Key Points
Question
Is mosaic BRCA1 promoter methylation in normal tissue associated with the risk of incident high-grade serous ovarian cancer (HGSOC) and triple-negative breast cancer (TNBC)?
Findings
In this nested case-control study including 637 women developing TNBC and 511 developing HGSOC, white blood cell BRCA1 promoter methylation was associated with a significantly elevated risk of developing both cancer forms (hazard ratio for HGSOC of 1.93 and TNBC of 2.35). The results remained significant in a subgroup analysis of women who received a diagnosis of cancer more than 5 years after blood sampling (hazard ratio of 1.82 and 2.52, respectively).
Meaning
The study results may serve as proof of concept for early life constitutional methylation as a cancer risk factor.
Abstract
Importance
About 25% of all triple-negative breast cancers (TNBCs) and 10% to 20% of high-grade serous ovarian cancers (HGSOCs) harbor BRCA1 promoter methylation. While constitutional BRCA1 promoter methylation has been observed in normal tissues of some individuals, the potential role of normal tissue methylation as a risk factor for incident TNBC or HGSOC is unknown.
Objective
To assess the potential association between white blood cell BRCA1 promoter methylation and subsequent risk of incident TNBC and HGSOC.
Design, Setting, and Participants
This case-control study included women who were participating in the Women’s Health Initiative study who had not received a diagnosis of either breast or ovarian cancer before study entrance. A total of 637 women developing incident TNBC and 511 women developing incident HGSOC were matched with cancer-free controls (1841 and 2982, respectively) in a nested case-control design. Cancers were confirmed after central medical record review. Blood samples, which were collected at entry, were analyzed for BRCA1 promoter methylation by massive parallel sequencing. The study was performed in the Mohn Cancer Research Laboratory (Bergen, Norway) between 2019 and 2022.
Main Outcomes and Measures
Associations between BRCA1 methylation and incident TNBC and incident HGSOC were analyzed by Cox proportional hazards regression.
Results
Of 2478 cases and controls in the TNBC group and 3493 cases and controls in the HGSOC group, respectively, 7 (0.3%) and 3 (0.1%) were American Indian or Alaska Native, 46 (1.9%) and 30 (0.9%) were Asian, 1 (0.04%) and 1 (0.03%) was Native Hawaiian or Pacific Islander, 326 (13.2%) and 125 (3.6%) were Black or African, 56 (2.3%) and 116 (3.3%) were Hispanic, 2046 (82.6%) and 3257 (93.2%) were White, and 35 (1.4%) and 35 (1.0%) were multiracial. Median (range) age at entry was 62 (50-79) years, with a median interval to diagnosis of 9 (TNBC) and 10 (HGSOC) years. Methylated BRCA1 alleles were present in 194 controls (5.5%). Methylation was associated with risk of incident TNBC (12.4% methylated; HR, 2.35; 95% CI, 1.70-3.23; P < .001) and incident HGSOC (9.4% methylated; HR, 1.93; 95% CI, 1.36-2.73; P < .001). Restricting analyses to individuals with more than 5 years between sampling and cancer diagnosis yielded similar results (TNBC: HR, 2.52; 95% CI, 1.75-3.63; P < .001; HGSOC: HR, 1.82; 95% CI, 1.22-2.72; P = .003). Across individuals, methylation was not haplotype-specific, arguing against an underlying cis-acting factor. Within individuals, BRCA1 methylation was observed on the same allele, indicating clonal expansion from a single methylation event. There was no association found between BRCA1 methylation and germline pathogenic variant status.
Conclusions and Relevance
The results of this case-control suggest that constitutional normal tissue BRCA1 promoter methylation is significantly associated with risk of incident TNBC and HGSOC, with potential implications for prediction of these cancers. These findings warrant further research to determine if constitutional methylation of tumor suppressor genes are pancancer risk factors.
This case-control study examines the potential association between white blood cell BRCA1 promoter methylation and subsequent risk of incident triple-negative breast cancer and high-grade serous ovarian cancer.
Introduction
Women with breast cancer type 1 susceptibility gene (BRCA1) germline pathogenic variants are at high risk of triple-negative breast cancer (TNBC) and high-grade serous ovarian cancer (HGSOC).1 BRCA1, like BRCA2 and several other genes, is required for homologous recombination repair of double-stranded DNA breaks.2 While most TNBCs and HGSOCs have gene expression profiles indicating homologous recombination deficiency (HRD),3,4,5 in many cases, no genetic alteration explains the HRD-positive status.
An alternative mechanism to HRD-positive status is BRCA1 promoter methylation, which causes downregulation of transcription from the BRCA1 gene.6,7 BRCA1 methylation has been detected in tumors of 25% to 30% of TNBCs and 10% to 20% of HGSOCs and has been associated with HRD mutational and gene expression signatures, such as those in cancers in BRCA1 germline pathogenic variant carriers.3,4,5,8 While BRCA1 methylation is commonly considered to have somatic origin,9 the HRD mutational signatures indicate that BRCA1 methylation may occur early in tumor evolution, potentially even as a first triggering event in certain TNBC and HGSOC cases.
Constitutional epimutations are aberrant normal tissue methylation events occurring in utero.10 While several studies have assessed white blood cell (WBC) BRCA1 methylation as a potential risk factor for TNBC or HGSOC,11,12,13,14,15,16,17 apart from 1 study in TNBC18 and 1 in HGSOC,19 these studies have provided mixed results because of a limited number of cases and lack of statistical power. However, the main limitation, which was shared by all prior studies, was blood sample collection after cancer diagnosis with potential for disease-related confounding. Consequently, to our knowledge, associations between constitutional BRCA1 methylation and incident TNBC and HGSOC have not been definitively assessed.
In this article, we assessed the potential association of antecedent WBC BRCA1 promoter methylation with incident TNBC and HGSOC among Women’s Health Initiative (WHI) study participants in a nested case-control study. These tumor forms were selected based on their association with BRCA1 germline pathogenic variants and previous findings of elevated BRCA1 methylation among individuals with a diagnosis of HGSOC but not among other ovarian cancers.19
Methods
Study Design
The WHI study design details were reported previously.20 Briefly, 161 808 postmenopausal women aged 50 to 79 years with an anticipated survival of longer than 3 years were recruited from 40 US clinical centers between 1993 and 1998 to participate in either an observational study or 1 or more of 4 clinical trials (Supplement 1). At study entry, self-administered questionnaires were used to collect demographic characteristics and medical, reproductive, and family history. Race and ethnicity were determined by participant self-report against fixed categories. Entry blood samples were obtained after at least 12 hours of fasting via a prespecified protocol standardized across all study sites. Samples were shipped on dry ice and stored at −80 °C at Fisher Bioservices (Rockville, Maryland).
