Abstract
Resolution of chromosome dimers, by site-specific recombination between dif sites, is carried out in Escherichia coli by XerCD recombinase in association with the FtsK protein. We show here that a variety of altered FtsK polypeptides, consisting of the N-terminal (cell division) domain alone or with deletions in the proline-glutamine-rich part of the protein, or polypeptides consisting of the C-terminal domain alone are all unable to carry out dif recombination. Alteration of the putative nucleotide-binding site also abolishes the ability of FtsK to carry out recombination between dif sites.
The ftsK gene of Escherichia coli encodes a very large protein (1,330 amino acids; ∼147 kDa) that is essential both for cell division and for resolution of chromosome dimers by site-specific recombination at the dif site (1, 10). The predicted amino acid sequence shows three distinct regions: the N-terminal 260 amino acids are predicted to form a series of transmembrane α-helices, the next 556 amino acids are rich in proline and glutamine (the PQ-rich domain), and the C-terminal 514 amino acids include a nucleotide-binding consensus sequence (1). Certain mutations in the N-terminal region abolish cell division at 42°C but do not affect chromosome segregation (1). Expression of a polypeptide consisting of the N-terminal ∼200 amino acids alone is sufficient to allow cell division, and this part of the protein therefore forms a functional domain (1, 5, 14). Within the central PQ-rich segment there is also a remarkable set of six consecutive repeats of a 10-amino-acid sequence: the “PQ repeat motif” (PQQPV[A/P]PQ[P/Q]Q) of unknown function. Deletion of the whole ftsK gene is lethal, but expression of the N-terminal polypeptide restores cell division and viability (5).
The N-terminal domain has sequence and predicted topological similarities to a number of FtsK-like proteins from other bacterial species (1). The C-terminal 514 amino acids show very strong sequence homology to all of these proteins and also to a number of smaller plasmid- or transposon-encoded polypeptides that do not possess the initial hydrophobic N-terminal sequence (1). In contrast, the ∼556-amino-acid PQ-rich sequence has been found only in FtsK from E. coli and Salmonella enterica serovar Typhi (http://www.Sanger.ac.uk/DataSearch).
Strains expressing the N-terminal domain alone, although mostly consisting of normal cells, show a proportion of filaments and chains of cells (4) in which DNA is localized abnormally at septa (5, 6). Diez et al. (4) showed that such mutant cultures were induced for the “universal” stress response protein, Usp, and Liu et al. (6) showed that the SOS response was also partly induced in such cultures. Liu et al. (6) also showed that cell division was necessary for SOS induction and concluded that some sort of chromosome damage resulted from cell division in a proportion of the cells. Steiner et al. (10) showed that FtsK (together with the site-specific recombinases XerC and XerD [2]) is required for monomerization of chromosome dimers (formed by homologous recombination between replicated parts of the circular E. coli chromosome) during cell division. They also showed that, although the N-terminal domain of the protein alone is sufficient for cell division, it is insufficient for resolution of chromosome dimers. Our hypothesis is that induction of Usp and the SOS response and the formation of chains in strains expressing only the N-terminal domain of FtsK are the results of chromosome dimers being trapped in closing septa.
Construction of deletion clones.
