Abstract
The reading frame rule suggests that Duchenne muscular dystrophy (DMD) results from DMD mutations causing an out-of-frame transcript, whereas the milder Becker muscular dystrophy results from mutations causing an in-frame transcript. However, predicted nonsense mutations may instead result in altered splicing and an in-frame transcript. Here we report a 10-year-old boy with a predicted nonsense mutation in exon 42 who had a 6-minute walk time of 157% of that of age matched DMD controls, characterized as intermediate muscular dystrophy. RNA sequencing analysis from a muscle biopsy revealed only 6.0–9.8% of DMD transcripts were in-frame, excluding exon 42, and immunoblot demonstrated only 3.2% dystrophin protein expression. Another potential genetic modifier noted was homozygosity for the protective IAAM LTBP4 haplotype. This case suggests that very low levels of DMD exon skipping and dystrophin protein expression may result in amelioration of skeletal muscle weakness, a finding relevant to current dystrophin-restoring therapies.
Keywords: dystrophin, nonsense mutation, RNA Sequencing, intermediate muscular dystrophy
1. INTRODUCTION
Duchenne and Becker muscular dystrophy (DMD and BMD) are X-linked allelic disorders that represent the most common type of muscular dystrophy. The phenotype is caused by reduced or absent dystrophin, a subsarcolemmal protein that links actin to the extracellular matrix. Normal levels of dystrophin are required for appropriate muscle membrane integrity. Reduced or absent levels result in an unstable muscle membrane leading to muscle breakdown which causes weakness. Absence of dystrophin results in the more severe DMD, marked by progressive muscle weakness leading, in the pre-steroid treatment era, to loss of ambulation before age 12; death due to cardiac and respiratory failure typically occurs in the third decade. The milder BMD phenotype results from the expression of a partially functional dystrophin protein, and is classically defined by preservation of ambulation past the age of 16. Many clinicians use the term intermediate muscular dystrophy (IMD) to describe those boys who lose ambulation between ages 12 and 16. There is a clinical spectrum seen within each category, but a much broader spectrum is seen in BMD, with some patients losing ambulation at age 16 and others remaining ambulatory lifelong with muscle cramping and fatigue as their only symptoms[1].
The reading-frame rule predicts phenotype with ~90% specificity in DMD patients[2] stating that mutations that maintain an open reading frame to allow translation of functional amino- and carboxy-terminal domains are associated with BMD, and mutations that truncate the reading frame result in DMD[3]. Around two-thirds of DMD mutations are exon deletions or duplications, which commonly follow this rule. On the basis of this rule, nonsense mutations, which cause ~15% of all dystrophinopathies,[4] are generally predicted to cause a severe phenotype[4]. However, milder phenotypes have been associated with predicted nonsense mutations that instead alter exon definition elements, resulting in alternate splicing with exclusion of all or a portion of the nonsense-containing exon from the mature transcript[5–11].
Detection of such altered splicing generally requires analysis of muscle-derived mRNA. Massive parallel RNA sequencing (RNA-Seq) is a relatively new tool that allows for assessment of gene expression, relative transcript abundance, and detection of alternative splice sites and their usage. Recent RNA-Seq analysis of the splicing pattern in wild type DMD noted that most of the 79 exons were constitutively spliced and only 12 alternative splicing events were identified[12]. Here we utilize RNA-Seq analysis to identify and quantify splicing alterations resulting from the predicted nonsense mutation, demonstrating that low levels of both altered mRNA splicing and dystrophin protein expression can be associated with a markedly ameliorated clinical phenotype.
