Abstract
The survival of most reef-building corals is dependent upon a symbiosis between the coral and the community of Symbiodiniaceae. Montipora capitata, one of the main reef-building coral species in Hawai'i, is known to host a diversity of symbionts, but it remains unclear how they change spatially and whether environmental factors drive those changes. Here, we surveyed the Symbiodiniaceae community in 600 M. capitata colonies from 30 sites across Kāne'ohe Bay and tested for host specificity and environmental gradients driving spatial patterns of algal symbiont distribution. We found that the Symbiodiniaceae community differed markedly across sites, with M. capitata in the most open-ocean (northern) site hosting few or none of the genus Durusdinium, whereas individuals at other sites had a mix of Durusdinium and Cladocopium. Our study shows that the algal symbiont community composition responds to fine-scale differences in environmental gradients; depth and temperature variability were the most significant predictor of Symbiodiniaceae community, although environmental factors measured in the study explained only about 20% of observed variation. Identifying and mapping Symbiodiniaceae community distribution at multiple scales is an important step in advancing our understanding of algal symbiont diversity, distribution and evolution and the potential responses of corals to future environmental change.
Keywords: Symbiodiniaceae, spatial pattern, coral reef
1. Introduction
Coral reefs are among the most biologically diverse and productive ecosystems on Earth and provide valuable ecosystem services as sources of tourism, coastal protection, natural products and nutrition [1–3]. The symbiotic interaction with an exceptionally diverse dinoflagellate (family Symbiodiniaceae) is inherently linked to the health and success of reef-building corals because they provide a large proportion of the coral energy requirement [4–7]. There are 11 described genera [8] of Symbiodiniaceae (and possibly many more yet to be described), each with different physiological characteristics that impact the nutrient provisioning and thermal tolerance of the coral host [9–16]. Cladocopium (previously Symbiodinium clade C) and Durusdinium (previously clade D) are the two genera most commonly hosted by corals in the Pacific [17]. Cladocopium is a generalist symbiont and also the most speciose genus [18,19] while Durusdinium is usually found in shallow corals exposed to elevated light, sea surface temperature or areas with high temperature variability [20] and is associated with increased resilience to thermal stress [9,16,19–22].
Thermal stress is the main threat affecting corals worldwide [23–27]. Sea temperatures in many tropical regions have increased by almost 1°C over the past 100 years and are currently increasing at approximately 1–2°C per century [27–30]. Temperature stress disrupts coral-dinoflagellate symbiosis, leading to algal symbiont loss and consequent paling, a phenomenon known as coral bleaching [31,32]. Mass coral bleaching events are increasing in frequency and duration, resulting in significant losses of live coral in many parts of the world [27,33–35]. Coral susceptibility to heat stress and bleaching is dependent on a wide range of factors, including the algal symbiont community they host [9,22,24,32,36]. Bleaching may also represent an opportunity for corals to rapidly change their current algal symbiont community composition to more resilient types [37,38]. However, this Adaptive Bleaching Hypothesis remains controversial because many coral taxa are algal symbiotic specialists (but see [39]), hosting a single algal symbiont taxon and although corals can sometimes change their Symbiodiniaceae symbionts, many also recover to the same algal symbiont community they had prior to bleaching [40–44], although a few studies have reported that corals were able to maintain the symbionts even after 2 years [45]. Furthermore, hosting the stress-tolerant Durusdinium often comes at an energetic cost, as it decreases the growth and metabolite exchange rate of the host [46–52].
Symbiodiniaceae assemblage structure in corals tends to be shaped by many factors, including the host species [18,53], large-scale factors like geography [54,55], and local scale factors like depth [36,56], habitat [21], and environmental factors such as light [57] and temperature [21,55]. Here, we investigated the local-scale environmental drivers of Symbiodiniaceae assemblage structure in a common reef-building coral Montipora capitata, which is one of the most abundant corals in Kāne‘ohe Bay and may harbour a community of Cladocopium (C), Durusdinium (D) or both symbiont genera [58–60]. Previous studies of the species in Hawai‘i have reported associations with C31 [58,61,62], C17, C21 [61,62] and Durusdinium glynnii (formerly, Symbiodinium glynii [ITS2 Type D161,62] and D 4-6 [58]).
Here, we used high-throughput sequencing of the internal transcribed spacer region (ITS2) to identify the Symbiodiniaceae assemblage and the local-scale environmental drivers of symbiont community composition for 600 colonies of the common reef-building coral Montipora capitata collected from across 30 sites in Kāne‘ohe Bay. This fine-scale sampling of symbionts from corals across Kāne‘ohe Bay included the more environmentally extreme northern and southern regions not sampled by previous studies. Our comprehensive sampling provides a baseline for symbiont communities across the full environmental gradient of the bay against which future consequences of coral bleaching can be compared and allows us to investigate the role of the environmental gradient in shaping the symbiont community of Montipora capitata.
2. Material and methods
2.1. Site selection and tagging
Details of the stratified random sampling design and environmental data are detailed by Caruso et al. [63]. Briefly, the bay was divided into five hydrodynamically defined regions (blocks) from South to North based on the water flow regimes and water residence time [64,65] and six sites were selected within each block following a stratified random sampling design [63]. Within each of these six sites, 20 visually healthy M. capitata were tagged, for a total of 600 tagged colonies distributed across 30 sites spread equally across the five hydrodynamically defined blocks. In early 2018, about 1 cm2 was sampled from each of 600 tagged Montipora capitata colonies. Sampled fragments were immediately preserved in 70% ethanol and stored at −20°C until processed. DNA was extracted using the Nucleospin Tissue Kits (Macherey-Nagel, Düren, Germany) following manufacturer instructions and quantified by fluorimetry (Quant-it HS dsDNA kit, Thermo-Fisher). Only corals that appeared healthy were sampled for this study.
Data loggers (Hobo Pendant or Water Temp Pro v. 2 loggers, Onset Computer Corp., Bourne, MA) recorded temperature and sediment traps were collected regularly from each site [63] and used to calculate variation in temperature and sedimentation rate (standard deviation, maximum, minimum and range for each). Degree heating weeks (DHW) per site was calculated as the number of weeks when temperature exceeded the bleaching threshold of 28.5°C (MMM +1°C; [59,66]) (figure 1).
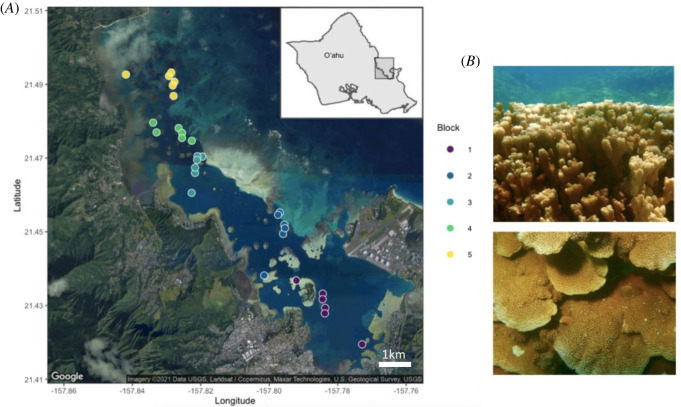
Figure 1.
(A) Sites and blocks in Kāne‘ohe Bay. Each point is a randomly selected site within blocks represented by colours (Site IDs in electronic supplementary material, table S1). Blocks go from the south bay (block 1) to north bay (block 5). (B) Montipora capitata, the rice coral. The figures illustrate the morphological plasticity of this species, which can be branching (top) or plating (lower picture). Site IDs consist of the digit corresponding to the block in which the site is contained, followed by the site number (e.g. 1_10, with six sites per block, but not necessarily in consecutive order). Map done in the R package ggmap [67].