Clinical outcomes were ascertained annually from enrollment in the observational study and every 6 months for clinical trial participants during the 8.5-year intervention period and annually thereafter. Self-reported cancers were initially confirmed with medical record review at the clinical centers by trained physician adjudicators, with final confirmation at the clinical coordinating center.
Participants provided written informed consent and study protocols were approved at all clinical centers. The current study was additionally approved by the regional ethical committee of the Western Norwegian Health Region.
For the current study, all WHI participants with incident TNBC or HGSOC were included (Supplement 1). Women reporting a history of breast or ovarian cancer at baseline were excluded. For the TNBC study, women with a previous unilateral or bilateral mastectomy for any reason were excluded, while women reporting previous oophorectomy were excluded from the HGSOC study.
The study was conducted as 2 nested case-control studies. Based on up-front statistical power calculations (eMethods in Supplement 2), women with incident TNBC and controls were matched at a 1:3 ratio, whereas women with incident HGSOC and controls were matched at a 1:6 ratio. Controls were matched for age at entry, hormone therapy use, race and ethnicity, and DNA extraction method. In addition, TNBC cases and controls were matched based on age at prior bilateral oophorectomy. Further, controls were required to be alive and disease free at the time of the case diagnosis (eMethods in Supplement 2). The statistical analyses included samples with BRCA1 promoter methylation determinations from incident TNBC (n = 637), incident HGSOC (n = 511), and matched cancer-free controls (n = 1841 and 2982, respectively), as depicted in Figure 1. To limit the required sample number, 1274 of the controls (35.9%) were included in the TNBC and HGSOC groups. Controls were first drawn for the HGSOC comparison and were then eligible to be selected as controls for the TNBC cases. Five cases had TNBC and HGSOC and were included in the risk assessments for both cancers. The study was conducted according to Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) reporting guidelines.
Figure 1. Flowchart Depicting Samples Drawn and Successfully Analyzed From Patients and Controls.
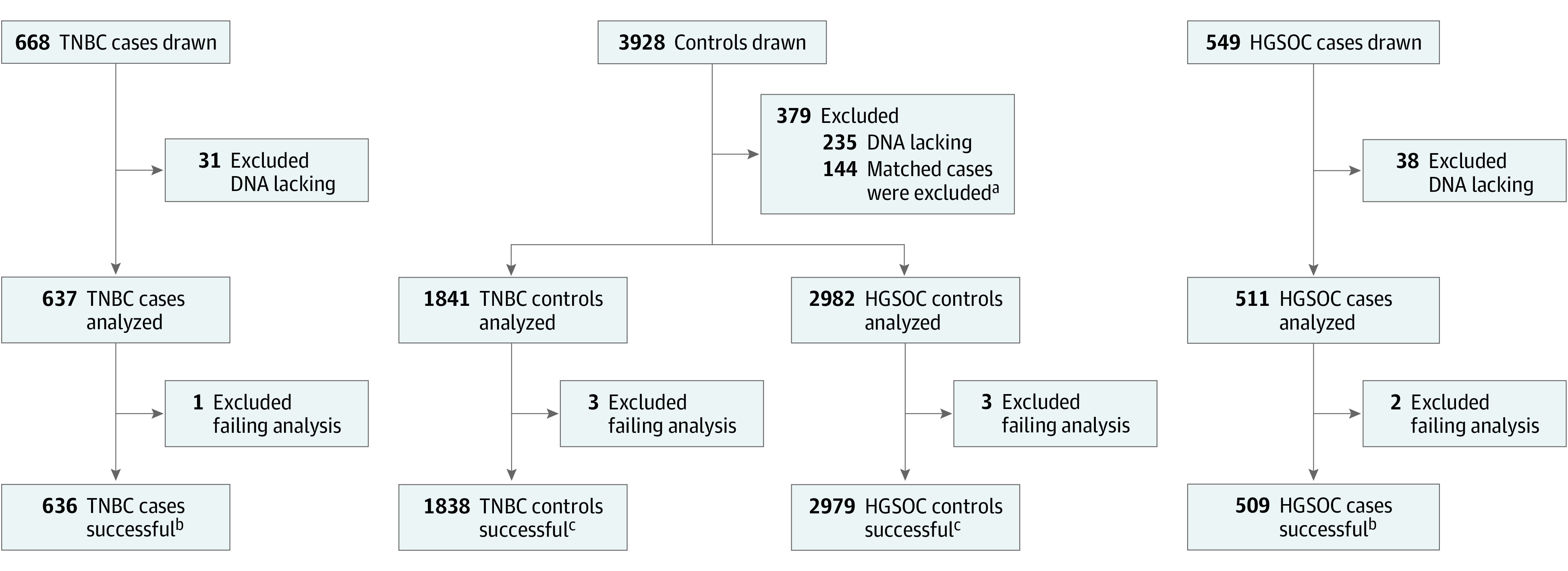
aA total of 144 controls were excluded because their matched cases had too low DNA concentration.
bFive cases had triple-negative breast cancer (TNBC) and high-grade serous ovarian cancer (HGSOC) and were included as cases in the hazard ratio estimates for TNBC and HGSOC.
cA total of 1272 controls were included as controls in the hazard ratio estimates for TNBC and HGSOC.
BRCA1 Promoter Methylation Analysis
A detailed description of the methylation analysis is given in eFigure 1 and eTables 1 and 2 in Supplement 2. In brief, WBC genomic DNA was bisulfite converted, and 4 overlapping regions covering the BRCA1 promoter area were amplified, pooled, indexed, and sequenced to a very high depth (>20 000×) using the Illumina MiSeq System. Methylation status was scored by predefined criteria as variant epiallele frequency (VEF), referring to the frequency of hypermethylated epialleles (eFigures 2 and 3 in Supplement 2).21
All samples were analyzed masked to case-control status. The main cutoff value for methylation positivity was computationally defined based on the assay sensitivity and VEF probability density across the entire sample set, which was masked to case-control status (eFigure 4 in Supplement 2).
Determining Allele Specificity of Methylation
The region covered by sequencing contains a highly prevalent single-nucleotide polymorphism (SNP), rs799905, which is located in the BRCA1 gene body (eFigure 1 in Supplement 2). By analyzing methylation in sequencing reads covering this SNP, we assessed the potential allele specificity of methylation in individuals heterozygous for the SNP.
