Abstract
The outer membrane protein contents of Salmonella enterica serovar Typhimurium strains with PhoP/PhoQ regulon mutations were compared by two-dimensional gel electrophoresis. At least 26 species of outer membrane proteins (OMPs) were identified as being regulated by PhoP/PhoQ activation. One PhoP/PhoQ-activated OMP was identified by semiautomated tandem mass spectrometry coupled with electronic database searching as PgtE, a member of the Escherichia coli OmpT and Yersinia pestis Pla family of outer membrane proteases. Salmonella PgtE expression promoted resistance to alpha-helical cationic antimicrobial peptides (α-CAMPs). Strains expressing PgtE cleaved C18G, an 18-residue α-CAMP present in culture medium, indicating that protease activity is likely to be the mechanism of OmpT-mediated resistance to α-CAMPs. PhoP/PhoQ did not regulate the transcription or export of PgtE, indicating that another PhoP/PhoQ-dependent mechanism is required for PgtE outer membrane localization. PgtE is a posttranscriptionally regulated component of the PhoP/PhoQ regulon that contributes to Salmonella resistance to innate immunity.
Innate immunity is a mechanism by which animals sense and control invading microbes, including bacteria. Cationic antimicrobial peptides (CAMPs) are a major component of innate immunity (55). CAMPs are released at mucosal and skin surfaces and are part of the phagocytic vacuole microbicidal mechanism. CAMPs have a variety of amphipathic structures that function to kill bacteria by permeabilization of lipid bilayers.
Upon host colonization, gram-negative bacteria can increase their resistance to innate host defenses, including CAMPs, by changing the structure, immunogenic properties, and permeability of their surfaces. The PhoP/PhoQ regulatory system of Salmonella enterica serovar Typhimurium is a host defense mechanism by which bacteria respond to environmental signals and induce changes in the bacterial outer membrane that promote CAMP resistance (20). These changes include addition of aminoarabinose and palmitate to the lipid A moiety of lipopolysaccharide (LPS) (16, 19, 21), which promotes resistance to polymyxin (15, 19), alpha-helical antimicrobial peptides, and protegrin, a β-sheet antimicrobial peptide (21). Besides regulating the expression of enzymes involved in LPS modifications, PhoP/PhoQ regulates the expression of several secreted and membrane proteins (4, 44) that could be important for resistance to bactericidal agents. To identify additional members of the PhoP/PhoQ regulon that are involved in resistance to CAMPs, outer membrane protein (OMP) profiles of Salmonella strains with phoP/phoQ mutations were compared.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids used in this study are described in Table 1. Bacterial cultures were grown at 37°C with aeration in Luria broth (LB) (31), tryptic soy broth (Difco), Mueller-Hinton broth (Difco), or N minimal medium supplemented with 0.1% casamino acids, 38 mM glycerol, and 8 μM MgCl2 (13). Antibiotics were used at the following concentrations: kanamycin, 45 μg/ml; ampicillin, 50 to 100 μg/ml; and streptomycin, 1,000 μg/ml.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or propertya | Source or referenceb |
|---|---|---|
| Serovar Typhimurium | ||
| CS015 | phoP102::Tn10d-Cam | 32 |
| CS401 | phoN2 zxx-6251::Tn10d-Cam rpsL (Strr) | 21 |
| CS093 | 14028s; wild type | ATCC |
| CS022 | pho-24 (PhoP constitutive); a mutation in phoQ that results in increased net phosphorylation of PhoP and unregulated expression of PhoP-activated genes (18) | 33 |
| TG59 | CS401 ΔpgtE::Kan | This work |
| TG61 | CS022 ΔpgtE::Kan | This work |
| TG71 | TG61 with pBluescript KS+ | This work |
| TG73 | TG61 with pTG73 | This work |
| CS435 | pho-24 pagP::Tn10d-Tet phoN2 zxx-6251::Tn10d-Cam (also LG069) | 21 |
| TG66 | CS435 ΔpgtE::Kan | This work |
| TG172 | CS401 with pTG171 | This work |
| TG173 | CS015 with pTG171 | This work |
| TG174 | CS022 with pTG171 | This work |
| TG200 | CS022 pagA::MudJ | This work |
| CS404 | CS022 pagA::MudJ pagP::Tn10d-Tet | This work |
| E. coli | ||
| SM10 λpir | thi-1 thr-1 leuB6 supE44 tonA21 lacY1 recA::RP4-2-Tc::Mu | |
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 endA1 recA1 hsdR17 deoR thi-1 supE441 gyrA96 relA1 | Gibco-BRL |
| CC118 | araD139 Δ(ara,leu)7697 ΔlacX74 phoAΔ20 galE galK thi rpsE rpoB argE(Am) recA1 | 28 |
| Plasmids and transposons | ||
| pKAS32 | Ampr Strs (rpsL); a low-copy allelic exchange vector | 41 |
| pTG51 | Ampr Kanr; pgtE with an inserted Kanr cassette (39) replacing a portion of pgtE is ligated to KpnI-SacI fragment of pKAS32 | This work |
| pBluescript KS+ | Ampr; a high-copy cloning vector | Stratagene |
| pTG73 | Ampr; contains pgtE and 904 bp of upstream region (nt 671 to 2671; GenBank accession no. M21279) cloned into XbaI site of pBluescript KS+ | This work |
| pGPLFR03 | Ampr; firefly (f-luc) and vanilla (v-luc) double luciferase suicide vector | 30 |
| pTG171 | Ampr; contains a transcriptional fusion of pgtE (nt 5266 to 5641; GenBank no. M21279) to f-luc; ligated with an EcoRI-KpnI fragment of pGPLFR03 | This work |
| pWSK129 | Kanr; low-copy cloning vector | 51 |
| pTG82 | Kanr; contains pgtE and 904 bp of upstream region (nt 671 to 2671; GenBank no. M21279) cloned into XbaI site of pWSK129 | This work |
| pTG85 | Kanr; contains TnphoA/in insertion at codon 112 of pgtE in pTG82 | This work |
| pTG86 | Kanr; contains TnphoA/in insertion at codon 152 of pgtE in pTG82 | This work |
| pTG87 | Kanr; contains TnphoA/in insertion at codon 190 of pgtE in pTG82 | This work |
| TnphoA/in | Tn5 derivative; generates translational fusions to alkaline phosphatase (phoA) | 27 |
nt, nucleotide.
ATCC, American Type Culture Collection.
Cloning of pgtE and construction of mutant strains.
DNA sequences flanking pgtE were amplified by PCR using appropriate oligonucleotide primers and cloned into the XbaI and HindIII sites of pBluescript KS+. The resulting plasmid contained DNA sequences deleted for most of the pgtE open reading frame (nucleotides 3912 to 4785 and 5594 to 5916; GenBank accession no. M21279). A kanamycin resistance cassette was then inserted into the EcoRI site within ΔpgtE. The ΔpgtE::Kan cassette was then cloned into the pKAS32 suicide vector (41) digested with KpnI and SacI (pTG51), and SM10λpir cells were transformed. Allelic exchange was performed by a conjugative transfer of pTG51 into CS401, a wild-type strain of serovar Typhimurium containing a recessive streptomycin sensitivity allele. Single homologous recombination derivatives were isolated by selection for ampicillin and kanamycin resistance and then selected on streptomycin for loss of plasmid sequences. The presence of the ΔpgtE::Kan cassette on the chromosome was determined by PCR amplification. The mutated locus was transferred into other Salmonella strains by P22HTint phage-mediated transduction.
