Abstract
Millions of breast imaging exams are performed each year in an effort to reduce the morbidity and mortality of breast cancer. Breast imaging exams are performed for cancer screening, diagnostic work-up of suspicious findings, evaluating extent of disease in recently diagnosed breast cancer patients, and determining treatment response. Yet, the interpretation of breast imaging can be subjective, tedious, time-consuming, and prone to human error. Retrospective and small reader studies suggest that deep learning (DL) has great potential to perform medical imaging tasks at or above human-level performance, and may be used to automate aspects of the breast cancer screening process, improve cancer detection rates, decrease unnecessary callbacks and biopsies, optimize patient risk assessment, and open up new possibilities for disease prognostication. Prospective trials are urgently needed to validate these proposed tools, paving the way for real-world clinical use. New regulatory frameworks must also be developed to address the unique ethical, medicolegal, and quality control issues that DL algorithms present. In this article, we review the basics of DL, describe recent DL breast imaging applications including cancer detection and risk prediction, and discuss the challenges and future directions of artificial intelligence-based systems in the field of breast cancer.
Introduction
Artificial intelligence (AI) has exciting potential to transform the field of medical imaging. Recent advances in computer algorithms, increased availability of computing power, and more widespread access to big data are fueling this revolution. AI algorithms can be taught to extract patterns from large data sets, including data sets containing a vast amount of medical images, and are able to meet, and even exceed, human-level performance in a variety of repetitive well-defined tasks. 1–6
Breast imaging is particularly well suited for AI algorithm development since the diagnostic question is straightforward and there is widespread availability of data. Most breast imaging exams are binary classification problems (e.g. malignant vs benign), and almost all studies have an accepted ground truth (e.g. histopathology or negative imaging follow-up) that is commonly available for use during algorithmic development. Furthermore, there is widespread availability of standard imaging data due to population-wide screening programs, and the American College of Radiology (ACR) Breast Imaging and Reporting Data System (BI-RADS) system enforces structured reporting and assessments.
To date, retrospective and small reader studies show that AI tools increase diagnostic accuracy, 7–9 improve breast cancer risk assessment, 10–12 and predict response to cancer therapy, 13,14 among other tasks. AI is also being applied to improve the image reconstruction process more generally, so that high quality images may be obtained with lower radiation dose in mammography and digital breast tomosynthesis (DBT), and with shorter scan times in MRI. 15–17
AI is uniquely poised to help breast imagers at both ends of the interpretive spectrum. On the one hand, AI can be used to automate simple tasks (e.g. removing completely normal exams from the radiology worklist, which relieves radiologists to tackle more challenging cases). Computer algorithms do not suffer from fatigue or distraction, and thus are uniquely suited for basic repetitive tasks that humans may find tedious or boring. On the other hand, AI has the potential to extend the frontiers of our practice of medicine. AI can identify complex patterns in imaging data that are not appreciated by the human eye, 10 adding a wealth of information to enable more sophisticated disease modelling and more individualized treatment planning. However, it is important to note that almost all studies to date have been either retrospective trials or small reader studies, which limits the generalizability of results. Prospective studies are now needed to more fully evaluate the performance of these AI tools, and are prerequisite to responsible clinical translation.
In this article, we will review the basics of AI and deep learning, describe some AI applications in clinical breast imaging, and discuss challenges and future directions.
Artificial intelligence and DL
Traditional machine learning, a subfield of AI, was used in the 1990s and 2000s to develop computer-aided detection (CAD) software for mammography. In initial studies, 18 CAD improved diagnostic accuracy, it received FDA approval in 1998, and became widely utilized over the next 18 years. 19 However, more recently, larger studies demonstrated that CAD generates large numbers of false positives and does not improve diagnostic accuracy, and therefore it has largely fallen out of favor. 20
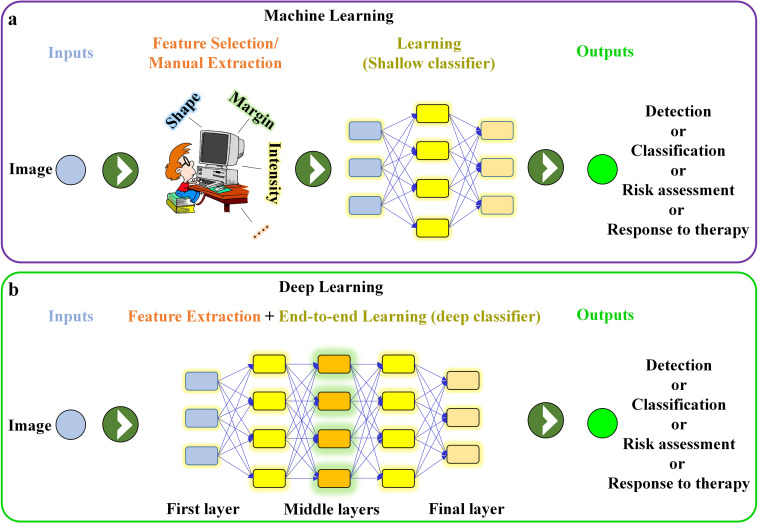
Deep learning (DL), a new type of representative machine learning, first gained widespread attention in 2012 when AlexNet won the ImageNet Large Scale Visual Recognition Challenge by a large margin. 21 Since 2016, there has been an explosion of effort in applying DL to diagnostic radiology, and to breast imaging in particular. 22,23 DL models not only classify input images as positive or negative, but they figure out which imaging features are needed to perform this classification, without expert input. 24 This is in contrast to traditional machine learning techniques (e.g. CAD), which rely on hand-crafted features (e.g. shape, margin) to perform the classification (Figure 1a). This major difference explains why new DL algorithms outperform traditional machine learning techniques (if there are sufficient data available). 25
Figure 1.
Schematic illustrating (a) feature-based (human-engineered) machine learning network (e.g. conventional CAD software) and (b) end-to-end deep learning network. CAD, computer-aided design.
Most DL algorithms for medical imaging use convolutional neural networks (CNN). CNNs have millions of weights (i.e. variables to be optimized) and multiple layers of processing designed to extract hierarchical patterns in data. Most DL models for medical imaging use a supervised learning technique, which means that training is performed using many labeled examples. Data labeling can be done on the exam level (e.g. a whole mammogram exam is labeled as benign or malignant), breast level (e.g. the left breast is labeled benign and the right breast is labeled malignant), pixel level (e.g. the area of malignancy is circled), or somewhere in between. Pixel-level labeling gives the most information and reduces training set size requirements, although it is costly to generate.
During CNN training, a general purpose learning procedure 24 is used to perform feature selection and classification simultaneously and without expert input (Figure 1b). During CNN training, large numbers of labeled medical images are fed directly to a CNN. The first layer learns small simple features (e.g. location and orientation of edges), the next layers learn particular combinations of those simpler features, and the deeper layers learn even more complex arrangements of those earlier patterns. The final layers use these imaging features or representations to classify the image or to detect other patterns of interest (Figure 1b). Once a CNN is fully trained, its performance is tested using a held-out test set not used during the training. Ideally, CNN performance is further validated using a data set from an outside institution (i.e. external validation).
DL methods are data-driven and results generally improve as the data set size increases. There is no specific formula to calculate the data set size needed to train a model for a given task, although training data sets must be large enough and diverse enough to encompass the range of phenotypes of the categories that they seek to classify. When it is not possible to curate a data set of sufficient size to train a CNN from scratch (a frequent occurrence in medical imaging), CNN weights may be initialized with weights learned for some other task (e.g. classifying cats versus dogs). This transfer learning technique substantially reduces data set size requirements for CNN training. 24,26
Since DL networks learn complex representations of images not appreciated by the human eye, they have the potential to identify new unseen patterns in data, transcending our current knowledge of disease diagnosis and treatment, although this aspect of DL is still at a pilot stage and warrants further exploration.
