Abstract
Drug resistance is an important factor for treatment failure of gastric cancer. N6‐methyladenosine (m6A) is the predominant mRNA internal modification in eukaryotes. The roles of m6A modification in drug resistance of gastric cancer remains unclear. In the present study, the m6A methylated RNA level was significantly decreased while the expression of m6A demethylase fat mass and obesity‐associated protein (FTO) was obviously elevated in cisplatin‐resistant (SGC‐7901/DDP) gastric cancer cells. Knockdown of FTO reversed cisplatin resistance of SGC‐7901/DDP cells both in vitro and in vivo, which was attributed to the inhibition of Unc‐51‐like kinase 1 (ULK1)‐mediated autophagy. Mechanistically, ULK1 expression was regulated in an FTO‐m6A‐dependent and YTHDF2‐mediated manner. Collectively, our findings indicate that the FTO/ULK1 axis exerts crucial roles in cisplatin resistance of gastric cancer.
Keywords: autophagy, cisplatin resistance, FTO, gastric cancer, ULK1
Knockdown of FTO reversed cisplatin resistance of SGC‐7901/DDP cells both in vitro and in vivo, which were attributed to the inhibition of ULK1 mediated‐autophagy. ULK1 expression was regulated in the FTO‐m6A dependent and YTHDF2‐mediated manner.
1. INTRODUCTION
Gastric cancer is one of the most common tumors that poses a potential threat to human health and quality of life. 1 , 2 At present, platinum‐based chemotherapy is commonly used for patients with gastric cancer, which can improve prognosis and prolong the survival of patients. Unfortunately, poor response to platinum‐based chemotherapy is often observed in gastric cancer patients due to the primary or obtained resistance, which becomes the major cause of treatment failure. 3 , 4 Over the past few decades, scientists have studied the mechanisms of cisplatin resistance. It is a complex cellular process that involves multiple pathways, including DNA repair, autophagy, and glutamine metabolism. 5 , 6 , 7 Consequently, the understanding of molecular mechanisms for platinum resistance and investigating potential therapeutic targets are urgent.
Autophagy is a system of cellular degradation, which is a prevalent element of drug resistance to chemotherapy in tumor cells. 8 , 9 Chemotherapeutic drugs could induce autophagy, which avoids cancer cell apoptosis, thereby producing drug resistance. 10 Previous studies have shown that 5‐fluorouracil induced autophagic death of gastric cancer cells and inhibited cell proliferation by upregulating Beclin1 expression by inhibiting microRNA‐30. 11 It was found that autophagy, which was involved in the formation and maintenance of chemotherapy resistance, inhibited the killing effect of imatinib on tumor cells in BCR‐ABL positive chronic myelogenous leukemia. 12 , 13 , 14 Some studies have found that paclitaxel increased the expression of BECN1 and decreased P62, inducing autophagy, in both cases. 15 , 16 Unc‐51‐like kinase 1 (ULK1) is a cytoplasmic kinase, which recruits the autophagy proteins to mediate the initiation of autophagy. 17 A selective inhibitor of ULK1 (SBI0206965) significantly inhibits the progress of autophagy in cisplatin‐resistant non‐small‐cell lung cancer (NSCLC) cells. 18 In addition, knockdown of ULK1 in NSCLC cells makes cells more sensitive to cisplatin. 19
N6‐methyladenosine (m6A), in which the sixth nitrogen (N) atom of adenine is methylated, is the most frequent modification on mRNAs in eukaryotes. Modification of m6A is regulated by methyltransferase complex (“writers”), demethylases (“erasers”), and RNA‐binding proteins (“readers”). 20 The main discovered components of the methyltransferases include methyltransferase‐like 3 (METTL3) and METTL14, while fat mass and obesity‐associated protein (FTO) and α‐ketoglutarate‐dependent dioxygenase homolog 5 (ALKBH5) have been identified as demethylated enzymes. The main known “reader” proteins include YTH domain families (YTHDF1, YTHDF2, and YTHDC1), which mediate mRNA stabilization. 21 , 22 In the current study, we aimed to clarify the effects and mechanisms of m6A RNA modification on autophagy‐related cisplatin resistance in gastric cancer cells.
Fat mass and obesity‐associated protein, the first identified m6A demethylase, has important roles in various types of disease. 23 , 24 , 25 The present study found that FTO was obviously elevated in the cisplatin‐resistant (SGC‐7901/DDP) gastric cancer cells. Knockdown of FTO reversed cisplatin resistance both in vitro and in vivo, which was attributed to the inhibition of ULK1‐mediated autophagy. Furthermore, ULK1 expression was regulated in the FTO‐m6A dependent and YTHDF2‐mediated manner.
2. MATERIALS AND METHODS
2.1. Cell culture
The human gastric epithelial cell line GES‐1 and the human gastric cancer cell line SGC‐7901 were obtained from the Health Science Research Resources Bank and used in our previous study. 26 Cells were cultured in RPMI‐1640 medium (Gibco) supplemented with 10% FBS (Lonsera). The cisplatin‐resistant cell line (SGC‐7901/DDP) was produced from parental SGC‐7901 cells. In brief, SGC‐7901 cells were subjected to increasing concentrations of cisplatin from 0.1 μM (Sigma‐Aldrich) until the cells acquired resistance to 1 μM cisplatin. The cells were additionally treated with 0.5 μM cisplatin to maintain the cisplatin‐resistant phenotype. The cisplatin in the medium was removed 2 weeks before other experiments.
2.2. Cell transfection
Lentivirus (LV) carrying FTO shRNA (shFTO) or negative control shRNA (shNC) and plasmid (pCDNA3.1) carrying FTO or YTHDF2 were purchased from HanBio. The ULK1 siRNA (siULK1), YTHDF2 siRNA (siYTHDF2), and negative control siRNA (siNC) were purchased from RiboBio. Cells in 6‐well culture plates were infected with lentivirus carrying shRNA for 72 h. Cell transfection with plasmid and siRNA was undertaken with Lipofectamine 3000 (Invitrogen) for 48 h. The procedures were carried out strictly in accordance with the manufacturer's instructions. The FTO shRNA sequences were: #1, 5′‐GGATGACTCTCATCTCGAAGG‐3′; and #2, 5′‐AAGAGCAGAGCAGCATACAACGTAA‐3′. The ULK1 siRNA sequences were: #1, 5′‐CGCCTGTTCTACGAGAAGA‐3′; and #2, 5′‐ACCAGCGCATTGAGCGAAA‐3′. The YTHDF2 siRNA sequence was 5′‐GACCAAGAATGGCATTGCA‐3′ and NC sequence was 5′‐UUCUCCGAACGUGUCACGUTT‐3′.
2.3. Cell viability assay
Cell viability assays were performed using the CCK‐8 method. Cells at log‐phase growth were seeded into 96‐well plates at 5000 cells/well. After treatment, 10 μl CCK‐8 solution (Beyotime) was added into each well, followed by an additional incubation for 2 h at 37°C. Finally, the 450 nm absorbance (OD) was determined using a microplate reader (Bio‐Tek). The percentage cell viability was calculated. Cells were treated with different concentrations of cisplatin (0, 1, 2, 4, 8, 16, and 32 μM) for 24 h. The cell viability was measured by CCK‐8 after cisplatin treatment. The IC50 values of cisplatin were calculated.