Isolation of bacterial outer membranes and analysis of OMPs by 2-D PAGE.
Outer membranes were isolated by a modified protocol of Osborn et al. (36). One liter of LB medium was inoculated from an overnight culture at a dilution factor of 1:100. Cultures were grown until late log phase (optical density at 600 nm [OD600] = 0.8 to 1.0), when cells were collected by centrifugation at 8,000 × g for 15 min at 4°C. Bacterial spheroplasts were generated by cold osmotic shock in 0.5 M sucrose–10 mM Tris-Cl (pH 7.8)–60 μg of lysozyme per ml and subsequent addition of an equal volume of ice-cold 1 μM EDTA. Spheroplasts were broken by French press at 16,000 lb/in2, unbroken cells were removed by centrifugation at 6,000 × g for 15 min at 4°C, and the bacterial extract was separated into fractions by centrifugation at 200,000 × g for 1 h at 4°C. The pellet fraction containing total bacterial membranes was homogenized in 20% sucrose and subjected to sucrose density gradient centrifugation at 180,000 × g for 12 to 16 h at 4°C. The outer membrane fraction was separated as the band of highest buoyant density in the sucrose gradient.
For more efficient separation during isoelectric focusing (IEF), aliquots containing 250 μg of total OMP were washed in deionized water and solubilized by boiling for 10 minutes in 1% (wt/vol) sodium dodecyl sulfate (SDS). Protein was subsequently precipitated in 10 volumes of ice-cold acetone. Removal of the bulk of the outer membrane lipid and LPS aggregates by this technique significantly improved protein separation by IEF. The protein precipitate was resuspended in solubilization buffer containing 9 M urea, 2% Triton X-100, 2% Pharmalyte pH 3-10 (Pharmacia), 2% β-mercaptoethanol, and the protease inhibitors pepstatin (2 μg/ml), aprotinin (2 μg/ml), and leupeptin (2 μg/ml). After 2 h of incubation at 37°C, insoluble material was removed by sedimentation at 14,000 × g for 10 min at room temperature. Solubilized protein was first separated by IEF using a Pharmacia Multiphor II electrophoresis unit with immobilized pH gradients (pH 4 to 7) and then on SDS–12% polyacrylamide gels. Protein spots were visualized by staining with Coomassie brilliant blue. Gels were scanned using an UMAX Astra 1200S scanner, and two-dimensional (2-D) profiles of outer membrane proteomes were compared.
Peptide isolation, sequencing by tandem mass spectrometry, and identification of PgtE.
Protein spots of interest were excised from Coomassie-stained 2-D SDS-polyacrylamide gels and digested in situ with trypsin as described below (obtained as a personal communication from Michael Kinter, University of Virginia). Polyacrylamide gel slices were fragmented, destained during an overnight incubation in 50% (vol/vol) methanol, and dehydrated by incubation in acetonitrile for 10 min. Excess liquid was removed under vacuum, and gel fragments were rehydrated in 50 μl of sequencing-grade modified trypsin (Promega) at the concentration of 20 μg/ml. After a 30-min incubation on ice, excess trypsin was removed, 20 μl of 50 mM ammonium bicarbonate was added, and the mixture was incubated overnight at 37°C. Resulting peptides were eluted from the gel with several changes of extraction buffer (200 μl of 50% acetonitrile–5% formic acid) and dried by evaporation. Peptides were solubilized in 5% acetonitrile–0.5% acetic acid for further analysis.
LC/MS/MS analysis (liquid chromatography combined with tandem mass spectrometry) of tryptic peptides was carried out with a Finnigan/Thermoquest TSQ 7000 triple quadrupole mass spectrometer (San Jose, Calif.), coupled with a microcapillary high-pressure liquid chromatography (HPLC) apparatus built in-house. The details of our modifications of a Shimadzu HPLC system (Shimadzu Scientific Instruments, Inc., Columbia, Md.) for use with capillary HPLC have been described previously (50). The Finnigan API (atmospheric pressure ionization) electrospray source was modified with a capacitive sprayer assembly (49). A linear binary gradient of solvents at a flow rate of 200 ml/min (Shimadzu LC-10AD pumps) was split precolumn to give a 1:1,000 split ratio, as measured at the beginning of the gradient. A fused-silica capillary column was packed in-house, 75 μm inner diameter by 12 cm (Magic C18; 5-μm packing; 100 Å pore size; Michrom Bioresources, Auburn, Calif.), and connected to the splitter. The column was eluted with a gradient of 0 to 75% acetonitrile in the presence of 0.4% acetic acid over 25 min. The temperature of the heated capillary inlet to the mass spectrometer was set at 180°C. When the main beam ion peaks for a peptide reached a preset threshold (40,000 counts), the ion of interest was automatically selected for collision-induced dissociation (CID) using the data-dependent scanning capabilities built into the TSQ, which are accessed using ICL (instrument control language) procedures (11). Protein identification was accomplished by use of the SEQUEST computer program (53).
Bacterial sensitivity to antimicrobial peptides.
Standard MICs of CAMPs were determined as described (45), except that bacterial cultures were grown overnight in N minimal medium with low (8 μM) magnesium or in tryptic soy broth and diluted to 2 × 105 bacteria per ml in N minimal medium with low magnesium. Complementation experiments were performed in Mueller-Hinton broth. Test peptides were assayed at final concentrations of 0.15 to 40.0 μg/ml in 96-well polypropylene microtiter plates (Costar). The MIC was determined as the lowest concentration of the peptide that did not allow visible bacterial growth after 24 h (for assays performed in rich medium) or after 48 h (for assays that were performed in minimal medium). C18G was a gift of Richard Darveau; LL-37 and CRAMP were a gift of Robert Lehrer. Resistance to defensins HNP-1 (a gift of Thomas Ganz), cryptidin 2 (a gift of Michael Selsted), and NP-1 and protegrin PG-1 (a gift of Robert Lehrer) were analyzed as above and in radial diffusion assays as described previously (23). Peptide-killing assays were performed with mid-log-phase bacterial cultures grown in rich medium (LB) as described in Miller et al. (34). All experiments were performed two or three times.
To determine the specificity of the protease action in pgtE-mediated CAMP resistance, 2 × 105 bacterial cells were incubated with 2.5 μg of C18G peptide per ml for 16 h in 0.5% tryptone. Cells were sedimented by centrifugation, and peptide-containing supernatants were collected. Relative amounts of cleaved and uncleaved peptide were determined by separating the supernatants on a reversed-phase column (250-mm by 1.00-mm Jupiter column; 5-μm particle size; 300-Å pore size; C-18 bonded silica; Pharmacia, Inc., Kalamazoo, Mich.) used with the Shimadzu LC-10AD VP liquid chromatography system. A linear gradient of 5 to 95% acetonitrile in H2O was applied over a period of 30 min. The aqueous and organic buffers contained 0.1 and 0.08% trifluoroacetic acid, respectively. The sensitivity of the UV detector at 214 nm was set at 0.1 absorbance unit full scale.
DNA techniques.
Bacterial chromosomal DNA was isolated as previously described (17). Plasmid DNA was isolated using kits from Promega and Qiagen. PCR was performed with Pfu Turbo DNA polymerase (Stratagene) and Taq DNA polymerase (Gibco-BRL) according to the manufacturers' instructions.
Luciferase assays.