2. CASE REPORT
2.1. Clinical Features
The patient (Bx103) was born at full term with a birth weight of 3,515 grams. He had normal fine motor, speech/language, and social development. He walked at 16 months of age, and was noted to run on his toes and to have large calves. Heel cord tightness was noted at age 7, for which he saw a physical therapist; at age 9, when seen by an orthopedist, a CK was obtained and was elevated at 15,370 U/L, leading to a referral to a neurologist. He was first examined in our clinic at 10 years 4 months of age, and was noted to have mild calf hypertrophy and heel cords reducible to just 90 degrees. Muscle strength was symmetric throughout, and was noted to be MRC grade 4 in neck flexion, shoulder abduction, elbow flexion, hip flexion, ankle dorsiflexion; grade 4+ in elbow extension, wrist flexion & extension, knee extension & flexion; and grade 5 in all remaining muscles. Mild lordosis was present, and he arose from the floor with a modified Gowers’ maneuver (one hand on his knee). Functional testing included a 6 minute walk test (6MWT) of 575 meters (630 meters is normal for a healthy 10–11 year old[13] and 366 meters is the median for a 9.5–10.5 year old DMD patient[14]) and a NSAA score of 30 (out of maximum score of 34). At this time of this examination, he had never received corticosteroids. His exam was judged to be atypical for an untreated patient with typical DMD and he was given a diagnosis of, at most, an IMD phenotype. Follow up examination 7 months later after 3 months on deflazacort was notable only for slight decline in NSAA score (28/34) and 6 minute walk test distance (537 meters), without changes in MRC scores. EKG and ECHO were within normal limits.
2.2. Molecular Analysis
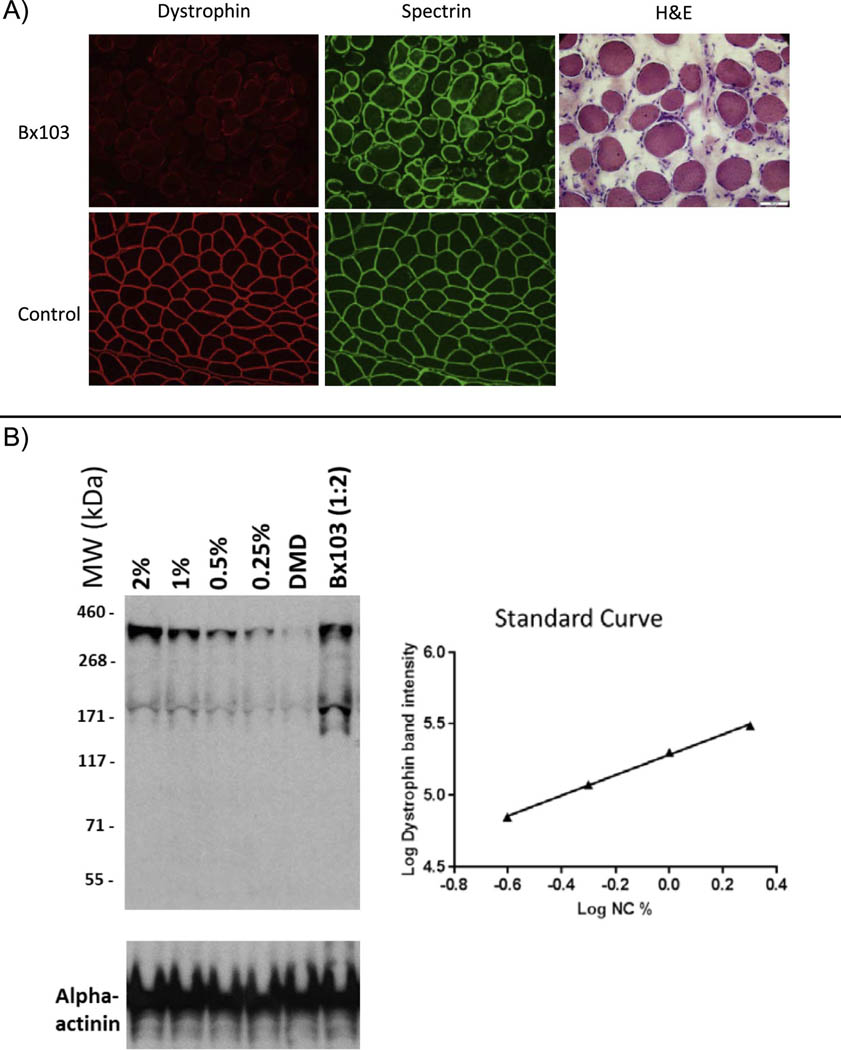
Genetic testing was performed prior to referral to our clinic and revealed a predicted nonsense mutation within exon 42 of the DMD gene (c.6115A>T; p.Lys2039Ter [p.K2039X]). Given his relatively mild phenotype, a left quadriceps muscle biopsy was subsequently performed at another hospital and frozen tissue was sent to our laboratory for RT-PCR and dystrophin expression analysis. Preparation artifact at the