Association Between BRCA1 Methylation Status and Cancer Risk Genes
We tested for the association of BRCA1 methylation status with germline pathogenic variants in a subgroup of 234 participants (5.0%) from the study (173 cases and 61 controls) who had previously been tested for germline pathogenic variants in BRCA1/2 and in 26 additional cancer risk genes (eTable 7 in Supplement 2) as part of another WHI ancillary study.22
Statistical Methods
The potential associations between BRCA1 methylation and incident TNBC and HGSOC were assessed by estimating hazard ratios (HRs) and 95% CIs. The HRs were determined using Cox proportional hazards regression in matched case-control groups, including age, race and ethnicity, previous hormone use, DNA extraction method, and (for TNBC) previous oophorectomy as covariates. In addition, we performed hypothesis-generating supportive subgroup analyses.
Power estimates based on previous results of methylation frequency among HGSOC cases and controls19 are outlined in the eMethods in Supplement 2. In brief, a conservative assumption of 600 TNBC cases and 400 HGSOC cases was made. Matching 600 TNBC with 1800 control samples in a nested design provides a power of 0.88 to detect an HR of 2.0. Similarly, for HGSOC, comparing 400 patients with 2400 controls provides a power of 0.80.
In sensitivity analyses, we analyzed HR associated with BRCA1 methylation status in different promoter subregions, age groups, cutoff levels for methylation positivity, and methylation positivity assessed by methylation beta values (ratio of methylated to total number of cytosines; eMethods in Supplement 2). As oophorectomy has been associated with a reduced risk of TNBC in BRCA1 germline pathogenic variant carriers,23 we also performed subgroup analysis assessing HRs for TNBC among cases and controls who were not undergoing an oophorectomy. Analysis were conducted using R, version 4.0.3 (R Foundation).
Results
The demographic characteristics and flowchart of TNBC and HGSOC cases and controls are presented in the Table and Figure 1. There was no systemic difference between individuals excluded from analysis because of lack of DNA and final participants. BRCA1 methylation status was not associated with a self-reported family history of either breast or ovarian cancer (eTable 3 in Supplement 2). Among controls, across all age groups, 194 (5.5%) had methylated BRCA1 alleles (eTable 4 in Supplement 2), contrasting with 79 (12.4%) and 48 (9.4%) in TNBC and HGSOC cases, respectively. For cases and controls, most alleles were either fully methylated or unmethylated (eFigures 2 and 3 in Supplement 2).
Table. Demographic Characteristics of Patients and Controls Included in the Trial.
| Characteristic | No. (%) | |||
|---|---|---|---|---|
| TNBC | HGSOC | |||
| Cases (n = 637) | Controls (n = 1841)a | Cases (n = 511) | Controls (n = 2982)a | |
| Age, y | ||||
| Mean (SD) | 62.1 (6.81) | 62.1 (6.74) | 62.3 (6.42) | 62.7 (6.82) |
| Median (range) | 62.0 (50.0-78.0) | 62.0 (50.0-79.0) | 62.0 (50.0-79.0) | 62.0 (50.0-79.0) |
| IQR (Q1-Q3) | 11.0 (57.0-68.0) | 10.0 (57.0-67.0) | 9.50 (57.5-67.0) | 11.0 (57.0-68.0) |
| Ethnicity | ||||
| Not Hispanic or Latino | 621 (97.5) | 1795 (97.5) | 493 (96.5) | 2880 (96.6) |
| Hispanic or Latino | 13 (2.0) | 43 (2.3) | 16 (3.1) | 100 (3.4) |
| Unknown or not reported | 3 (0.5) | 3 (0.2) | 2 (0.4) | 2 (0.1) |
| Race | ||||
| American Indian or Alaska Native | 1 (0.2) | 6 (0.3) | 2 (0.4) | 1 (0.0) |
| Asian | 13 (2.0) | 33 (1.8) | 5 (1.0) | 25 (0.8) |
| Native Hawaiian or other Pacific Islander | 0 | 1 (0.1) | 0 | 1 (0.0) |
| Black or African American | 81 (12.7) | 245 (13.3) | 18 (3.5) | 107 (3.6) |
| White | 525 (82.4) | 1521 (82.6) | 479 (93.7) | 2778 (93.2) |
| Multiracial | 12 (1.9) | 23 (1.2) | 3 (0.6) | 32 (1.1) |
| Unknown or not reported | 5 (0.8) | 12 (0.7) | 4 (0.8) | 38 (1.3) |
| Years from DNA sampling to diagnosis | ||||
| Mean (SD) | 10.1 (5.23) | NA | 10.4 (5.80) | NA |
| Median (range) | 9.00 (0-23.0) | NA | 10.0 (0-23.0) | NA |
| IQR (Q1-Q3) | 8.00 (6.00-14.0) | NA | 9.00 (6.00-15.0) | NA |
| Missing | 0 | 1841 (100) | 0 | 2982 (100) |
| Bilateral oophorectomy | ||||
| No | 509 (79.9) | 1493 (81.1) | 511 (100) | 2982 (100) |
| Yes | 118 (18.5) | 320 (17.4) | 0 | 0 |
| Missing | 10 (1.6) | 28 (1.5) | 0 | 0 |
| Family history of breast cancer | ||||
| No | 158 (24.8) | 519 (28.2) | 137 (26.8) | 857(28.7) |
| Yes | 154 (24.2) | 294 (16.0) | 94 (18.4) | 514 (17.2) |
| Missing | 325 (51.0) | 1028 (55.8) | 280 (54.8) | 1611 (54.0) |
| Family history of ovarian cancer | ||||
| No | 289 (45.4) | 723 (39.3) | 209 (40.9) | 1252 (42.0) |
| Yes | 18 (2.8) | 42 (2.3) | 13 (2.5) | 61 (2.0) |
| Do not know | 16 (2.5) | 64 (3.5) | 17 (3.3) | 86 (2.9) |
| Missing | 314 (49.3) | 1012 (55.0) | 272 (53.2) | 1583 (53.1) |
| High-risk germline variants | ||||
| BRCA1 | 2 (0.3) | 0 | 1 (0.2) | 0 |
| BRCA2 | 1 (0.2) | 0 | 1 (0.2) | 0 |
| No variant detected | 101 (15.9) | 25 (1.4) | 4 (0.8) | 36 (1.2) |
| Missing | 533 (83.7) | 1816 (98.6) | 505 (98.8) | 2946 (98.8) |
Abbreviations: HGSOC, high-grade serous ovarian cancer; NA, not applicable; Q, quartile; TNBC, triple-negative breast cancer.
