metabolites and their usage numbers in ADTree1 and ADTree2 are depicted (Figure S2). The top three metabolites (N‐acetylputrescine, 4‐methyl‐2‐oxopentanoate, and 5‐oxoproline) were used in both models.
FIGURE 4.
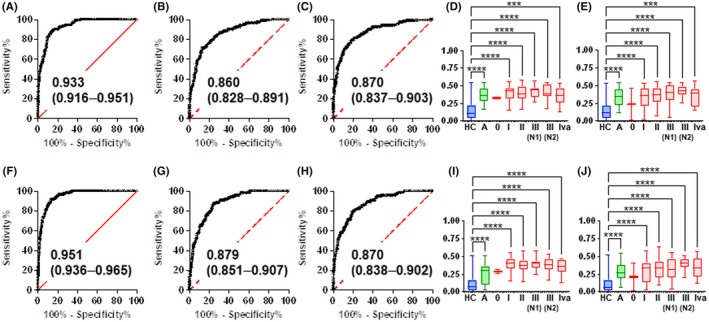
Discriminability of machine learning (ML) models. Receiver‐operating characteristic (ROC) curves of all data (A) as well as the cross‐validation (CV) of the training (B) and validation (C) datasets by alternative decision tree (ADTree)1. The ADTree1 prediction probability for adenoma (AD) + colorectal cancer (CRC) using the training (D) and validation (E) datasets. ADTree2 ROC curves for all data (F) as well as the CV of the training (G) and validation (H) datasets. ADTree2 prediction probability for CRC using the training (I) and validation (J) datasets. A, B, E, F, All area under ROC curves (AUC) values are presented with a 95% confidence interval (CI) between parentheses. The values were statistically significant (p < 0.0001). D, E, I, J, Asterisks indicate the P value of Dunn's post‐test after the Kruskal‐Wallis test. ***p < 0.01 and ****p < 0.0001. The y‐axis indicates the prediction probability
3.4. Multiple logistic regression models
To compare the discrimination ability of the ADTree and MLR models, we developed two MLR models for the comparisons (1) and (2) (Figure 1C). We developed MLR1 to discriminate AD + CRC from HC, and MLR2 to discriminate CRC from AD + HC. The ROC curves using training and validation datasets and stage‐specific prediction probabilities are shown in Figure S3. MLR1 and MLR2 included three metabolites, and N‐acetylputrescine was selected in both models (Table S1). All AUC values yielded by both MLR models showed a p < 0.0001 (Table S2). However, comparing the results following validation, the AUC values of ML were higher than those of MLR models.
3.5. Comparisons with tumor markers
Carcinoembryonic antigen and CA19‐9 of AD and CRC subjects were measured, and the comparisons of sensitivities among the tumor markers (TMs), ML, and MLR models were summarized (Table S3). For TMs, the subjects showing a CEA > 5.0 ng/ml or CA19‐9 > 37 U/ml3 were counted as positive. For MLR and ML, the optimal cutoff value was defined to maximize the sensitivity and specificity using the training dataset, and validated using the validation dataset. The predicted probabilities higher than these cutoff values were counted as positive. In the validation dataset, the sensitivity to CEA and CA19‐9 from CRC subjects were 30.5% and 16.9%, respectively. Those of ADTree1, ADTree2, MLR1, and MLR2 were 77.1%, 70.4%, 77.1%, and 76.3%, respectively. All TM values for AD were negative. ADTree1 and MLR1 indicated 68% sensitivities, while ADTree2 and MLR2 indicated 60% sensitivities.
The correlations between predicted probabilities and TM are listed in Table S4. In the validation dataset, the correlations (R, Spearman correlation) with ADTree1 were 0.174 (CEA) and 0.133 (CA19‐9). Those with ADTree2 were 0.216 (CEA) and 0.169 (CA19‐9). Only the comparison of MLR1 and CEA showed no significance. The correlations values with MLR1 were 0.114 (CEA) and 0.0900 (CA19‐9), and those with MLR2 were 0.117 (CEA) and 0.0375 (CA19‐9). All correlations produced by the MLR showed no significance.