2.2. Symbiodiniaceae ITS2 amplicon sequencing library preparation
Symbiodiniaceae amplicon library preparation and sequencing followed the protocol outlined in [68]. Briefly, the ITS2 region of Symbiodiniaceae ribosomal DNA was targeted for sequencing using Symbiodiniaceae primers 454-ITSinfor2 (5′-GAATTGCAGAACTCCGTG-3′), ITSD (5′-GTGAATTGCAGAACTCCGTG-3′) and 454-ITS2-reverse (5′-GGGATCCATATGCTTAAGTTCAGCGGGT-3′) and its2rev2 (5′-CCTCCGCTTACTTATATGCTT-3′) modified from Arif et al. [69] to include Nextera indexes to allow multiplexing (see electronic supplementary material). Library products were sequenced on the Illumina MiSeq platform (v. 3 2 × 300 bp PE). Each library plate included a negative control (wells to which no template DNA was added). In addition, to ensure repeatability and test for the effect of sequencing run on the detection of symbionts, 192 samples (two 96 well plates) were sequenced more than one time in different runs. We found no differences in either community composition or symbiont identity that would alter our results or interpretation, and so the replicated control samples were simply pooled and analysed as a single sample for that individual.
2.3. Amplicon sequencing analysis with symportal
Raw sequences were first demultiplexed using Cutadapt [70]. Demultiplexed forward and reverse reads were submitted to SymPortal [71], a platform for genetically identifying Symbiodiniaceae using high throughput ITS2 sequence data that differentiates intra- and intergenomic sources of ITS2 sequence variance (electronic supplementary material, figure S1).
Many terms have been used to describe the Symbiodiniaceae unity of resolution. To avoid ambiguity, we restrict our use to ‘Symbiodiniaceae type’ and ‘Symbiodiniaceae profile’. A type refers to Symbiodiniaceae taxa that have a specific sequence as their most abundant sequence. A Symbiodiniaceae profile is a set of ITS2 sequences that have been found in a sufficient number of independent samples to be identified as a ‘defining intragenomic variant’ (DIV). For example, C17 is a Symbiodiniaceae type and C17d/C31-C21-C17e-C21ac-C17f-C17 g is a Symbiodiniaceae profile with C17d present in higher abundance than the other types.
2.4. Statistical analysis
All analyses were done in the R statistical environment [72]. Bray-Curtis dissimilarity of relative abundance of the Symbiodiniaceae community composition was tested by permutational multivariate analysis of variance (PERMANOVA) in the function adonis (for effect of block, with site nested within block), and pairwise.adonis (for pairwise PERMANOVA) in the vegan package [73], each with 999 permutations. To better visualize the similarity among blocks, R2 from the PERMANOVA was plotted in a dendrogram using the function pheatmap in the package gplots [74]. The function metaMDS was used in the R package vegan [73] to generate non-metric multidimensional scaling visualizations using Bray-Curtis dissimilarities of algal symbiont community per block.
To investigate the effect of the environmental data in driving the Symbiodiniaceae community, we performed distance-based redundancy analysis (dbRDA) using the function capscale. After running an ANOVA to check the significance of the constrains axis as well as the significance of the environmental variables (electronic supplementary material, table S1), we visualized the significant variables in the dbRDA. Samples were considered to have majority Cladocopium (C) or Durusdinium (D) if the proportion of either algal symbiont exceeded 80% in the sample (modified from [60]). All remaining samples with Cladocopium and Durusdinium were designated as CD, corresponding to corals with neither algal symbiont genus achieving greater than 80% dominance. Correlation among the environmental factors was calculated using the function cor.test in R. Data and R code to execute and reproduce all the analyses and figures presented in the manuscript are archived at Zenodo [75].
3. Results
3.1. Symbiodiniaceae ITS2 sequences and ITS2 type profiles
550 Montipora capitata samples returned high-quality reads. Prior to quality filtering, these samples included 3 285 481 sequences which were reduced to 1 632 505 following quality control and minimum entropy decomposition with SymPortal, an average of 2968 sequences per sample. 11 386 sequences had not been previously curated by SymPortal and were unique to this dataset. A total of 283 Symbiodiniacae types were identified, 85% belonging to the genus Cladocopium, and 15% belonging to the genus Durusdinium. Twenty-six ITS2 type profiles were identified across all samples, 23 of which were from the genus Cladocopium, with the remaining three belonging to the genus Durusdinium. Overall, 43% of Montipora capitata hosted Cladocopium only, 11% hosted Durusdinium only, and 46% hosted a combination of both genera. From those mixed colonies, 32.5% were dominated by Cladocopium (C greater than 80%), while 37% of colonies were dominated by Durusdinium (D > 80%). To confirm differences in read number do not change the results or interpretation, we excluded 20 samples whose number of reads were more than two standard deviations above or below the mean.
3.2. Biogeography of symbiodiniaceae in Kāne‘ohe Bay
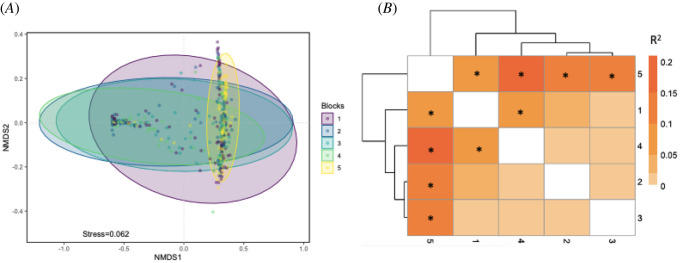
Symbiodiniaceae composition varied significantly among corals within each of the environmentally delineated blocks (PERMANOVA, F25 = 6.5632, p = 0.001; electronic supplementary material, table S2). Pairwise comparisons revealed that blocks 1 and 5 were significantly different, whereas blocks 2–4 were rarely significantly different from one another (electronic supplementary material, table S2, figure 2a,b).
Figure 2.
Similarity of Symbiodiniaceae communities detected in 600 Montipora capitata colonies collected across five hydrodynamically defined blocks in Kāne‘ohe Bay (block 1 is furthest south, while block 5 is furthest north in the bay). (A) nMDS of Symbiodiniaceae per block. Each point represents the symbiont community in a colony. Ellipses are 95% confidence intervals. (B) Dendrogram and heat map of R2 of the PERMANOVA of Symbiodiniaceae per block highlighting the similarity among blocks in the center of the bay that have been sampled in previous studies relative to block 1 and 5.