Mammography and digital breast tomosynthesis
Cancer detection and classification studies
Each year, more than 300,000 cases of breast cancer are diagnosed in the United States alone. While screening mammography decreases breast cancer mortality by 20–35%, it is not a perfect tool. 27 The diagnostic accuracy of mammography varies widely, even among breast imaging experts, with sensitivity and specificity ranging from 67 to 99% and 71 to 97%, respectively. 28 DL has the potential to improve these metrics, both increasing cancer detection rates and decreasing unnecessary callbacks. Several retrospective and reader studies have already shown AI model performance at or beyond the level of expert radiologists 9,22,29–31 (see Supplementary Material 1). Reader studies have used a combination of fellowship-trained breast imagers, general radiologists, and sometimes even trainees, which is an important consideration when making claims about the superiority of AI tools compared to human readers. Of note, while initial DL studies were based on 2D mammography, more recent work has focused on DBT, 7,29,32,33 which is a more complex technical task but which has potential to improve AI performance even further. DBT increases the radiologist’s interpretation time by approximately 50% compared to 2D mammography, 34 and so AI tools for DBT are being developed not just to find more cancers, but with an eye towards clinical efficiency.
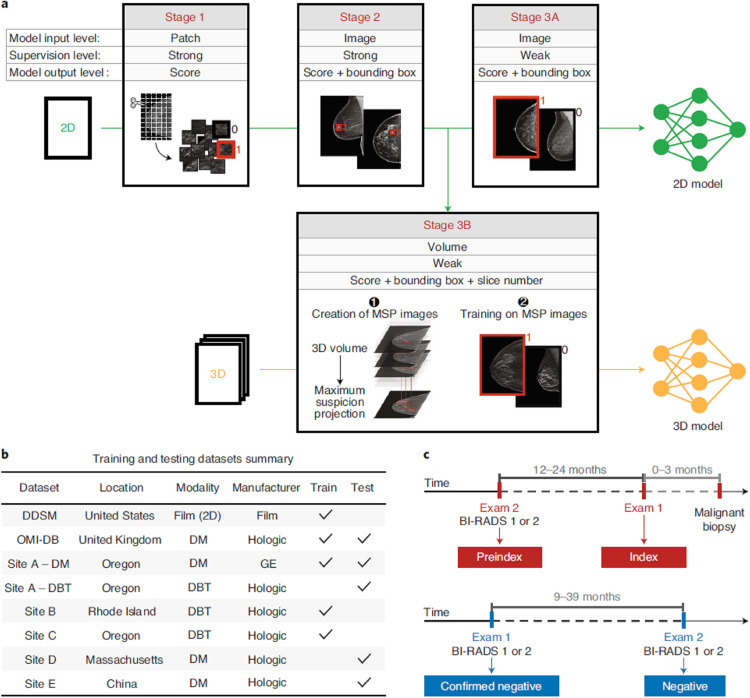
In one of the most important AI mammography/DBT studies to date, Lotter et al presented a DL model for cancer detection that yielded state-of-the-art performance for mammographic classification, showing an area under the curve (AUC) of 0.945 in their retrospective study. 29 The DL model outperformed five expert breast imagers in a reader study, worked for both 2D digital mammography and 3D DBT, and was externally validated using imaging data from several national and one international site, demonstrating good generalizability (Figure 2). Another key retrospective study, from Salim et al, independently evaluated performance of three commercial AI systems for mammography screening using a single standardized data set. 35 One of the three AI algorithms outperformed human readers, and combining that algorithm with a human reader outperformed two human readers.
Figure 2.
An illustration of multistage deep learning model training and data summary. 29 (a) Illustration of model training in stages. Stage 1 illustrates patch-level classification. Stage 2 demonstrates an intermediate step where a detection-based model was trained on full mammography images to identify bounding boxes and malignancy likelihood scores. Stage 3A re-trains the detection-based model using multi-instance learning approach wherein maximum score over all bounding boxes in each full mammography image was computed for classification of cancer or no cancer. In Stage 3B, an analogous detection-based model was trained for DBT using maximum suspicion projection images. (b) Summary of multi-institution training and testing data sets. (c) Illustration of exam definitions used in the study. 29 Reprinted by permission from Springer Nature: Nature Medicine, 29 copyright 2021.
To pave the way for clinical translation, prospective clinical trials are needed. Several such trials are currently recruiting, most of which are evaluating performance of the Transpara (ScreenPoint Medical) and INSIGHT MMG (Lunit) commercial AI software, in different clinical and geographic settings. For example, the AITIC trial (NCT04949776) will evaluate if Transpara can reduce the workload of a breast screening program by 50% with non-inferior cancer detection and recall rate, whereas the ScreenTrustCAD trial (NCT04778670) will compare the Lunit software to single and double readings by radiologists.
Calcifications
Calcifications, a common finding on mammogram, are conventionally classified as suspicious, probably benign, or benign using the ACR BI-RADS lexicon. 36 However, more than half of calcifications classified as suspicious yield benign pathology, 37 and so there is much interest in developing DL tools to improve the classification process and avoid unnecessary biopsies. Some groups have demonstrated increases in diagnostic accuracy with DL algorithms, although data set sizes are small and larger validation studies are needed. 38–40
AI for mammography workflow optimization
In the coming years, DL seems poised to transcend its role as a mammography decision-support tool and instead to serve as an independent reader of “ultra-normal” mammograms. Over 20 million 22 screening mammograms are performed in the United States each year, and over 99% of them are completely normal. If an independent AI reader signed-off on even a fraction of these studies without radiologist input, there could be significant cost savings and impacts on workflow. Several studies 41–43 have investigated this, with results suggesting that AI could remove up to 20% of the lowest-likelihood-of-cancer screening mammograms from the worklist without missing cancers. Larger trials are warranted to validate these promising findings. Ethical, medicolegal, and regulatory aspects of standalone AI warrant further consideration prior to clinical translation. For example, it will be important to develop AI-specific quality controls, including a schedule for continuous algorithmic assessment and fine-tuning to ensure that AI performance does not drift over time.