A fusion of the pgtE promoter region to the transcriptional reporter f-luc was integrated into the chromosome of different Salmonella phoP strains using the suicide vector pGPLFRO3 (30) (Table 1). Correct chromosomal localization of the pgtE-f-luc fusion was confirmed by Southern blotting and hybridization and also genetically by allelic replacement of a kanamycin resistance marker inserted into pgtE. Bacteria were grown in LB medium or in N minimal medium containing different concentrations of Mg2+. Luciferase assays were performed throughout the growth curve of each strain as previously described (30). Aliquots (20 μl) of bacterial cultures were lysed by freezing and thawing, followed by sonication for 20 s. Luciferase activity of cell lysates was determined using the Luciferase Reporter assay system (Promega). Units were recorded in a Berthold LB9501 luminometer.
Nucleotide sequence accession numbers.
The revised nucleotide sequence of pgtE has been deposited in GenBank under accession number AF239770. Salmonella genome sequence data were produced by the Salmonella typhi Sequencing Group at the Sanger Centre, Cambridge, U.K., and by the Genomes Sequencing Center at Washington University, St. Louis, Mo., and can be obtained from ftp://ftp.sanger.ac.uk/pub/pathogens/st/ and ftp://genome.wustl.edu/pub/gsc1/sequence/st.louis/bacterial/salmonella/, respectively.
RESULTS
PhoP/PhoQ regulates OMPs.
A variety of studies have identified PhoP/PhoQ-mediated transcriptional regulation of genes that encode predicted envelope proteins and alter outer membrane structure through LPS modifications (4, 16, 20, 44). To further define PhoP/PhoQ-regulated changes in the protein composition of the Salmonella envelope, a method for high-resolution analysis of gram-negative OMPs by 2-D polyacrylamide gel electrophoresis (2-D PAGE) was developed. Salmonella outer membranes were purified by separation on a sucrose gradient. Then OMPs were separated from LPS aggregates by heating in the presence of an anionic detergent. Next, protein separation was accomplished by IEF (described in Materials and Methods). A large number of Salmonella OMPs were detected by Coomassie staining of 2-D polyacrylamide gels.
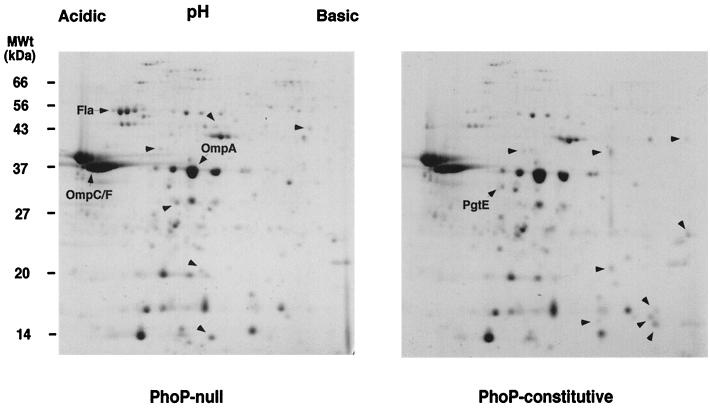
Twenty-six spots corresponding to OMPs that are members of the PhoP/PhoQ regulon were detected by comparing the 2-D PAGE profiles of strains with PhoP-null and PhoP-constitutive phenotypes (Fig. 1 and data not shown). Twelve OMP spots were unique to the PhoP-null strain (CS015), and 14 spots were unique to the strain with increased PhoQ kinase activity (PhoP constitutive, CS022). Thus, PhoP/PhoQ mediates significant alteration in the protein content of the outer membrane in addition to LPS structural changes.
FIG. 1.
2-D PAGE map of PhoP/PhoQ-regulated OMPs. OMPs were separated by IEF on a linear pH gradient of 4 to 7 and on SDS–12% PAGE gels. Proteins were visualized by staining in Coomassie brilliant blue. 2-D map positions of the porins OmpA, OmpC/OmpF, and PhoP-regulated phase I flagellin (Fla) were determined by comparison with previous studies (38). Unlabeled arrowheads point to protein species present exclusively in the absence (PhoP null) or presence (PhoP constitutive) of an active PhoP/PhoQ regulatory system. PgtE is a protein that was sequenced and analyzed in this study.
Identification of PgtE protease as a PhoP-regulated OMP.
One abundant protein species of approximately 32 kDa and isoelectric point 5.2 localized to the outer membrane of strains that expressed activated PhoP/PhoQ but not to the membrane of the PhoP-null strain (Fig. 1). To confirm that this protein was localized to the outer membrane as a result of PhoP/PhoQ activation, OMPs isolated from wild-type serovar Typhimurium grown in a PhoP-repressing (1 mM) or PhoP-inducing (8 μM) concentration of Mg2+ (13) were separated by 2-D PAGE. A 32-kDa protein of pI 5.2 was detected in the outer membranes of salmonellae that were grown in PhoP-activating conditions (8 μM Mg2+), but not in the outer membranes of salmonellae grown in the presence of a high concentration of Mg2+ (data not shown), further indicating that this protein was regulated by PhoP/PhoQ-activating conditions and was not specific to strains with this phoQ mutation.
To begin the characterization of PhoP-regulated OMPs, this protein was excised from the polyacrylamide gel and digested in situ with trypsin, and the resulting peptide mixture was eluted from the gel. An aliquot of this tryptic peptide mixture was analyzed by on-line microcapillary HPLC coupled with electrospray ionization-mass spectrometry (Fig. 2). Sequences of isolated tryptic peptides were determined by semiautomated tandem mass spectrometry coupled with protein database searching. Raw-product ion (MS2 or MS/MS) data were searched using the SEQUEST computer program, which is capable of identifying peptides from protein amino acid sequence data coded in FASTA format, such as that found in the OWL database (5), from uninterpreted MS/MS spectra (53). Three peptides (LSQLDWK, AGVTAGYQETR, and SIHPDTSVNYANEYDLN) had high SEQUEST cross-correlation scores (2.96, 3.80, and 7.18, respectively) with peptides originating from PgtE, the Salmonella OmpT protease homologue (14). The SEQUEST search results suggested that all three peptides originated from the same protein, based on the gene sequence found in the SWISS-PROT database (2), accession number P06185. Fragmentation of selected peptide ions by CID (24) resulted in sufficient partial sequence information to characterize the protein as PgtE (Fig. 3). Salmonella pgtE was originally identified as an open reading frame located downstream of the inducible pgtBCA operon, which is involved in phosphoglycerate transport (54).
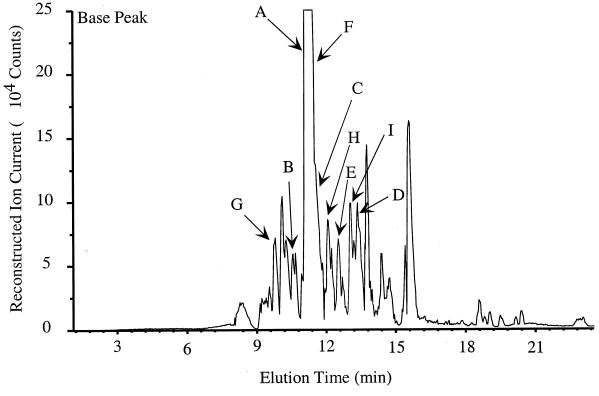
FIG. 2.