2.4. m6A RNA methylation quantification
Total RNA was extracted from GES‐1, SGC‐7901, and SGC‐7901/DDP cells using TRIzol reagent. The m6A content in the total RNA was detected using the m6A RNA Methylation Assay Kit (Abcam). The percentage of m6A in total RNA was carried out using the following formula: m6A% = {[(sample OD − negative control OD) ÷ sample amount] / [(positive control OD − NC OD) ÷ positive control amount]} × 100%.
2.5. Western blot analysis
Cells or tumor tissues were lysed in RIPA lysis buffer with protease and phosphatase inhibitors (Beyotime). Thirty micrograms of total protein was subjected to SDS‐PAGE gels and subsequently transferred to PVDF membranes. The membranes were blocked and then incubated with primary Abs overnight at 4°C. After washing three times, the membranes were incubated with the secondary Abs for 2 h at room temperature. Finally, protein bands were visualized by a chemiluminescence imaging system (Tanon) using western chemiluminescent HRP substrate (ECL, Millipore). The following Abs were purchased: METTL3, METTL14, FTO, ALKBH5, and YTHDF2 (Cat. No. ab195352, ab220030, ab126605, ab195377, and ab220163, respectively; Abcam); ULK1, LC3B, and P62/SQSTM1 (Cat. No. 8054, 3868, and 88588, respectively; Cell Signaling Technology); ATG13 pS318 (Cat. No. 600‐401‐C49; Rockland); actin (Cat. No. A5441; Sigma‐Aldrich); and anti‐rabbit HRP‐linked Ab and anti‐mouse HRP‐linked Ab (Cat. No. 7074 and 7076, respectively; Cell Signaling Technology).
2.6. Transmission electron microscopy
Autophagosomes refer to endogenous substances, including damaged organelles or excess glycogen stored in cells due to physiological or pathological reasons, which can be formed by wrapping membranes of the cells themselves (such as endoplasmic reticulum or Golgi complex membranes). Under transmission electron microscopy (TEM), autophagosomes were characterized as crescent‐shaped or cup‐shaped double‐ or multilayered autophagosomes with a tendency to enclose cytoplasmic components. Cells were centrifuged and the supernatant discarded. After centrifugation, the cell clumps were fixed in 2% glutaraldehyde overnight at 4°C, then incubated in 1% osmium tetroxide for 1 h at 4°C, dehydrated in graded ethanol, saturated in graded acetone, and cut into 50‐nm ultrathin sections. The autophagosomes in cells were viewed under a JEM‐1010 TEM (JEOL).
2.7. Immunofluorescence analysis
Cells were fixed with 4% paraformaldehyde and then permeabilized with Triton X‐100 (Beyotime) at room temperature. Cells were incubated with anti‐LC3B Abs (Abcam) at 4°C overnight after blocking with the Immunol staining blocking buffer (Beyotime) for 60 min. After washing three times with TBST solution, cells were incubated with Cy3 goat anti‐rabbit IgG (Abclonal) at room temperature for 1 h. Cell nuclei were stained with DAPI (Beyotime) for 10 min at room temperature. Cells were observed under fluorescence microscope and images were collected after adding antifluorescence attenuation sealer.
2.8. Quantitative real‐time PCR
TRIzol reagent (Invitrogen) was used to extract total RNA from cells, and a reverse transcription kit (Tiangen) was used for reverse transcription. cDNA was generated according to the manufacturer's instructions. In addition, quantitative real‐time PCR (qPCR) was carried out to analyze gene expression using the SYBR Green PCR Kit (Tiangen). The PCR data were normalized to human GAPDH expression. The relative gene expression was calculated using the comparative cycle threshold (2−ΔΔCt) method. The primer sequences for genes are shown as follows:
GAPDH forward, 5′‐CAGGAGGCATTGCTGATGAT‐3′ and reverse, 5′‐GAAGGCTGGGGCTCATTT‐3′; ATG5 forward, 5′‐GCAGATGGACAGTTGCACACAC‐3′ and reverse, 5′‐GAGGTGTTTCCAACATTGGCTCA‐3′; ATG7 forward, 5′‐CGTTGCCCACAGCATCATCTTC‐3′ and reverse, 5′‐CACTGAGGTTCACCATCCTTGG‐3′; ULK1 forward, 5′‐GCAAGGACTCTTCCTGTGACAC‐3′ and reverse 5′‐CCACTGCACATCAGGCTGTCTG‐3′; and Beclin1 forward, 5′‐CTGGACACTCAGCTCAACGTCA‐3′ and reverse, 5′‐CTCTAGTGCCAGCTCCTTTAGC‐3′.
2.9. N6‐methyladenosine RIP qPCR assay
This procedure was undertaken with the m6A RIP (MeRIP) kit (Bersinbio) according to the manufacturer's instructions. Briefly, total RNA was extracted from 2 × 107 SGC‐7901/DDP cells using TRIzol reagent, and RNA was further fragmentated using ultrasound. The processed fragment size was approximately 300 bp. After fragmentation, 50 μl RNA samples were stored as Input samples at −80°C; the remaining RNA samples were immunoprecipitated with anti‐m6A Ab (Abcam) or anti‐IgG Ab and washed with immunoprecipitation (IP) buffer. Protein A/G magnetic beads were incubated with RNA‐Ab hybridization solution for 1 h at 4°C in a vertical mixer. The beads were washed three times with IP buffer and then digested with proteinase K at 55°C for 45 min. Supernatant was transferred to new RNase‐free tubes, and RNA was purified, followed by qRT‐PCR. Primer sequences were:
ULK1 (site 1) forward, 5′‐TCTGTGCCTGACCTTTCTGG‐3′ and reverse, 5′‐GCTCAGCACCAGCGATCA‐3′; ULK1 (site 2) forward, 5′‐ATGGCGCTGATCGCTGG‐3′ and reverse, 5′‐AATTCTGTGATCCCCACCTGT‐3′; and ULK1 (site 3) forward, 5′‐AGCCTCTCCCTCCGAGATAC‐3′ and reverse, 5′‐TGTACAAACACCAGGCCACT‐3′.
2.10. RNA immunoprecipitation PCR
RNA immunoprecipitation was undertaken with the RIP kit (Bersinbio) according to the manufacturer's instructions. Briefly, total RNA was extracted from 2 × 107 cells using TRIzol reagent. RNA samples were divided into three: the IP group, IgG group and Input group. The Input samples were stored at −80°C. Anti‐FTO, ‐YTHDF2, or ‐IgG Abs were conjugated to protein A/G magnetic beads in IP buffer for 16 h at 4°C in a vertical mixer. The RNA was eluted from the beads with proteinase K at 55°C for 45 min. Finally, the precipitated RNA and input RNA were detected by qRT‐PCR.
2.11. mRNA stability analysis
Cells were transfected with either the NC siRNA or YTHDF2 siRNA for 24 h and then treated with 5 μg/ml actinomycin D (MCE) to inhibit mRNA transcription. The cells were collected and total RNA was extracted by TRIzol reagent after treatment for the indicated times. The levels of mRNA were detected by qRT‐PCR.
2.12. Animals and treatments
Five‐week‐old male BALB/c nude mice were purchased from Hangzhou Medical College Laboratory Animal Co., Ltd. The mice were raised in pathogen‐free conditions, and all the experiments were implemented according to the Guide for the Care and Use of Laboratory Animals published by the US NIH.