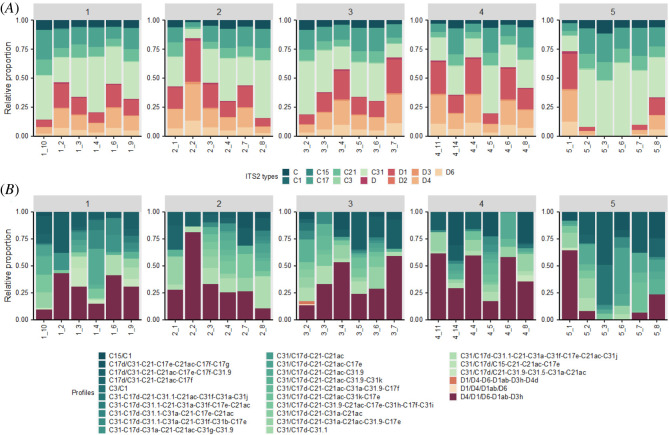
Most sites had corals with both Cladocopium and Durusdinium present, except two sites in block 5 (sites 5_3 and 5_6) which did not have any samples in which Durusdinium was detected (figure 3a,b; electronic supplementary material, figure S1). It is noteworthy that sites 5_3 and 5_6 were also the least diverse of all we sampled, and unlike other sites, some colonies hosted only a single type (C3) of algal symbiont (electronic supplementary material, figure S1). Corals within block 5 hosted significantly less Durusdinium symbionts than any other site, with all sites including corals hosting a majority of Cladocopium. Site 2_2 had the highest relative proportion of Durusdinium and site 3_2 was the only site to have three profiles of Durusdinium.
Figure 3.
(A) Major Symbiodiniaceae types by site and block. (B) Symbiodiniaceae profiles by site. Block 1 is the southernmost in Kāne‘ohe Bay, while block 5 is the most northern and similar to offshore conditions. Symbiodiniaceae ITS2 subtypes were summarized to the major subtype to facilitate visualization in the bar charts (i.e. C31a and C31b were summarized as C31). Due to the wide diversity of ITS2 available in the SymPortal database, not all sequences are given names. Only those sequences that are used in the definition of ITS2 type profiles (i.e. DIVs) are named. Unnamed Cladocopium and Durusdinium sequences were combined for visualization and represented as summed ‘C’ and ‘D’ types, respectively.
3.3. Environmental drivers of symbiodiniaceae community composition
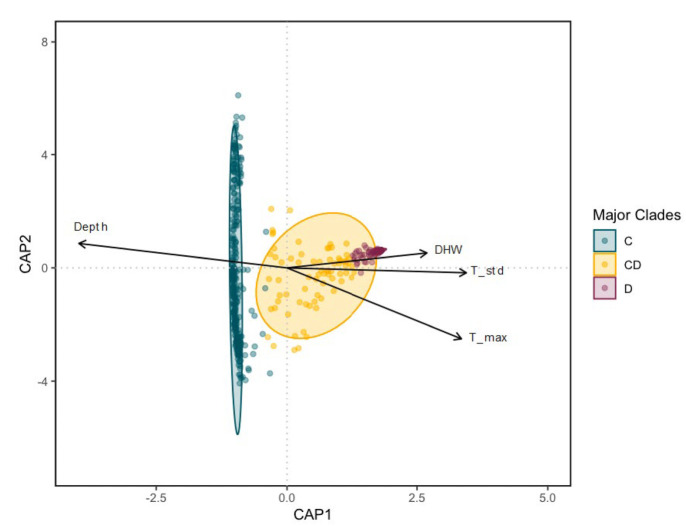
Environmental data from each site is provided in Supplemental materials (electronic supplementary material, tables S1 & S3). dbRDA showed that M. capitata hosting majority Cladocopium are distinct from M. capitata hosting majority Durusdinium or mixed C & D (figure 4). PERMANOVA of the environmental factors in the dbRDA (electronic supplementary material, table S4) showed that depth, DHW, maximum mean daily temperature recorded at the site and mean daily standard deviation were all significant drivers of Symbiodiniaceae community composition. The environmental factors in the study explained only 20% of the Symbiodiniaceae variation, with depth having the greatest relative contribution (61%) followed by daily temperature standard deviation (19%), maximum temperature (9.9%) and DHW (4.8%) (electronic supplementary material, table S4).
Figure 4.
Distance based redundancy analysis (dbRDA) for environmental drivers of the Symbiodiniaceae communities in Montipora capitata sampled throughout Kāne‘ohe Bay. Each point represents an individual colony sampled irrespective of site. Samples were considered as majority Cladocopium (C) if they contain greater than 80%C, and majority Durusdinium (D) if greater than 80% D. Only vectors for the environmental factors contributing significantly to the algal symbiont diversity are plotted. Each arrow signifies the multiple partial correlation of the environmental driver in the RDA whose length and direction can be interpreted as indicative of its contribution to the proportion of variation explained.
4. Discussion
Here, we contribute to a more nuanced understanding of coral algal symbiosis by examining the associations of environmental drivers across a small spatial scale (approx. 10 km) on the community composition of Symbiodiniaceae in one of the dominant reef-building corals in the region, Montipora capitata. Previous surveys of the Symbiodiniaceae community structure of M. capitata across Kāne‘ohe Bay [60,61] found algal symbiont structure differed at the level of site and colony, but did not find significant differences in community composition of symbionts among regions of the bay. Here we report significant differences in symbiont community composition across the environmental gradient of Kāne‘ohe Bay that is not seen in these previous surveys. This difference may result from the increased resolution possible with high-throughput methods in our study. Stat et al. [61] used cloning and Sanger sequencing of ITS2 to identify algal symbionts, with fewer total colonies sampled (52), each with far less resolution (5–7 clones per colony) and in a smaller portion of the bay (Blocks 1 and 2) than is possible with the amplicon approach using high-throughput sequencing employed here. Innis et al. [60]) sampled a large number of colonies (707) using a qPCR assay targeting Cladocopium and Durusdinium but did not have the resolution to identify the diversity of subtypes of Cladocopium and Durusdinium as in this study. However, it is not the additional resolution so much as the geographic range of sampling that appears to underlie the difference, because Innis et al. [60] had no samples from either the far north or far south of the bay (blocks 1 & 5 in our study). Innis et al. [60] concluded that Symbiodiniaceae community variability may arise from either holobiont phenotypic plasticity or differential survival of Symbiodiniaceae across light gradients and recommended additional study into whether algal symbiont communities were stable or plastic within individuals across these gradients. Our findings are concordant with both Stat et al. [61] and Innes et al. [76] in that there is little differentiation among symbiont communities across the central portion of Kāne‘ohe Bay (blocks 2–4), but without blocks 1 and 5, in which we found the greatest difference in symbiont composition, they did not sample the ends of the environmental gradient over which the community composition is observed to shift significantly in our study.
Using a high-throughput metabarcoding approach, our study shows that the Symbiodiniaceae community composition varied at all spatial scales examined, both among the sites within each region and among hydrodynamically defined blocks across the bay. Interestingly, the relative proportion of algal symbiont types is relatively consistent among the colonies (i.e. when Cladocopium is present, C31 is typically most abundant followed by C17 and C21, whereas for Durusdinium, D1 is the most abundant, followed by D4 and D6), no matter the location (figure 3; electronic supplementary material, figure S1). In total, seven types of Cladocopium (C1, C3, C15, C17, C21, C31) are detected among colonies across Kāne‘ohe Bay, with C31, C17 and C1 being the most common. Similarly, we detected six types of Durusdinium (D, D1, D2, D3, D4, D6) with D1, D4, and D6 being the most prevalent. These results are consistent with multiple studies reporting strong Symbiodiniaceae specificity with type C31 being the most common symbiont associated with Montipora capitata in Hawai‘i [58,61,62], but the distinctly structured communities found in the northern and southern extremes of the bay were undetected by previous studies.