DL for breast cancer risk assessment
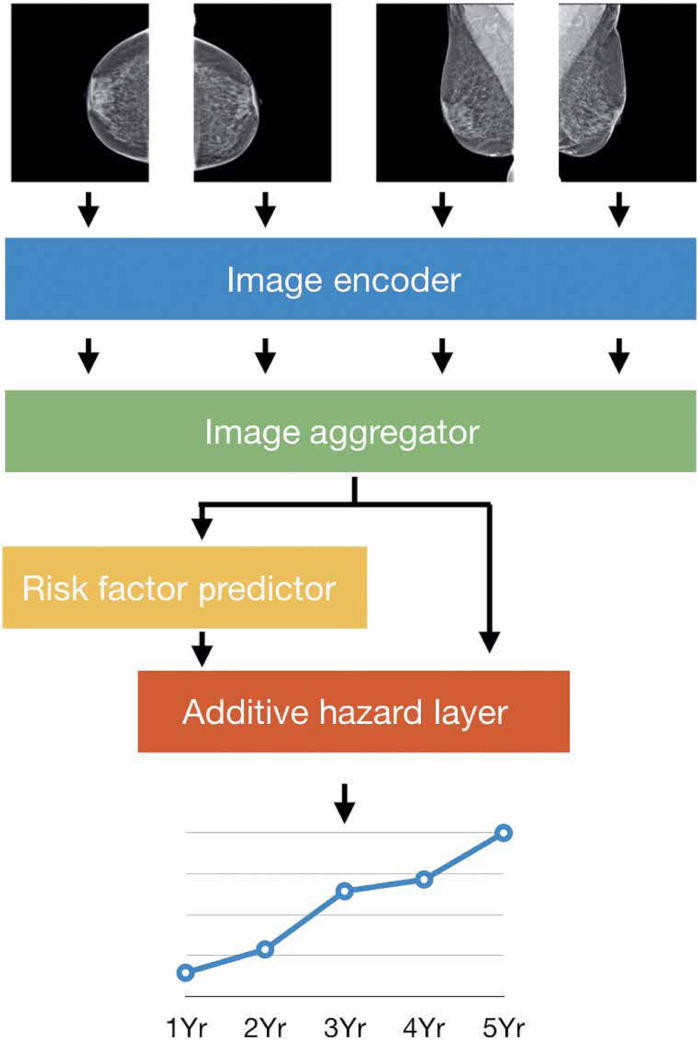
In one of the most significant AI-for-medical-imaging developments to date, DL has been used in an effort to optimize breast cancer screening practices. At present, conventional risk assessment models such as the Tyrer–Cuzick model are used to determine whether a woman is at high risk of breast cancer, a status which warrants supplemental screening with contrast-enhanced MRI in addition to standard-of-care annual screening mammography. Yala et al and others (see Supplementary Material 1), have developed DL models using mammograms 10,44,45 (or MR images 12 ) that outperform the Tyrer–Cuzick model (Figure 3), and that have been externally validated on large and diverse data sets from the United States, Europe and Asia. 10 A prospective trial (ScreenTrustMRI, NCT04832594) is currently recruiting, which will evaluate use of one commercial AI tool and one in-house academic tool, to predict future breast cancer risk based on mammography images, and thereby optimize the triage of women to supplemental screening with breast MRI.
Figure 3.
A schematic illustration of breast cancer risk-prediction DL model. 10 The model input is four standard views of an individual mammogram. The image encoder and image aggregator together provide a combined single vector for all mammogram views. Standard clinical variables (e.g. age, family history) are incorporated into the DL model, and if any of these clinical variables are unavailable, a risk factor predictor module is used to fill in the missing pieces. Finally, the additive hazard layer combines the imaging and clinical data to predict breast cancer risk for five consecutive years. Reprinted by permission from The American Association for the Advancement of Science: Science Translational Medicine, 10 copyright 2021. DL, deep learning.
Going one step further, Manley et al 46 demonstrated that their DL breast cancer risk score tool is modifiable, and that chemoprevention can decrease risk. DL tools have also been developed to automate the assessment of mammographic breast density, and have been clinically implemented at both academic and clinical radiology centers. 47,48
Ultrasound
Breast ultrasound can be used as both a supplemental screening modality (where it increases the cancer detection rate over mammography alone, particularly in women with dense breasts), and as a diagnostic tool in the work-up of mammographic or clinical findings. 49 Unfortunately, in many cases, ultrasound demonstrates low specificity and prompts unnecessary biopsies. It also has high interreader variability. 50 In an effort to boost the diagnostic accuracy of ultrasound, DL methods have been developed for breast ultrasound lesion segmentation, lesion detection, and lesion classification, for both automated and handheld ultrasound. Automated breast ultrasound generates thousands of images per patient exam, and so DL tools are particularly needed for lesion detection and to reduce interpretation time. 51 DL-based segmentation methods are state of the art, outperforming conventional computerized methods. 51–53 DL has also been applied to lesion detection and classification, 54–62 with several reader studies reporting DL models that are equivalent or superior to radiologists, 49,62,63 although in most of these studies, DL models were compared against general radiologists without subspecialty training in breast imaging, small data set sizes were used, and data were from a single institution. 58,62 As such, more work is needed to demonstrate the generalizability of these models.
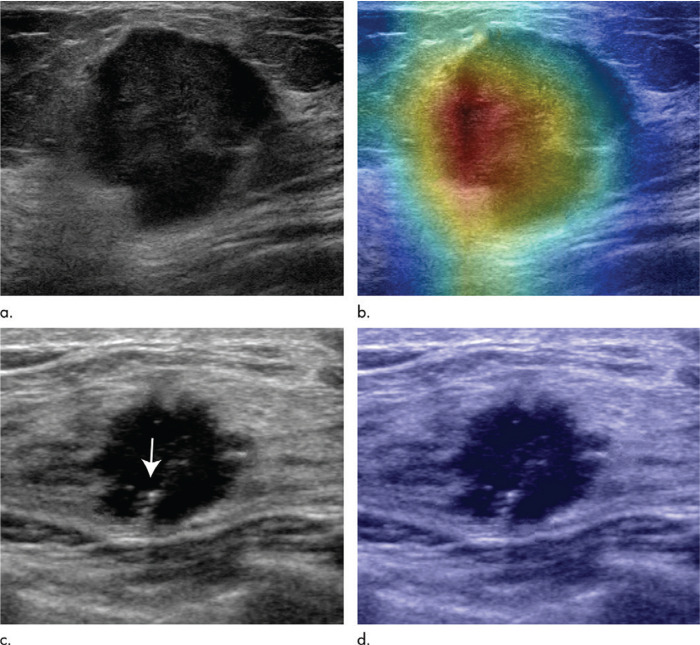
DL for breast ultrasound has also been explored as a prognostication tool. Zhou et al 58 and Zheng et al 64 used ultrasound images of primary breast tumors to predict the presence of axillary node metastases, with AUCs up to 0.90. Figure 4 illustrates the use of DL to predict axillary nodal metastases using ultrasound images of primary breast cancer.
Figure 4.
2D visualization of primary breast cancer in ultrasound images, and DL-aided prediction of clinically positive and negative lymph node metastasis. 58 Here, DL enabled accurate prediction of positive lymph node metastasis in 67-year-old females (a, b) and negative lymph node metastasis in 46-year-old females (c, d). Reprinted by permission from Radiology, 58 copyright Radiological Society of North America 2020. DL, deep learning.
Magnetic resonance imaging
Breast MRI is the most sensitive tool currently available for breast cancer detection, 65 and is an indispensible tool in the screening of high risk women, in evaluating extent of disease, in assessing treatment response, and as a problem solving tool in challenging diagnostic situations. However, its use is often limited by high price tag and long exam time. High resolution dynamic contrast-enhanced MRI is an information-rich imaging modality with different imaging sequences (e.g. T 1W, T 2W, DWI, dynamic pre- and post-contrast imaging) reflecting different aspects of the underlying pathophysiology (e.g. water content, vascular permeability, etc.). This data-richness gives DL real potential not just to automate simple breast MR interpretation tasks, but to learn new patterns that uncover new connections between imaging and disease, opening new avenues for personalized medicine. To date, DL has been applied to breast MR image segmentation, lesion detection, risk prediction, and treatment response. 66
Segmentation, lesion detection and lesion classification
Today, DL is considered the state-of-the-art method for 3D segmentation of breast MRI images, including segmentation of the whole breast, 67,68 fibroglandular tissue (FGT), 69,70 and mass lesions 71,72 (see Supplementary Material 1).