Base peak capillary HPLC mass chromatogram from the tryptic digest of Salmonella PgtE. In this type of HPLC trace, the signal intensity of the most abundant ion is plotted for each scan. Each scan of the mass spectrometer takes about 1 s. Peptides identified by SEQUEST are indicated by arrows and contain the following partial sequences: (A) ELVYDTDTGR; (B) ELVYDTDGRK; (C) KLSQLDWK; (D) GWLLQGDNYK; (E) FSWTAR; (F) YIGNFPHGVR; (G) GIGYSQR; (H) YSDWVNAHDNDEHYMR; and (I) IFAEFAYSK.
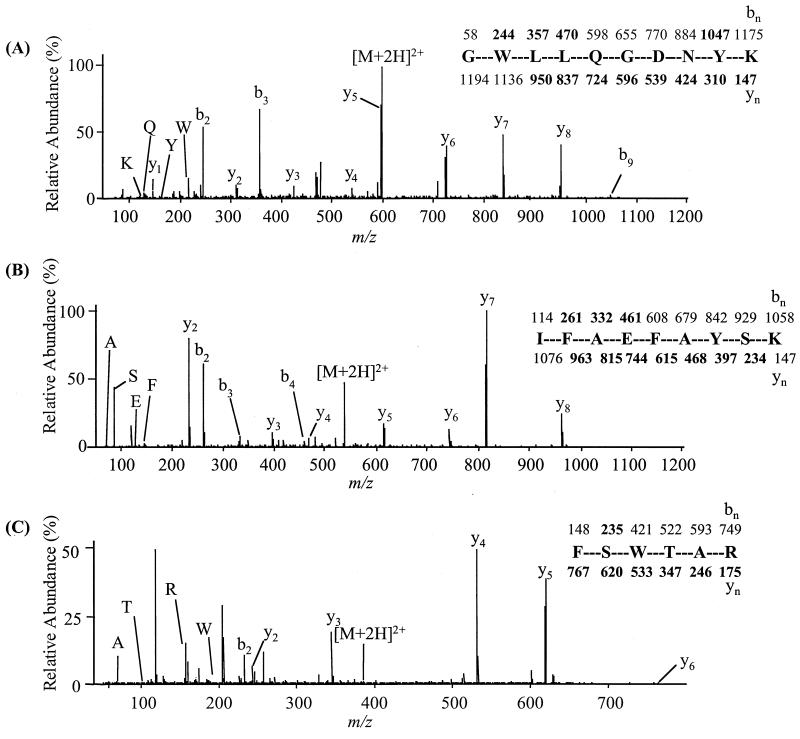
FIG. 3.
Peptide sequencing by tandem mass spectrometry and identification of Salmonella PgtE. Fragment ions observed are indicated in boldface above (b-type ions) and below (y-type ions) the peptide sequence. (A) CID mass spectrum of GWLLQGDNYK. (B) CID mass spectrum of IFAEFAYSK. (C) CID mass spectrum of FSWTAR. The nomenclature used for b and y peptide fragment ions has been described by Biemann (4a). Low-mass ions that are indicative of amino acid composition but not sequence are described by the amino acid single-letter code.
Nucleotide sequence analysis and correction of PgtE sequence.
The genomic region of serovar Typhimurium that contains pgtE was cloned into a plasmid vector (pTG73, Table 1), and its nucleotide sequence was analyzed and compared to sequences in GenBank and to sequences available from the Salmonella genome sequencing projects. Plasmid-derived pgtE sequence was identical to regions of two S. enterica LT2 genomic fragments (gnl/WUGSC 99287/stmlt2-.Contig1497 and gnl/WUGSC 99287/stmlt2-A2A.Contig104, respectively) and highly similar to genomic fragments of serovars Typhi (gnl/Sanger 601/S. typhi Contig427) and Paratyphi (gnl/WUGSC 32027/spara B SPA.0.1882). The nucleotide sequences derived from plasmid pTG73 and from the above-mentioned Salmonella genome fragments contain several frameshifts within the pgtE open reading frame with respect to a previously reported pgtE sequence (GenBank accession number M21279). The newly assigned PgtE sequence exhibits higher similarity to other OmpT-like proteases than the previously reported one (72% identity and 83% similarity to Y. pestis Pla, 46% identity and 65% similarity to E. coli OmpT and OmpP, and 38% identity and 59% similarity to Shigella flexneri SopA) throughout the length of the protein. Revised PgtE contains a C-terminal phenylalanine residue that is essential for outer membrane insertion of trimeric OMPs (46). E. coli contains two highly homologous genes encoding OmpT-like proteases: ompT on the chromosome (15) and ompP (25) on plasmid F (29). OmpP and OmpT have 87% sequence identity (25). A pgtE-specific probe hybridized to a single Salmonella genomic fragment on a Southern blot (data not shown).
PhoP/PhoQ does not regulate pgtE transcription and export into the periplasm.
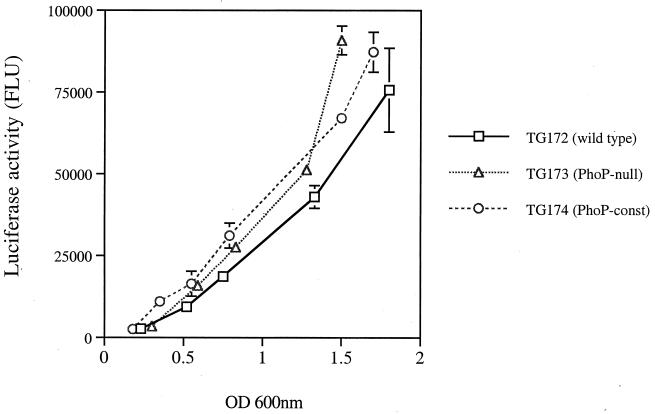
Adams et al. (1) have reported the activity of a PhoP-regulated OmpT-like protease in serovar Typhimurium cellular extracts (1). The 2-D PAGE analysis described in this study suggested that PhoP/PhoQ might regulate the transcription of pgtE. Single-copy transcriptional fusions of pgtE to the firefly luciferase (f-luc) gene were constructed by using the pGPLFR03 suicide vector (Table 1). The resulting gene fusion contained f-luc inserted into the last third of the pgtE. Expression of f-luc was measured throughout the growth curve in bacterial cultures grown in LB and in minimal medium supplemented with a high (PhoP-repressing) or low (PhoP-inducing) concentration of Mg2+. Surprisingly, similar levels of pgtE-f-luc expression were detected in the PhoP-null (TG172), wild-type (TG173), and PhoP-constitutive (TG174) strains. Expression of the pgtE-f-luc fusion increased steadily in all strains during logarithmic growth (Fig. 4). These results indicated that transcription of pgtE is constitutive and it is not dependent on PhoP/PhoQ. Therefore, localization of PgtE to the outer membrane is mediated by a PhoP/PhoQ-dependent mechanism that acts after the transcription of the pgtE gene.
FIG. 4.
pgtE expression does not require PhoP. The expression of pgtE was measured throughout the growth of bacterial cultures by quantitating the amount of luciferase activity produced by strains containing the pgtE-f-luc transcriptional fusion. Bacterial cultures were grown in rich medium (LB). The graph depicts one experiment performed in triplicate and is representative of several experiments. Error bars represent the standard deviation (SD); no bars indicate that the SD is insignificant. FLU, firefly luciferase light units. const, constitutive.