3.6. Effect of age on the metabolomic profile
Age showed a significant effect on the metabolic profile. Therefore, age‐matched data were generated by eliminating HC subjects of lower ages. The MLR1 and MLR2 model's coefficients without feature selection were trained using training datasets and evaluated using validation datasets. In short, these models used already selected metabolites (Table S2). We conducted feature selection using the age‐matched data and developed MLR1 and MLR2 (Table S5). Both models with/without feature selection showed high AUC values (p < 0.0001) in both training and validation datasets (Table S6).
4. DISCUSSION
In this study, we investigated the use of metabolomics to discover salivary‐based biomarkers and discriminate among CRC, AD, and HC. As described in the heatmap (Figure 2), both training and validation datasets were highly similar. All metabolites in the map were elevated in CRC (Figure S1). Among them, end products of glycolysis, such as lactate and pyruvate, were elevated only in CRC. Lactate, also an end product of glycolysis, was observed in various reports, including our oral cancer saliva data. 35 It can be inferred that the Warburg effect 48 and glutaminolysis, 49 , 50 which are characteristic of cancer metabolism, might underlie the observed characteristics. In addition, several amino acids, such as isoleucine, valine, lysine, and alanine, were elevated in both AD and CRC. The intermediate metabolites associated with these energy and amino acid pathways were frequently reported. 15 , 51
In both ML models, acetylated polyamines were selected as predictive features. We previously reported the high polyamines in the urine collected from CRC patients as a valuable biomarker. 20 , 22 The synthesis of polyamines is attributed to various pathways, with the activation of ornithine decarboxylase that converts ornithine to putrescine in cancer cells largely affecting the polyamine contents. 52 The activation of acetylation of spermine and spermidine resulted in high level of their acetylated forms, which spread to the scrounging biofluid (Figure S4). 53 As frequently reported, we also previously confirmed that N 1,N 12‐diacetylspermine showed the most clearly elevated levels in CRC urine. 22 , 54 , 55 However, other forms of acetylated polyamines, such as N 1‐acetylspermine and N 8‐diacetylspermidine, were also elevated, consistent with other studies. 56 , 57 In the current saliva data, N 1‐acetylspermine and N 1,N 8‐diacetylspermidine showed higher discriminability than N 1,N 12‐diacetylspermine (Figures S5 and S6). The differences in AUC values between all stages and early stages (0, I, and II) of CRC were small. These data are beneficial in enhancing the capacity to detect early‐stage CRC subjects. The prediction probabilities calculated by the two ML models were also successfully utilized. We observed significant differences between each CRC stage and HC, even in the early stages, although stage 0 showed no significance because of the small sample numbers (Figure 4). These metabolites also showed similar discriminability for AD (Figures S5 and S6). We previously confirmed the similar aberrant metabolomic profiles of AD and CRC, which were shifted by the activation of MYC genes observed in AD. 16 MYC induced the activation of ODC, resulting in the polyamine synthesis activation (Figure S4). The prediction probabilities of AD by the developed models were significantly higher compared with HC, even though AD was grouped as negative data in ADTree2 (Figure 4I,J) and MLR2 (Figure S3G,H). These data indicate the usefulness of detecting AD and CRC, whereas the differentiation between AD and CRC is not satisfactory.
We compared the sensitivity of ADTree models with those of CEA and CA19‐9 in AD and CRC data. Both ADTree models showed better sensitivity compared with these two TM. TM did not detect AD subjects; however, the ADTree and MLR models showed more than 60% sensitivity (Table S3). Furthermore, R = 0.216 between CEA and ADTree2 was the highest positive correlation between the ML model and TM in the validation data (Table S4). Therefore, the complimentary use of ADTree models and TM would benefit the screening of AD and CRC.
Right‐sided CRC has a higher mortality rate and worse prognosis than left‐sided CRC, and both genetic and metabolomic differences between these two sides have been reported. 58 , 59 , 60 Therefore, we evaluated the difference between the left and right colon on prediction accuracy. There was no significant difference between tumor locations even in the training and validation data (Figure S7). This trend was observed for both MLR1 and MLR2. Therefore, the prediction accuracies of the developed models were not affected by tumor location.