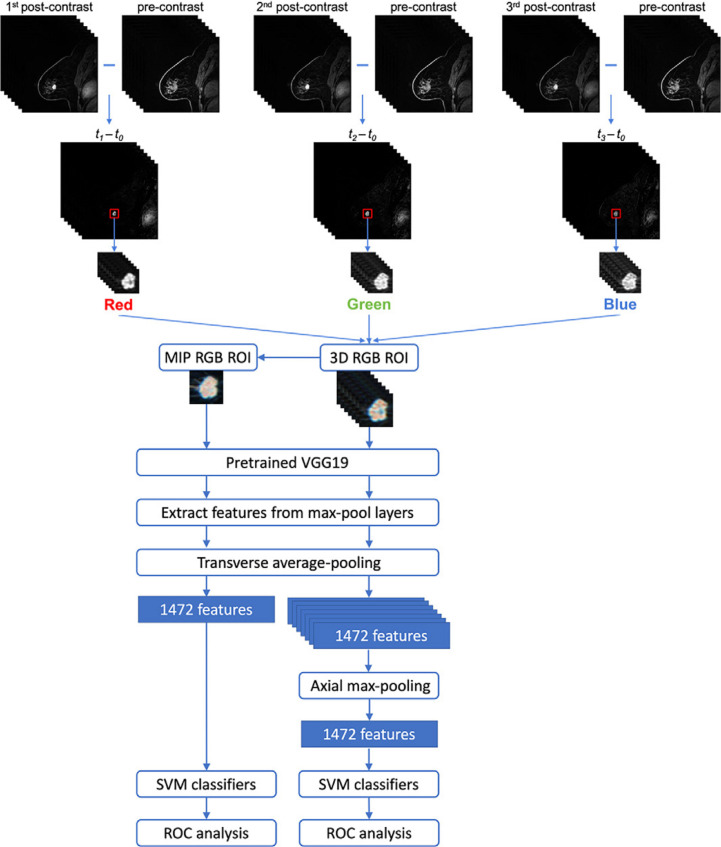
Several groups have harnessed DL for both the detection and classification of lesions on breast MRI 73–81 (see Supplementary Material 1 for details). The large size of a 4D breast MRI dataset makes en mass model training computationally difficult. Hence, breast MRI DL models hinge on imaging pre-processing pipelines that distill clinically relevant spatial and temporal information. The most popular (and easiest) approach is to collapse a 4D data set into a 2D maximum intensity projection (MIP) of the subtraction image (post-contrast minus pre-contrast), enabling the use of standard 2D CNN architectures for model training. 73–76,78,82 Other groups have experimented with different approaches, including: (i) using a “MIP” of CNN features, 73,74 (ii) using 3D lesion ROIs, not whole images, thus reducing data size requirements and enabling use of a 3D CNN, 76 (iii) incorporating multiparametric information (e.g. T2, DWI), 76–78 and (iv) fusing DL feature extraction methods with traditional machine learning classifiers. 74,76,77 Figure 5 depicts a fused architecture integrating DL feature extraction and machine learning classifiers for lesion classification from subtraction images of dynamic contrast-enhanced (DCE) MRI sequences. 74 Several papers show that DL methods for breast MRI outperform traditional machine learning methods, particularly as training data set sizes increase. 25 Of the many studies published on DL for breast MRI, only a few include reader studies (two showing that DL models performed similarly to humans, 75,76 and one showing that the DL model was inferior 71 ). Of note, readers in these studies are not always radiologists, let alone fellowship-trained breast radiologists, and data set sizes are small. Additional larger and multicenter reader studies are needed to determine how DL models for breast MRI compares with human experts. In fact, small data set size is a limitation of all breast MRI DL papers to date, with the largest studies including only a couple thousand breast MRI exams. Breast MRI exams are also notoriously hard to curate, given the myriad sequences and the variations in protocol parameters and naming conventions even within the same institution.
Figure 5.
An illustration of combined DL feature extraction methods with traditional machine learning classifiers for lesion classification from cropped ROI’s of MIP and 3D RGB of subtraction images of DCE MRI sequences. 74 The top layer illustrates the construction of the cropped ROI’s from the DCE MRI sequences. MIP and 3D RGB features were integrated with max-pooling and then passed to a machine learning classifier. Reprinted by permission from Radiology: Artificial Intelligence, 74 copyright Radiological Society of North America 2021. DCE, dynamic contrast-enhanced; DL, deep learning; MIP, maximum intensity projection; ROI, region of interest.
Risk prediction and treatment response
Background parenchymal enhancement (BPE) is a qualitative measure of normal breast tissue enhancement after intravenous contrast administration. Similar to breast density, BPE is included in a radiologist’s breast MRI report both to give information about whether the sensitivity for cancer detection is limited by BPE, and because BPE is a risk factor for breast cancer. 83 Radiologists demonstrate significant interreader variability in categorizing BPE. DL has been applied both to segment 84 and to classify BPE on breast MRI, 85 enabling full automation and standardization of this process.
Additionally, in work analogous to that done with mammography, DL has also been used to predict 5-year breast cancer risk from the breast MRI MIP image directly, 12 outperforming the standard-of-care Tyrer–Cuzick model.
The richness of breast MRI data makes it particularly well-suited for more complex DL-based prognostication. To create a virtual biopsy tool, DL with breast MRI images has been developed to predict breast tumor molecular subtypes using pathology ground truth of luminal A, luminal B, HER2+, and basal subtypes. 86–88 Other groups have used DL to predict Oncotype Dx Recurrence Score using breast MRI data. 89 DL has also been applied to the prediction of axillary nodal status using breast MR images of the primary tumor, with high cross-validation accuracy. 90–92
Finally, DL techniques to predict patient response to neoadjuvant chemotherapy is an area of high interest. The landmark I-SPY 2 trial found that combinations of MRI features can predict pathologic treatment response using basic logistic regression analysis with AUCs of 0.81. 93 Several groups are now working to improve predictive power using newer machine learning techniques, and with a combination of pre- and post- neoadjuvant chemotherapy MRI images. 13,14,91,94 Much of this work is still preliminary, with small data sets from single institutions, although over the coming years, larger efforts in this area could personalize and optimize cancer therapy.
Challenges and future directions
Since 2016, there has been exponential growth in the application of DL methods to all aspects of breast imaging. Still, more work is needed in several key areas.
First, large multi-institutional prospective trials, conducted by independent third parties, are needed to assess whether AI tools will work as expected in clinic. Retrospective and small reader studies show that AI mammography tools perform at or beyond the level of expert radiologists, and several AI decision-support products have already gained FDA approval. 95 But prior to responsible clinical use, rigorous evaluation of these tools in a prospective setting is imperative.
AI mammography tools have been retrospectively validated in a few large multi-institutional external validation studies, but similar validation work is needed for ultrasound and especially for MRI. AI model development for breast MRI has been encouraging, but it is important to note that almost all published studies used small data sets and were from a single institution without external validation. DL breast MRI projects can be especially challenging since the breast MRI protocols are so variable across institutions, and even within the same institution over time. Still, there is a wealth of information within a breast MRI exam, and it remains a promising area of inquiry with potential to identify new and better ways to personalize the management of breast cancer patients to maximize therapeutic benefit and minimize harm.
In the realm of mammography, more technical work is needed to optimize AI tools for DBT. Radiologists detect more cancers and have fewer callbacks when using DBT compared to 2D mammography, and so it follows that AI-enhanced DBT should outperform AI-enhanced 2D mammography. But this is not yet the case. Developing state-of-the-art AI for DBT is a technically challenging pursuit. DBT exam sizes are much larger than 2D mammography, which translates into markedly increased computational costs during training which can pose technical limitations. Additionally, DBT image post-processing is even less standardized across vendors than 2D digital mammography, with significant variations in both acquisition technique (i.e. hardware) and in reconstruction technique (i.e. software). Available DBT data sets also tend to be smaller. Still, a number of recent studies have reported encouraging results, and also have an eye towards decreasing interpretation times.