Despite considerable variation in the composition of the communities of Symbiodiniaceae hosted by Montipora capitata sampled across Kāne‘ohe Bay, we find that the coral–algal symbiont association was strongly influenced by environmental gradients. M. capitata located at the environmental extremes (block 1 in the south and block 5 in the north) hosted Symbiodiniaceae communities that were significantly different from blocks in the centre of the bay. We find four factors (depth, DHW, maximum and variability in temperature) that are significant drivers of Symbiodiniaceae community composition (electronic supplementary material, table S4), although these four significant factors combined explained less than 20% of the variation in symbiont community composition. Although 80% of the variation remains unexplained, meta-analyses show that environment generally explains about 20% of the variation in community structure across terrestrial, freshwater and marine ecosystems [77]. Other factors such as light [57], nutrients [78,79] and pH [80] were unmeasured here, but have also been found to influence the symbiont community in other systems. It is important to note that in our study, we cannot determine if these correlated environmental factors are the direct drivers of algal symbiont diversity. Until future studies determine the underlying mechanisms driving variation in algal symbiont community composition, we remain cautious in our interpretation of these environmental drivers.
4.1. Depth
Consistent with previous studies [18,57,60,81,82], depth appears to be the strongest environmental driver of symbiont community composition measured in this study. The deepest block (5) hosted the lowest proportion of Durusdinium, with most colonies hosting only Cladocopium (figure 3; electronic supplementary material, figure S1). A similar pattern is found when looking at individual sites surveyed across all blocks, with the deepest sites (1_3, 1_4, 1_9, 1_10, 2_4 and 2_8) being mostly dominated by Cladocopium. While Innis et al. [60] also found depth to be the primary driver of algal symbiont clades in Kāne‘ohe Bay they concluded the mechanism was most likely light attenuation in deeper reefs as opposed to depth per se. It is well known that light attenuates with depth in water, but the relationship is complicated in coral reef environments where a variety of other factors can alter penetration of light to deeper corals [83–85]. Likewise, an entire suite of environmental parameters other than light also covary with depth, such that it is often difficult to know exactly which environmental or biological factors drive changes in community structure with depth [86–89]. Depth is simple to measure and is well-correlated with changes in coral reef community structure in many studies (reviewed by [86,90,91]) but is much harder to isolate as an environmental factor to determine the quantitative relative contribution in studies such as this (electronic supplementary material, table S5).
4.2. Sedimentation
Suspended sediments can both impact corals directly and alter their light penetration and irradiance. Sedimentation can also impact corals negatively through increased nutrient input, damaging the coral surface and by making it harder for corals to feed and photosynthesize [92–94]. Sediment particles in the water increase turbidity, which attenuates light similarly as does depth. In our study, however, none of the sediment parameters were significant predictors of symbiont community composition (electronic supplementary material, table S4). The lack of significance may result from the complex interaction of turbidity and depth on light because deeper low sediment reefs (like most sites in block 5) may experience higher irradiance than shallower sites with higher turbidity.
4.3. Temperature
The role of temperature in mediating the symbiosis between the coral host and their algal endosymbionts has been widely studied [31,32,95,96], so it is not surprising that several aspects of temperature come out as significant drivers of community composition. Degree heating weeks (DHW) and maximum temperature are widely established to be major predictors of the breakdown of symbiosis between the partners and result in coral bleaching [27,97]. Unsurprisingly, both were significant in our analysis of environmental drivers, although neither explained much of the variation in algal symbiont community structure among sites. By contrast, variability in temperature, in this case, mean daily temperature standard deviation, had the second largest contribution to the observed variation in algal symbiont community structure. A number of studies highlight the importance of daily temperature fluctuations for coral acclimatization to higher temperatures [98,99], suggesting that temperature fluctuations encourage greater thermal tolerance by exposing the corals to short periods of thermal stress without causing mortality. Blocks in the centre of the bay had higher temperature variation (higher mean average temperature standard deviation and higher mean daily range) and our data shows that the existing community structure of algal symbionts responds to such variability (figure 3). Blocks in the centre of the bay (blocks 2–4) had higher proportions of Durusdinium, while block 1 and block 5 (extreme north and south, respectively) had the lowest proportion, suggesting that blocks at the extreme ends of the bay may be more vulnerable to bleaching events. This prediction is consistent with coral surveys during the 2015 bleaching event in Kāne‘ohe Bay which found the highest levels of bleaching and paling in the north (70%) and south (60%) of the bay [100].
Here we advance previous work by showing that algal symbiont communities within a single species of coral in a single embayment are finely tuned to their environmental conditions. Whether community response results from selection for Symbiodiniaceae types living under different environments, adaptive shuffling of Symbiodiniaceae communities in response to environmental conditions, or both, remains to be determined. Comparing microscale environmental variability [101,102] to algal symbiont community structure might explain much of the variability we see, because algal symbionts in colonies may be adaptively responding to fine-scale variability at the same site within the broad regional differences we compared here. A more detailed understanding of the relationship between adaptive tuning of algal symbiont communities to local environmental conditions will require fine-scale environmental measurements coupled with long-term monitoring of corals in the field to determine whether and how algal symbiont communities within individual colonies respond through time.
5. Conclusion
Fine-scale sampling of 600 M. capitata colonies across a relatively small spatial gradient (approx. 10 km) within Kāne‘ohe Bay showed that algal symbiont community structure is associated with depth and temperature. This fine-scale variation in algal symbiont community composition across local environmental gradients suggests that algal symbiont communities can adaptively match the environmental conditions surrounding the holobiont. Previous studies of Symbiodiniaceae in the Bay focused on the three central regions that exhibit the least environmental variability among sites across the environmental gradient from north to south. Here we extend the sampling to include the full gradient across the bay and overturned previous conclusions that algal symbiont structure did not differ significantly among regions of the bay. Our results support that conclusion for symbiont communities in the central portion of the bay, but show that both environmental extremes of the far northern and southern regions of Kāne‘ohe Bay sampled here for the first time differ significantly from those in the central region. These results imply that the community composition responds to the conditions under which the holobiont is living, setting the stage for understanding the role of environmental conditions in how Symbiodiniaceae communities are distributed in space and time.
Our study highlights the complex interactions among environmental factors and algal symbiont diversity in the reef-building coral Montipora capitata. While depth was the main factor driving algal symbiont community composition in our study, several aspects of temperature (DHW, max temp & mean standard deviation) likewise appear to be drivers of algal symbiont community composition. We also note that many factors correlate with depth, such as light, temperature, sedimentation rate and water flow, such that fine-scale measurements of the full range of environmental factors surrounding individual colonies through time will be needed to pinpoint the most important environmental drivers of algal symbiont community structure. Regardless of the ultimate suite of parameters that drive algal symbiont community structure in corals, our study shows that Symbiodiniaceae communities are attuned to fine-scale environmental gradients and that understanding these complex interactions across the heterogeneous mosaic of coral reef environments is needed to better predict spatial patterns in biological responses such as bleaching susceptibility.
Acknowledgements
All members of the Coral Resilience Lab (the legacy of Ruth Gates) and ToBo Lab who assisted with collections, molecular work, or bioinformatics (special thanks to Kira Hughes, Joshua Hancock, Janaya Bruce, Dennis Conneta, Caroline Hobbs, Valerie Kahkejian and Christian Marin). This is SOEST contribution number 11553 and HIMB contribution number 1899.
Ethics
Montipora capitata colonies were tagged and collected under Special Activity Permit 2018-03 to HIMB from Hawaii Department of Aquatic Resources.
Data accessibility
All materials, code and data are available for download on Zenodo (75; https://zenodo.org/record/5670832#.YvBKpC1h2iM).
The data are provided in electronic supplementary material [103].