Recently developed liquid biopsies for CRCs include methylation and abnormal levels of circulating tumor DNA and noncoding RNA, mainly micro‐RNA, as markers; however, most of these markers are detected in plasma and stool. 61 In saliva samples, MiR‐21 has shown CRC and HC discrimination ability. 62 A single marker that shows high specificity for a disease is beneficial for developing simple and reasonable assays as compared with simultaneous analyses of multiple markers for the detection of diseases. The analysis of volatile compounds in saliva has also shown potential to detect CRC. 63 The current study measured only hydrophilic metabolites, and more comprehensive analyses could explore the accurate biomarkers of CRC.
Several limitations need to be acknowledged. First, although the sample size is relatively large, this is a case‐control study; in short, the proportion of the three groups does not reflect the actual prevalence of these diseases. Second, the current data has an age bias between HC and the other groups. Therefore, age‐matched subsets were randomly generated, and the models showing discrimination at a significant level were confirmed (Tables S5 and S6). However, evaluating the developed models using age‐matched data, including larger samples, is necessary for rigorous validations. Third, the comparison with other diseases, especially using other cancer types, was not performed. For example, salivary polyamines were elevated even in breast and pancreatic cancers. 36 , 37 , 64 The elevation of salivary amino acids was also reported in breast cancer. 65 Therefore, a single marker may not be enough for a disease‐specific index, and an ML capturing multiple metabolite patterns would enhance the specificity. To use the developed biomarkers for diagnostic purposes, rigorous validation is necessary, for example, comparison of clinical‐pathological features between HC and patients with CRC. The approach in the current study showed CRC detection abilities; however, room to improve the sensitivity and specificity of CRC detection still exists. In general, a lower threshold to determine the positive cases enhances the sensitivity and reduces the false‐negative cases for screening purposes. Meanwhile, a higher threshold is used to enhance the specificity and reduce the false‐positive cases for diagnostic purposes. Saliva metabolomics demonstrated in this study showed a high sensitivity, which is suitable for a screening test; however, the specificity is not enough for diagnostic purposes. The current result can encourage patients who show a higher risk of AD or CRC to undergo other diagnostic tests.
In conclusion, we analyzed the salivary metabolic profiles of CRC, AD, and HC. The data showed consistent profile patterns, including polyamines, with previous studies. The ensemble ADTree models successfully discriminated against these groups with high sensitivity and specificity. We also showed high generality using validation datasets. In addition, the models showed higher sensitivity compared with CEA and CA19‐9. The models could contribute to clinical screening for AD and CRC.
ACKNOWLEDGMENTS
The authors thank the members of Center for Health Surveillance and Preventive Medicine, Tokyo Medical University Hospital for collecting saliva samples.
DISCLOSURE
Tomoyoshi Soga is an editorial board member of Cancer Science. The authors have no conflict of interest.
ETHICAL APPROVAL
Approval of the research protocol by an Institutional Reviewer Board: This study was approved by the Ethics Committee of Tokyo Medical University (Nos. 2346 and 3405).
INFORMED CONSENT
All informed consent was obtained from the subjects.
Supporting information
Appendix S1
Kuwabara H, Katsumata K, Iwabuchi A, et al. Salivary metabolomics with machine learning for colorectal cancer detection. Cancer Sci. 2022;113:3234‐3243. doi: 10.1111/cas.15472
Funding information
This work was supported by JSPS KAKENHI (Grant Numbers 26462027, 15K08751, 16H05408, 16K10554, 20H05743, and 21K07228) and research grants from the Yamagata Prefectural Government and the City of Tsuruoka.