As more AI tools are developed with potential for clinical translation, it is essential to tackle the associated ethical, medicolegal and regulatory issues. This is particularly important for standalone AI tools that independently interpret breast imaging exams (i.e. where no human radiologist looks at the images). 81 On the ethical front, there are many unanswered questions. In which circumstances are clinicians obligated to inform patients about the use of AI tools in their clinical work-up? It might be particularly important in situations where AI acts as a “black box,” where clinicians act on an AI tool output but do not understand how the algorithm arrived at the conclusion. Who is liable when the AI tool misses a cancer? How much human oversight should be required? Algorithmic biases are also an ethical concern. AI models perform better on images that resemble images in the training data set, and so ongoing vigilance is needed to handle potentially underrepresented subgroups in the training data (e.g. racial groups, vendors, etc.). More work is also needed to improve the robustness of image normalization techniques, so that DL models can better generalize to data across institutions with different imaging hardware or imaging post-processing software. Finally, it is essential to develop new regulatory frameworks for rigorous AI quality assessment. This might include a regular schedule of AI algorithm quality control testing (similar to how imaging hardware undergoes regular quality control testing), as well as occasional fine-tuning of the algorithm prevent model performance from deteriorating over time.
Finally, one of the most striking aspects of this literature to date is the lack of improved algorithm performance when images at multiple prior timepoints are used. 30 It is well known that the diagnostic accuracy of breast imagers markedly improves with the availability of prior mammograms, and yet the algorithms developed to date are not able to show similar improvements. This is clearly an area ripe for technical development.
Conclusion
DL tools for breast imaging interpretation are being developed at a rapid pace and are likely to transform the clinical landscape of breast imaging over the coming years. Notably, DL mammography tools for breast cancer detection and breast cancer risk assessment demonstrate performance at or above human-level, and prospective trials are warranted to pave the way for clinical translation. Other work on DL for breast imaging opens up new possibilities for disease prognostication and personalized therapies. As DL tools are incorporated into clinical practice, however, regulatory oversight is needed to avoid algorithmic biases, prevent AI “performance drift”, and to address the unique ethical, medicolegal, and quality control issues that DL algorithms present.
Footnotes
Acknowledgment: Authors thanks Joanne Chin for her help with editing the manuscript.
Funding: The authors are supported in part through the NIH/NCI Cancer Center Support Grant P30 CA008748.
Contributor Information
Arka Bhowmik, Email: bhowmika@mskcc.org, arkabhowmik@yahoo.co.uk.
Sarah Eskreis-Winkler, Email: eskreiss@mskcc.org.
REFERENCES
- 1. Shen D, Wu G, Suk HI. Deep learning in medical image analysis. Annu Rev Biomed Eng 2017; 19: 221–48. doi: 10.1146/annurev-bioeng-071516-044442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, et al. A survey on deep learning in medical image analysis. Med Image Anal 2017; 42: 60–88. doi: 10.1016/j.media.2017.07.005 [DOI] [PubMed] [Google Scholar]
- 3. Chartrand G, Cheng PM, Vorontsov E, Drozdzal M, Turcotte S, Pal CJ, et al. Deep learning: A primer for radiologists. Radiographics 2017; 37: 2113–31. doi: 10.1148/rg.2017170077 [DOI] [PubMed] [Google Scholar]
- 4. Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017; 542: 115–18. doi: 10.1038/nature21056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Esteva A, Robicquet A, Ramsundar B, Kuleshov V, DePristo M, Chou K, et al. A guide to deep learning in healthcare. Nat Med 2019; 25: 24–29. doi: 10.1038/s41591-018-0316-z [DOI] [PubMed] [Google Scholar]
- 6. De Fauw J, Ledsam JR, Romera-Paredes B, Nikolov S, Tomasev N, Blackwell S, et al. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat Med 2018; 24: 1342–50. doi: 10.1038/s41591-018-0107-6 [DOI] [PubMed] [Google Scholar]
- 7. Rodriguez-Ruiz A, Lång K, Gubern-Merida A, Broeders M, Gennaro G, Clauser P, et al. Stand-alone artificial intelligence for breast cancer detection in mammography: comparison with 101 radiologists. J Natl Cancer Inst 2019; 111: 916–22. doi: 10.1093/jnci/djy222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu N, Phang J, Park J, Shen Y, Huang Z, Zorin M, et al. Deep neural networks improve radiologists’ performance in breast cancer screening. IEEE Trans Med Imaging 2020; 39: 1184–94. doi: 10.1109/TMI.2019.2945514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McKinney SM, Sieniek M, Godbole V, Godwin J, Antropova N, Ashrafian H, et al. International evaluation of an AI system for breast cancer screening. Nature 2020; 577: 89–94. doi: 10.1038/s41586-019-1799-6 [DOI] [PubMed] [Google Scholar]
- 10. Yala A, Mikhael PG, Strand F, Lin G, Smith K, Wan Y-L, et al. Toward robust mammography-based models for breast cancer risk. Sci Transl Med 2021; 13: 578. doi: 10.1126/scitranslmed.aba4373 [DOI] [PubMed] [Google Scholar]
- 11. Dembrower K, Liu Y, Azizpour H, Eklund M, Smith K, Lindholm P, et al. Comparison of a deep learning risk score and standard mammographic density score for breast cancer risk prediction. Radiology 2020; 294: 265–72. doi: 10.1148/radiol.2019190872 [DOI] [PubMed] [Google Scholar]
- 12. Portnoi T, Yala A, Schuster T, Barzilay R, Dontchos B, Lamb L, et al. Deep learning model to assess cancer risk on the basis of a breast MR image alone. AJR Am J Roentgenol 2019; 213: 227–33. doi: 10.2214/AJR.18.20813 [DOI] [PubMed] [Google Scholar]
- 13. El Adoui M, Drisis S, Benjelloun M. Multi-input deep learning architecture for predicting breast tumor response to chemotherapy using quantitative MR images. Int J Comput Assist Radiol Surg 2020; 15: 1491–1500. doi: 10.1007/s11548-020-02209-9 [DOI] [PubMed] [Google Scholar]
- 14. Ha R, Chin C, Karcich J, Liu MZ, Chang P, Mutasa S, et al. Prior to initiation of chemotherapy, can we predict breast tumor response? deep learning convolutional neural networks approach using a breast MRI tumor dataset. J Digit Imaging 2019; 32: 693–701. doi: 10.1007/s10278-018-0144-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Teuwen J, Moriakov N, Fedon C, Caballo M, Reiser I, Bakic P, et al. Deep learning reconstruction of digital breast tomosynthesis images for accurate breast density and patient-specific radiation dose estimation. Med Image Anal 2021; 71: 102061. doi: 10.1016/j.media.2021.102061 [DOI] [PubMed] [Google Scholar]