SGC‐7901/DDP cells expressing LV‐shFTO or LV‐shNC were resuspended in PBS solution. A total of 5 × 106 cells in 200 μl PBS were injected subcutaneously into the flanks of nude mice. After injection, cisplatin treatment was initiated on day 5. Mice were injected with 5 mg/kg cisplatin or PBS solution in the abdominal cavity once a week for 3 weeks. The tumor volume was measured and calculated. The formula volume = ½ (larger diameter) × (smaller diameter) × (smaller diameter). The animal experiments were approved by the Ethics Committee of The First Affiliated Hospital of Wannan Medical College.
2.13. Hematoxylin−eosin and immunohistochemistry staining
Tumor tissues of the mice were fixed in 4% paraformaldehyde and then embedded in paraffin. After embedding and sectioning, the tumor tissues were stained with H&E. Immunohistochemistry staining was carried out using an anti‐Ki‐67 mAb (Cell Signaling Technology). The protein expression levels of Ki‐67 in tumor tissues were observed under microscopy.
2.14. Statistical analysis
The data are presented as the mean ± SD. The statistical analyses were undertaken using a two‐tailed Student's t‐test or ANOVA, which was used to analyze the differences when there were more than two groups. The statistical analyses were carried out using SPSS 19.0 software. P values less than 0.05 were considered to be statistically significant.
3. RESULTS
3.1. Fat mass and obesity‐associated protein is a potential target of cisplatin resistance in gastric cancer cells
CCK‐8 assays were used to examine the effects of cisplatin on the viability of gastric cancer cells and cisplatin‐resistant cells. Compared with SGC‐7901 cells, cisplatin‐resistant cells (SGC‐7901/DDP) showed significant resistance to cisplatin treatment. The IC50 value of cisplatin in SGC‐7901/DDP cells was significantly higher than that in SGC‐7901 cells (SGC‐7901/DDP, 16.29 ± 3.58 μM vs. SGC‐7901, 3.18 ± 0.40 μM; Figure 1A,B). To clarify the relationship between m6A methylation modification and cisplatin resistance of gastric cancer, we first quantified the m6A methylation content in total RNAs and the protein expression of m6A methyltransferases and demethylases in GES‐1, SGC‐7901, and SGC‐7901/DDP cells. As shown in Figure 1C, the m6A levels in total RNAs were obviously decreased in SGC‐7901/DDP cells compared to that in GES‐1 and SGC‐7901 cells. Furthermore, FTO expression was dramatically upregulated in SGC‐7901/DDP cells, while the expression levels of METTL3, METTL14, FTO, and ALKBH5 were considerably less pronounced (Figure 1D,E). To further investigate the effects of FTO on cisplatin resistance, we knocked down FTO by shRNAs (shFTO‐1 and shFTO‐2) and overexpressed FTO by pcDNA3.1 plasmid in SGC‐7901/DDP and SGC‐7901 cells (Figure 1F–H). Knockdown or overexpression of FTO did not affect the cell viability of SGC‐7901 or SGC‐7901/DDP cells (Figure 1I,J). However, FTO silencing obviously enhanced the sensitivity of SGC‐7901/DDP cells to cisplatin (Figure 1K). We also observed that FTO overexpression significantly reduced the sensitivity of SGC‐7901 cells to cisplatin (Figure 1L). These results indicate that m6A demethylase enzyme FTO mediates cisplatin resistance of gastric cancer cells.
FIGURE 1.
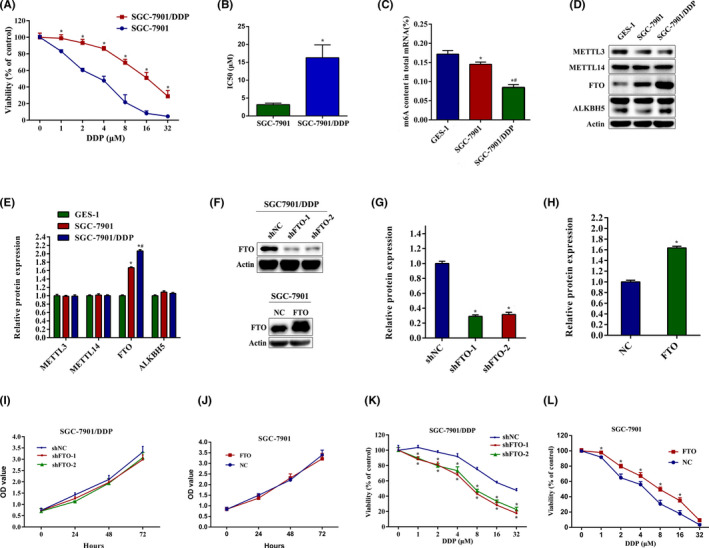
Fat mass and obesity‐associated protein (FTO) is the potential target of cisplatin (DDP) resistance in gastric cancer cells. (A) SGC‐7901 and SGC‐7901/DDP cells were treated with 0–32.0 μM DDP, and CCK‐8 assays were carried out to assess cell viability after 24 h. (B) IC50 values of cisplatin in SGC‐7901 and SGC‐7901/DDP cells were calculated. (C) m6A methylation levels in total RNAs in GES‐1, SGC‐7901, and SGC‐7901/DDP cells were assessed. (D, E) Expression of METTL3, METTL14, FTO, and ALKBH5 in GES‐1, SGC‐7901, and SGC‐7901/DDP cells was measured by western blotting. (F–H) SGC‐7901/DDP cells were transfected with lentivirus FTO shRNA. SGC‐7901 cells were transfected with pcDNA3.1‐FTO plasmid. Western blot analysis was used to detect the protein expression of FTO. (I, J) SGC‐7901/DDP and SGC‐7901 cells were transfected with FTO shRNA and FTO plasmid, and the cell viability was measured by CCK‐8 assays. (K, L) SGC‐7901/DDP and SGC‐7901 cells were transfected with FTO shRNA and FTO plasmid and the cell viability was measured by CCK‐8 assays after DDP treatment for 24 h. Results are presented as mean ± SD of n = 3–4 independent experiments. *p < 0.05 vs. SGC‐7901, GES‐1, negative control shRNA (shNC), or negative control (NC) group; # p < 0.05 vs. SGC‐7901 group. FTO, pcDNA3.1‐FTO plasmid; OD, optical density; shFTO, FTO shRNA
3.2. Fat mass and obesity‐associated protein promotes cisplatin resistance by facilitating autophagy in gastric cancer cells
Autophagy is a common cellular process in eukaryotic cells, which is largely involved in the cisplatin resistance of tumors. 6 , 27 , 28 During autophagy, LC3 I is converted to LC3 II through lipidation by a ubiquitin‐like system, and P62 can be degraded by the autophagosome. 29 , 30 Consequently, degradation of P62 and accumulation of LC3 II (bottom LC3 band) are indicators of autophagy induction. To explore whether FTO mediated autophagy in cisplatin‐resistant and cisplatin‐sensitive gastric cancer cells, protein levels of the autophagy markers LC3B and P62 were measured to determine the autophagy activation. The results showed that the expression of LCB3 II was significantly decreased while the P62 expression was obviously increased in SGC‐7901/DDP cells with FTO silencing (Figure 2A,B). However, overexpression of FTO by plasmid dramatically increased LCB3 II expression and reduced P62 level in SGC‐7901 cells (Figure 2C,D). In addition, immunofluorescence assays showed that knockdown of FTO significantly reduced the formation of LC3B puncta in SGC‐7901/DDP cells, whereas LC3B puncta was enhanced in FTO‐overexpressed SGC‐7901 cells (Figure 2E). The results of TEM images also revealed that FTO silencing reduced the number of autophagosomes in SGC‐7901/DDP cells. In contrast, overexpression of FTO increased the number of autophagosomes in SGC‐7901 cells (Figure 2F). To further confirm the roles of autophagy in cisplatin resistance, we used 3‐methyladenine and chloroquine to inhibit autophagy and found that inhibiting autophagy reversed the cisplatin resistance of SGC‐7901/DDP cells (Figure 2G–I).