A total of 1274 controls were included as controls for comparison with TNBC and HGSOC. Among these 1274, 2 failed analyses. Thus, 1272 controls were included in the final hazard ratio estimates for TNBC and HGSOC (Figure 1).
Risk of TNBC and HGSOC
For women with incident TNBC, the median (IQR) follow-up from sampling to diagnosis was 9 (8) years. The presence of methylated BRCA1 alleles in WBCs was significantly associated with incident TNBC risk (HR, 2.35; 95% CI, 1.70-3.23; P < .001; Figure 2). The HR was 1.83 (95% CI, 0.92-3.64; P = .08) for TNBC diagnosed 5 years before or less and 2.52 (95% CI, 1.75-3.63; P < .001) for those who received a diagnosis more than 5 years after blood sampling. There was no difference between subgroups defined by age at entry or methylation level (Figure 2).
Figure 2. Hazard Ratios for Triple-Negative Breast Cancer (TNBC) Associated With the Presence of Methylated BRCA1 Alleles in the Overall Cohort and Selected Subgroups.
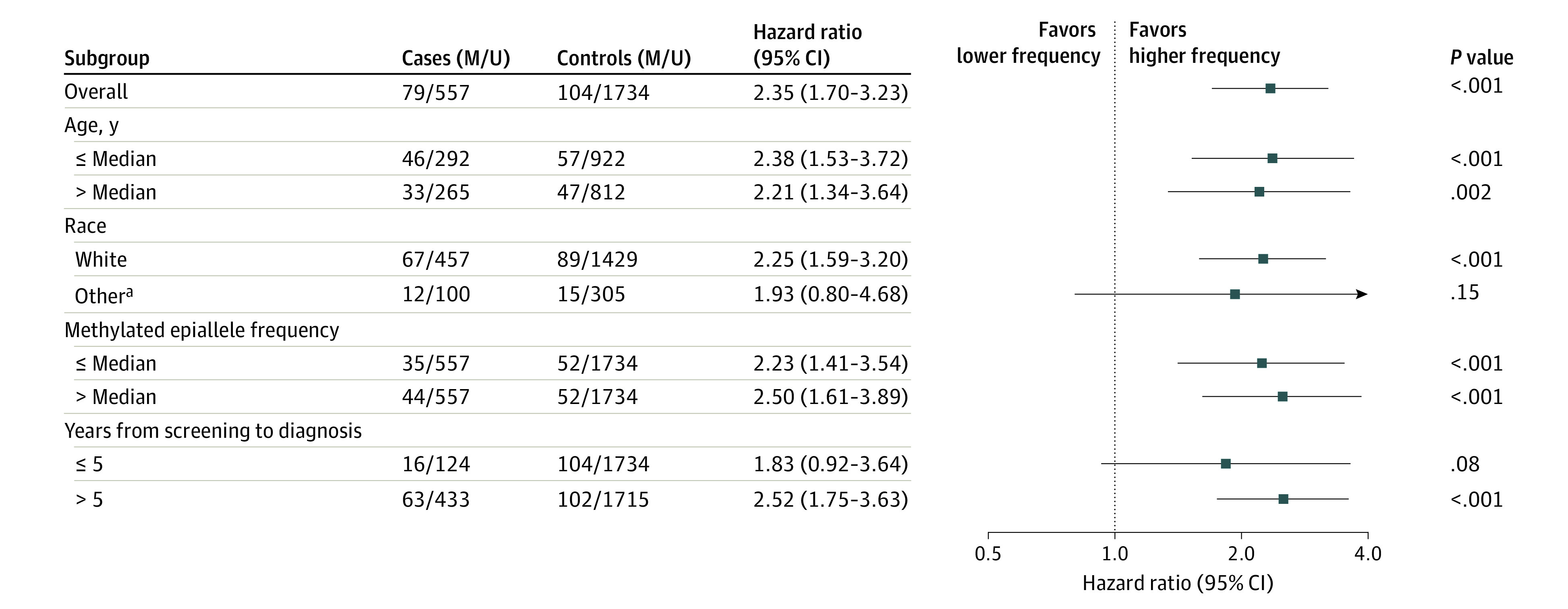
M and U represent the number of methylated and unmethylated samples, respectively. Median methylated epiallele frequency equals median variant epiallele frequency for samples classified as methylation-positive.
aOther race includes American Indian or Alaska Native, Asian, Native Hawaiian or Pacific Islander, Black or African American, multiracial, unknown, or not reported.
For women with incident HGSOC, the median (IQR) time from sampling to diagnosis was 10 (9) years. Like the findings for TNBC, the presence of methylated BRCA1 alleles in WBCs was significantly associated with incident HGSOC risk (HR, 1.93; 95% CI, 1.36-2.73; P < .001; Figure 3). The HR was significant for HGSOC diagnosed 5 years or less (HR, 2.28; 95% CI, 1.13-4.59; P = .02) or more than 5 years (HR, 1.82; 95% CI, 1.22-2.72; P = .003) after blood sampling. Like for TNBC, the association remained significant in subgroups stratified for age at inclusion and methylation level (Figure 3).
Figure 3. Hazard Ratios for High-grade Serous Ovarian Cancer (HGSOC) Associated With the Presence of Methylated BRCA1 Alleles in the Overall Cohort and Selected Subgroups.