Deletion clones of ftsK were made by PCR. For the internal deletions, primers were made with unique restriction sites for ftsK. For amplification of the N-terminal portions, the 5′ UP primer incorporated an EcoRI site, and the 3′ REV primers utilized an XbaI site. For the C-terminal region, the reverse was true, with the 5′ UP primer incorporating an XbaI site and the 3′ REV primer containing the EcoRI site (Fig. 1). The PCR products were cloned into pUC19 and sequenced automatically (data not shown) to ensure no errors were made during amplification. The three internal-deletion clones were then made by ligating the deletion fragments via the XbaI sites and simultaneously into the EcoRI site in pBAD18. pBADK-PQ, pBADK-1/2, and pBADK-ALL were made in this manner (Fig. 1). In clone pBADK-PQ, the PQ repeat motif (FtsK 727 to 822) was removed. In pBADK-1/2, the region coding for amino acids 509 to 822 was removed. In pBADK-ALL, residues 206 to 822 were removed. The N-terminal portion of ftsK in pBADK′ codes for only the first 206 residues of FtsK and was constructed by restricting pBADK-ALL with XbaI to excise the C-terminal region before religation (Fig. 1). pBADK3 was constructed by cloning the 4-kb HindIII fragment from pBADK (5), which codes for most of ftsK, into pBADK′. Restriction of pBADK3 with SphI removed the last 322 amino acids from the C terminus to create pBADK4 (Fig. 1). pETAB1 and pETAB2 were two C-terminal clones with novel NdeI sites at both ends after PCR-directed mutagenesis. They were cloned in frame to the N-terminal His tag in pET16b. The first fusion (encoded in pETAB1) started at the methionine residue at position 863. The second fusion (encoded in pETAB2) started at the methionine residue at position 825. The N-terminal His-tagged ′FtsK C-terminal fusions were subcloned into pJF118EH to create pJFAB1 and pJFAB2 (Fig. 1). FtsK contains consensus sequences to the Walker A and B nucleotide-binding sites (1) (Fig. 1 shows the corresponding region in ftsK). The Walker A site (GTTGSGKSV; positions 989 to 997) was mutagenized by changing residues K995 and S996 to I and P, respectively (Fig. 1). Changing SGKS to SGIS in the Walker A site of MalK (part of the maltose binding complex in Salmonella enterica serovar Typhimurium) abolished ATPase binding in vitro (13). Clone pETK1 was made by changing the start codon of ftsK to ATG via an NdeI site introduced by PCR-directed mutagenesis (Fig. 1). Thus, the whole ftsK gene was fused at the N terminus to the poly-His tag. Clones of the deletions for the radiolabeling of the truncated polypeptides (Fig. 2) were constructed by cloning the relevant HindIII fragments from the pBAD clones into pETK1 cut with the same enzyme.
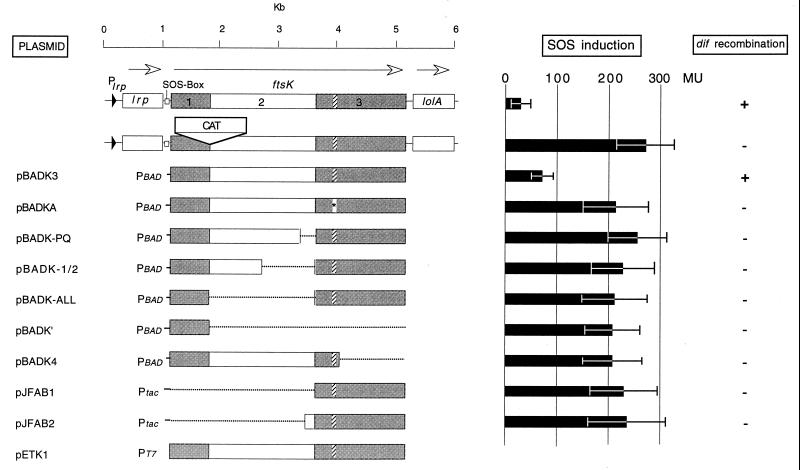
FIG. 1.
The chromosomal locus of ftsK is shown in the top line. The regions coding for the three proposed domains of FtsK are shaded and/or numbered. The hatched regions code for consensus sequences to the Walker A and B nucleotide-binding sites. The Walker A site (asterisk) was mutagenized in pBADKA (see the text). The second line shows the cat insertion in the ftsK::cat-1 allele. The remaining lines show the ftsK regions subcloned into various plasmids, together with the promoters from which they were expressed. Details of the construction of the deletion clones are given in the text. Either arabinose or glucose was added to the media at a final concentration of 0.1% (wt/vol). SOS induction levels are shown as β-galactosidase specific activities (MU, U of β-galactosidase [7]); the bars show mean values (±1 standard deviation) for log-phase cultures (5 to 10 samples each). Induction levels for the control strains DMG1(ftsK+)/pBAD18 and DMG2 (ftsK::cat-1)/pBAD18 are also shown. +, present; −, absent.
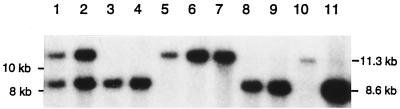
FIG. 2.