Authors' contributions
M.R.S.: conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, project administration, supervision, validation, visualization, writing—original draft, writing—review and editing; C.C.: conceptualization, data curation, methodology, writing—review and editing; L.R.: conceptualization, data curation, methodology, writing—review and editing; C.D.: data curation, funding acquisition, resources, validation, visualization, writing—review and editing; R.G.: conceptualization, funding acquisition, resources; R.T.: funding acquisition, resources, supervision, writing—original draft, writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Funding
This work was funded by a Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brasil (CAPES) fellowship, the National Science Foundation (OA-1416889 & BioOCE-1924604 to R.J.T.) and the Paul G. Allen Family Foundation.
References
- 1.Hicks CC, Graham NAJ, Cinner JE. 2013. Synergies and tradeoffs in how managers, scientists, and fishers value coral reef ecosystem services. Glob. Environ. Change 23, 1444-1453. ( 10.1016/j.gloenvcha.2013.07.028) [DOI] [Google Scholar]
- 2.Moberg F, Folke C. 1999. Ecological goods and services of coral reef ecosystems. Ecol. Econ. 29, 215-233. ( 10.1016/S0921-8009(99)00009-9) [DOI] [Google Scholar]
- 3.Woodhead AJ, Hicks CC, Norström AV, Williams GJ, Graham NAJ. 2019. Coral reef ecosystem services in the Anthropocene. Funct. Ecol. 33, 1023-1034. ( 10.1111/1365-2435.13331) [DOI] [Google Scholar]
- 4.Cantin NE, van Oppen MJH, Willis BL, Mieog JC, Negri AP. 2009. Juvenile corals can acquire more carbon from high-performance algal symbionts. Coral Reefs 28, 405-414. ( 10.1007/s00338-009-0478-8) [DOI] [Google Scholar]
- 5.Cunning R, Muller EB, Gates RD, Nisbet RM. 2017. A dynamic bioenergetic model for coral- Symbiodinium symbioses and coral bleaching as an alternate stable state. J. Theor. Biol. 431, 49-62. ( 10.1016/j.jtbi.2017.08.003) [DOI] [PubMed] [Google Scholar]
- 6.Davy SK, Allemand D, Weis VM. 2012. Cell biology of Cnidarian-Dinoflagellate symbiosis. Microbiol. Mol. Biol. Rev. 76, 229-261. ( 10.1128/MMBR.05014-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muscatine L, Porter JW. 1977. Reef corals: mutualistic symbioses adapted to nutrient-poor environments. BioScience 27, 454-460. ( 10.2307/1297526) [DOI] [Google Scholar]
- 8.Yorifuji M, et al. 2021. Unique environmental Symbiodiniaceae diversity at an isolated island in the northwestern Pacific. Mol. Phylogenet. Evol. 161, 107158. ( 10.1016/j.ympev.2021.107158) [DOI] [PubMed] [Google Scholar]
- 9.Berkelmans R, van Oppen MJH. 2006. The role of zooxanthellae in the thermal tolerance of corals: a ‘nugget of hope’ for coral reefs in an era of climate change. Proc. R. Soc. B 273, 2305-2312. ( 10.1098/rspb.2006.3567) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grégoire V, Schmacka F, Coffroth MA, Karsten U. 2017. Photophysiological and thermal tolerance of various genotypes of the coral endosymbiont Symbiodinium sp. (Dinophyceae). J. Appl. Phycol. 29, 1893-1905. ( 10.1007/s10811-017-1127-1) [DOI] [Google Scholar]
- 11.Hoadley KD, et al. 2015. Physiological response to elevated temperature and pCO2 varies across four Pacific coral species: understanding the unique host+symbiont response. Sci. Rep. 5, 18371. ( 10.1038/srep18371) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howells EJ, Beltran VH, Larsen NW, Bay LK, Willis BL, van Oppen MJH. 2012. Coral thermal tolerance shaped by local adaptation of photosymbionts. Nat. Clim. Change 2, 116-120. ( 10.1038/nclimate1330) [DOI] [Google Scholar]
- 13.McIlroy SE, Gillette P, Cunning R, Klueter A, Capo T, Baker AC, Coffroth MA. 2016. The effects of Symbiodinium (Pyrrhophyta) identity on growth, survivorship, and thermal tolerance of newly settled coral recruits. J. Phycol. 52, 1114-1124. ( 10.1111/jpy.12471) [DOI] [PubMed] [Google Scholar]
- 14.Quigley KM, Baker AC, Coffroth MA, Willis BL, van Oppen MJH. 2018. Bleaching resistance and the role of algal endosymbionts. In Coral bleaching (eds van Oppen MJH, Lough JM), pp. 111-151. Cham, Switzerland: Springer International Publishing. ( 10.1007/978-3-319-75393-5_6) [DOI] [Google Scholar]
- 15.Robison JD, Warner ME. 2006. Differential impacts of photoacclimation and thermal stress on the photobiology of four different phylotypes of Symbiodinium (Pyrrhophyta). J. Phycol. 42, 568-579. ( 10.1111/j.1529-8817.2006.00232.x) [DOI] [Google Scholar]
- 16.Stat M, Gates RD. 2011. Clade D Symbiodinium in Scleractinian corals: a ‘nugget’ of hope, a selfish opportunist, an ominous sign, or all of the above? J. Mar. Biol. 2011, 1-9. ( 10.1155/2011/730715) [DOI] [Google Scholar]
- 17.LaJeunesse TC. 2005. ‘Species’ radiations of symbiotic Dinoflagellates in the Atlantic and Indo-Pacific since the Miocene-Pliocene transition. Mol. Biol. Evol. 22, 570-581. ( 10.1093/molbev/msi042) [DOI] [PubMed] [Google Scholar]
- 18.Baker AC. 2003. Flexibility and specificity in coral-algal symbiosis: diversity, ecology, and biogeography of Symbiodinium. Ann. Rev. Ecol. Evol. Syst. 34, 661-689. ( 10.1146/annurev.ecolsys.34.011802.132417) [DOI] [Google Scholar]
- 19.LaJeunesse TC, Parkinson JE, Gabrielson PW, Jeong HJ, Reimer JD, Voolstra CR, Santos SR. 2018. Systematic revision of Symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr. Biol. 28, 2570-2580.e6. ( 10.1016/j.cub.2018.07.008) [DOI] [PubMed] [Google Scholar]
- 20.Oliver TA, Palumbi SR. 2011. Do fluctuating temperature environments elevate coral thermal tolerance? Coral Reefs 30, 429-440. ( 10.1007/s00338-011-0721-y) [DOI] [Google Scholar]
- 21.LaJeunesse TC, Pettay DT, Sampayo EM, Phongsuwan N, Brown B, Obura DO, Hoegh-Guldberg O, Fitt WK. 2010. Long-standing environmental conditions, geographic isolation and host-symbiont specificity influence the relative ecological dominance and genetic diversification of coral endosymbionts in the genus Symbiodinium. J. Biogeogr. 37, 785-800. ( 10.1111/j.1365-2699.2010.02273.x) [DOI] [Google Scholar]
- 22.Rowan R. 2004. Thermal adaptation in reef coral symbionts. Nature 430, 742-742. ( 10.1038/430742a) [DOI] [PubMed] [Google Scholar]