outside hospital resulted in severe artifactual changes, but hematoxylin and eosin staining revealed chronic and severe myopathic changes consistent with a muscular dystrophy, including fiber size variation, degenerating and necrotic fibers, phagocytosis, hypercontracted fibers, and significant endomysial fibrosis (Figure 1, left). Interpretation of immunofluorescent analysis using a standard panel of antibodies was questionable, given the poor condition of the specimen, but scattered fibers stain faintly with dystrophin (Figure 1, left). Immunoblot analysis, in contrast, showed low level expression of a full-length dystrophin, measured as 3.2% of normal, in comparison to a standard curve generated from pooled normal muscle specimens run on the same blot (Supplemental Methods; Figure 1, right).
Figure 1. Dystrophin expression analysis in patient muscle.
(A) Dystrophin (red) and spectrin (green) staining of quadriceps muscle biopsy specimens. Dystrophin expression appears minimal but is present in muscle from the patient (Bx103, top row); staining in control muscle is normal (bottom row). The generally poor state of the patient specimen is demonstrated by hematoxylin and eosin staining. (B) Western blot analysis of dystrophin expression. (Left) Western blot image showing dystrophin expression in muscle from pooled normal specimens diluted from 2% to 0.25% (Lanes 1–4), a DMD control (Lane 5) and the patient (Lane 6). As discussed in the text, quantification of the patient sample is 3.2% of the value in normal tissue. (Right) A four-point standard curve from pooled control muscle. Red = C-terminal dystrophin (Abcam 15277, Thermo, Inc.); Green = spectrin (NCL spec 1, Leica Biosystems)antibodies.
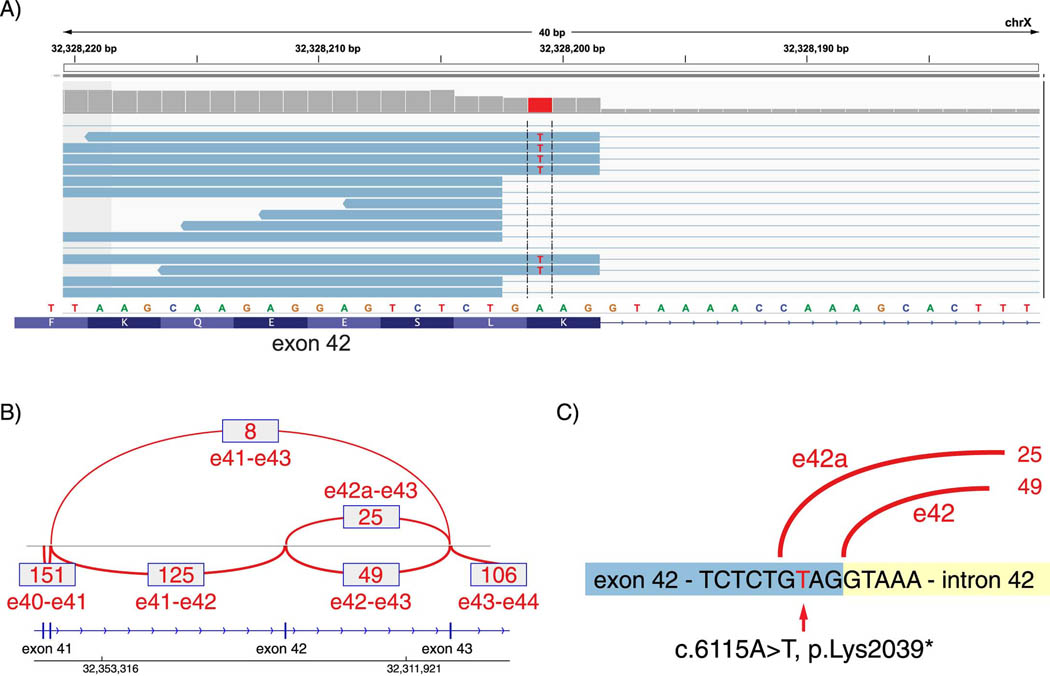
Despite the relatively low amount of dystrophin expressed, we predicted, using the Human Splice Finder database (www.umd.be/HSF3/HSF), that altered splicing resulting in the in-frame exclusion of exon 42 might account for his relatively mild phenotype. To assess splicing, RNA-Seq was performed using RNA extracted from patient muscle sections, and the splicing pattern observed across the exon 41, 42 and 43 region estimated by counting reads