Assay for recombination between duplicate dif sites. A Southern blot of the PK3302-based strains probed with the Ω cassette (8) showing dif restriction fragments at 8.6 and/or 11.3 kb (12). Lane 1, PK3302/pBAD18; lane 2, PK3302 ftsK::cat-Δ5/pBADK3; lane 3, PK3302 ftsK::cat-Δ5/pBADK-PQ; lane 4, PK3302 ftsK::cat-Δ5/pBADK-1/2; lane 5, PK3302 ΔCDK5/pBADK-ALL; lane 6, PK3302 ftsK::cat-Δ5/pBADK4; lane 7, PK3302 ftsK::cat-Δ5/pBADK′; lane 8, PK3302 ftsK::cat-Δ5/pBADKA; lane 9, PK3302 ftsK::cat-1/pJF118HE; lane 10, PK3302 ftsK::cat-1/pJFAB1; lane 11, PK3302 ftsK::cat-1/pJFAB2.
The altered ftsK alleles, expressed from inducible promoters on plasmids, were introduced into strain DMG2, which carries the nonlethal ftsK::cat-1 allele (5). DMG2 (ΔlacZYA) was lysogenized with λsfiA::lacZ to allow us to measure induction of the SOS response. The ability of the altered peptides to carry out cell division was tested by the abilities of the various plasmids to support the growth of C600 cells that had been transduced with the lethal deletion allele ftsK::cat-Δ5 (5).
Log-phase DMG1 (like DMG2, but ftsK+) cells (carrying the vector plasmid pBAD18) in Luria broth plus arabinose (0.1% [wt/vol]) show a low level of β-galactosidase (34 ± 9 MU [7]) expressed from the SOS-inducible promoter of the sfiA gene (Fig. 1). In contrast, DMG2 cells, carrying the ftsK::cat-1 allele, show an approximately eightfold induction (270 ± 55 MU [Fig. 1]), as was previously reported for another strain that expresses only the N-terminal peptide of FtsK (6). We have previously presented evidence that induction of the SOS response (in about 20% of the cells) is caused by damage to chromosome dimers during cell division (6, 10). Induction of the ftsK+ allele from the PBAD promoter in plasmid pBADK3 is sufficient to complement the lethal ftsK::cat-Δ5 allele (data not shown) and greatly reduces the level of SOS expression in DMG2 cells (76 ± 14 MU [Fig. 1]). In Luria broth plus glucose medium, pBADK3 allows a higher level of SOS induction (149 ± 20 MU) but can still restore viability to ftsK::cat-Δ5 cells.
In contrast, none of the other plasmids shown in Fig. 1 can significantly reduce the level of SOS induction in DMG2 cells, although all those that express the N-terminal domain of FtsK can restore viability to ftsK::cat-Δ5 cells (even in the absence of arabinose [5]). The plasmids (pJFAB1 and pJFAB2) that do not encode the N-terminal domain cannot complement the ftsK::cat-Δ5 allele, but they do express polypeptides, as indicated by the fact that addition of inducer (IPTG [isopropyl-β-d-thiogalactopyranoside]) causes a marked reduction in growth rate and the appearance of inclusion bodies in the cells.
Because the SOS regulon is sensitive to even small amounts of DNA damage, it was possible that some of the altered forms of FtsK retained some activity in dimer resolution. To test this, the ftsK::cat-Δ5 allele was transduced into PK3302 cells carrying each of the plasmids (except in the case of plasmids that do not express the essential N-terminal domain of FtsK, where the ftsK::cat-1 allele was introduced instead). PK3302 contains, in addition to the normal copy of dif in the terminus region, a second chromosomal copy at another location. This second copy is inverted with respect to the orientation of the normal dif sequence and also contains the spc gene (12). Site-specific recombination between the inverted dif sites causes inversion of the chromosome segment between them. The size of the EcoRV restriction fragment containing the spc-dif insertion (visualized by probing with 32P-labeled spc DNA) is either 11.3 or 8.6 kb, depending on the orientation of the intervening chromosome segment. Clones of xerC+ xerD+ ftsK+ cells always contain both orientations (Fig. 2, lane 1) because of the high frequency of recombination between the dif sites. If XerC, XerD, or the C-terminal domain of FtsK is absent, however, recombination cannot take place, and single-cell clones contain only one or the other of the two possible orientations (10, 12). Figure 2, lane 2, shows that, although pBADK3 (PBAD::ftsK+) clones also contain both orientations of this segment, single-cell clones of all of the other plasmids contained only one or the other orientation. Because each clone was isolated from a single cell and grown for many generations before being tested, this result shows that none of the altered forms of FtsK has even residual function in dif recombination.