REFERENCES
- 1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209‐249. [DOI] [PubMed] [Google Scholar]
- 2. Siegel RL, Miller KD, Goding Sauer A, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70:145‐164. [DOI] [PubMed] [Google Scholar]
- 3. Ahmed FE. Effect of diet, life style, and other environmental/chemopreventive factors on colorectal cancer development, and assessment of the risks. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2004;22:91‐147. [DOI] [PubMed] [Google Scholar]
- 4. Randi G, Edefonti V, Ferraroni M, La Vecchia C, Decarli A. Dietary patterns and the risk of colorectal cancer and adenomas. Nutr Rev. 2010;68:389‐408. [DOI] [PubMed] [Google Scholar]
- 5. Hewitson P, Glasziou P, Watson E, Towler B, Irwig L. Cochrane systematic review of colorectal cancer screening using the fecal occult blood test (hemoccult): an update. Am J Gastroenterol. 2008;103:1541‐1549. [DOI] [PubMed] [Google Scholar]
- 6. Simon JB. Fecal occult blood testing: clinical value and limitations. Gastroenterologist. 1998;6:66‐78. [PubMed] [Google Scholar]
- 7. de Wijkerslooth TR, Stoop EM, Bossuyt PM, et al. Immunochemical fecal occult blood testing is equally sensitive for proximal and distal advanced neoplasia. Am J Gastroenterol. 2012;107:1570‐1578. [DOI] [PubMed] [Google Scholar]
- 8. Maida M, Macaluso FS, Ianiro G, et al. Screening of colorectal cancer: present and future. Expert Rev Anticancer Ther. 2017;17:1131‐1146. [DOI] [PubMed] [Google Scholar]
- 9. Locker GY, Hamilton S, Harris J, et al. Asco 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J Clin Oncol. 2006;24:5313‐5327. [DOI] [PubMed] [Google Scholar]
- 10. Cancer Genome Atlas Network . Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330‐337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lao VV, Grady WM. Epigenetics and colorectal cancer. Nat Rev Gastroenterol Hepatol. 2011;8:686‐700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Warburg O. On the origin of cancer cells. Science. 1956;123:309‐314. [DOI] [PubMed] [Google Scholar]
- 13. Ashton TM, McKenna WG, Kunz‐Schughart LA, Higgins GS. Oxidative phosphorylation as an emerging target in cancer therapy. Clin Cancer Res. 2018;24:2482‐2490. [DOI] [PubMed] [Google Scholar]
- 14. Zhao Y, Zhao X, Chen V, et al. Colorectal cancers utilize glutamine as an anaplerotic substrate of the tca cycle in vivo. Sci Rep. 2019;9:19180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hirayama A, Kami K, Sugimoto M, et al. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time‐of‐flight mass spectrometry. Cancer Res. 2009;69:4918‐4925. [DOI] [PubMed] [Google Scholar]
- 16. Satoh K, Yachida S, Sugimoto M, et al. Global metabolic reprogramming of colorectal cancer occurs at adenoma stage and is induced by myc. Proc Natl Acad Sci USA. 2017;114:E7697‐E7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tevini J, Eder SK, Huber‐Schönauer U, et al. Changing metabolic patterns along the colorectal adenoma‐carcinoma sequence. J Clin Med. 2022;11(3):721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dalal N, Jalandra R, Sharma M, et al. Omics technologies for improved diagnosis and treatment of colorectal cancer: technical advancement and major perspectives. Biomed Pharmacother. 2020;131:110648. [DOI] [PubMed] [Google Scholar]
- 19. Uchiyama K, Yagi N, Mizushima K, et al. Serum metabolomics analysis for early detection of colorectal cancer. J Gastroenterol. 2017;52:677‐694. [DOI] [PubMed] [Google Scholar]
- 20. Sakurai T, Katsumata K, Udo R, et al. Validation of urinary charged metabolite profiles in colorectal cancer using capillary electrophoresis‐mass spectrometry. Metabolites. 2022;12:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Qiu Y, Cai G, Su M, et al. Urinary metabonomic study on colorectal cancer. J Proteome Res. 2010;9:1627‐1634. [DOI] [PubMed] [Google Scholar]