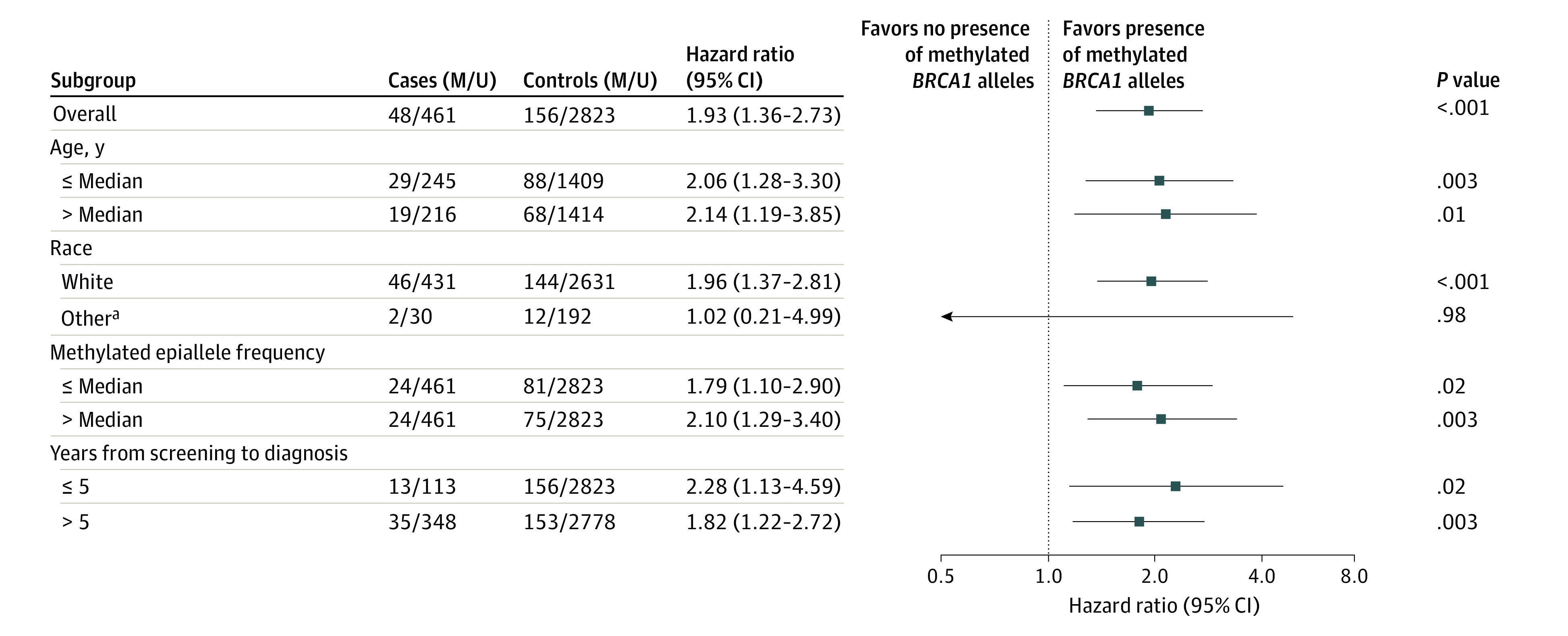
M and U represent the number of methylated and unmethylated samples, respectively. Median methylated epiallele frequency here equals median variant epiallele frequency for samples classified as methylation-positive.
aOther race includes American Indian or Alaska Native, Asian, Native Hawaiian or Pacific Islander, Black or African American, multiracial, unknown, or not reported.
Sensitivity Analysis
We assessed the potential effect of promoter area selection, analytical cutoff, and different methods for methylation classification. These analyses revealed results that confirmed the findings from the main analysis (eFigures 5-14 in Supplement 2). Subgroup analysis excluding TNBC cases and controls who underwent oophorectomy or the 5 cases who had TNBC and HGSOC after DNA collection had no significant association with HRs (eFigure 15 in Supplement 2).
Allele-Specific BRCA1 Methylation
Assessing methylation in the region harboring BRCA1 SNP rs799905, we found methylation frequencies to be similar among individuals carrying the different rs799905 alleles. This suggested that BRCA1 methylation is not associated with a cis-acting factor (factor located on the same allele as the methylation). However, in individuals who were heterozygous for rs799905 for whom allele-spesific methylation could be determined, the intraindividual methylation was strongly enriched at a single allele. In more than 90% of individuals, more than 95% of the methylation was associated with one of the rs799905 alleles, indicating that BRCA1 methylation may have occurred as a single, early event that was followed by clonal expansion of the methylated cell (eFigure 16 in Supplement 2).
Association of BRCA1 Methylation With Germline Pathogenic Variant Status
BRCA1 methylation was not associated with germline mutation status for either BRCA1 or BRCA2. Similarly, methylation was not associated with the germline status of any of the 26 other cancer risk genes analyzed (eTable 7 in Supplement 2).
Discussion
In a nested case-control design in postmenopausal WHI participants, including women with incident TNBC (n = 637), incident HGSOC (n = 511), and matched cancer-free controls (n = 1841 and 2982, respectively), WBC BRCA1 promoter methylation was significantly associated with higher risk of incident TNBC and incident HGSOC. The association was also significant in analyses restricted to cancers diagnosed more than 5 years after sampling. While an association between BRCA1 methylation in normal tissue and TNBC and HGSOC was established previously,11,18,19 these studies were performed on normal tissue obtained after the patients received their cancer diagnoses. Thus, this study’s findings represent a potential conceptual breakthrough, the results suggesting that BRCA1 normal tissue methylation in association with TNBC and HGSOC occurs before, and not because of, cancer development. The association of BRCA1 normal tissue methylation with higher risk for TNBC and HGSOC, the 2 major cancers associated with germline BRCA1 pathogenic variants, supports this conclusion. Given the frequency of BRCA1 methylation in TNBC and HGSOC, BRCA1 normal tissue methylation may be an underlying cause of a substantial fraction of TNBC and HGSOC cases.
Constitutional epimutations may be classified as primary epimutations (methylation in absence of any genetic aberration) or secondary epimutations that are associated with rare germline genetic variants.10,24 Secondary epimutations have been detected in only a few cases associated with the Lynch syndrome–associated MLH1 gene, but also BRCA1.24,25 In such cases, the fraction of methylated alleles is typically around 50% (ie, 100% of cells carrying the epigenetic variant in a heterozygous individual), and cancer penetrance is high. In contrast, primary BRCA1 normal tissue methylation assessed in WBCs is not a rare event; it occurs as a low-mosaic phenomenon in 4% to 10% of adult women and newborn girls without cancer.19,26 A potential pathogenic role of mosaic methylations may be paralleled with an elevated cancer risk associated with mosaic germline pathogenic variants in BRCA1 as well as other tumor suppressor genes.27,28,29,30,31 The finding that BRCA1 methylation was not associated with a family history of breast/ovarian cancer was expected, considering the magnitude of the HRs reported.
Recent trials have provided preliminary evidence suggesting that BRCA1 methylation may be associated with a more favorable response to chemotherapy, as well as polyadenosine diphosphate-ribose polymerase inhibition, in breast cancer.8,32 Neither trial distinguished somatic from constitutional methylation. Thus, it remains to be determined if constitutionally methylated tumors have the same phenotypical characteristics as those methylated somatically.
One may assume that the effect of BRCA1 methylation and BRCA1 pathogenic variants is similar in individual cells. In this article, we found the odds ratios for TNBC as well as HGSOC associated with WBC BRCA1 methylation to be modest as compared with the odds ratio of more than 50 for TNBC in BRCA1 germline pathogenic variant carriers.33 This likely reflects the fact that germline pathogenic variants affect all cells in the body, whereas only a small fraction, 0.1% to 10% of the cells, carry methylated alleles. The present study’s finding that BRCA1 methylation occurs independent of rs799905 genotype across individuals is consistent with previous findings,19 arguing against a cis-acting genetic factor, subject to mendelian inheritance, as the underlying cause of methylation.