- 23.Heron SF, Maynard JA, van Hooidonk R, Eakin CM. 2016. Warming trends and bleaching stress of the world's coral reefs 1985–2012. Sci. Rep. 6, 38402. ( 10.1038/srep38402) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoegh-Guldberg O, et al. 2007. Coral reefs under rapid climate change and ocean acidification. Science 318, 1737-1742. ( 10.1126/science.1152509) [DOI] [PubMed] [Google Scholar]
- 25.Hughes TP. 2003. Climate change, human impacts, and the resilience of coral reefs. Science 301, 929-933. ( 10.1126/science.1085046) [DOI] [PubMed] [Google Scholar]
- 26.Hughes TP, et al. 2017. Global warming and recurrent mass bleaching of corals. Nature 543, 373-377. ( 10.1038/nature21707) [DOI] [PubMed] [Google Scholar]
- 27.Sully S, Burkepile DE, Donovan MK, Hodgson G, van Woesik R. 2019. A global analysis of coral bleaching over the past two decades. Nat. Commun. 10, 1264. ( 10.1038/s41467-019-09238-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lough JM, Anderson KD, Hughes TP. 2018. Increasing thermal stress for tropical coral reefs: 1871–2017. Sci. Rep. 8, 6079. ( 10.1038/s41598-018-24530-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peñaflor EL, Skirving WJ, Strong AE, Heron SF, David LT. 2009. Sea-surface temperature and thermal stress in the Coral Triangle over the past two decades. Coral Reefs 28, 841-850. ( 10.1007/s00338-009-0522-8) [DOI] [Google Scholar]
- 30.IPCC. 2021 [Google Scholar]
- 31.Brown BE. 1997. Coral bleaching: causes and consequences. Coral Reefs 16, S129-S138. ( 10.1007/s003380050249) [DOI] [Google Scholar]
- 32.Douglas AE. 2003. Coral bleaching––how and why? Mar. Pollut. Bull. 46, 385-392. ( 10.1016/S0025-326X(03)00037-7) [DOI] [PubMed] [Google Scholar]
- 33.Donner SD, Skirving WJ, Little CM, Oppenheimer M, Hoegh-Guldberg O. 2005. Global assessment of coral bleaching and required rates of adaptation under climate change. Glob. Change Biol. 11, 2251-2265. ( 10.1111/j.1365-2486.2005.01073.x) [DOI] [PubMed] [Google Scholar]
- 34.Hoegh-Guldberg O. 1999. Climate change, coral bleaching and the future of the world's coral reefs. Mar. Freshw. Res. 50, 839-866. ( 10.1071/MF99078) [DOI] [Google Scholar]
- 35.Oliver JK, Berkelmans R, Eakin CM. 2018. Coral bleaching in space and time. In Coral bleaching: patterns, processes, causes and consequences (eds van Oppen MJH, Lough JM), pp. 27-49. Cham: Springer International Publishing. ( 10.1007/978-3-319-75393-5_3) [DOI] [Google Scholar]
- 36.Warner ME, LaJeunesse TC, Robison JD, Thur RM. 2006. The ecological distribution and comparative photobiology of symbiotic dinoflagellates from reef corals in Belize: Potential implications for coral bleaching. Limnol. Oceanogr. 51, 1887-1897. ( 10.4319/lo.2006.51.4.1887) [DOI] [Google Scholar]
- 37.Buddemeier RW, Fautin DG. 1993. Coral bleaching as an adaptive mechanism. BioScience 43, 320-326. ( 10.2307/1312064) [DOI] [Google Scholar]
- 38.Fautin DG, Buddemeier RW. 2004. Adaptive bleaching: a general phenomenon. Hydrobiologia 530/531, 459-467. ( 10.1007/s10750-004-2642-z) [DOI] [Google Scholar]
- 39.Mies M, Francini-Filho RB, Zilberberg C, Garrido AG, Longo GO, Laurentino E, Güth AZ, Sumida PY, Banha TN. 2020. South Atlantic coral reefs are major global warming refugia and less susceptible to bleaching. Front. Mar. Sci. 7, 514. ( 10.3389/fmars.2020.00514) [DOI] [Google Scholar]
- 40.Baird A, Cumbo V, Leggat W, Rodriguez-Lanetty M. 2007. Fidelity and flexibility in coral symbioses. Mar. Ecol. Prog. Ser. 347, 307-309. ( 10.3354/meps07220) [DOI] [Google Scholar]
- 41.Goulet T. 2006. Most corals may not change their symbionts. Mar. Ecol. Prog. Ser. 321, 1-7. ( 10.3354/meps321001) [DOI] [Google Scholar]
- 42.Goulet T. 2007. Most scleractinian corals and octocorals host a single symbiotic zooxanthella clade. Mar. Ecol. Prog. Ser. 335, 243-248. ( 10.3354/meps335243) [DOI] [Google Scholar]
- 43.Goulet T, Coffroth M. 2003. Stability of an octocoral-algal symbiosis over time and space. Mar. Ecol. Prog. Ser. 250, 117-124. ( 10.3354/meps250117) [DOI] [Google Scholar]
- 44.Hoegh-Guldberg O, Jones RJ, Ward S, Loh WK. 2002. Is coral bleaching really adaptive? Nature 415, 601-602. ( 10.1038/415601a) [DOI] [PubMed] [Google Scholar]
- 45.Grottoli AG, Warner ME, Levas SJ, Aschaffenburg MD, Schoepf V, McGinley M, Baumann J, Matsui Y. 2014. The cumulative impact of annual coral bleaching can turn some coral species winners into losers. Glob. Change Biol. 20, 3823-3833. ( 10.1111/gcb.12658) [DOI] [PubMed] [Google Scholar]
- 46.Baker DM, Andras JP, Jordán-Garza AG, Fogel ML. 2013. Nitrate competition in a coral symbiosis varies with temperature among Symbiodinium clades. ISME J. 7, 1248-1251. ( 10.1038/ismej.2013.12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cunning R, Silverstein RN, Baker AC. 2018. Symbiont shuffling linked to differential photochemical dynamics of Symbiodinium in three Caribbean reef corals. Coral Reefs 37, 145-152. ( 10.1007/s00338-017-1640-3) [DOI] [Google Scholar]
- 48.Jones A, Berkelmans R. 2010. Potential costs of acclimatization to a warmer climate: growth of a reef coral with heat tolerant vs. sensitive symbiont types. PLoS ONE 5, e10437. ( 10.1371/journal.pone.0010437) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jones AM, Berkelmans R. 2011. Tradeoffs to thermal acclimation: energetics and reproduction of a reef coral with heat tolerant Symbiodinium type-D. J. Mar. Biol. 2011, 1-12. ( 10.1155/2011/185890) [DOI] [Google Scholar]
- 50.Matthews JL, Oakley CA, Lutz A, Hillyer KE, Roessner U, Grossman AR, Weis VM, Davy SK. 2018. Partner switching and metabolic flux in a model cnidarian–dinoflagellate symbiosis. Proc. R. Soc. B 285, 20182336. ( 10.1098/rspb.2018.2336) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sproles AE, Oakley CA, Krueger T, Grossman AR, Weis VM, Meibom A, Davy SK. 2020. Sub-cellular imaging shows reduced photosynthetic carbon and increased nitrogen assimilation by the non-native endosymbiont Durusdinium trenchii in the model cnidarian Aiptasia. Environ. Microbiol. 22, 3741-3753. ( 10.1111/1462-2920.15142) [DOI] [PubMed] [Google Scholar]