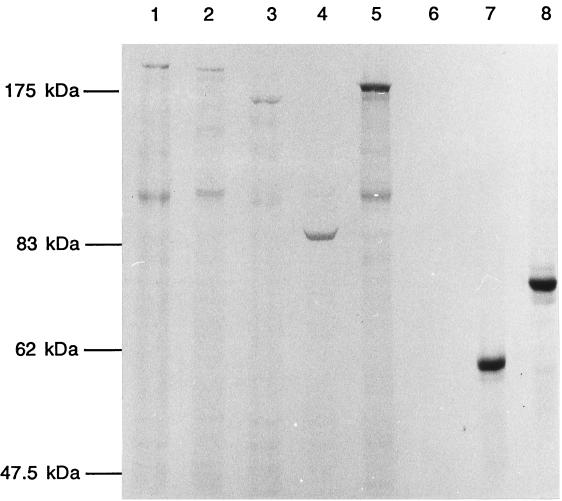
Our tests showed that each of the plasmids that were expected to express the N-terminal cell division domain were indeed able to complement the lethal ftsK::cat-Δ5 deletion allele and therefore expressed stable functional polypeptides. It could be inferred that the remaining two plasmids, which did not encode this domain, also expressed polypeptides, because induction of the Ptac promoter caused the appearance of inclusion bodies and reduced the growth rate. To check whether polypeptides of the expected kinds were actually produced by each of these constructs, ftsK and its different deletion alleles were subcloned into the expression vector pET16b, and radiolabeled polypeptides were detected directly by sodium dodecyl sulfate (SDS)–8% polyacrylamide gel electrophoresis (PAGE) (Fig. 3). All FtsK peptides, with the exception of the smallest clone expressing only the NH domain, showed anomalous mobility on SDS-PAGE, with apparent molecular masses higher than predicted. Figure 3 shows that FtsK (predicted mass, 147 kDa) migrates aberrantly in SDS-PAGE, with an apparent molecular mass of 190 kDa, as previously reported by Wang and Lutkenhaus (14). All the altered FtsK peptides showed mobilities proportional to their predicted sizes, but all migrated more slowly than their calculated mobilities. Figure 3 also shows the breakdown peptides (apparent mass, 90 to 100 kDa), as reported by Wang and Lutkenhaus (14), in lanes with FtsK peptides that contained at least the N-terminal domain and part of the PQ-rich domain. The exception was the entire PQ deletion from pBADK-ALL (Fig. 1); therefore, we conclude that the cleavage site lies in the first half of this central domain.
FIG. 3.
SDS-PAGE analysis of radiolabeled polypeptides from the deletion clones. The deletions were cloned into pET16b or pETK1 to make N-terminal fusions, which were then overproduced, radiolabeled, and electrophoresed through SDS–8% PAGE. Mass markers are on the left. Lane 1, pETK1 (147/200 kDa [calculated mass/observed mass]); lane 2, pETK-PQ (132/190 kDa); lane 3, pETK-1/2 (107/160 kDa); lane 4, pETK-ALL (74/85 kDa); lane 5, pETK4 (114/180 kDa); lane 6, pETK′ (23 kDa/not applicable); lane 7, pETAB1 (54/55 kDa); lane 8, pETAB2 (58/75 kDa). Lanes 1, 2, 3, and 5 show a breakdown product of His-FtsK at ∼100 kDa.
Diez et al. (4) reported the construction of a plasmid that expressed a C-terminal peptide of FtsK which abolished the “chaining” phenotype of their ftsK mutant. Analysis of the locations of the chromosomes in the chains described by Diez et al. (4) revealed that they were located at the cell midpoint. In our similar mutant strain (ftsK::cat-1), chaining of cells is thought to result from chromosome trapping at septa (6), but neither of our plasmids expressing C-terminal peptides had any effect on SOS induction (also supposed to arise from chromosome trapping) or was able to support dif recombination (Fig. 3).