- 22. Nakajima T, Katsumata K, Kuwabara H, et al. Urinary polyamine biomarker panels with machine‐learning differentiated colorectal cancers, benign disease, and healthy controls. Int J Mol Sci. 2018;19:756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Leichtle AB, Nuoffer JM, Ceglarek U, et al. Serum amino acid profiles and their alterations in colorectal cancer. Metabolomics. 2012;8:643‐653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Farshidfar F, Weljie AM, Kopciuk KA, et al. A validated metabolomic signature for colorectal cancer: exploration of the clinical value of metabolomics. Br J Cancer. 2016;115:848‐857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hyvärinen E, Savolainen M, Mikkonen JJW, Kullaa AM. Salivary metabolomics for diagnosis and monitoring diseases: Challenges and possibilities. Metabolites. 2021;11:587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ishikawa S, Sugimoto M, Kitabatake K, et al. Effect of timing of collection of salivary metabolomic biomarkers on oral cancer detection. Amino Acids. 2017;49:761‐770. [DOI] [PubMed] [Google Scholar]
- 27. Sugimoto M, Saruta J, Matsuki C, et al. Physiological and environmental parameters associated with mass spectrometry‐based salivary metabolomic profiles. Metabolomics. 2013;9:454‐463. [Google Scholar]
- 28. Okuma N, Saita M, Hoshi N, et al. Effect of masticatory stimulation on the quantity and quality of saliva and the salivary metabolomic profile. PLoS One. 2017;12:e0183109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kawanishi N, Hoshi N, Masahiro S, et al. Effects of inter‐day and intra‐day variation on salivary metabolomic profiles. Clin Chim Acta. 2019;489:41‐48. [DOI] [PubMed] [Google Scholar]
- 30. Tomita A, Mori M, Hiwatari K, et al. Effect of storage conditions on salivary polyamines quantified via liquid chromatography‐mass spectrometry. Sci Rep. 2018;8:12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sugimoto M, Ota S, Kaneko M, Enomoto A, Soga T. Quantification of salivary charged metabolites using capillary electrophoresis time‐of‐flight‐mass spectrometry. Bio Protoc. 2020;10:e3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sugimoto M. Salivary metabolomics for cancer detection. Expert Rev Proteomics. 2020;17:639‐648. [DOI] [PubMed] [Google Scholar]
- 33. Sugimoto M, Wong DT, Hirayama A, Soga T, Tomita M. Capillary electrophoresis mass spectrometry‐based saliva metabolomics identified oral, breast and pancreatic cancer‐specific profiles. Metabolomics. 2010;6:78‐95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ishikawa S, Sugimoto M, Konta T, et al. Salivary metabolomics for prognosis of oral squamous cell carcinoma. Front Oncol. 2021;11:789248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ishikawa S, Sugimoto M, Kitabatake K, et al. Identification of salivary metabolomic biomarkers for oral cancer screening. Sci Rep. 2016;6:31520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Asai Y, Itoi T, Sugimoto M, et al. Elevated polyamines in saliva of pancreatic cancer. Cancers. 2018;10:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Murata T, Yanagisawa T, Kurihara T, et al. Salivary metabolomics with alternative decision tree‐based machine learning methods for breast cancer discrimination. Breast Cancer Res Treat. 2019;177:591‐601. [DOI] [PubMed] [Google Scholar]
- 38. Takada M, Sugimoto M, Naito Y, et al. Prediction of axillary lymph node metastasis in primary breast cancer patients using a decision tree‐based model. BMC Med Inform Decis Mak. 2012;12:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Takada M, Sugimoto M, Ohno S, et al. Predictions of the pathological response to neoadjuvant chemotherapy in patients with primary breast cancer using a data mining technique. Breast Cancer Res Treat. 2012;134:661‐670. [DOI] [PubMed] [Google Scholar]
- 40. Sobin LH, Gospodarowicz MK, Wittekind C. TNM classification of malignant tumours. 7th ed. Wiley‐Blackwell; 2009. [Google Scholar]
- 41. Sugimoto M, Kawakami M, Robert M, Soga T, Tomita M. Bioinformatics tools for mass spectroscopy‐based metabolomic data processing and analysis. Curr Bioinform. 2012;7:96‐108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wold S, Sjöström M, Eriksson L. Pls‐regression: a basic tool of chemometrics. Chemom Intell Lab Syst. 2001;58:109‐130. [Google Scholar]
- 43. Gromski PS, Muhamadali H, Ellis DI, et al. A tutorial review: metabolomics and partial least squares‐discriminant analysis—a marriage of convenience or a shotgun wedding. Anal Chim Acta. 2015;879:10‐23. [DOI] [PubMed] [Google Scholar]