FIGURE 2.
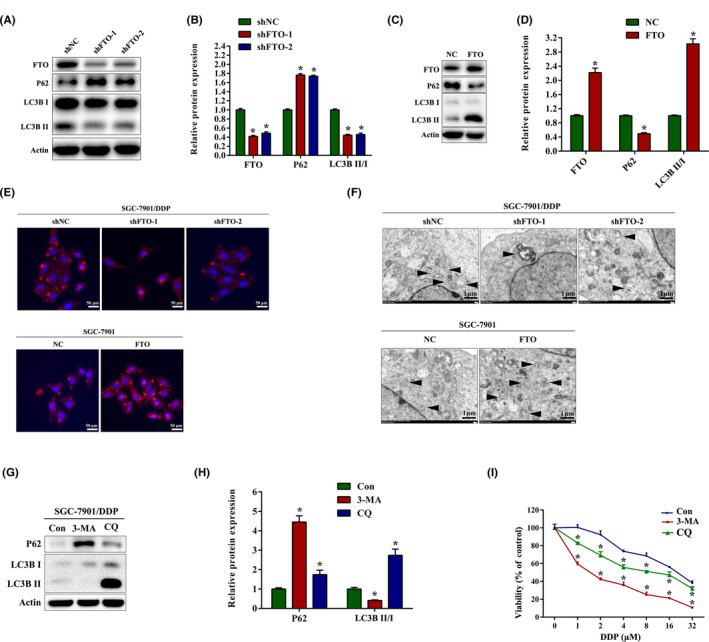
Fat mass and obesity‐associated protein (FTO) promotes cisplatin (DDP) resistance by facilitating autophagy in gastric cancer cells. (A, B) Western blot analysis of FTO, P62, and LC3B expression in SGC‐7901/DDP cells with FTO knockdown. (C, D) Western blot analysis of FTO, P62, and LC3B expression in SGC‐7901 cells with FTO overexpression. (E) Immunofluorescence images of LC3B puncta in SGC‐7901/DDP and SGC‐7901 cells transfected with FTO shRNA and FTO overexpression plasmids. (F) Transmission electron microscopy analysis of autophagosomes in SGC‐7901/DDP and SGC‐7901 cells transfected with FTO shRNA and FTO overexpression plasmids. Arrows indicate autophagosomes. (G, H) Western blot analysis of FTO, P62, and LC3B expression in SGC‐7901/DDP cells after treatment with 5 μM 3‐methyladenine (3‐MA) or 10 μM chloroquine (CQ) for 24 h. (I) SGC‐7901/DDP cells were pretreated with 3‐MA or CQ for 1 h, and the cell viability was measured by CCK‐8 assay after DDP treatment for 24 h. Results are presented as mean ± SD of n = 3–4 independent experiments. *p < 0.05 vs. negative control shRNA (shNC), negative control (NC), or control (Con) group. FTO, pcDNA3.1‐FTO plasmid; shFTO, FTO shRNA
3.3. Loss of FTO attenuates expression of ULK1
To identify potential target genes of FTO in autophagy, we undertook qRT‐PCR assays to determine the mRNA expression changes of autophagy‐related genes following FTO knockdown in SGC‐7901/DDP cells. Similar to the results obtained from cervical cancer HeLa cells, 24 the level of ULK1 mRNA was significantly downregulated after FTO knockdown, while mRNA levels of ATG5, ATG7, and BECLIN1 were not evidently changed (Figure 3A). As shown in Figure 3B–D, under‐ and overexpression of FTO markedly decreased and increased protein levels of ULK1, respectively, in SGC‐7901/DDP and SGC‐7901 cells. To further confirm the effects of ULK1 on autophagy, we transfected SGC‐7901/DDP cells with ULK1 siRNA, and discovered that knockdown of ULK1 significantly increased P62 expression and decreased LCB3 II protein level. In addition, the protein expression of phosphorylation of ATG13 S318 (P‐ATG13) was downregulated, indicating that ULK1 activity decreased 31 , 32 (Figure 3E,F). Moreover, knockdown of ULK1 obviously promoted the inhibitory effects of cisplatin on growth in SGC‐7901/DDP cells (Figure 3G). These results indicate that ULK1 is functionally important for autophagy and cisplatin resistance in gastric cancer cells.
FIGURE 3.
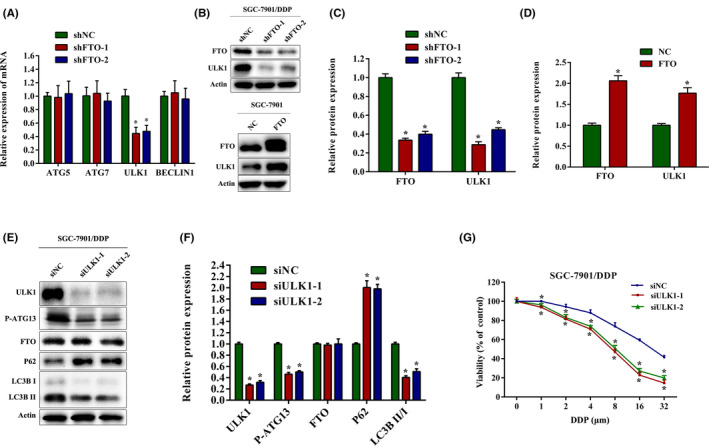
Loss of fat mass and obesity‐associated protein (FTO) attenuates the expression of Unc‐51‐like kinase 1 (ULK1). (A) mRNA expression of autophagy‐related genes (ATG5, ATG7, ULK1, and BECLIN1) was detected by quantitative real‐time PCR in SGC‐7901/DDP cells with FTO knockdown. (B–D) Western blot analysis of FTO and ULK1 expression in SGC‐7901/DDP and SGC‐7901 cells with FTO shRNA and FTO overexpression plasmid. (E, F) Western blot analysis of FTO, ULK1, P‐ATG13, P62, and LC3B expression in SGC‐7901/DDP cells after transfection with ULK1 siRNA. (G) SGC‐7901/DDP cells were transfected with ULK1 siRNA and the cell viability was measured by CCK‐8 assays after DDP treatment for 24 h. Results are presented as mean ± SD of n = 3–4 independent experiments. *p < 0.05 vs. negative control shRNA (shNC), negative control (NC), or negative control siRNA (siNC) group. DDP, cisplatin; FTO, pcDNA3.1‐FTO plasmid; shFTO, FTO shRNA; siULK1, ULK1 siRNA
3.4. Fat mass and obesity‐associated protein regulates autophagy and cisplatin resistance through targeting ULK1 in an m6A‐dependent manner
To confirm whether FTO influenced autophagy and cisplatin resistance through targeting ULK1, we undertook rescue experiments and observed that ULK1 inhibition reversed the upregulated LC3B II, P‐ATG13 and increased the downregulated P62 in SGC‐7901/DDP cells with FTO overexpression (Figure 4A,B). In addition, immunofluorescence assays showed that the elevated LC3B puncta induced by FTO overexpression was compromised in SGC‐7901/DDP cells when treated with siULK1 (Figure 4C ). Overexpression of FTO significantly attenuated the inhibitory effects of cisplatin on growth in SGC‐7901/DDP cells. Transfection with siULK1 obviously reversed these effects (Figure 4D). To further explore the potential underlying mechanisms of m6A in autophagy regulation, MeRIP‐qPCR and RIP‐qPCR assays were carried out. As shown in Figure 4E, silencing of FTO significantly increased the m6A levels on mRNA of ULK1 at three sites. Furthermore, RIP‐qPCR analysis revealed that ULK1 was a direct target of FTO (Figure 4F). These results indicate that FTO regulates the expression of ULK1 in an m6A‐dependent manner.