To further analyze PhoP-mediated regulation, pgtE and a 904-bp upstream region were cloned downstream of the inducible lac promoter on a low-copy-number plasmid (pTG82, Table 1). Random translational fusions of pgtE to the alkaline phosphatase gene (phoA) were generated in E. coli by utilizing transposon TnphoA/in delivered by the method of Manoil and Bailey (27). Plasmids containing transposon insertions in pgtE were isolated and analyzed. Three plasmids, pTG85, pTG86, and pTG87, contained exported PhoA fusions to residues 112, 152, and 190 of PgtE, respectively. These plasmids were transferred to appropriate Salmonella strains, and the alkaline phosphatase activity of the fusion proteins in the presence and absence of phoP expression was measured as previously described (6). Rates of PgtE-PhoA synthesis and export were similar in all strains (data not shown), indicating that the translation initiation and likely Sec-dependent translocation of PgtE across the inner membrane were independent of PhoP/PhoQ.
The results of this study suggested that localization at the outer membrane could be important for the protease activity of PgtE. It has been demonstrated that proper folding, oligomerization, and insertion of some E. coli OMPs into the outer membrane are dependent on interaction with LPS molecules or specific LPS structures (9, 10, 40). Therefore, it is possible that PhoP/PhoQ-mediated modifications of LPS could affect the insertion and localization of some Salmonella OMPs, including PgtE. To explore this possibility, OMPs from PhoP-constitutive Typhimurium strains containing a mutation in pagP, pagA, or both genes (CS435, TG200, and CS404, respectively), were analyzed by 2-D PAGE for the presence of PgtE. PhoP-regulated pagP and pagA mediate additions of palmitate and aminoarabinose, respectively, to Salmonella lipid A (19, 21). The absence of these two lipid A modifications did not affect the localization of PgtE to the outer membrane, as determined by 2-D PAGE (data not shown), indicating that these modifications are not essential for localization of PgtE to the outer membrane.
PgtE promotes resistance to antimicrobial peptides.
PgtE belongs to the family of outer membrane endopeptidases (42) that specifically cleave between paired basic residues and after a basic residue that is followed by a nonpolar amino acid (48). Therefore, CAMPs are potential targets of PgtE. Recently, Stumpe et al. (48) have demonstrated that expression of OmpT increases survival of E. coli grown in the presence of protamine, a CAMP isolated from salmon sperm (47). To test whether PgtE plays a similar role in serovar Typhimurium, pgtE mutants were generated by allelic exchange and their ability to survive in the presence of CAMPs that contain predicted OmpT (PgtE) cleavage sites was determined. The ability of pgtE strains to resist the bactericidal action of alpha-helical peptide C18G (8) was determined in a growth inhibition (MIC) assay. The pgtE deletion strain TG61 showed increased sensitivity to C18G (Table 2). The sensitivity of this mutant was increased when bacterial cultures were grown in N minimal medium prior to their incubation with C18G peptide. A previously described pagP mutant (CS435) that is sensitive to alpha-helical CAMPs (α-CAMPs) (21) had a higher survival rate than TG61 under the same assay conditions. A strain containing mutations of pagP and pgtE (TG66) showed greater sensitivity to C18G than strains containing single pagP or pgtE mutations (Table 2). pgtE mutants also displayed increased sensitivity to human CAMP LL-37 (26) and mouse CRAMP (3), other naturally occurring alpha-helical CAMPs that contain predicted OmpT cleavage sites (Table 2).
TABLE 2.
MICs of cationic peptidesa
| Straina | MIC (μg/ml)
|
|||||
|---|---|---|---|---|---|---|
| C18G
|
LL-37, N-minimal | CRAMP, N-minimal | NP-1, N-minimal | PG-1, N-minimal | ||
| N-minimal | TSB | |||||
| CS015 (PhoP null) | ND | 0.62 | ND | ND | ND | ND |
| CS022 (PhoPC) | 2.50 | 2.50 | 2.50 | 2.50 | 0.62 | 0.31 |
| TG61 (PhoPCompT) | 0.31 | 1.25 | 1.25 | 1.25 | 0.62 | 0.31 |
| CS435 (PhoPCpagP) | 0.62 | 1.25 | 2.50 | 1.25 | 0.62 | 0.31 |
| TG66 (PhoPCpagP ompT) | 0.15 | 0.62 | 1.25 | 0.62 | 0.62 | 0.31 |
PhoPC, PhoP constitutive. Bacterial strains were grown either in N minimal medium containing 8 μM Mg2+ or in tryptic soy broth (TSB) prior to incubation with the peptides. Bacterial cells were diluted in N minimal medium when incubated with alpha-helical peptides C18G, LL-37, and CRAMP and in 1% TSB in 10 mM sodium phosphate buffer (pH 7.4) when incubated with defensin NP-1 and protegrin PG-1. ND, not determined. N minimal medium (13) does not support growth of CS015.
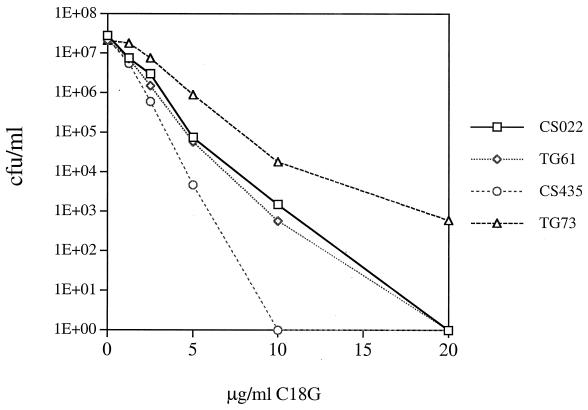
To observe if the phenotype conferred by deletion of pgtE could be complemented, a high-copy-number plasmid expressing pgtE (pTG73) or a control vector was introduced into mutant strain TG61. Bacterial cultures were grown in rich medium prior to incubation with CAMP to ensure high levels of pgtE expression. As shown in Table 3, high-level expression of pgtE greatly increases the survival of serovar Typhimurium in the presence of C18G compared to strain CS022 (PhoP constitutive). In a peptide-killing assay, the number of surviving CFU was determined after 2 × 105 bacteria were exposed to varied amounts of C18G for 2 h. Although the pgtE mutant (TG61) did not display sensitivity to C18G under these conditions, expression of pgtE from a high-copy vector increased the resistance of serovar Typhimurium to C18G (Fig. 5).
TABLE 3.
High-copy expression of PgtE protease increases Salmonella survival upon exposure to C18G and LL-37 (strains were grown in Mueller-Hinton broth)a
| Strain | MIC (μg/ml)
|
|
|---|---|---|
| C18G | LL-37 | |
| CS022 (PhoPC) | 5.00 | 5.00 |
| TG71 (PhoPCpgtE + vector) | 1.25 | 2.50 |
| TG73 (PhoPCpgtE + pTG73) | 40.0 | 40.0 |
See Table 2 footnote a.
FIG. 5.
High levels of pgtE expression increase Salmonella survival in the presence of C18G. Mid-log-phase bacterial cultures were incubated with the indicated concentrations of C18G, and the number of CFU was determined. Expression of pgtE from a high-copy plasmid (in TG73) increased survival in the presence of C18G compared to the parental strain (CS022), while the strain with the pgtE deletion (TG61) did not display significant sensitivity to C18G. A pagP mutant (CS435) was sensitive to C18G.