The expression of an FtsK truncation, with only the II and III domains, was previously shown to be unable to resolve dimers via dif in a plasmid-based recombination assay (9). We conclude that peptides consisting of the cytoplasmic domain alone are nonfunctional, probably because they are not correctly localized with respect to the closing septum and the dif sites in dimeric chromosomes that are trapped there. However, in view of the results of Diez et al. (4), we cannot rule out the possibility that some other form of FtsK can carry out this function, at least to some extent.
Our experiments therefore show that, perhaps surprisingly, deletion of any part of the central PQ-rich domain abolishes the dif recombination activity of FtsK, as also, more predictably, does deletion of all or part of the C-terminal domain or alteration of the nucleotide-binding motif. Thus, the central domain plays an essential role in the coresolvase activity of FtsK in E. coli, although this domain is absent in homologous proteins from most other bacterial species. Possibly this part of the protein is required to position the active C-terminal domain at a sufficient distance from the cell membrane to allow it to interact productively with the chromosome, XerC, and/or XerD. If so, it may be possible to replace it with a different “spacer” peptide, although the peculiar PQ repeat region may have some special additional function.
Acknowledgments
We thank Peter Kuempel for strain PK3302.
We thank the European Commission for funding.
REFERENCES
- 1.Begg K J, Dewar S J, Donachie W D. A new Escherichia coli cell division gene, ftsK. J Bacteriol. 1995;177:6211–6222. doi: 10.1128/jb.177.21.6211-6222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blakely G, May G, McCulloch R, Grant K, Neilson L, Sheratt D J. Two related recombinases are required for site specific recombination at dif and cer in E. coli K12. Cell. 1993;75:351–361. doi: 10.1016/0092-8674(93)80076-q. [DOI] [PubMed] [Google Scholar]
- 3.D'Ari R, Huisman O. Novel mechanism of cell division inhibition associated with the SOS response in Escherichia coli. J Bacteriol. 1983;156:243–250. doi: 10.1128/jb.156.1.243-250.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diez A A, Farewell A, Nannmark U, Nyström T. A mutation in the ftsK gene of Escherichia coli affects cell separation, stationary-phase survival, stress adaptation, and expression of the gene encoding the stress protein UspA. J Bacteriol. 1997;179:5878–5883. doi: 10.1128/jb.179.18.5878-5883.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Draper G C, McLennan N F, Begg K J, Masters M, Donachie W D. Only the N-terminal domain of FtsK functions in cell division. J Bacteriol. 1998;180:4621–4627. doi: 10.1128/jb.180.17.4621-4627.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu G, Draper G C, Donachie W D. FtsK is a bifunctional protein involved in cell division and chromosome localisation in Escherichia coli. Mol Microbiol. 1998;29:893–903. doi: 10.1046/j.1365-2958.1998.00986.x. [DOI] [PubMed] [Google Scholar]
- 7.Miller J F. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. β-Galactosidase assay; pp. 72–77. [Google Scholar]
- 8.Prentki P, Kirsch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1986;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 9.Recchia G D, Aroyo M, Wolf D, Blakely G, Sherratt D J. FtsK-dependent and -independent pathways of Xer site-specific recombination. EMBO J. 1999;18:5724–5734. doi: 10.1093/emboj/18.20.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steiner W, Liu G, Donachie W D, Kuempel P L. The FtsK cell division protein is required for resolution of chromosome dimers in Escherichia coli. Mol Microbiol. 1999;31:579–583. doi: 10.1046/j.1365-2958.1999.01198.x. [DOI] [PubMed] [Google Scholar]
- 11.Studier W F, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 12.Tecklenburg M, Naumer A, Olagappan N, Kuempel P L. The dif resolvase locus of the Escherichia coli chromosome can be replaced by a 33-bp sequence, but function depends on location. Proc Natl Acad Sci USA. 1995;92:1352–1356. doi: 10.1073/pnas.92.5.1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walter C, Wilken S, Schneider E. Large-scale purification, nucleotide binding properties, and ATPase activity of the MalK subunit of Salmonella typhimurium maltose transport complex. J Biol Chem. 1992;268:23685–23696. [PubMed] [Google Scholar]
- 14.Wang L, Lutkenhaus J F. FtsK is an essential cell division protein that is localized to the septum and induced as part of the SOS response. Mol Microbiol. 1998;29:731–740. doi: 10.1046/j.1365-2958.1998.00958.x. [DOI] [PubMed] [Google Scholar]