FIGURE 4.
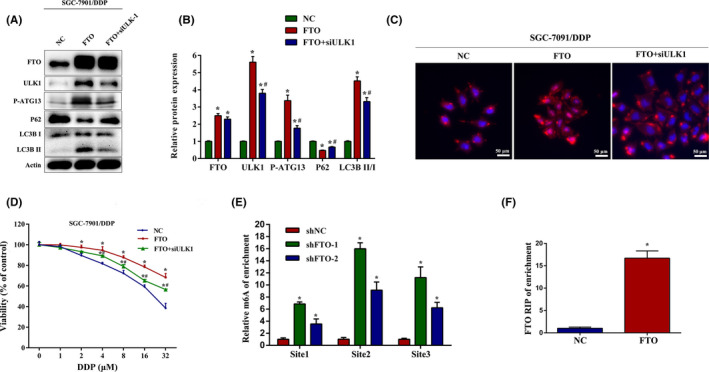
Fat mass and obesity‐associated protein (FTO) regulates autophagy and cisplatin (DDP) resistance through targeting Unc‐51‐like kinase 1 (ULK1) in an N6‐methyladenosine (m6A)‐dependent manner. (A, B) Western blot analysis of FTO, ULK1, P‐ATG13, P62, and LCB3 protein expression in FTO‐overexpressed SGC‐7901/DDP cells which were transfected with or without ULK1 siRNA. (C) Immunofluorescence images of LC3B puncta in SGC‐7901/DDP cells treated in the same method as described above. (D) FTO‐overexpressed SGC‐7901/DDP cells were transfected with or without ULK1 siRNA, and the cell viability was measured by CCK‐8 assays after DDP treatment for 24 h. (E) m6A levels of ULK1 mRNA were detected by methylated RNA immunoprecipitation (MeRIP)–quantitative PCR (qPCR) assays in SGC‐7901/DDP cells transfected with shFTO. (F) RIP‐qPCR assay was carried out using FTO Ab, and the enrichment of ULK1 was measured in SGC‐7901/DDP cells transfected with FTO plasmid. Results are presented as mean ± SD of n = 3–4 independent experiments. *p < 0.05 vs. negative control (NC) or negative control shRNA (shNC) group; # p < 0.05 vs. pcDNA3.1‐FTO plasmid (FTO) group. shFTO, FTO shRNA; siULK1, ULK1 siRNA
3.5. Fat mass and obesity‐associated protein modulates ULK1 expression through YTHDF2
It is well known that m6A methylation mediates targeted mRNAs by specific RNA‐binding proteins. YTHDF2, a major m6A reader, is reported to selectively bind and destabilize m6A‐modified mRNAs. 23 , 33 To investigate whether the expression of ULK1 was affected by YTHDF2, we treated SGC‐7901/DDP cells with YTHDF2 siRNA and found that YTHDF2 silencing significantly increased the protein level of ULK1 and P‐ATG13 (Figure 5A,B). The RIP‐qPCR assays indicated the direct binding within YTHDF2 and ULK1 mRNA (Figure 5C). In addition, mRNA stability analysis showed that the decay of ULK1 mRNA was inhibited in SGC‐7901/DDP cells transfected with YTHDF2 siRNA, when the transcription was halted with actinomycin D (Figure 5D). To confirm whether FTO regulated ULK1‐mediated autophagy through YTHDF2, we undertook rescue experiments and observed that knockdown of YTHDF2 could reverse the downregulation of ULK1, LC3B II, and P‐ATG13, and decrease the upregulation of P62 in SGC‐7901/DDP cells with FTO knockdown (Figure 5E,F). Moreover, CCK‐8 experiments revealed that siYTHDF2 transfection markedly restrained the inhibitory effects of cisplatin on growth in FTO‐downregulated SGC‐7901/DDP cells (Figure 5G). Together, our data suggest that FTO regulates ULK1 in a YTHDF2‐dependent manner.
FIGURE 5.
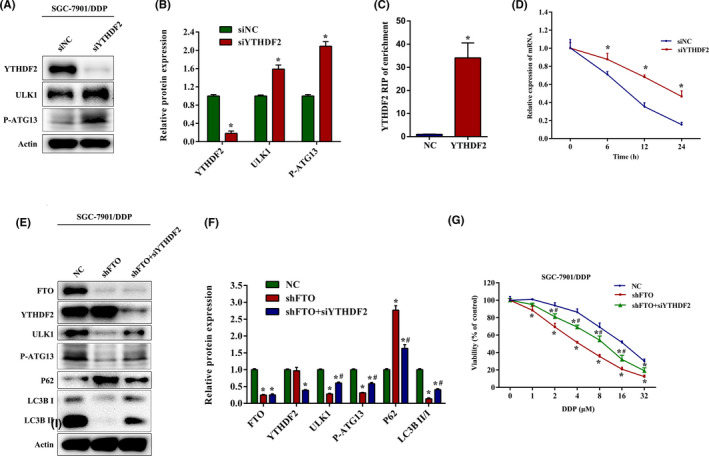
Fat mass and obesity‐associated protein (FTO) modulates Unc‐51‐like kinase 1 (ULK1) expression levels by YTHDF2. (A, B) Western blot analysis of YTHDF2, ULK1, and P‐ATG13 expression in SGC‐7901/DDP cells transfected with YTHDF2 siRNA. (C) RIP–quantitative PCR assay was carried out using YTHDF2 Ab, and the enrichment of ULK1 was measured in SGC‐7901/DDP cells transfected with YTHDF2 plasmid. (D) SGC‐7901/DDP cells transfected with YTHDF2 siRNA were treated with 5 μg/ml actinomycin D for the indicated times. mRNA expression of ULK1 was quantified by quantitative PCR. (E, F) Western blot analysis of FTO, YTHDF2, ULK1, P‐ATG13, P62, and LCB3 expression in FTO‐silenced SGC‐7901/DDP cells transfected with or without YTHDF2 siRNA. (G) FTO‐silenced SGC‐7901/DDP cells were transfected with or without YTHDF2 siRNA, and the cell viability was measured by CCK‐8 assays after cisplatin (DDP) treatment for 24 h. Results are presented as mean ± SD of n = 3–4 independent experiments. *p < 0.05 vs. negative control siRNA (siNC) or negative control (NC) group; # p < 0.05 vs. FTO shRNA (shFTO) group. siYTHDF2, YTHDF2 siRNA
3.6. Knockdown of FTO improves the sensitivity of cisplatin‐resistant gastric cancer cells to cisplatin in vivo
We finally explored the effects of FTO knockdown on tumor formation using nude mice xenograft experiments. Either control or FTO‐silenced SGC‐7901/DDP cells were injected subcutaneously in the flanks of male nude mice. Mice were injected with cisplatin or PBS solution in the abdominal cavity. In nude mice xenografts, the volume of tumors that carried FTO shRNA showed significantly slower growth than in the shNC group when the mice treated with DDP (Figure 6A,B). Although the data from H&E staining had no obvious difference between groups, knockdown of FTO inhibited cell proliferation in DDP‐treated nude mice as evidenced by the decreased Ki‐67 staining on the xenograft (Figure 6C,D). Consistent with the data of the in vitro study, FTO knockdown alleviated protein expression of ULK1 and LC3B II while elevating P62 protein abundance (Figure 6E,F). Taken together, these results suggest that FTO silencing could increase the sensitivity of cisplatin through inactivating ULK1‐dependent autophagy in vivo.