- 52.Sproles AE, Oakley CA, Matthews JL, Peng L, Owen JG, Grossman AR, Weis VM, Davy SK. 2019. Proteomics quantifies protein expression changes in a model cnidarian colonized by a thermally tolerant but suboptimal symbiont. ISME J. 13, 2334-2345. ( 10.1038/s41396-019-0437-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Finney JC, Pettay DT, Sampayo EM, Warner ME, Oxenford HA, LaJeunesse TC. 2010. The relative significance of host–habitat, depth, and geography on the ecology, endemism, and speciation of coral endosymbionts in the genus Symbiodinium. Microb. Ecol. 60, 250-263. ( 10.1007/s00248-010-9681-y) [DOI] [PubMed] [Google Scholar]
- 54.LaJeunesse T, Thornhill D, Cox E, Stanton F, Fitt W, Schmidt G. 2004. High diversity and host specificity observed among symbiotic dinoflagellates in reef coral communities from Hawaii. Coral Reefs 23, 596-603. ( 10.1007/s00338-004-0428-4) [DOI] [Google Scholar]
- 55.Tonk L, Sampayo EM, Weeks S, Magno-Canto M, Hoegh-Guldberg O. 2013. Host-specific interactions with environmental factors shape the distribution of Symbiodinium across the Great Barrier Reef. PLoS ONE 8, e68533. ( 10.1371/journal.pone.0068533) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bongaerts P, Ridgway T, Sampayo EM, Hoegh-Guldberg O. 2010. Assessing the ‘deep reef refugia’ hypothesis: Focus on Caribbean reefs. Coral Reefs 29, 309-327. ( 10.1007/s00338-009-0581-x) [DOI] [Google Scholar]
- 57.Rowan R, Knowlton N, Baker A, Jara J. 1997. Landscape ecology of algal symbionts creates variation in episodes of coral bleaching. Nature 388, 265-269. ( 10.1038/40843) [DOI] [PubMed] [Google Scholar]
- 58.Cunning R, Ritson-Williams R, Gates R. 2016. Patterns of bleaching and recovery of Montipora capitata in Kāne‘ohe Bay, Hawai‘i, USA. Mar. Ecol. Prog. Ser. 551, 131-139. ( 10.3354/meps11733) [DOI] [Google Scholar]
- 59.Dilworth J, Caruso C, Kahkejian VA, Baker AC, Drury C. 2021. Host genotype and stable differences in algal symbiont communities explain patterns of thermal stress response of Montipora capitata following thermal pre-exposure and across multiple bleaching events. Coral Reefs 40, 151-163. ( 10.1007/s00338-020-02024-3) [DOI] [Google Scholar]
- 60.Innis T, Cunning R, Ritson-Williams R, Wall CB, Gates RD. 2018. Coral color and depth drive symbiosis ecology of Montipora capitata in Kāne‘ohe Bay, O‘ahu, Hawai‘i. Coral Reefs 37, 423-430. ( 10.1007/s00338-018-1667-0) [DOI] [Google Scholar]
- 61.Stat M, et al. 2011. Variation in Symbiodinium ITS2 sequence assemblages among coral colonies. PLoS ONE 6, e15854. ( 10.1371/journal.pone.0015854) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stat M, Pochon X, Franklin EC, Bruno JF, Casey KS, Selig ER, Gates RD. 2013. The distribution of the thermally tolerant symbiont lineage (Symbiodinium clade D) in corals from Hawaii: Correlations with host and the history of ocean thermal stress. Ecol. Evol. 3, 1317-1329. ( 10.1002/ece3.556) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Caruso C, et al. 2021. Genetic patterns in Montipora capitata across an environmental mosaic in Kāne'ohe Bay. BioRxiv ( 10.1101/2021.10.07.463582) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lowe, et al. 2009. A numerical study of circulation in a coastal reef-lagoon system. J. Geophys. Res. 114, C06022. ( 10.1029/2008JC005081) [DOI] [Google Scholar]
- 65.Lowe, et al. 2009. Wave-driven circulation of a coastal reef–lagoon system. Journal of Physical Oceanography 39, 873-893. ( 10.1175/2008JPO3958.1) [DOI] [Google Scholar]
- 66.Wyatt ASJ, Leichter JJ, Toth LT, Miyajima T, Aronson RB, Nagata T. 2020. Heat accumulation on coral reefs mitigated by internal waves. Nat. Geosci. 13, 28-34. ( 10.1038/s41561-019-0486-4) [DOI] [Google Scholar]
- 67.Kahle D, Wickham H. 2013. ggmap: spatial visualization with ggplot2. R J 5, 144. ( 10.32614/RJ-2013-014) [DOI] [Google Scholar]
- 68.Jacobs KP, Hunter CL, Forsman ZH, Pollock AL, de Souza MR, Toonen RJ. 2021. A phylogenomic examination of Palmyra Atoll's corallimorpharian invader. Coral Reefs 41, 673-685. ( 10.1007/s00338-021-02143-5) [DOI] [Google Scholar]
- 69.Arif, et al. 2014. Assessing Symbiodinium diversity in scleractinian corals via next-generation sequencing-based genotyping of the ITS2 rDNA region. Mol. Ecol. 23, 4418-4433. ( 10.1111/mec.12869) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 3. ( 10.14806/ej.17.1.200) [DOI] [Google Scholar]
- 71.Hume BCC, et al. 2019. SymPortal: a novel analytical framework and platform for coral algal symbiont next-generation sequencing ITS2 profiling. Mol. Ecol. Resour. 19, 1063-1080. ( 10.1111/1755-0998.13004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.R Core Team 2020. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. (https://www.R-project.org/) [Google Scholar]
- 73.Dixon 2003. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14, 927-930. ( 10.1111/j.1654-1103.2003.tb02228.x) [DOI] [Google Scholar]
- 74.Warnes, et al. 2020. gplots: Various R programming tools for plotting data. R package version. [Google Scholar]
- 75.de Souza MR, Caruso C, Ruiz-Jones L, Dury C, Gates R, Toonen RJ. 2021. Community composition of coral associated Symbiodiniaceae differs across fine scale environmental gradients in Kāne‘ohe Bay. Zenodo. ( 10.1111/zenodo.org/record/5670832#.YvBKpC1h2iM) [DOI] [PMC free article] [PubMed]
- 76.Innis, et al. 2018. Coral color and depth drive symbiosis ecology of Montipora capitata in Kane‘ohe Bay, O‘ahu, Hawai‘i. Coral Reefs 37, 423-430. ( 10.1007/s00338-018-1667-0) [DOI] [Google Scholar]
- 77.Cottenie K. 2005. Integrating environmental and spatial processes in ecological community dynamics: Meta-analysis of metacommunities. Ecol. Lett. 8, 1175-1182. ( 10.1111/j.1461-0248.2005.00820.x) [DOI] [PubMed] [Google Scholar]
- 78.Claar DC, McDevitt-Irwin JM, Garren M, Vega Thurber R, Gates RD, Baum JK. 2020. Increased diversity and concordant shifts in community structure of coral-associated Symbiodiniaceae and bacteria subjected to chronic human disturbance. Mol. Ecol. 29, 2477-2491. ( 10.1111/mec.15494) [DOI] [PubMed] [Google Scholar]
- 79.Wiedenmann J, D'Angelo C, Smith EG, Hunt AN, Legiret F-E, Postle AD, Achterberg EP. 2013. Nutrient enrichment can increase the susceptibility of reef corals to bleaching. Nat. Clim. Change 3, 160-164. ( 10.1038/nclimate1661) [DOI] [Google Scholar]
- 80.Wee HB, Kurihara H, Reimer JD. 2019. Reduced Symbiodiniaceae diversity in Palythoa tuberculosa at a heavily acidified coral reef. Coral Reefs 38, 311-319. ( 10.1007/s00338-019-01776-x) [DOI] [Google Scholar]