An important question is to what extent WBC BRCA1 methylation represents BRCA1 methylation in normal tissue across other organs, like the ovaries, fallopian tubes, breasts, and others. Global DNA methylation pattern may vary across different tissue compartments and even between WBC subfractions.34,35 However, we previously found WBC BRCA1 promoter methylation to be strongly associated with methylation status in other benign tissues.19 This finding, in concert with the observation that BRCA1 mosaic promoter methylation occurs across all age groups, including newborns, supports the hypothesis that this methylation is constitutive,10 thus affecting different tissue compartments derived from all the embryonic germ layers.
General methylation patterns may change with age.36 However, in a previous study,19 we found WBC BRCA1 methylation among females across all age groups, with a slight drop in frequency during lifetime. In the present study, we found BRCA1 methylation to be predominantly monoallelic, nearly completely restricted to the same allele across affected cells within the same individual, as previously indicated by Hansman and colleagues13 in a small group of patients. While this finding does not fully exclude the possibility of dynamic modulations of BRCA1 methylation through life, it supports the hypothesis that normal tissue BRCA1 methylation has a clonal origin, arising in a single cell at an early embryonic stage.
An important question is whether BRCA1 methylation could be a secondary event to germline pathogenic variants in BRCA1 or other tumor suppressor genes. Our subgroup analysis revealed no evidence for such a covariance. Regarding BRCA1 pathogenic variants, in a previous study analyzing more than 250 individuals harboring BRCA1 germline pathogenic variants who had received a diagnosis of HGSOC, we found constitutional methylation frequency to be lower compared with individuals harboring either germline BRCA2 variants or being wild type for both genes.19
Limitations
All WHI participants were postmenopausal, with a median age of 62 years, with 813 (17%) being 70 years or older at entry. In general, TNBC is more common in younger women. Thus, while TNBC constitutes about 15% of all US breast cancers, the percentage of TNBC in WHI is 7%.37 Moreover, the prevalence of tumor BRCA1 methylation is higher in younger compared with older women with TNBC.4 Thus, in our previous study assessing BRCA1 WBC methylation status in women at all ages who already received a diagnosis of HGSOC19 the odds ratio associated with HGSOC varied between 2.2 to 2.9 and was higher among younger compared with older individuals. This indicates that the risk of TNBC and HGSOC associated with WBC BRCA1 methylation may be underestimated in the present study. While information on BRCA1 germline pathogenic variant status was not available for all patients in the current study, in the subgroup from whom BRCA1 methylation could be compared with BRCA1/2 germline pathogenic variant status, we detected no association between germline variant status and BRCA1 methylation. Notably, BRCA1 promoter methylation and BRCA1 germline pathogenic variants have been reported to be mutually exclusive in breast cancer tissue4 as well as in WBC collected from patients with a diagnosis of HGSOC.19 The present study was not powered to assess different racial and ethnic groups, precluding conclusions for risk in African American individuals, for whom the incidence of TNBC is known to be elevated.
For cancer risk studies, the findings should be confirmed in independent cohorts. To our knowledge, there are few population-based cohorts that are enrolling sufficient numbers of participants, including follow-ups with regular health assessment, that are adequate to confirm our findings. However, our study contains an indirect independent validation because it suggests that there is an association between BRCA1 methylation and TNBC and HGSOC, the 2 cancer forms most strongly associated with germline BRCA1 pathogenic variants.
Conclusions
In this case-control study, we found normal tissue (WBC) BRCA1 promoter methylation to be associated with an elevated HR for incident TNBC and HGSOC (also when restricting analysis to cancers developing more than 5 years after WBC sampling). The study findings have 2 major implications. First, WBC BRCA1 methylation may help identify women at elevated risk of TNBC and HGSOC. This raises the question whether methylation carriers should be offered breast cancer screening at a younger age compared with the general population. While multimodal screening for HGSOC has not been recommended for women without deleterious germline BRCA1/BRCA2 pathogenic variants,38 the study findings add evidence to future discussion of potential screening strategies for this serious cancer diagnosis. Second, the current study findings, in concert with previous data revealing WBC BRCA1 methylation to occur in women across all age groups, even newborns,19 point to BRCA1 methylation as an early embryonic event that is followed by clonal expansion. Prenatal events have been associated with risk of breast cancer, as well as other cancers.39,40,41 Thus, this study’s findings should trigger further research assessing the potential association of constitutional methylation of other genes with the risk of other cancer types and research into the cause(s) of constitutional normal tissue methylation of tumor suppressor genes.
Trial protocol
eMethods.
eResults.
eTable 1. GRCh38 genomic coordinates of PCR amplicons and individual CpGs
eTable 2. Primer sequences and amplification conditions for PCR amplicons
eTable 3. Fractions of methylated samples by case-control status and family history of cancer
eTable 4. Fractions of methylated controls by age at screening
eTable 5. Fractions of methylated BRCA1 epiallele carriers by hormone therapy group
eTable 6. Sample counts in relation to rs799905 SNP genotype.
eTable 7. Distribution of germline mutations in shared case and control samples
eFigure 1. The genomic structure of the BRCA1 promoter region
eFigure 2. Observed versus expected methylation levels in control samples
eFigure 3. Empirical cumulative density function (eCDF) plots
eFigure 4. Histogram of log10-transformed VEF values for region CpG14–34
eFigure 5. Risk of incident TNBC or HGSOC applying increasing methylation cutoff values
eFigure 6. Risk of incident TNBC or HGSOC calculated using optimized cutoffs
eFigure 7. Risk of incident TNBC or HGSOC calculated for different sets of CpGs
eFigure 8. Risk of incident TNBC or HGSOC calculated for different PCR amplicons
eFigure 9. Risk (OR) of incident TNBC or HGSOC calculated using 2x2 contingency tables
eFigure 10. Risk (OR) of incident TNBC or HGSOC calculated using multivariate logistic regression
eFigure 11. Scatter plot of methylation beta and VEF values
eFigure 12. Risk of incident TNBC or HGSOC calculated using beta instead of VEF values
eFigure 13. Risk of incident TNBC or HGSOC in subgroups based on hormone therapy exposure
eFigure 14. Risk of incident TNBC or HGSOC accounting for potential interaction between methylation and hormone therapy
eFigure 15. Risk of incident TNBC or HGSOC calculated for subsets of samples
eFigure 16. Allele specificity of methylation
eReferences.