FIGURE 6.
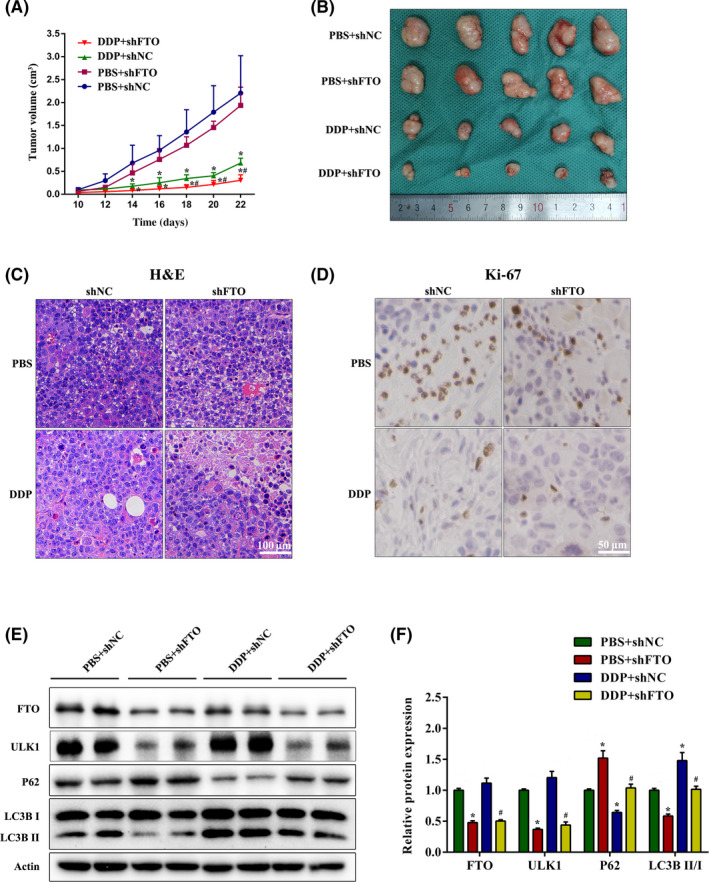
Knockdown of fat mass and obesity‐associated protein (FTO) improves the sensitivity of drug‐resistant gastric cancer cells to cisplatin (DDP) in vivo. (A) SGC7901/DDP cells transfected with FTO shRNA (shFTO) were injected in the flanks of BALB/c nude mice. Mice were injected with 5 mg/kg i.p. cisplatin or PBS once a week. Tumor volume was measured every other day. (B) Representative photographs of xenograft tumors in nude mice. (C, D) Representative images of (C) H&E staining and (D) Ki‐67 immunohistochemical staining. (E, F) Western blot analysis of FTO, Unc‐51‐like kinase 1 (ULK1), P62, and LCB3 expression in xenograft tumors of nude mice. *p < 0.05 vs. PBS + negative control shRNA (shNC) group; # p < 0.05 vs. DDP + shNC group
4. DISCUSSION
The m6A modification is one of the most crucial RNA modifications in human malignant diseases. 34 , 35 N6‐methyladenosine modification plays a crucial role in regulating gene expression through RNA splicing, stability, translocation, and translation. 36 , 37 The expression levels of m6A regulators, including writers, erasers, and readers, are usually dysregulated in cancers, which contributes to drug resistance and cancer relapse. 38 , 39 , 40 Expression of METTL3 is downregulated in sorafenib‐resistant hepatocellular carcinoma and triggered degradation of FOXO3, which elevates autophagy‐induced sorafenib resistance. 41 The FTO‐mediated m6A demethylation regulates mRNA stability of MERTK, then controls TKI resistance. 42 However, few studies have focused on the roles of m6A modification in regulating drug resistance for gastric cancer.
In the present study, we first revealed the relationship between m6A RNA modification and cisplatin resistance. The m6A levels in total RNAs were obviously decreased in SGC7901/DDP cells, which was attributed to the upregulation of FTO. Furthermore, knockdown of FTO in cisplatin‐resistant SGC7901/DDP cells increased the reactivity to cisplatin, whereas overexpression of FTO in cisplatin‐sensitive SGC7901 cells decreased the cisplatin sensitivity. These results indicate the direct roles of FTO‐mediated m6A modification in cisplatin resistance of gastric cancer.
Previous studies have indicated that FTO positively regulates autophagy in cervical cancer HeLa cells 24 and mouse pre‐adipocyte 3T3‐L1 cells. 23 Autophagy functions as a protective factor in resistance of cancer cells exposed to anticancer drugs. 28 , 43 Our data determined a similar modulatory role of FTO in mediating autophagy, and FTO‐dependent cisplatin resistance in gastric cancer was regulated by promoting autophagy. Silencing FTO reduced the number of autophagosomes and accumulation of LC3B II in gastric cancer cells, which indicated that suppression of autophagy might reverse the m6A‐dependent resistance to cisplatin.
To elucidate the mechanisms by which FTO mediated autophagy, we identified the target genes of autophagy following FTO knockdown. ATG5 and ATG7 are significantly attenuated following FTO knockdown in 3T3‐L1 cells. 16 Knockdown of FTO positively regulates the expression of ULK1 in HeLa cells. 17 These results suggested that target genes involved in the FTO‐mediated autophagy might be species‐ and cell‐specific. We identified ULK1, but not ATG5 or ATG7, as a primary regulator connecting m6A modification with autophagy in gastric cancer cells. ULK1 is essential for the recruitment of other autophagy‐related proteins to initiate the autophagosome formation, linking cellular nutrient status to downstream events in autophagy. 44 ULK1 forms a complex with ATG101, ATG13, and FIP200. 45 Following autophagy induction, ULK1 can phosphorylate serine‐318 (P‐S318) in ATG13, which serves as an indicator for ULK1 activity. 31 The expression of ULK1 protects tumor cells from excessive autophagy. 46 , 47 , 48 Knockdown of ULK1 makes cells more sensitive to cisplatin in NSCLC cells. 19 MicroRNA‐489 directly targets ULK1 and negatively regulates its expression, and thus overcomes resistance to doxorubicin, tamoxifen, and cisplatin by inhibiting autophagy in breast cancer cells. 48 , 49 Consistently, our results suggested that knockdown of ULK1 significantly inhibited autophagy and promoted the inhibitory effects on growth in SGC‐7901/DDP cells treated with cisplatin, which indicated that ULK1 is functionally important for autophagy and cisplatin resistance in gastric cancer cells.