- 16. Kaye EA, Aherne EA, Duzgol C, Häggström I, Kobler E, Mazaheri Y, et al. Accelerating prostate diffusion-weighted MRI using a guided denoising convolutional neural network: retrospective feasibility study. Radiol Artif Intell 2020; 2: e200007. doi: 10.1148/ryai.2020200007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zeng G, Guo Y, Zhan J, Wang Z, Lai Z, Du X, et al. A review on deep learning MRI reconstruction without fully sampled k-space. BMC Med Imaging 2021; 21(1): 195. doi: 10.1186/s12880-021-00727-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Freer TW, Ulissey MJ. Screening mammography with computer-aided detection: prospective study of 12,860 patients in a community breast center. Radiology 2001; 220: 781–86. doi: 10.1148/radiol.2203001282 [DOI] [PubMed] [Google Scholar]
- 19. Keen JD, Keen JM, Keen JE. Utilization of computer-aided detection for digital screening mammography in the united states, 2008 to 2016. J Am Coll Radiol 2018; 15: 44–48. doi: 10.1016/j.jacr.2017.08.033 [DOI] [PubMed] [Google Scholar]
- 20. Lehman CD, Wellman RD, Buist DSM, Kerlikowske K, Tosteson ANA, Miglioretti DL, et al. Diagnostic accuracy of digital screening mammography with and without computer-aided detection. JAMA Intern Med 2015; 175: 1828–37. doi: 10.1001/jamainternmed.2015.5231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Krizhevsky A, Sutskever I, GEJAinips H. Imagenet classification with deep convolutional neural networks. Neural Information Processing Systems 2012. [Google Scholar]
- 22. Geras KJ, Mann RM, Moy L. Artificial intelligence for mammography and digital breast tomosynthesis: current concepts and future perspectives. Radiology 2019; 293: 246–59. doi: 10.1148/radiol.2019182627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Morgan MB, Mates JL. Applications of artificial intelligence in breast imaging. Radiol Clin North Am 2021; 59: 139–48. doi: 10.1016/j.rcl.2020.08.007 [DOI] [PubMed] [Google Scholar]
- 24. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015; 521: 436–44. doi: 10.1038/nature14539 [DOI] [PubMed] [Google Scholar]
- 25. Truhn D, Schrading S, Haarburger C, Schneider H, Merhof D, Kuhl C. Radiomic versus convolutional neural networks analysis for classification of contrast-enhancing lesions at multiparametric breast MRI. Radiology 2019; 290: 290–97. doi: 10.1148/radiol.2018181352 [DOI] [PubMed] [Google Scholar]
- 26. Boström H, Knobbe A, Soares C, Papapetrou P. (n.d.). Advances in Intelligent Data Analysis XV. In: 15th International Symposium, IDA 2016. Cham: Springer International Publishing. doi: 10.1007/978-3-319-46349-0 [DOI] [Google Scholar]
- 27. Elmore JG, Armstrong K, Lehman CD, Fletcher SW. Screening for breast cancer. JAMA 2005; 293: 1245–56. doi: 10.1001/jama.293.10.1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lehman CD, Arao RF, Sprague BL, Lee JM, Buist DSM, Kerlikowske K, et al. National performance benchmarks for modern screening digital mammography: update from the breast cancer surveillance consortium. Radiology 2017; 283: 49–58. doi: 10.1148/radiol.2016161174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lotter W, Diab AR, Haslam B, Kim JG, Grisot G, Wu E, et al. Robust breast cancer detection in mammography and digital breast tomosynthesis using an annotation-efficient deep learning approach. Nat Med 2021; 27: 244–49. doi: 10.1038/s41591-020-01174-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schaffter T, Buist DSM, Lee CI, Nikulin Y, Ribli D, Guan Y, et al. Evaluation of combined artificial intelligence and radiologist assessment to interpret screening mammograms. JAMA Netw Open 2020; 3: e200265. doi: 10.1001/jamanetworkopen.2020.0265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shen L, Margolies LR, Rothstein JH, Fluder E, McBride R, Sieh W. Deep learning to improve breast cancer detection on screening mammography. Sci Rep 2019; 9(1): 12495. doi: 10.1038/s41598-019-48995-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Conant EF, Toledano AY, Periaswamy S, Fotin SV, Go J, Boatsman JE, et al. Improving accuracy and efficiency with concurrent use of artificial intelligence for digital breast tomosynthesis. Radiol Artif Intell 2019; 1: e180096. doi: 10.1148/ryai.2019180096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pinto MC, Rodriguez-Ruiz A, Pedersen K, Hofvind S, Wicklein J, Kappler S, et al. Impact of artificial intelligence decision support using deep learning on breast cancer screening interpretation with single-view wide-angle digital breast tomosynthesis. Radiology 2021; 300: 529–36. doi: 10.1148/radiol.2021204432 [DOI] [PubMed] [Google Scholar]
- 34. Dang PA, Freer PE, Humphrey KL, Halpern EF, Rafferty EA. Addition of tomosynthesis to conventional digital mammography: effect on image interpretation time of screening examinations. Radiology 2014; 270: 49–56. doi: 10.1148/radiol.13130765 [DOI] [PubMed] [Google Scholar]
- 35. Salim M, Wåhlin E, Dembrower K, Azavedo E, Foukakis T, Liu Y, et al. External evaluation of 3 commercial artificial intelligence algorithms for independent assessment of screening mammograms. JAMA Oncol 2020; 6: 1581–88. doi: 10.1001/jamaoncol.2020.3321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.. D’Orsi C. Breast imaging reporting and data system (BI-RADS). In: Lee CI, Lehman C, Bassett L, editors. Breast Imaging. New York: Oxford University Press; 2018. [Google Scholar]
- 37. Farshid G, Sullivan T, Downey P, Gill PG, Pieterse S. Independent predictors of breast malignancy in screen-detected microcalcifications: biopsy results in 2545 cases. Br J Cancer 2011; 105: 1669–75. doi: 10.1038/bjc.2011.466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang J, Yang Y. A context-sensitive deep learning approach for microcalcification detection in mammograms. Pattern Recognit 2018; 78: 12–22. doi: 10.1016/j.patcog.2018.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang J, Yang X, Cai H, Tan W, Jin C, Li L. Discrimination of breast cancer with microcalcifications on mammography by deep learning. Sci Rep 2016; 6: 27327. doi: 10.1038/srep27327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu H, Chen Y, Zhang Y, Wang L, Luo R, Wu H, et al. A deep learning model integrating mammography and clinical factors facilitates the malignancy prediction of BI-RADS 4 microcalcifications in breast cancer screening. Eur Radiol 2021; 31: 5902–12. doi: 10.1007/s00330-020-07659-y [DOI] [PubMed] [Google Scholar]
- 41. Rodriguez-Ruiz A, Lång K, Gubern-Merida A, Teuwen J, Broeders M, Gennaro G, et al. Can we reduce the workload of mammographic screening by automatic identification of normal exams with artificial intelligence? A feasibility study. Eur Radiol 2019; 29: 4825–32. doi: 10.1007/s00330-019-06186-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yala A, Schuster T, Miles R, Barzilay R, Lehman C. A deep learning model to triage screening mammograms: A simulation study. Radiology 2019; 293: 38–46. doi: 10.1148/radiol.2019182908 [DOI] [PubMed] [Google Scholar]
- 43. Lång K, Dustler M, Dahlblom V, Åkesson A, Andersson I, Zackrisson S. Identifying normal mammograms in a large screening population using artificial intelligence. Eur Radiol 2021; 31: 1687–92. doi: 10.1007/s00330-020-07165-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yala A, Lehman C, Schuster T, Portnoi T, Barzilay R. A deep learning mammography-based model for improved breast cancer risk prediction. Radiology 2019; 292: 60–66. doi: 10.1148/radiol.2019182716 [DOI] [PubMed] [Google Scholar]