The effect of pgtE deletion on resistance to several beta-sheet CAMPs was also examined by peptide-killing assays, MIC assays, and radial diffusion assays. Strains deleted of pgtE did not exhibit increased sensitivity or resistance to HNP-1, NP-1, cryptidin-2, or protegrin PG-1 under any of the conditions tested (Table 2 and data not shown). Therefore, pgtE is essential for Salmonella resistance to alpha-helical CAMPs but not to peptides of beta-sheet structure. A pagP mutant (CS435) did not exhibit sensitivity to protegrin PG-1 in MIC assays (Table 2), although it was shown to be sensitive to PG-1 in a peptide-killing assay (22).
Evidence that protease activity is the mechanism of PgtE-mediated CAMP resistance.
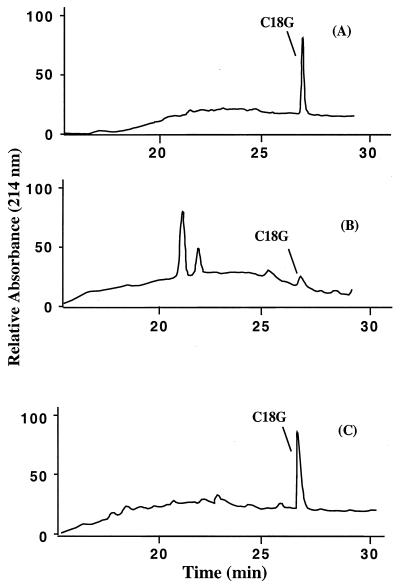
To determine if peptide cleavage promotes resistance to CAMP, Salmonella strains expressing pgtE were assayed for cleavage of C18G. C18G is an 18-residue CAMP (ALYKKLLKKLLKSAKKLG) that contains at least three putative PgtE cleavage sites. In this experiment, supernatants of cultures grown in the presence of C18G were collected, and their contents were separated on a reversed-phase HPLC column. C18G was completely degraded when incubated with strain TG73, which expresses high levels of PgtE, while no peptide degradation was observed when it was incubated with the pgtE-null strain TG61 (Fig. 6). An intermediate amount (approximately 40%) of C18G was cleaved when incubated with parental strain CS022 (PhoP constitutive) (data not shown). These results indicated that peptide cleavage correlates with the serovar Typhimurium CAMP resistance phenotype.
FIG. 6.
Salmonella cells expressing PgtE cleave C18G peptide present in the culture supernatants. Culture supernatants were collected after an MIC experiment, and their contents were analyzed by reversed-phase HPLC as described in Materials and Methods. A strain expressing pgtE from a high-copy plasmid (TG73) efficiently cleaved C18G (B), while a strain carrying a mutation in ompT (TG61) did not cleave C18G (C). The control sample contained C18G in buffer without bacteria present (A).
DISCUSSION
Previous studies have demonstrated that PhoP/PhoQ regulates the transcription of over one dozen genes encoding S. enterica envelope and secreted proteins, including ones such as PagC that were predicted to locate to the outer membrane (4, 37). PhoP/PhoQ also regulates changes in structure of the lipid A component of LPS, which constitutes the outer leaflet of the Salmonella outer membrane (16, 19, 20). In recent years, analyses of proteomes by 2-D gel electrophoresis and tandem mass spectrometry have allowed characterization of complex biological processes (52). In this work the technique of 2-D proteome mapping was utilized to study PhoP/PhoQ regulation of Salmonella outer membrane proteins. This work indicates that PhoP/PhoQ regulates a significant number of OMP species. Fourteen species of OMPs that are positively regulated by PhoP/PhoQ were detected, and 12 species were repressed. Therefore, it appears that in addition to alteration of LPS, a major function of PhoP/PhoQ is to regulate extensive structural changes in both the lipid and protein components of the outer membrane.
A surprising observation of this work was that PgtE is an abundant component of the outer membrane upon PhoP/PhoQ activation, even though pgtE transcription and PgtE export into the bacterial periplasm are not regulated by PhoP/PhoQ. This finding indicates the utility of searching for posttranscriptionally regulated factors localized to the bacterial envelope. The results of this study suggested that PgtE insertion into the outer membrane is dependent upon transcription of another PhoP/PhoQ-activated factor(s). It is possible that PhoP/PhoQ-mediated modifications of LPS could affect the insertion and localization of some OMPs. Though localization of PgtE was not affected by mutations in pagP and pagA, which mediate some Salmonella PhoP-activated LPS modifications, other regulated LPS modifications or factors might be important for localization of PgtE.
In this study, maximal resistance to CAMPs was observed in strains expressing PgtE, indicating that PgtE is part of the resistance to innate immunity regulated by PhoP/PhoQ. The PgtE contribution to inducible resistance was significant when bacteria were exposed to peptides with an alpha-helical structure, such as C18G and LL-37. Such resistance correlated with the ability of bacteria to digest this peptide in the culture medium, indicating that PgtE cleavage of such peptides is the likely mechanism of resistance. The sensitivity of the pgtE mutant was the most obvious in the microbroth dilution (MIC) assays, when the bacteria were incubated with the peptide for a longer period of time (24 to 48 h). The rate of PgtE-mediated peptide hydrolysis is likely to be the limiting factor for the survival of the bacteria. Expression of PgtE is likely advantageous for a small number of bacteria that survive the initial exposure to α-CAMPs. Bacteria expressing PgtE can slowly digest the remaining unbound α-CAMP that was not bound to the bacterial membranes and replicate more efficiently than the bacteria lacking the protease. In support of this, the contribution of PgtE to bacterial resistance in a peptide-killing assay was significant only in the presence of high-copy pgtE. The PgtE protease was not demonstrated to contribute to Salmonella resistance to defensins or protegrin, CAMPs with an amphipathic beta-sheet structure stabilized through intramolecular disulfide bonds (35). Defensin structure could prevent access of PgtE to cleavage sites predicted by amino acid sequence of defensins. In contrast to α-CAMPs, which are produced throughout the animal kingdom, defensins have been found only in higher vertebrates, mammals (22), and birds (7). Higher vertebrates might have evolved defensins as an additional component of innate immunity to combat microbial pathogens which acquired the ability to resist peptides of a less complex structure through PgtE (OmpT) production.
In this work, mutation of both pgtE and pagP resulted in greater α-CAMP sensitivity than in strains containing a single mutation in either gene. Though mutation of pagP has a minor effect on the beta-sheet CAMP protegrin, it also has been found to have a greater effect on antimicrobial α-CAMP resistance. pagP encodes a PhoP/PhoQ-activated acyltransferase which catalyzes the addition of palmitate to lipid A and promotes a decrease in the permeability of the Salmonella outer membrane (21). The envelope-preserving function of pagP could contribute to the survival of salmonellae exposed to a variety of environmental stresses or to antimicrobial compounds and CAMPs within the intestinal lumen or phagocyte vacuoles. The action of a surface protease such as PgtE could further protect bacteria by lowering the concentration of α-CAMP to sublethal doses prior to insertion of the cationic peptide into the bacterial envelope. Synergistic action of PgtE and PagP could significantly decrease the amount of α-CAMP inserted into the cytoplasmic membrane and allow increased survival of bacteria during exposure to CAMPs.
Interestingly, the E. coli and Y. pestis homologues of PgtE have been implicated in virulence. E. coli ompT has been associated with the ability to cause urinary tract infections (12). Additionally, the Y. pestis homologue Pla is essential for virulence in mice when injected subcutaneously but not intravenously (43). It is possible that these effects are a result of OmpT-mediated resistance to innate immunity.