References
- 1.Kuchenbaecker KB, Hopper JL, Barnes DR, et al. ; BRCA1 and BRCA2 Cohort Consortium . Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA. 2017;317(23):2402-2416. doi: 10.1001/jama.2017.7112 [DOI] [PubMed] [Google Scholar]
- 2.Mateo J, Lord CJ, Serra V, et al. A decade of clinical development of PARP inhibitors in perspective. Ann Oncol. 2019;30(9):1437-1447. doi: 10.1093/annonc/mdz192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies H, Glodzik D, Morganella S, et al. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat Med. 2017;23(4):517-525. doi: 10.1038/nm.4292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glodzik D, Bosch A, Hartman J, et al. Comprehensive molecular comparison of BRCA1 hypermethylated and BRCA1 mutated triple negative breast cancers. Nat Commun. 2020;11(1):3747. doi: 10.1038/s41467-020-17537-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Konstantinopoulos PA, Ceccaldi R, Shapiro GI, D’Andrea AD. Homologous recombination deficiency: exploiting the fundamental vulnerability of ovarian cancer. Cancer Discov. 2015;5(11):1137-1154. doi: 10.1158/2159-8290.CD-15-0714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rice JC, Ozcelik H, Maxeiner P, Andrulis I, Futscher BW. Methylation of the BRCA1 promoter is associated with decreased BRCA1 mRNA levels in clinical breast cancer specimens. Carcinogenesis. 2000;21(9):1761-1765. doi: 10.1093/carcin/21.9.1761 [DOI] [PubMed] [Google Scholar]
- 7.Bell D, Berchuck A, Birrer M, et al. ; Cancer Genome Atlas Research Network . Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609-615. doi: 10.1038/nature10166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eikesdal HP, Yndestad S, Elzawahry A, et al. Olaparib monotherapy as primary treatment in unselected triple negative breast cancer. Ann Oncol. 2021;32(2):240-249. doi: 10.1016/j.annonc.2020.11.009 [DOI] [PubMed] [Google Scholar]
- 9.Mazor T, Pankov A, Johnson BE, et al. DNA methylation and somatic mutations converge on the cell cycle and define similar evolutionary histories in brain tumors. Cancer Cell. 2015;28(3):307-317. doi: 10.1016/j.ccell.2015.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lønning PE, Eikesdal HP, Løes IM, Knappskog S. Constitutional mosaic epimutations—a hidden cause of cancer? Cell Stress. 2019;3(4):118-135. doi: 10.15698/cst2019.04.183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Snell C, Krypuy M, Wong EM, Loughrey MB, Dobrovic A; kConFab investigators . BRCA1 promoter methylation in peripheral blood DNA of mutation negative familial breast cancer patients with a BRCA1 tumour phenotype. Breast Cancer Res. 2008;10(1):R12. doi: 10.1186/bcr1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong EM, Southey MC, Fox SB, et al. Constitutional methylation of the BRCA1 promoter is specifically associated with BRCA1 mutation-associated pathology in early-onset breast cancer. Cancer Prev Res (Phila). 2011;4(1):23-33. doi: 10.1158/1940-6207.CAPR-10-0212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansmann T, Pliushch G, Leubner M, et al. Constitutive promoter methylation of BRCA1 and RAD51C in patients with familial ovarian cancer and early-onset sporadic breast cancer. Hum Mol Genet. 2012;21(21):4669-4679. doi: 10.1093/hmg/dds308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iwamoto T, Yamamoto N, Taguchi T, Tamaki Y, Noguchi S. BRCA1 promoter methylation in peripheral blood cells is associated with increased risk of breast cancer with BRCA1 promoter methylation. Breast Cancer Res Treat. 2011;129(1):69-77. doi: 10.1007/s10549-010-1188-1 [DOI] [PubMed] [Google Scholar]
- 15.Bosviel R, Michard E, Lavediaux G, Kwiatkowski F, Bignon YJ, Bernard-Gallon DJ. Peripheral blood DNA methylation detected in the BRCA1 or BRCA2 promoter for sporadic ovarian cancer patients and controls. Clin Chim Acta. 2011;412(15-16):1472-1475. doi: 10.1016/j.cca.2011.04.027 [DOI] [PubMed] [Google Scholar]
- 16.Kontorovich T, Cohen Y, Nir U, Friedman E. Promoter methylation patterns of ATM, ATR, BRCA1, BRCA2 and p53 as putative cancer risk modifiers in Jewish BRCA1/BRCA2 mutation carriers. Breast Cancer Res Treat. 2009;116(1):195-200. doi: 10.1007/s10549-008-0121-3 [DOI] [PubMed] [Google Scholar]
- 17.Azzollini J, Pesenti C, Pizzamiglio S, et al. Constitutive BRCA1 promoter hypermethylation can be a predisposing event in isolated early-onset breast cancer. Cancers (Basel). 2019;11(1):E58. doi: 10.3390/cancers11010058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prajzendanc K, Domagała P, Hybiak J, et al. BRCA1 promoter methylation in peripheral blood is associated with the risk of triple-negative breast cancer. Int J Cancer. 2020;146(5):1293-1298. doi: 10.1002/ijc.32655 [DOI] [PubMed] [Google Scholar]
- 19.Lønning PE, Berge EO, Bjørnslett M, et al. White blood cell BRCA1 promoter methylation status and ovarian cancer risk. Ann Intern Med. 2018;168(5):326-334. doi: 10.7326/M17-0101 [DOI] [PubMed] [Google Scholar]
- 20.Anderson G, Cummings S, Freedman LS, et al. ; Women’s Health Initiative Study Group . Design of the Women’s Health Initiative clinical trial and observational study. Control Clin Trials. 1998;19(1):61-109. doi: 10.1016/S0197-2456(97)00078-0 [DOI] [PubMed] [Google Scholar]
- 21.Nikolaienko O, Lønning PE, Knappskog S. epialleleR: an R/BioC package for sensitive allele-specific methylation analysis in NGS data. bioRxiv 2022: 2022.06.30.498213. doi: 10.1101/2022.06.30.498213 [DOI] [PMC free article] [PubMed]
- 22.Kurian AW, Bernhisel R, Larson K, et al. Prevalence of pathogenic variants in cancer susceptibility genes among women with postmenopausal breast cancer. JAMA. 2020;323(10):995-997. doi: 10.1001/jama.2020.0229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Domchek SM, Friebel TM, Singer CF, et al. Association of risk-reducing surgery in BRCA1 or BRCA2 mutation carriers with cancer risk and mortality. JAMA. 2010;304(9):967-975. doi: 10.1001/jama.2010.1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hitchins MP. Constitutional epimutation as a mechanism for cancer causality and heritability? Nat Rev Cancer. 2015;15(10):625-634. doi: 10.1038/nrc4001 [DOI] [PubMed] [Google Scholar]