We further validated ULK1 as a direct target of FTO in an m6A‐dependent manner in gastric cancer cells. Knockdown of FTO markedly reduced ULK1 expression at both mRNA and protein levels. Furthermore, by using MeRIP‐qPCR assays with gene‐specific primers according to the published data of the m6A‐sequence, 24 we found that FTO overexpression elevated m6A levels of ULK1 mRNA. The RIP‐qPCR assay revealed a strong signal for ULK1 mRNA following overexpression of FTO, suggesting that ULK1 was the target of FTO. Previous study showed that m6A‐modified mRNA tended to be less stable, which was largely attributed to the YTHDF2‐mediated mRNA degradation. 50 We treated SGC‐7901/DDP cells with YTHDF2 siRNA and found that YTHDF2 silencing markedly increased ULK1 expression. The RIP‐qPCR assay validated ULK1 as a direct target of YTHDF2. In addition, mRNA stability analysis indicated that the decay of ULK1 mRNA was inhibited when transfected with YTHDF2 siRNA. To confirm whether FTO regulated ULK1‐mediated autophagy through YTHDF2, we undertook rescue experiments and observed that knockdown of YTHDF2 could reverse the downregulation of ULK1 and the inhibitory effects of cisplatin on growth in SGC‐7901/DDP cells with FTO silencing.
In summary, the present study reveals a critical role of FTO in mediating cisplatin resistance of gastric cancer cells. Upregulated FTO promotes autophagy‐induced cisplatin resistance by regulating YTHDF2‐related ULK1 expression in cisplatin‐resistant gastric cancer cells. Our findings clarify the underlying mechanisms through which m6A mRNA methylation functions in cisplatin‐resistant gastric cancer cells, suggesting a potential therapeutic target.
AUTHOR CONTRIBUTIONS
W.‐P.D. and F.‐Y.D. designed the experiment and supervised the project. Y.Z., L.‐X.G., W.W., and T.Z. performed the experiments. F.‐Y.D. and W.‐P.D. analyzed the data and drafted the manuscript. Y.Z. and W.‐P.D. were responsible for critical reading, editing, and revising the manuscript. All authors read and approved the final manuscript.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ETHICS STATEMENT
Approval of the research protocol by an institutional review board: N/A.
Informed consent: N/A.
Registry and registration no. of the study/trial: N/A.
Animal studies: The animal experiments were approved by the Ethics Committee of The First Affiliated Hospital of Wannan Medical College.
ACKNOWLEDGMENTS
This study was supported by the grants from the Key University Science Research Project of Anhui Province (KJ2020A0594 and KJ2021A0827) and the Key Project Research Fund of Wannan Medical College (WK2020ZF19).
Zhang Y, Gao L‐x, Wang W, Zhang T, Dong F‐y, Ding W‐p. M6A demethylase fat mass and obesity‐associated protein regulates cisplatin resistance of gastric cancer by modulating autophagy activation through ULK1 . Cancer Sci. 2022;113:3085‐3096. doi: 10.1111/cas.15469
Yan Zhang and Ling‐xi Gao contributed equally to this work.
Contributor Information
Fang‐yi Dong, Email: dfy01c15@rjh.com.cn.
Wen‐ping Ding, Email: dingwenping99@163.com.
DATA AVAILABILITY STATEMENT
The datasets generated for this study are available on request to the corresponding author.
REFERENCES
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7‐34. [DOI] [PubMed] [Google Scholar]
- 2. Iwatsuki M, Yamamoto H, Miyata H, et al. Effect of hospital and surgeon volume on postoperative outcomes after distal gastrectomy for gastric cancer based on data from 145,523 Japanese patients collected from a nationwide web‐based data entry system. Gastric Cancer. 2019;22(1):190‐201. [DOI] [PubMed] [Google Scholar]
- 3. Yuan L, Xu ZY, Ruan SM, Mo S, Qin JJ, Cheng XD. Long non‐coding RNAs towards precision medicine in gastric cancer: early diagnosis, treatment, and drug resistance. Mol Cancer. 2020;19(1):96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Biagioni A, Skalamera I, Peri S, et al. Update on gastric cancer treatments and gene therapies. Cancer Metastasis Rev. 2019;38(3):537‐548. [DOI] [PubMed] [Google Scholar]
- 5. Yimit A, Adebali O, Sancar A, Jiang Y. Differential damage and repair of DNA‐adducts induced by anti‐cancer drug cisplatin across mouse organs. Nat Commun. 2019;10:309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kim M, Jung JY, Choi S, et al. GFRA1 promotes cisplatin‐induced chemoresistance in osteosarcoma by inducing autophagy. Autophagy. 2017;13(1):149‐168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guo J, Satoh K, Tabata S, Mori M, Tomita M, Soga T. Reprogramming of glutamine metabolism via glutamine synthetase silencing induces cisplatin resistance in A2780 ovarian cancer cells. BMC Cancer. 2021;21:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bahar E, Kim JY, Yoon H. Chemotherapy resistance explained through endoplasmic reticulum stress‐dependent signaling. Cancers (Basel). 2019;11(3):338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lin TY, Chan HH, Chen SH, et al. BIRC5/Survivin is a novel ATG12‐ATG5 conjugate interactor and an autophagy‐induced DNA damage suppressor in human cancer and mouse embryonic fibroblast cells. Autophagy. 2020;16(7):1296‐1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang X, Zeng X, Liang X, et al. The chemotherapeutic potential of PEG‐b‐PLGA copolymer micelles that combine chloroquine as autophagy inhibitor and docetaxel as an anti‐cancer drug. Biomaterials. 2014;35(33):9144‐9154. [DOI] [PubMed] [Google Scholar]
- 11. Yang C, Pan Y. Fluorouracil induces autophagy‐related gastric carcinoma cell death through Beclin‐1 upregulation by miR‐30 suppression. Tumour Biol. 2015;37:15489‐15494. [DOI] [PubMed] [Google Scholar]
- 12. Zeng X, Zhao H, Li Y, et al. Targeting hedgehog signaling pathway and autophagy overcomes drug resistance of BCR‐ABL‐positive chronic myeloid leukemia. Autophagy. 2015;11:355‐372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kumar A, Singh UK, Chaudhary A. Targeting autophagy to overcome drug resistance in cancer therapy. Future Med Chem. 2015;7(12):1535‐1542. [DOI] [PubMed] [Google Scholar]
- 14. Li LQ, Xie WJ, Pan D, Chen H, Zhang L. Inhibition of autophagy by bafilomycin A1 promotes chemosensitivity of gastric cancer cells. Tumour Biol. 2016;37(1):653‐659. [DOI] [PubMed] [Google Scholar]
- 15. Xi G, Hu X, Wu B, et al. Autophagy inhibition promotes paclitaxel‐induced apoptosis in cancer cells. Cancer Lett. 2011;307:141‐148. [DOI] [PubMed] [Google Scholar]
- 16. Yu YF, Hu PC, Wang Y, et al. Paclitaxel induces autophagy in gastric cancer BGC823 cells. Ultrastruct Pathol. 2017;41:284‐290. [DOI] [PubMed] [Google Scholar]