ACKNOWLEDGMENTS
We thank Robert Lehrer, Thomas Ganz, Mike Selsted, and Mike Giblin for gifts of antimicrobial peptides and useful comments and suggestions. We also thank members of the Miller lab for critical comments and suggestions. Kheng B. Lim assisted with the tandem mass spectrometry experiments.
This work was supported by R01 AI30479 to Samuel I. Miller from the National Institutes of Health and funds provided to Murray Hackett by the Department of Medicinal Chemistry and School of Pharmacy, University of Washington.
REFERENCES
- 1.Adams P, Fowler R, Howell G, Kinsella N, Skipp P, Coote P, O'Connor C D. Defining protease specificity with proteomics: a protease with a dibasic amino acid recognition motif is regulated by a two-component signal transduction system in Salmonella. Electrophoresis. 1999;20:2241–2247. doi: 10.1002/(SICI)1522-2683(19990801)20:11<2241::AID-ELPS2241>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 2.Bairoch A, Apweiler R. The SWISS-PROT protein sequence data bank and its supplement TrEMBL in 1999. Nucleic Acids Res. 1999;27:49–54. doi: 10.1093/nar/27.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bals R, Wang X, Zasloff M, Wilson J M. The peptide antibiotic LL-37/hCAP-18 is expressed in epithelia of the human lung where it has broad antimicrobial activity at the airway surface. Proc Natl Acad Sci USA. 1998;95:9541–9546. doi: 10.1073/pnas.95.16.9541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belden W J, Miller S I. Further characterization of the PhoP regulon: identification of new PhoP-activated virulence loci. Infect Immun. 1994;62:5095–5101. doi: 10.1128/iai.62.11.5095-5101.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.Biemann K. Contributions of mass spectrometry to peptide and protein structure. Biomed Environ Mass Spectrom. 1988;16:99–111. doi: 10.1002/bms.1200160119. [DOI] [PubMed] [Google Scholar]
- 5.Bleasby A J, Akrigg D, Attwood T K. OWL—a non-redundant composite protein sequence database. Nucleic Acids Res. 1994;22:3574–3577. [PMC free article] [PubMed] [Google Scholar]
- 6.Brickman E, Beckwith J. Analysis of the regulation of Escherichia coli alkaline phosphatase synthesis using deletions and phi80 transducing phages. J Mol Biol. 1975;96:307–316. doi: 10.1016/0022-2836(75)90350-2. [DOI] [PubMed] [Google Scholar]
- 7.Brockus C W, Jackwood M W, Harmon B G. Characterization of beta-defensin prepropeptide mRNA from chicken and turkey bone marrow. Anim Genet. 1998;29:283–289. doi: 10.1046/j.1365-2052.1998.00338.x. [DOI] [PubMed] [Google Scholar]
- 8.Darveau R P, Blake J, Seachord C L, Cosand W L, Cunningham M D, Cassiano-Clough L, Maloney G. Peptides related to the carboxyl terminus of human platelet factor IV with antibacterial activity. J Clin Investig. 1992;90:447–455. doi: 10.1172/JCI115880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Cock H, Brandenburg K, Wiese A, Holst O, Seydel U. Non-lamellar structure and negative charges of lipopolysaccharides required for efficient folding of outer membrane protein PhoE of Escherichia coli. J Biol Chem. 1999;274:5114–5119. doi: 10.1074/jbc.274.8.5114. [DOI] [PubMed] [Google Scholar]
- 10.Diedrich D L, Stein M A, Schnaitman C A. Associations of Escherichia coli K-12 OmpF trimers with rough and smooth lipopolysaccharides. J Bacteriol. 1990;172:5307–5311. doi: 10.1128/jb.172.9.5307-5311.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ducret A, Oostven I V, Eng J K, Yates J R, Aebersold R. High throughoutput protein characterization by automated reversed-phase chromatography/electrospray tandem mass spectrometry. Protein Sci. 1998;7:706–719. doi: 10.1002/pro.5560070320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foxman B, Zhang L, Tallman P, Palin K, Rode C, Bloch C, Gillespie B, Marrs C F. Virulence characteristics of Escherichia coli causing first urinary tract infection predict risk of second infection. J Infect Dis. 1995;172:1536–1541. doi: 10.1093/infdis/172.6.1536. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Vescovi E, Soncini F C, Groisman A A. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell. 1996;84:165–174. doi: 10.1016/s0092-8674(00)81003-x. [DOI] [PubMed] [Google Scholar]
- 14.Grodberg J, Dunn J J. Comparison of Escherichia coli K-12 outer membrane protease OmpT and Salmonella typhimurium E protein. J Bacteriol. 1989;171:2903–2905. doi: 10.1128/jb.171.5.2903-2905.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grodberg J, Lundrigan M D, Toledo D L, Mangel W F, Dunn J J. Complete nucleotide sequence and deduced amino acid sequence of the ompT gene of Escherichia coli K-12. Nucleic Acids Res. 1988;16:1209. doi: 10.1093/nar/16.3.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Groisman E A, Kayser J, Soncini F C. Regulation of polymyxin resistance and adaptation to low-Mg2+ environments. J Bacteriol. 1997;179:7040–7045. doi: 10.1128/jb.179.22.7040-7045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gunn J S, Alpuche-Aranda C A, Loomis W P, Miller S I. Characterization of the Salmonella typhimurium pagC/pagD chromosomal region. J Bacteriol. 1995;177:5040–5047. doi: 10.1128/jb.177.17.5040-5047.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gunn J S, Hohmann E L, Miller S I. Transcriptional regulation of Salmonella virulence: a PhoQ periplasmic domain mutation results in increased net phosphotransfer to PhoP. J Bacteriol. 1996;178:6369–6373. doi: 10.1128/jb.178.21.6369-6373.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gunn J S, Lim K B, Krueger J, Kim K, Guo L, Hackett M, Miller S I. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol Microbiol. 1998;27:1171–1182. doi: 10.1046/j.1365-2958.1998.00757.x. [DOI] [PubMed] [Google Scholar]
- 20.Guo L, Lim K, Gunn J S, Bainbridge B, Darveau R, Hackett M, Miller S I. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science. 1997;276:250–253. doi: 10.1126/science.276.5310.250. [DOI] [PubMed] [Google Scholar]
- 21.Guo L, Lim K B, Poduje C M, Daniel M, Gunn J S, Hackett M, Miller S I. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell. 1998;95:189–198. doi: 10.1016/s0092-8674(00)81750-x. [DOI] [PubMed] [Google Scholar]
- 22.Hancock R E W, Falla T, Brown M. Cationic bactericidal peptides. In: Poole R K, editor. Advances in microbial physiology. London, U.K: Academic Press; 1995. pp. 135–175. [DOI] [PubMed] [Google Scholar]
- 23.Harwig S S, Ganz T, Lehrer R I. Neutrophil defensins: purification, characterization, and antimicrobial testing. Methods Enzymol. 1994;236:160–172. doi: 10.1016/0076-6879(94)36015-4. [DOI] [PubMed] [Google Scholar]