- 25.Evans DGR, van Veen EM, Byers HJ, et al. A dominantly inherited 5′ UTR variant causing methylation-associated silencing of BRCA1 as a cause of breast and ovarian cancer. Am J Hum Genet. 2018;103(2):213-220. doi: 10.1016/j.ajhg.2018.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Al-Moghrabi N, Al-Showimi M, Al-Yousef N, et al. Methylation of BRCA1 and MGMT genes in white blood cells are transmitted from mothers to daughters. Clin Epigenetics. 2018;10(1):99. doi: 10.1186/s13148-018-0529-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steinke-Lange V, de Putter R, Holinski-Feder E, Claes KBM. Somatic mosaics in hereditary tumor predisposition syndromes. Eur J Med Genet. 2021;64(12):104360. doi: 10.1016/j.ejmg.2021.104360 [DOI] [PubMed] [Google Scholar]
- 28.Friedman E, Efrat N, Soussan-Gutman L, et al. Low-level constitutional mosaicism of a de novoBRCA1 gene mutation. Br J Cancer. 2015;112(4):765-768. doi: 10.1038/bjc.2015.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J, Walsh MF, Wu G, et al. Germline mutations in predisposition genes in pediatric cancer. N Engl J Med. 2015;373(24):2336-2346. doi: 10.1056/NEJMoa1508054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evans DG, Ramsden RT, Shenton A, et al. Mosaicism in neurofibromatosis type 2: an update of risk based on uni/bilaterality of vestibular schwannoma at presentation and sensitive mutation analysis including multiple ligation-dependent probe amplification. J Med Genet. 2007;44(7):424-428. doi: 10.1136/jmg.2006.047753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pareja F, Ptashkin RN, Brown DN, et al. Cancer-causative mutations occurring in early embryogenesis. Cancer Discov. 2022;12(4):949-957. doi: 10.1158/2159-8290.CD-21-1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stefansson OA, Hilmarsdottir H, Olafsdottir K, et al. BRCA1 promoter methylation status in 1031 primary breast cancers predicts favorable outcomes following chemotherapy. JNCI Cancer Spectr. 2019;4(2):pkz100. doi: 10.1093/jncics/pkz100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walsh T, Gulsuner S, Lee MK, et al. Inherited predisposition to breast cancer in the Carolina Breast Cancer Study. NPJ Breast Cancer. 2021;7(1):6. doi: 10.1038/s41523-020-00214-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teschendorff AE, Menon U, Gentry-Maharaj A, et al. An epigenetic signature in peripheral blood predicts active ovarian cancer. PLoS One. 2009;4(12):e8274. doi: 10.1371/journal.pone.0008274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones MJ, Goodman SJ, Kobor MS. DNA methylation and healthy human aging. Aging Cell. 2015;14(6):924-932. doi: 10.1111/acel.12349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chlebowski RT, Aragaki AK, Prentice RL. Dietary moderation and deaths from breast cancer. J Clin Oncol. 2020;38(26):3071-3072. doi: 10.1200/JCO.20.01218 [DOI] [PubMed] [Google Scholar]
- 38.Sroczynski G, Gogollari A, Kuehne F, et al. A systematic review on cost-effectiveness studies evaluating ovarian cancer early detection and prevention strategies. Cancer Prev Res (Phila). 2020;13(5):429-442. doi: 10.1158/1940-6207.CAPR-19-0506 [DOI] [PubMed] [Google Scholar]
- 39.Denholm R, De Stavola B, Hipwell JH, et al. Pre-natal exposures and breast tissue composition: findings from a British pre-birth cohort of young women and a systematic review. Breast Cancer Res. 2016;18(1):102. doi: 10.1186/s13058-016-0751-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qiu L, Onoyama S, Low HP, et al. Effect of preeclampsia on umbilical cord blood stem cells in relation to breast cancer susceptibility in the offspring. Carcinogenesis. 2015;36(1):94-98. doi: 10.1093/carcin/bgu231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swerdlow AJ, De Stavola BL, Swanwick MA, Maconochie NES. Risks of breast and testicular cancers in young adult twins in England and Wales: evidence on prenatal and genetic aetiology. Lancet. 1997;350(9093):1723-1728. doi: 10.1016/S0140-6736(97)05526-8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Trial protocol
eMethods.
eResults.
eTable 1. GRCh38 genomic coordinates of PCR amplicons and individual CpGs
eTable 2. Primer sequences and amplification conditions for PCR amplicons
eTable 3. Fractions of methylated samples by case-control status and family history of cancer
eTable 4. Fractions of methylated controls by age at screening
eTable 5. Fractions of methylated BRCA1 epiallele carriers by hormone therapy group
eTable 6. Sample counts in relation to rs799905 SNP genotype.
eTable 7. Distribution of germline mutations in shared case and control samples
eFigure 1. The genomic structure of the BRCA1 promoter region
eFigure 2. Observed versus expected methylation levels in control samples
eFigure 3. Empirical cumulative density function (eCDF) plots
eFigure 4. Histogram of log10-transformed VEF values for region CpG14–34
eFigure 5. Risk of incident TNBC or HGSOC applying increasing methylation cutoff values
eFigure 6. Risk of incident TNBC or HGSOC calculated using optimized cutoffs
eFigure 7. Risk of incident TNBC or HGSOC calculated for different sets of CpGs
eFigure 8. Risk of incident TNBC or HGSOC calculated for different PCR amplicons
eFigure 9. Risk (OR) of incident TNBC or HGSOC calculated using 2x2 contingency tables
eFigure 10. Risk (OR) of incident TNBC or HGSOC calculated using multivariate logistic regression
eFigure 11. Scatter plot of methylation beta and VEF values
eFigure 12. Risk of incident TNBC or HGSOC calculated using beta instead of VEF values
eFigure 13. Risk of incident TNBC or HGSOC in subgroups based on hormone therapy exposure
eFigure 14. Risk of incident TNBC or HGSOC accounting for potential interaction between methylation and hormone therapy
eFigure 15. Risk of incident TNBC or HGSOC calculated for subsets of samples
eFigure 16. Allele specificity of methylation
eReferences.