- 17. Tamargo‐Gómez I, Mariño G AMPK: Regulation of metabolic dynamics in the context of autophagy. Int J Mol Sci 2018. 19(12):3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tang F, Hu P, Yang Z, et al. SBI0206965, a novel inhibitor of Ulk1, suppresses non‐small cell lung cancer cell growth by modulating both autophagy and apoptosis pathways. Oncol Rep. 2017;37(6):3449‐3458. [DOI] [PubMed] [Google Scholar]
- 19. Zhao Z, Li J, Jiang Y, Xu W, Li X, Jing W. CLDN1 increases drug resistance of non‐small cell lung cancer by activating autophagy via up‐regulation of ULK1 phosphorylation. Med Sci Monit. 2017;23:2906‐2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol. 2019;20(10):608‐624. [DOI] [PubMed] [Google Scholar]
- 21. Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6‐methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014;16(2):191‐198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Berulava T, Buchholz E, Elerdashvili V, et al. Changes in m6A RNA methylation contribute to heart failure progression by modulating translation. Eur J Heart Fail. 2020;22(1):54‐66. [DOI] [PubMed] [Google Scholar]
- 23. Wang X, Wu R, Liu Y, et al. m6A mRNA methylation controls autophagy and adipogenesis by targeting Atg5 and Atg7. Autophagy. 2020;16(7):1221‐1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jin S, Zhang X, Miao Y, et al. m6A RNA modification controls autophagy through upregulating ULK1 protein abundance. Cell Res. 2018;28(9):955‐957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhou S, Bai ZL, Xia D, et al. FTO regulates the chemo‐radiotherapy resistance of cervical squamous cell carcinoma (CSCC) by targeting β‐catenin through mRNA demethylation. Mol Carcinog. 2018;57(5):590‐597. [DOI] [PubMed] [Google Scholar]
- 26. Ma M, Zhang Y, Weng M, et al. lncRNA GCAWKR promotes gastric cancer development by scaffolding the chromatin modification factors WDR5 and KAT2A. Mol Ther. 2018;26(11):2658‐2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guo WJ, Zhang YM, Zhang L, et al. Novel monofunctional platinum (II) complex mono‐Pt induces apoptosis‐independent autophagic cell death in human ovarian carcinoma cells, distinct from cisplatin. Autophagy. 2013;9(7):996‐1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Peng L, Sang H, Wei S, et al. circCUL2 regulates gastric cancer malignant transformation and cisplatin resistance by modulating autophagy activation via miR‐142‐3p/ROCK2. Mol Cancer. 2020;19(1):156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ichimura Y, Kirisako T, Takao T, et al. A ubiquitin‐like system mediates protein lipidation. Nature. 2000;408:488‐492. [DOI] [PubMed] [Google Scholar]
- 30. Komatsu M, Waguri S, Koike M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy‐deficient mice. Cell. 2007;131:1149‐1163. [DOI] [PubMed] [Google Scholar]
- 31. Shoemaker CJ, Huang TQ, Weir NR, Polyakov NJ, Schultz SW, Denic V. CRISPR screening using an expanded toolkit of autophagy reporters identifies TMEM41B as a novel autophagy factor. PLoS Biol. 2019;17:e2007044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang C, Wang H, Zhang D, et al. Phosphorylation of ULK1 affects autophagosome fusion and links chaperone‐mediated autophagy to macroautophagy. Nat Commun. 2018;9:3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu Y, You Y, Lu Z, et al. N 6‐methyladenosine RNA modification‐mediated cellular metabolism rewiring inhibits viral replication. Science. 2019;365(6458):1171‐1176. [DOI] [PubMed] [Google Scholar]
- 34. Choe J, Lin S, Zhang W, et al. mRNA circularization by METTL3‐eIF3h enhances translation and promotes oncogenesis. Nature. 2018;561(7724):556‐560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Weng H, Huang H, Wu H, et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m6A modification. Cell Stem Cell. 2018;22(2):191‐205.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Min KW, Zealy RW, Davila S, et al. Profiling of m6A RNA modifications identified an age‐associated regulation of AGO2 mRNA stability. Aging Cell. 2018;17(3):e12753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ma S, Chen C, Ji X, et al. The interplay between m6A RNA methylation and noncoding RNA in cancer. J Hematol Oncol. 2019;12(1):121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Feng ZY, Gao HY, Feng TD. Immune infiltrates of m6A RNA methylation‐related lncRNAs and identification of PD‐L1 in patients with primary head and neck squamous cell carcinoma. Front Cell Dev Biol. 2021;9:672248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zheng W, Dong X, Zhao Y, et al. Multiple functions and mechanisms underlying the role of METTL3 in human cancers. Front Oncol. 2019;9:1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lan Q, Liu PY, Haase J, Bell JL, Hüttelmaier S, Liu T. The critical role of RNA m6A methylation in cancer. Cancer Res. 2019;79(7):1285‐1292. [DOI] [PubMed] [Google Scholar]
- 41. Lin Z, Niu Y, Wan A, et al. RNA m6 a methylation regulates sorafenib resistance in liver cancer through FOXO3‐mediated autophagy. EMBO J. 2020;39(12):e103181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yan F, Al‐Kali A, Zhang Z, et al. A dynamic N6‐methyladenosine methylome regulates intrinsic and acquired resistance to tyrosine kinase inhibitors. Cell Res. 2018;28(11):1062‐1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang J, Zhou JY, Kho D, Reiners JJ Jr, Wu GS. Role for DUSP1 (dual‐specificity protein phosphatase 1) in the regulation of autophagy. Autophagy. 2016;12(10):1791‐1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lin MG, Hurley JH. Structure and function of the ULK1 complex in autophagy. Curr Opin Cell Biol. 2016;39:61‐68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hosokawa N, Sasaki T, Iemura S, Natsume T, Hara T, Mizushima N. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy. 2009;5:973‐979. [DOI] [PubMed] [Google Scholar]
- 46. Lechauve C, Keith J, Khandros E, et al. The autophagy‐activating kinase ULK1 mediates clearance of free α‐globin in β‐thalassemia. Sci Transl Med. 2019;11(506):eaav4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Quan Y, Lei H, Wahafu W, Liu Y, Ping H, Zhang X. Inhibition of autophagy enhances the anticancer effect of enzalutamide on bladder cancer. Biomed Pharmacother. 2019;120:109490. [DOI] [PubMed] [Google Scholar]
- 48. Liu L, Yan L, Liao N, Wu WQ, Shi JL. A review of ULK1‐mediated autophagy in drug resistance of cancer. Cancers (Basel). 2020;12(2):352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Soni M, Patel Y, Markoutsa E, et al. Autophagy, cell viability, and chemoresistance are regulated by miR‐489 in breast cancer. Mol Cancer Res. 2018;16(9):1348‐1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang X, Lu Z, Gomez A, et al. N6‐methyladenosine‐dependent regulation of messenger RNA stability. Nature. 2014;505(7481):117‐120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.