mapped to exon:exon junctions (Figure 2a,b). The c.6115A>T mutation was clearly visible in the RNA-Seq alignment, at its predicted location. Notably, this variant occurs at the −3 position relative to the end of exon 42; it results in a new “GT” splice donor site motif, utilization of which results in a four nucleotide deletion from the end of exon 42, and an out-of-frame transcript (termed “e42a”, Figure 2). As a result, three different transcripts are possible. Two of these truncate the reading frame, including (1) a normally-spliced but reading-frame disrupted transcript (due to the nonsense codon within the standard reading frame) (termed “e42” in Figure 2), and (2) an out-of-frame transcript utilizing the cryptic splice site at the end of exon 42 (e42a). The remaining transcript excludes exon 42 entirely, resulting in preservation of the reading frame, without a frameshift or nonsense codon.
Figure 2.
Splice junction read counts from RNA-Sequencing mapped to DMD. (A) Read alignments to exon 42 splice donor sites, (B) Splice junction read counts from DMD exon 40 through exon 44, and (C) the exon 42 splice donor region from patient showing the novel splice donor site created by the c.6115A>T nonsense mutation. Quantification of these junctions is summarized in the Table.
The RNA-Seq data allows quantification of each of these transcripts, as summarized in the Table. Junctional reads from the 3’ end of exon 41 splice to exon 42 in 125 of 133 sequences (94.0%), and to exon 43–the in-frame transcript–in only 8 of 133 (6.0%). These same 8 reads make up 9.8% of the 82 reads in which a junction of the 5’ end of exon 43 is detected; the remaining exon 43 junctional reads include 49 utilizing the standard exon 42 splicing site (59.8%), and 25 utilizing the novel cryptic splice donor site (30.5%). These results allow us to place boundaries on an estimate that between 6.0% and 9.8% of the DMD transcript maintains an open reading frame. Further RNA-Seq analysis demonstrated the IAAM haplotype in the patient (Supplemental Figure 1), which has been reported to be a beneficial modifier of DMD[15].
Table.
Quantification of mRNA junctions observed from RNA-Sequencing analysis.