- 24.Hunt D F, Yates J R, 3rd, Shabanowitz J, Winston S, Hauer C R. Protein sequencing by tandem mass spectrometry. Proc Natl Acad Sci USA. 1986;83:6233–6237. doi: 10.1073/pnas.83.17.6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaufmann A, Stierhof Y D, Henning U. New outer membrane-associated protease of Escherichia coli K-12. J Bacteriol. 1994;176:359–367. doi: 10.1128/jb.176.2.359-367.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larrick J W, Hirata M, Balint R F, Lee J, Zhong J, Wright S C. Human CAP18: a novel antimicrobial lipopolysaccharide-binding protein. Infect Immun. 1995;63:1291–1297. doi: 10.1128/iai.63.4.1291-1297.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manoil C, Bailey J. A simple screen for permissive sites in proteins: analysis of Escherichia coli lac permease. J Mol Biol. 1997;267:250–263. doi: 10.1006/jmbi.1996.0881. [DOI] [PubMed] [Google Scholar]
- 28.Manoil C, Beckwith J. TnphoA: a transposon probe for protein export signals. Proc Natl Acad Sci USA. 1985;82:8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsuo E, Sampei G, Mizobuchi K, Ito K. The plasmid F OmpP protease, a homologue of OmpT, as a potential obstacle to E. coli-based protein production. FEBS Lett. 1999;461:6–8. doi: 10.1016/s0014-5793(99)01418-0. [DOI] [PubMed] [Google Scholar]
- 30.Miao E A, Scherer C A, Tsolis R M, Kingsley R A, Adams L G, u. AJ B, Miller S I. Salmonella typhimurium leucine-rich repeat proteins are targeted to the SPI1 and SPI2 type III secretion systems. Mol Microbiol. 1999;34:850–864. doi: 10.1046/j.1365-2958.1999.01651.x. [DOI] [PubMed] [Google Scholar]
- 31.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 32.Miller S I, Kukral A M, Mekalanos J J. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc Natl Acad Sci USA. 1989;86:5054–5058. doi: 10.1073/pnas.86.13.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller S I, Mekalanos J J. Constitutive expression of the PhoP regulon attenuates Salmonella virulence and survival within macrophages. J Bacteriol. 1990;172:2485–2490. doi: 10.1128/jb.172.5.2485-2490.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller S I, Pulkkinen W S, Selsted M E, Mekalanos J J. Characterization of defensin resistance phenotypes associated with mutations in phoP virulence regulon of Salmonella typhimurium. Infect Immun. 1990;58:3706–3710. doi: 10.1128/iai.58.11.3706-3710.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicolas P, Mor A. Peptides as weapons against microorganisms in the chemical defense system of vertebrates. Annu Rev Microbiol. 1995;49:277–304. doi: 10.1146/annurev.mi.49.100195.001425. [DOI] [PubMed] [Google Scholar]
- 36.Osborn M J, Gander J E, Parisi E, Carson J. Mechanism of assembly of the outer membrane of Salmonella typhimurium: isolation and characterization of cytoplasmic and outer membrane. J Biol Chem. 1972;247:3962–3972. [PubMed] [Google Scholar]
- 37.Pulkkinen W S, Miller S I. A Salmonella typhimurium virulence protein is similar to a Yersinia enterocolitica invasion protein and a bacteriophage lambda outer membrane protein. J Bacteriol. 1991;173:86–93. doi: 10.1128/jb.173.1.86-93.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qi S Y, Moir A, O'Connor C D. Proteome of Salmonella typhimurium SL1344: identification of novel abundant cell envelope proteins and assignment to a two-dimensional reference map. J Bacteriol. 1996;178:5032–5038. doi: 10.1128/jb.178.16.5032-5038.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruther U. Construction and properties of a new cloning vehicle, allowing direct screening for recombinant plasmids. Mol Gen Genet. 1980;178:475–477. doi: 10.1007/BF00270503. [DOI] [PubMed] [Google Scholar]
- 40.Sen K, Nikaido H. Lipopolysaccharide structure required for in vitro trimerization of Escherichia coli OmpF porin. J Bacteriol. 1991;173:926–928. doi: 10.1128/jb.173.2.926-928.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skorupski K, Taylor R K. Positive selection vectors for allelic exchange. Gene. 1996;169:47–52. doi: 10.1016/0378-1119(95)00793-8. [DOI] [PubMed] [Google Scholar]
- 42.Sodeinde O A, Goguen J D. Nucleotide sequence of the plasminogen activator gene of Yersinia pestis: relationship to ompT of Escherichia coli and gene E of Salmonella typhimurium. Infect Immun. 1989;57:1517–1523. doi: 10.1128/iai.57.5.1517-1523.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sodeinde O A, Subrahmanyam Y V, Stark K, Quan T, Bao Y, Goguen J D. A surface protease and the invasive character of plague. Science. 1992;258:1004–1007. doi: 10.1126/science.1439793. [DOI] [PubMed] [Google Scholar]
- 44.Soncini F C, Garcia Vescovi E, Solomon F, Groisman E A. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J Bacteriol. 1996;178:5092–5099. doi: 10.1128/jb.178.17.5092-5099.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Steinberg D A, Hurst M A, Fuji C A, Kung A H C, Ho J F, Cheng F-C, Loury D J, Fiddes J C. Protegrin-1: a broad, rapidly microbicidal peptide with in vivo activity. Antimicrob Agents Chemother. 1997;41:1739–1742. doi: 10.1128/aac.41.8.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Struyve M, Moons M, Tommassen J. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J Mol Biol. 1991;218:141–148. doi: 10.1016/0022-2836(91)90880-f. [DOI] [PubMed] [Google Scholar]
- 47.Stumpe S, Schmid R, Stephens D L, Georgiou G, Bakker E P. Identification of OmpT as the protease that hydrolyzes the antimicrobial peptide protamine before it enters growing cells of Escherichia coli. J Bacteriol. 1998;180:4002–4006. doi: 10.1128/jb.180.15.4002-4006.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sugimura K, Nishihara T. Purification, characterization, and primary structure of Escherichia coli protease VII with specificity for paired basic residues: identity of protease VII and OmpT. J Bacteriol. 1988;170:5625–5632. doi: 10.1128/jb.170.12.5625-5632.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang H, Hackett M. Ionization within a cylindrical capacitor: electrospray without an externally applied high voltage. Anal Chem. 1998;70:205–212. [Google Scholar]
- 50.Wang H, Lim K B, Lawrence R F, Howald W N, Taylor J A, Ericsson L H, Walsh K A, Hackett M. Stability enhancement for peptide analysis by electrospray using the triple quadrupole mass spectrometer. Anal Biochem. 1997;250:162–168. doi: 10.1006/abio.1997.2214. [DOI] [PubMed] [Google Scholar]
- 51.Wang R F, Kushner S R. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 52.Yates J R., 3rd Mass spectrometry and the age of the proteome. J Mass Spectrom. 1998;33:1–19. doi: 10.1002/(SICI)1096-9888(199801)33:1<1::AID-JMS624>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 53.Yates J R, 3rd, Eng J K, McCormack A L, Schieltz D. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal Chem. 1995;67:1426–1436. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
- 54.Yu G Q, Hong J S. Identification and nucleotide sequence of the activator gene of the externally induced phosphoglycerate transport system of Salmonella typhimurium. Gene. 1986;45:51–57. doi: 10.1016/0378-1119(86)90131-9. [DOI] [PubMed] [Google Scholar]
- 55.Zasloff M. Antibiotic peptides as mediators of innate immunity. Curr Opin Immunol. 1992;4:3–7. doi: 10.1016/0952-7915(92)90115-u. [DOI] [PubMed] [Google Scholar]