- 45. Ha R, Chang P, Karcich J, Mutasa S, Pascual Van Sant E, Liu MZ, et al. Convolutional neural network based breast cancer risk stratification using a mammographic dataset. Acad Radiol 2019; 26: 544–49. doi: 10.1016/j.acra.2018.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Manley H, Mutasa S, Chang P, Desperito E, Crew K, Ha R. Dynamic changes of convolutional neural network-based mammographic breast cancer risk score among women undergoing chemoprevention treatment. Clin Breast Cancer 2021; 21: e312-18. doi: 10.1016/j.clbc.2020.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lehman CD, Yala A, Schuster T, Dontchos B, Bahl M, Swanson K, et al. Mammographic breast density assessment using deep learning: clinical implementation. Radiology 2019; 290: 52–58. doi: 10.1148/radiol.2018180694 [DOI] [PubMed] [Google Scholar]
- 48. Chang K, Beers AL, Brink L, Patel JB, Singh P, Arun NT, et al. Multi-institutional assessment and crowdsourcing evaluation of deep learning for automated classification of breast density. J Am Coll Radiol 2020; 17: 1653–62. doi: 10.1016/j.jacr.2020.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kim J, Kim HJ, Kim C, Kim WH. Artificial intelligence in breast ultrasonography. Ultrasonography 2021; 40: 183–90. doi: 10.14366/usg.20117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mango VL, Sun M, Wynn RT, Ha R. Should we ignore, follow, or biopsy? impact of artificial intelligence decision support on breast ultrasound lesion assessment. AJR Am J Roentgenol 2020; 214: 1445–52. doi: 10.2214/AJR.19.21872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yap MH, Pons G, Marti J, Ganau S, Sentis M, Zwiggelaar R, et al. Automated breast ultrasound lesions detection using convolutional neural networks. IEEE J Biomed Health Inform 2018; 22: 1218–26. doi: 10.1109/JBHI.2017.2731873 [DOI] [PubMed] [Google Scholar]
- 52. Yap MH, Edirisinghe EA, Bez HE. A novel algorithm for initial lesion detection in ultrasound breast images. J Appl Clin Med Phys 2008; 9: 181–99. doi: 10.1120/jacmp.v9i4.2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Virmani J, Kumar V, Kalra N, Khandelwal N. Neural network ensemble based CAD system for focal liver lesions from B-mode ultrasound. J Digit Imaging 2014; 27: 520–37. doi: 10.1007/s10278-014-9685-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lei Y, He X, Yao J, Wang T, Wang L, Li W, et al. Breast tumor segmentation in 3D automatic breast ultrasound using mask scoring R-CNN. Med Phys 2021; 48: 204–14. doi: 10.1002/mp.14569 [DOI] [PubMed] [Google Scholar]
- 55. Zhou Y, Chen H, Li Y, Liu Q, Xu X, Wang S, et al. Multi-task learning for segmentation and classification of tumors in 3D automated breast ultrasound images. Med Image Anal 2021; 70: 101918. doi: 10.1016/j.media.2020.101918 [DOI] [PubMed] [Google Scholar]
- 56. Byra M, Galperin M, Ojeda-Fournier H, Olson L, O’Boyle M, Comstock C, et al. Breast mass classification in sonography with transfer learning using a deep convolutional neural network and color conversion. Med Phys 2019; 46: 746–55. doi: 10.1002/mp.13361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Han S, Kang H-K, Jeong J-Y, Park M-H, Kim W, Bang W-C, et al. A deep learning framework for supporting the classification of breast lesions in ultrasound images. Phys Med Biol 2017; 62: 7714–28. doi: 10.1088/1361-6560/aa82ec [DOI] [PubMed] [Google Scholar]
- 58. Zhou L-Q, Wu X-L, Huang S-Y, Wu G-G, Ye H-R, Wei Q, et al. Lymph node metastasis prediction from primary breast cancer US images using deep learning. Radiology 2020; 294: 19–28. doi: 10.1148/radiol.2019190372 [DOI] [PubMed] [Google Scholar]
- 59. Costa MGF, Campos JPM, de Aquino E Aquino G, de Albuquerque Pereira WC, Costa Filho CFF. Evaluating the performance of convolutional neural networks with direct acyclic graph architectures in automatic segmentation of breast lesion in US images. BMC Med Imaging 2019; 19(1): 85. doi: 10.1186/s12880-019-0389-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hu Y, Guo Y, Wang Y, Yu J, Li J, Zhou S, et al. Automatic tumor segmentation in breast ultrasound images using a dilated fully convolutional network combined with an active contour model. Med Phys 2019; 46: 215–28. doi: 10.1002/mp.13268 [DOI] [PubMed] [Google Scholar]
- 61. Zhuang Z, Li N, Joseph Raj AN, Mahesh VGV, Qiu S. An RDAU-NET model for lesion segmentation in breast ultrasound images. PLoS One 2019; 14(8): e0221535. doi: 10.1371/journal.pone.0221535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Qian X, Pei J, Zheng H, Xie X, Yan L, Zhang H, et al. Prospective assessment of breast cancer risk from multimodal multiview ultrasound images via clinically applicable deep learning. Nat Biomed Eng 2021; 5: 522–32. doi: 10.1038/s41551-021-00711-2 [DOI] [PubMed] [Google Scholar]
- 63. Kim S-Y, Choi Y, Kim E-K, Han B-K, Yoon JH, Choi JS, et al. Deep learning-based computer-aided diagnosis in screening breast ultrasound to reduce false-positive diagnoses. Sci Rep 2021; 11(1): 395. doi: 10.1038/s41598-020-79880-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zheng X, Yao Z, Huang Y, Yu Y, Wang Y, Liu Y, et al. Deep learning radiomics can predict axillary lymph node status in early-stage breast cancer. Nat Commun 2020; 11(1): 1236. doi: 10.1038/s41467-020-15027-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mann RM, Cho N, Moy L. Breast MRI: state of the art. Radiology 2019; 292: 520–36. doi: 10.1148/radiol.2019182947 [DOI] [PubMed] [Google Scholar]
- 66. Sheth D, Giger ML. Artificial intelligence in the interpretation of breast cancer on MRI. J Magn Reson Imaging 2020; 51: 1310–24. doi: 10.1002/jmri.26878 [DOI] [PubMed] [Google Scholar]
- 67. Zhang L, Mohamed AA, Chai R, Guo Y, Zheng B, Wu S. Automated deep learning method for whole-breast segmentation in diffusion-weighted breast MRI. J Magn Reson Imaging 2020; 51: 635–43. doi: 10.1002/jmri.26860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Piantadosi G, Sansone M, Fusco R, Sansone C. Multi-planar 3D breast segmentation in MRI via deep convolutional neural networks. Artif Intell Med 2020; 103: 101781. doi: 10.1016/j.artmed.2019.101781 [DOI] [PubMed] [Google Scholar]
- 69. Dalmış MU, Litjens G, Holland K, Setio A, Mann R, Karssemeijer N, et al. Using deep learning to segment breast and fibroglandular tissue in MRI volumes. Med Phys 2017; 44: 533–46. doi: 10.1002/mp.12079 [DOI] [PubMed] [Google Scholar]