- 44. Xia J, Psychogios N, Young N, Wishart DS. Metaboanalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009;37:W652‐W660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Freund YML. The alternating decision tree learning algorithm. ICML. 1999;1999:124‐133. [Google Scholar]
- 46. Che D, Liu Q, Rasheed K, Tao X. Decision tree and ensemble learning algorithms with their applications in bioinformatics. Adv Exp Med Biol. 2011;696:191‐199. [DOI] [PubMed] [Google Scholar]
- 47. Pang Z, Chong J, Zhou G, et al. Metaboanalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021;49:W388‐W396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pascale RM, Calvisi DF, Simile MM, Feo CF, Feo F. The Warburg effect 97 years after its discovery. Cancers. 2020;12:2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yang L, Venneti S, Nagrath D. Glutaminolysis: a hallmark of cancer metabolism. Annu Rev Biomed Eng. 2017;19:163‐194. [DOI] [PubMed] [Google Scholar]
- 50. Yoo HC, Yu YC, Sung Y, Han JM. Glutamine reliance in cell metabolism. Exp Mol Med. 2020;52:1496‐1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yusof HM, Ab‐Rahim S, Suddin LS, Saman MSA, Mazlan M. Metabolomics profiling on different stages of colorectal cancer: a systematic review. Malays J Med Sci. 2018;25:16‐34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gerner EW, Meyskens FL Jr. Polyamines and cancer: old molecules, new understanding. Nat Rev Cancer. 2004;4:781‐792. [DOI] [PubMed] [Google Scholar]
- 53. Soda K. The mechanisms by which polyamines accelerate tumor spread. J Exp Clin Cancer Res. 2011;30:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hiramatsu K, Takahashi K, Yamaguchi T, et al. N 1, N 12‐diacetylspermine as a sensitive and specific novel marker for early‐ and late‐stage colorectal and breast cancers. Clin Cancer Res. 2005;11:2986‐2990. [DOI] [PubMed] [Google Scholar]
- 55. Kawakita M, Hiramatsu K, Yanagiya M, Doi Y, Kosaka M. Determination of N 1, N 12‐diacetylspermine in urine: a novel tumor marker. Methods Mol Biol. 2011;720:367‐378. [DOI] [PubMed] [Google Scholar]
- 56. Loser C, Folsch UR, Paprotny C, Creutzfeldt W. Polyamines in colorectal cancer. Evaluation of polyamine concentrations in the colon tissue, serum, and urine of 50 patients with colorectal cancer. Cancer. 1990;65:958‐966. [DOI] [PubMed] [Google Scholar]
- 57. Kawakita M, Hiramatsu K. Diacetylated derivatives of spermine and spermidine as a novel promising tumor markers. J Biochem. 2006;139:315‐322. [DOI] [PubMed] [Google Scholar]
- 58. Deng K, Han P, Song W, et al. Plasma metabolomic profiling distinguishes right‐sided from left‐sided colon cancer. Clin Chim Acta. 2018;487:357‐362. [DOI] [PubMed] [Google Scholar]
- 59. Su MW, Chang CK, Lin CW, et al. Genomic and metabolomic landscape of right‐sided and left‐sided colorectal cancer: potential preventive biomarkers. Cell. 2022;11:527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jiang Y, Yan X, Liu K, et al. Discovering the molecular differences between right‐ and left‐sided colon cancer using machine learning methods. BMC Cancer. 2020;20:1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Palanca‐Ballester C, Rodriguez‐Casanova A, Torres S, et al. Cancer epigenetic biomarkers in liquid biopsy for high incidence malignancies. Cancers. 2021;13:3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sazanov AA, Kiselyova E, Zakharenko A, Romanov MN, Zaraysky M. Plasma and saliva mir‐21 expression in colorectal cancer patients. J Appl Genet. 2017;58:231‐237. [DOI] [PubMed] [Google Scholar]
- 63. Bel'skaya LV, Sarf EA, Shalygin SP, Postnova TV, Kosenok VK. Identification of salivary volatile organic compounds as potential markers of stomach and colorectal cancer: a pilot study. J Oral Biosci. 2020;62:212‐221. [DOI] [PubMed] [Google Scholar]
- 64. DeFelice BC, Fiehn O. Rapid lc‐ms/ms quantification of cancer related acetylated polyamines in human biofluids. Talanta. 2019;196:415‐419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cheng F, Wang Z, Huang Y, Duan Y, Wang X. Investigation of salivary free amino acid profile for early diagnosis of breast cancer with ultra performance liquid chromatography‐mass spectrometry. Clin Chim Acta. 2015;447:23‐31. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1