| exon junction observed | reads per junction | total reads observed | percent | |
|---|---|---|---|---|
| Analyzed by exit of exons 41 | 41–42 | 125 | 133 | 94.0% |
| 41–43 | 8 | 6.0% | ||
|
| ||||
| Analyzed by exit of exons 42/42a | 42–43 | 40 | 65 | 61.5% |
| 42a-43 | 25 | 38.5% | ||
|
| ||||
| Analyzed by entry to exon 43 | 41–43 | 8 | 82 | 9.8% |
| 42–43 | 49 | 59.8% | ||
| 42a-43 | 25 | 30.5% | ||
3. DISCUSSION:
The reading frame rule has a positive predictive value of nearly 90% in cases of DMD, but may be as low as 50% in cases of BMD[16]. The principle that predicted nonsense alleles may in fact alter pre-mRNA splicing, resulting in a milder than expected clinical phenotype, is now well-described[16–18] although arguably under-recognized in clinical practice. Such mutations typically affect exon definition elements, and may ablate exon splice enhancer or create exon splice suppressor sequences[8, 18–23]. In nearly all cases, the predicted nonsense alleles are found in a “zero-frame” context, in which exclusion of the mutation-containing exon results in an open reading frame, and clinical severity typically inversely correlates with the degree of efficiency exon exclusion. The large block of exons from 23–42 is the largest contiguous zero-frame block in DMD, and BMD-associated nonsense mutations are most frequently found in this region[16].
In the case presented here, the predicted nonsense mutation is found within this permissive region, in exon 42. Analysis of the c.6115A>T mutation using the Human Splice Finder tools predicted that the splicing pattern would be affected, with potential activation of an exonic cryptic donor site, creation of an exonic ESS site or alteration of an exonic ESE site. Significant exon 42 skipping would be expected to result in levels of a functional protein sufficient to explain the amelioration of his phenotype, with his well-preserved ambulation, relatively mild muscle weakness, and functional measures (such as 6MWT and NSAA) that are not typical of DMD subjects at the same age.
What is particularly informative about his muscle biopsy–and arguably, surprising–is the low level of in-frame transcript and the low level of dystrophin protein that appear to be sufficient for significant functional improvement. RNA-Seq analysis revealed 6.0–9.8% production of an in-frame transcript and protein analysis revealed only 3.2% relative dystrophin expression in this patient. Given his symptom severity, these are surprisingly low but do prove that expression of essentially normal length dystrophin is present. One may conclude that <10% production of a dystrophin transcript may be sufficient to ameliorate the DMD phenotype. There are several caveats this conclusion, however. First, although the patient has been classified as no more severe than an IMD phenotype at present, and appears significantly stronger than DMD patients of the same age based upon DMD-normed data, he has not yet reached the ages at which preserved ambulation define clinical phenotypes (12 years for IMD, and 16 years for BMD). Second, our quantification of dystrophin may underrepresent the actual amount; given the artifact revealed by H&E staining, and the associated poor laminin α2 staining, the amount of dystrophin protein recoverable for immunoblot analysis may be low. However, the actinin control on the immunoblot shows abundant signal (arguing against significant degradation despite the findings seen on H&E). Third, this patient is homozygous for the latent TGFβ binding protein 4 (LTBP4) IAAM haplotype that has been associated with prolonged ambulation in multiple cohorts[15, 24, 25], although determining whether the LTBP4 haplotype is particularly influential on the phenotype in the presence of low levels of dystrophin will require identification of additional patients with similar molecular mechanisms.
The finding that such low levels of an in-frame transcript and dystrophin expression of may affect phenotype is consistent with recent observations in clinical trials. The FDA has recently approved the exon 51-skipping agent eteplirsen, which showed an improvement in 6MWT distances despite a minimal yet significant increase dystrophin expression in muscle in treated DMD boys[26]. Similarly, treatment with ataluren, a nonsense mutation readthrough agent, showed minimal but measurable increases in dystrophin expression[27] and a trend toward improvement in 6MWT distances[28]; although equivocal, these results led to conditional approval in Europe. Consistent with these clinical trials, the patient we report provides evidence that a robust improvement in dystrophin expression may not be needed for significant clinical and functional amelioration of the DMD phenotype. While this is an interesting and noteworthy case, it is unlikely that results of exon skipping of one exon can be extrapolated to all exons due to likely variability in protein stability. However, extrapolation may be considered reasonable for exons located in the central rod domain that do not significantly shorten protein length. Continued reporting of these variations will help us to determine the best candidates for exon skipping and further therapies, in addition to helping elucidate the significantly complex genotype-phenotype correlations. Gathering this data will hopefully allow us to one day provide better prognostication and treatment for patients and their families.