- 81.Rowan R, Knowlton N. 1995. Intraspecific diversity and ecological zonation in coral-algal symbiosis. Proc. Natl Acad. Sci. USA 92, 2850-2853. ( 10.1073/pnas.92.7.2850) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Toller WW, Rowan R, Knowlton N. 2001. Repopulation of Zooxanthellae in the Caribbean corals Montastraea annularis and M. faveolata following experimental and disease-associated bleaching. Biol. Bull. 201, 360-373. ( 10.2307/1543614) [DOI] [PubMed] [Google Scholar]
- 83.Edmunds PJ, Tsounis G, Boulon R, Bramanti L. 2018. Long-term variation in light intensity on a coral reef. Coral Reefs 37, 955-965. ( 10.1007/s00338-018-1721-y) [DOI] [Google Scholar]
- 84.Hochberg EJ, Peltier SA, Maritorena S. 2020. Trends and variability in spectral diffuse attenuation of coral reef waters. Coral Reefs 39, 1377-1389. ( 10.1007/s00338-020-01971-1) [DOI] [Google Scholar]
- 85.Storlazzi CD, Norris BK, Rosenberger KJ. 2015. The influence of grain size, grain color, and suspended-sediment concentration on light attenuation: Why fine-grained terrestrial sediment is bad for coral reef ecosystems. Coral Reefs 34, 967-975. ( 10.1007/s00338-015-1268-0) [DOI] [Google Scholar]
- 86.Kahng SE, Garcia-Sais JR, Spalding HL, Brokovich E, Wagner D, Weil E, Hinderstein L, Toonen RJ. 2010. Community ecology of mesophotic coral reef ecosystems. Coral Reefs 29, 255-275. ( 10.1007/s00338-010-0593-6) [DOI] [Google Scholar]
- 87.Lesser MP, Slattery M, Leichter JJ. 2009. Ecology of mesophotic coral reefs. J. Exp. Mar. Biol. Ecol. 375, 1-8. ( 10.1016/j.jembe.2009.05.009) [DOI] [Google Scholar]
- 88.Morgan KM, Moynihan MA, Sanwlani N, Switzer AD. 2020. Light limitation and depth-variable sedimentation drives vertical reef compression on turbid coral reefs. Front. Mar. Sci. 7, 571256. ( 10.3389/fmars.2020.571256) [DOI] [Google Scholar]
- 89.Van den Hoek C, Breeman AM, Bak RPM, Van Buurt G. 1978. The distribution of algae, corals and gorgonians in relation to depth, light attenuation, water movement and grazing pressure in the fringing coral reef of Curaçao, Netherlands Antilles. Aquat. Bot. 5, 1-46. ( 10.1016/0304-3770(78)90045-1) [DOI] [Google Scholar]
- 90.Baldwin CC, Tornabene L, Robertson DR. 2018. Below the mesophotic. Sci. Rep. 8, 4920. ( 10.1038/s41598-018-23067-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lesser MP, Slattery M, Mobley CD. 2018. Biodiversity and functional ecology of mesophotic coral reefs. Ann. Rev. Ecol. Evol. Syst. 49, 49-71. ( 10.1146/annurev-ecolsys-110617-062423) [DOI] [Google Scholar]
- 92.Duckworth A, Giofre N, Jones R. 2017. Coral morphology and sedimentation. Mar. Pollut. Bull. 125, 289-300. ( 10.1016/j.marpolbul.2017.08.036) [DOI] [PubMed] [Google Scholar]
- 93.Jones R, Fisher R, Bessell-Browne P. 2019. Sediment deposition and coral smothering. PLoS ONE 14, e0216248. ( 10.1371/journal.pone.0216248) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Weber M, De Beer D, Lott C, Polerecky L, Kohls K, Abed RM, Ferdelman TG, Fabricius KE. 2012. Mechanisms of damage to corals exposed to sedimentation. Proc. Natl Acad. Sci. USA 109, E1558-E1567. ( 10.1073/pnas.1100715109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fitt W, Brown B, Warner M, Dunne R. 2001. Coral bleaching: interpretation of thermal tolerance limits and thermal thresholds in tropical corals. Coral Reefs 20, 51-65. ( 10.1007/s003380100146) [DOI] [Google Scholar]
- 96.Gates RD, Baghdasarian G, Muscatine L. 1992. Temperature stress causes host cell detachment in symbiotic cnidarians: implications for coral bleaching. Biol. Bull. 182, 324-332. ( 10.2307/1542252) [DOI] [PubMed] [Google Scholar]
- 97.Thompson DM, van Woesik R. 2009. Corals escape bleaching in regions that recently and historically experienced frequent thermal stress. Proc. R. Soc. B 276, 2893-2901. ( 10.1098/rspb.2009.0591) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Barshis DJ, Stillman JH, Gates RD, Toonen RJ, Smith LW, Birkeland C. 2010. Protein expression and genetic structure of the coral Porites lobata in an environmentally extreme Samoan back reef: does host genotype limit phenotypic plasticity? Mol. Ecol. 19, 1705-1720. ( 10.1111/j.1365-294X.2010.04574.x) [DOI] [PubMed] [Google Scholar]
- 99.Safaie A, Silbiger NJ, McClanahan TR, Pawlak G, Barshis DJ, Hench JL, Rogers JS, Williams GJ, Davis KA. 2018. High frequency temperature variability reduces the risk of coral bleaching. Nat. Commun. 9, 1671. ( 10.1038/s41467-018-04074-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bahr KD, Rodgers KS, Jokiel PL. 2017. Impact of three bleaching events on the reef resiliency of Kāne‘ohe Bay, Hawai‘i. Front. Mar. Sci. 4, 398. ( 10.3389/fmars.2017.00398) [DOI] [Google Scholar]
- 101.Gorospe KD, Karl SA. 2011. Small-scale spatial analysis of in situ sea temperature throughout a single coral patch reef. J. Mar. Biol. 2011, 1-12. ( 10.1155/2011/719580) [DOI] [Google Scholar]
- 102.Gorospe KD, Karl SA. 2015. Depth as an organizing force in Pocillopora damicornis: intra-reef genetic architecture. PLoS ONE 10, e0122127. ( 10.1371/journal.pone.0122127) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.de Souza MR, Caruso C, Ruiz-Jones L, Dury C, Gates R, Toonen RJ. 2022. Community composition of coral-associated Symbiodiniaceae differs across fine-scale environmental gradients in Kāne‘ohe Bay. FigShare. ( 10.6084/m9.figshare.c.6167440) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- de Souza MR, Caruso C, Ruiz-Jones L, Dury C, Gates R, Toonen RJ. 2021. Community composition of coral associated Symbiodiniaceae differs across fine scale environmental gradients in Kāne‘ohe Bay. Zenodo. ( 10.1111/zenodo.org/record/5670832#.YvBKpC1h2iM) [DOI] [PMC free article] [PubMed]
- de Souza MR, Caruso C, Ruiz-Jones L, Dury C, Gates R, Toonen RJ. 2022. Community composition of coral-associated Symbiodiniaceae differs across fine-scale environmental gradients in Kāne‘ohe Bay. FigShare. ( 10.6084/m9.figshare.c.6167440) [DOI] [PMC free article] [PubMed]
Data Availability Statement
All materials, code and data are available for download on Zenodo (75; https://zenodo.org/record/5670832#.YvBKpC1h2iM).
The data are provided in electronic supplementary material [103].