- 70. Zhang Y, Chen J-H, Chang K-T, Park VY, Kim MJ, Chan S, et al. Automatic breast and fibroglandular tissue segmentation in breast MRI using deep learning by a fully-convolutional residual neural network U-net. Acad Radiol 2019; 26: 1526–35. doi: 10.1016/j.acra.2019.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zhang Y, Chan S, Park VY, Chang K-T, Mehta S, Kim MJ, et al. Automatic detection and segmentation of breast cancer on MRI using mask R-CNN trained on non-fat-sat images and tested on fat-sat images. Acad Radiol 2022; 29 Suppl 1: S135-44. doi: 10.1016/j.acra.2020.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. van der Velden BHM, Janse MHA, Ragusi MAA, Loo CE, Gilhuijs KGA. Volumetric breast density estimation on MRI using explainable deep learning regression. Sci Rep 2020; 10(1): 18095. doi: 10.1038/s41598-020-75167-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Antropova N, Abe H, Giger ML. Use of clinical MRI maximum intensity projections for improved breast lesion classification with deep convolutional neural networks. J Med Imaging (Bellingham) 2018; 5: 014503. doi: 10.1117/1.JMI.5.1.014503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Hu Q, Whitney HM, Li H, Ji Y, Liu P, Giger ML. Improved classification of benign and malignant breast lesions using deep feature maximum intensity projection MRI in breast cancer diagnosis using dynamic contrast-enhanced MRI. Radiol Artif Intell 2021; 3: e200159. doi: 10.1148/ryai.2021200159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Fujioka T, Yashima Y, Oyama J, Mori M, Kubota K, Katsuta L, et al. Deep-learning approach with convolutional neural network for classification of maximum intensity projections of dynamic contrast-enhanced breast magnetic resonance imaging. Magn Reson Imaging 2021; 75: 1–8. doi: 10.1016/j.mri.2020.10.003 [DOI] [PubMed] [Google Scholar]
- 76. Dalmiş MU, Gubern-Mérida A, Vreemann S, Bult P, Karssemeijer N, Mann R, et al. Artificial intelligence-based classification of breast lesions imaged with a multiparametric breast MRI protocol with ultrafast DCE-MRI, T2, and DWI. Invest Radiol 2019; 54: 325–32. doi: 10.1097/RLI.0000000000000544 [DOI] [PubMed] [Google Scholar]
- 77. Parekh VS, Macura KJ, Harvey SC, Kamel IR, Ei-Khouli R, Bluemke DA, et al. Multiparametric deep learning tissue signatures for a radiological biomarker of breast cancer: preliminary results. Med Phys 2020; 47: 75–88. doi: 10.1002/mp.13849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hu Q, Whitney HM, Giger ML. A deep learning methodology for improved breast cancer diagnosis using multiparametric MRI. Sci Rep 2020; 10(1): 10536. doi: 10.1038/s41598-020-67441-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zhou J, Luo L-Y, Dou Q, Chen H, Chen C, Li G-J, et al. Weakly supervised 3D deep learning for breast cancer classification and localization of the lesions in MR images. J Magn Reson Imaging 2019; 50: 1144–51. doi: 10.1002/jmri.26721 [DOI] [PubMed] [Google Scholar]
- 80. Liu MZ, Swintelski C, Sun S, Siddique M, Desperito E, Jambawalikar S, et al. Weakly supervised deep learning approach to breast MRI assessment. Acad Radiol 2022; 29 Suppl 1: S166-72. doi: 10.1016/j.acra.2021.03.032 [DOI] [PubMed] [Google Scholar]
- 81. Hickman SE, Baxter GC, Gilbert FJ. Adoption of artificial intelligence in breast imaging: evaluation, ethical constraints and limitations. Br J Cancer 2021; 125: 15–22. doi: 10.1038/s41416-021-01333-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Adachi M, Fujioka T, Mori M, Kubota K, Kikuchi Y, Xiaotong W, et al. Detection and diagnosis of breast cancer using artificial intelligence based assessment of maximum intensity projection dynamic contrast-enhanced magnetic resonance images. Diagnostics (Basel) 2020; 10(5): E330. 10.3390/diagnostics10050330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Watt GP, Sung J, Morris EA, Buys SS, Bradbury AR, Brooks JD, et al. Association of breast cancer with MRI background parenchymal enhancement: the IMAGINE case-control study. Breast Cancer Res 2020; 22: 138. doi: 10.1186/s13058-020-01375-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ha R, Chang P, Mema E, Mutasa S, Karcich J, Wynn RT, et al. Fully automated convolutional neural network method for quantification of breast MRI fibroglandular tissue and background parenchymal enhancement. J Digit Imaging 2019; 32: 141–47. doi: 10.1007/s10278-018-0114-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Eskreis-Winkler S, Sutton EJ, D’Alessio D, Gallagher K, Saphier N, Stember J, et al. (n.d.). Breast MRI background parenchymal enhancement (BPE) categorization using deep learning: outperforming the radiologist. J Magn Reson Imaging. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Zhang Y, Chen J-H, Lin Y, Chan S, Zhou J, Chow D, et al. Prediction of breast cancer molecular subtypes on DCE-MRI using convolutional neural network with transfer learning between two centers. Eur Radiol 2021; 31: 2559–67. doi: 10.1007/s00330-020-07274-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Liu W, Cheng Y, Liu Z, Liu C, Cattell R, Xie X, et al. Preoperative prediction of ki-67 status in breast cancer with multiparametric MRI using transfer learning. Acad Radiol 2021; 28: e44-53. doi: 10.1016/j.acra.2020.02.006 [DOI] [PubMed] [Google Scholar]
- 88. Ha R, Mutasa S, Karcich J, Gupta N, Pascual Van Sant E, Nemer J, et al. Predicting breast cancer molecular subtype with MRI dataset utilizing convolutional neural network algorithm. J Digit Imaging 2019; 32: 276–82. doi: 10.1007/s10278-019-00179-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ha R, Chang P, Mutasa S, Karcich J, Goodman S, Blum E, et al. Convolutional neural network using a breast MRI tumor dataset can predict oncotype dx recurrence score. J Magn Reson Imaging 2019; 49: 518–24. doi: 10.1002/jmri.26244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Ha R, Chang P, Karcich J, Mutasa S, Fardanesh R, Wynn RT, et al. Axillary lymph node evaluation utilizing convolutional neural networks using MRI dataset. J Digit Imaging 2018; 31: 851–56. doi: 10.1007/s10278-018-0086-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ha R, Chang P, Karcich J, Mutasa S, Van Sant EP, Connolly E, et al. Predicting post neoadjuvant axillary response using a novel convolutional neural network algorithm. Ann Surg Oncol 2018; 25: 3037–43. doi: 10.1245/s10434-018-6613-4 [DOI] [PubMed] [Google Scholar]
- 92. Cooper K, Meng Y, Harnan S, Ward S, Fitzgerald P, Papaioannou D, et al. Positron emission tomography (PET) and magnetic resonance imaging (MRI) for the assessment of axillary lymph node metastases in early breast cancer: systematic review and economic evaluation. Health Technol Assess 2011; 15: 1–134. doi: 10.3310/hta15040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Li W, Newitt DC, Gibbs J, Wilmes LJ, Jones EF, Arasu VA, et al. Predicting breast cancer response to neoadjuvant treatment using multi-feature MRI: results from the I-SPY 2 TRIAL. NPJ Breast Cancer 2020; 6: 63. doi: 10.1038/s41523-020-00203-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Choi JH, Kim H-A, Kim W, Lim I, Lee I, Byun BH, et al. Early prediction of neoadjuvant chemotherapy response for advanced breast cancer using PET/MRI image deep learning. Sci Rep 2020; 10(1): 21149. doi: 10.1038/s41598-020-77875-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Tadavarthi Y, Vey B, Krupinski E, Prater A, Gichoya J, Safdar N, et al. The state of radiology AI: considerations for purchase decisions and current market offerings. Radiol Artif Intell 2020; 2: e200004. doi: 10.1148/ryai.2020200004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Eskreis-Winkler S, Onishi N, Pinker K, Reiner J, Kaplan J, Morris EA, et al. Using deep learning to improve nonsystemic viewing of breast cancer on MRI. Journal of Breast Imaging 2021; 3: 201–7. [DOI] [PubMed] [Google Scholar]