Supplementary Material
Highlights.
Nonsense dystrophin mutations do not always result in a severe phenotype
Predicted nonsense mutations may instead affect exon splicing
Very low-level dystrophin expression may be sufficient to significantly attenuate phenotype
ACKNOWLEDGMENTS
The authors would like to thank the patient and his parents for permission to report his case.
FUNDING
This work was supported by the National Institutes of Health grant # 5T32NS077984–04 (MAW).
Footnotes
CONFLICTS OF INTEREST
MAW, FG, RBW, KMF: none related to this work.
SEH and DEF: Full-time employees of Sarepta Therapeutics, Inc.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Flanigan KM. Duchenne and Becker muscular dystrophies. Neurol Clin 2014;32:671–88, viii. [DOI] [PubMed] [Google Scholar]
- [2].Aartsma-Rus A, Van Deutekom JC, Fokkema IF, Van Ommen GJ, Den Dunnen JT. Entries in the Leiden Duchenne muscular dystrophy mutation database: an overview of mutation types and paradoxical cases that confirm the reading-frame rule. Muscle Nerve 2006;34:135–44. [DOI] [PubMed] [Google Scholar]
- [3].Monaco AP, Bertelson CJ, Liechti-Gallati S, Moser H, Kunkel LM. An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics 1988;2:90–5. [DOI] [PubMed] [Google Scholar]
- [4].Dent KM, Dunn DM, von Niederhausern AC, et al. Improved molecular diagnosis of dystrophinopathies in an unselected clinical cohort. Am J Med Genet A 2005;134:295–8. [DOI] [PubMed] [Google Scholar]
- [5].Shiga N, Takeshima Y, Sakamoto H, et al. Disruption of the splicing enhancer sequence within exon 27 of the dystrophin gene by a nonsense mutation induces partial skipping of the exon and is responsible for Becker muscular dystrophy. J Clin Invest 1997;100:2204–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Tuffery-Giraud S, Saquet C, Thorel D, et al. Mutation spectrum leading to an attenuated phenotype in dystrophinopathies. Eur J Hum Genet 2005;13:1254–60. [DOI] [PubMed] [Google Scholar]
- [7].Flanigan KM, Dunn DM, von Niederhausern A, et al. Nonsense mutation-associated Becker muscular dystrophy: interplay between exon definition and splicing regulatory elements within the DMD gene. Hum Mutat 2011;32:299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Trabelsi M, Beugnet C, Deburgrave N, et al. When a mid-intronic variation of DMD gene creates an ESE site. Neuromuscul Disord 2014;24:1111–7. [DOI] [PubMed] [Google Scholar]
- [9].Banihani R, Baskin B, Halliday W, et al. A Novel Mutation in DMD (c.10797+5G>A) Causes Becker Muscular Dystrophy Associated with Intellectual Disability. J Dev Behav Pediatr 2016;37:239–44. [DOI] [PubMed] [Google Scholar]
- [10].Ginjaar IB, Kneppers AL, v d Meulen JD, et al. Dystrophin nonsense mutation induces different levels of exon 29 skipping and leads to variable phenotypes within one BMD family. Eur J Hum Genet 2000;8:793–6. [DOI] [PubMed] [Google Scholar]
- [11].Melis MA, Muntoni F, Cau M, et al. Novel nonsense mutation (C-->A nt 10512) in exon 72 of dystrophin gene leading to exon skipping in a patient with a mild dystrophinopathy. Hum Mutat 1998;Suppl 1:S137–8. [DOI] [PubMed] [Google Scholar]
- [12].Bouge AL, Murauer E, Beyne E, et al. Targeted RNA-Seq profiling of splicing pattern in the DMD gene: exons are mostly constitutively spliced in human skeletal muscle. Sci Rep 2017;7:39094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Goemans N, Klingels K, van den Hauwe M, et al. Six-minute walk test: reference values and prediction equation in healthy boys aged 5 to 12 years. PLoS One 2013;8:e84120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Goemans N, van den Hauwe M, Wilson R, van Impe A, Klingels K, Buyse G. Ambulatory capacity and disease progression as measured by the 6-minute-walk-distance in Duchenne muscular dystrophy subjects on daily corticosteroids. Neuromuscul Disord 2013;23:618–23. [DOI] [PubMed] [Google Scholar]
- [15].Flanigan KM, Ceco E, Lamar KM, et al. LTBP4 genotype predicts age of ambulatory loss in Duchenne muscular dystrophy. Ann Neurol 2013;73:481–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Flanigan KM, Dunn DM, von Niederhausern A, et al. Mutational spectrum of DMD mutations in dystrophinopathy patients: application of modern diagnostic techniques to a large cohort. Hum Mutat 2009;30:1657–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Disset A, Bourgeois CF, Benmalek N, Claustres M, Stevenin J, Tuffery-Giraud S. An exon skipping-associated nonsense mutation in the dystrophin gene uncovers a complex interplay between multiple antagonistic splicing elements. Hum Mol Genet 2006;15:999–1013. [DOI] [PubMed] [Google Scholar]
- [18].Flanigan KM, Dunn D, Larsen CA, Medne L, Bonnemann CB, Weiss RB. Becker muscular dystrophy due to an inversion of exons 23 and 24 of the DMD gene. Muscle Nerve 2011;44:822–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Martone J, Briganti F, Legnini I, et al. The lack of the Celf2a splicing factor converts a Duchenne genotype into a Becker phenotype. Nat Commun 2016;7:10488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Tuffery-Giraud S, Chambert S, Demaille J, Claustres M. Point mutations in the dystrophin gene: evidence for frequent use of cryptic splice sites as a result of splicing defects. Hum Mutat 1999;14:359–68. [DOI] [PubMed] [Google Scholar]
- [21].Juan-Mateu J, Gonzalez-Quereda L, Rodriguez MJ, et al. Interplay between DMD point mutations and splicing signals in Dystrophinopathy phenotypes. PLoS One 2013;8:e59916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Niba ETE, Nishida A, Tran VK, et al. Cryptic splice activation but not exon skipping is observed in minigene assays of dystrophin c.9361+1G>A mutation identified by NGS. J Hum Genet 2017;62:531–537. [DOI] [PubMed] [Google Scholar]
- [23].Deburgrave N, Daoud F, Llense S, et al. Protein- and mRNA-based phenotype-genotype correlations in DMD/BMD with point mutations and molecular basis for BMD with nonsense and frameshift mutations in the DMD gene. Hum Mutat 2007;28:183–95. [DOI] [PubMed] [Google Scholar]
- [24].Bello L, Kesari A, Gordish-Dressman H, et al. Genetic modifiers of ambulation in the Cooperative International Neuromuscular Research Group Duchenne Natural History Study. Ann Neurol 2015;77:684–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].van den Bergen JC, Hiller M, Bohringer S, et al. Validation of genetic modifiers for Duchenne muscular dystrophy: a multicentre study assessing SPP1 and LTBP4 variants. J Neurol Neurosurg Psychiatry 2015;86:1060–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Mendell JR, Rodino-Klapac LR, Sahenk Z, et al. Eteplirsen for the treatment of Duchenne muscular dystrophy. Ann Neurol 2013;74:637–47. [DOI] [PubMed] [Google Scholar]
- [27].Finkel RS, Flanigan KM, Wong B, et al. Phase 2a study of ataluren-mediated dystrophin production in patients with nonsense mutation Duchenne muscular dystrophy. PLoS One 2013;8:e81302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Bushby K, Finkel R, Wong B, et al. Ataluren treatment of patients with nonsense mutation dystrophinopathy. Muscle Nerve 2014;50:477–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.