Summary
Spinocerebellar ataxia type 10 (SCA10) is an autosomal-dominant disorder caused by an expanded pentanucleotide repeat in the ATXN10 gene. This repeat expansion, when fully penetrant, has a size of 850–4,500 repeats. It has been shown that the repeat composition can be a modifier of disease, e.g., seizures. Here, we describe a Mexican kindred in which we identified both pure (ATTCT)n and mixed (ATTCT)n-(ATTCC)n expansions in the same family. We used amplification-free targeted sequencing and optical genome mapping to decipher the composition of these repeat expansions. We found a considerable degree of mosaicism of the repeat expansion. This mosaicism was confirmed in skin fibroblasts from individuals with ATXN10 expansions with RNAScope in situ hybridization. All affected family members with the mixed ATXN10 repeat expansion showed typical clinical signs of spinocerebellar ataxia and epilepsy. In contrast, individuals with the pure ATXN10 expansion present with Parkinson's disease or are unaffected, even in individuals more than 20 years older than the average age at onset for SCA10. Our findings suggest that the pure (ATTCT)n expansion is non-pathogenic, while repeat interruptions, e.g., (ATTCC)n, are necessary to cause SCA10. This mechanism has been recently described for several other repeat expansions including SCA31 (BEAN1), SCA37 (DAB1), and three loci for benign adult familial myoclonic epilepsy BAFME (SAMD12, TNRC6A, RAPGEF2). Therefore, long-read sequencing and optical genome mapping of the entire genomic structure of repeat expansions are critical for clinical practice and genetic counseling, as variations in the repeat can affect disease penetrance, symptoms, and disease trajectory.
Key words: spinocerebellar ataxia type 10, SCA10, ATAXIN10, ATXN10, penetrance, long-read sequencing, amplification-free targeted sequencing, optical genome mapping
Spinocerebellar ataxia type 10 (SCA10) is a rare autosomal-dominant pentanucleotide repeat expansion disorder of the ATXN10 gene. We used advanced amplification-free targeted long-read sequencing and optical genome mapping to decipher the repeat composition. Our findings suggest that the pure (ATTCT)n expansion is non-pathogenic, while repeat interruptions are causal for SCA10.
Introduction
Complex pentanucleotide repeats within intron 9 of the ATAXIN 10 (ATXN10 [MIM:611150]) gene cause spinocerebellar ataxia type 10 (SCA10 [MIM:603516]), a rare autosomal-dominant disease that is most prevalent in Middle and South America.1 These repeats can comprise several thousand when clinically fully penetrant.2,3 The normal ATTCT repeat allele size is 10–32 repeats, whereas intermediate alleles with reduced penetrance contain 280–850 repeats. Complete penetrance of SCA10 typically has between 850 and 4,500 repeats.2,3
Clinically, SCA10 presents with a symptom complex of spinocerebellar ataxia, gait ataxia, dysarthria, nystagmus, epilepsy, cognitive decline, and non-motor symptoms.4 These symptoms affect quality of life measures, particularly “physical functioning” and “physical role.”5 In addition, non-motor symptoms include increased fatigue (32% of SCA10 versus 3.6% control group [p = 0.005]),6 changes in rapid eye movement (REM) sleep patterns (more extended REM periods and more REM arousals), and a higher respiratory disturbance index (RDI).7 Assessment of changes in the sense of smell or cancer risk shows no significant differences compared with controls.8,9 High-resolution magnetic resonance imaging (MRI) in 18 patients with SCA10 revealed white matter atrophy starting in the posterior/floculonodular cerebellum and gray matter degeneration in the cerebellum and brainstem thalamus. The putamen was more pronounced in patients with severe disease. Additionally, patients with SCA10 with epilepsy had lower gray matter intensity in the thalamus and a loss of integrity of the white matter in lobule VI of the cerebellum.10
Diagnostically, genetic sequencing analysis of ATXN10 expansion length has been challenging due to its repetitive nature. Furthermore, southern blotting has inherent problems with accurate sizing and considerable variability between clinical diagnostic laboratories. PCR for repetitive regions shows reduced amplification efficiency, allele bias, allele dropout,11 and replication slippage effects.12 Therefore, the somatic variability of repeat elements is challenging to assess because of replication errors.
Besides the repeat length, the ATXN10 pentanucleotide repeat can have variable genomic structure configurations: pure (ATTCT)n repeats and mixed (ATTCT)n-(ATTCC)n or (ATTCT)n-(ATCCT)n-(ATCCC)n repeats with additional interruptions of hepta- or octanucleotide repeats.3 However, current clinical genetic diagnostics cannot determine the ATXN10 pentanucleotide repeat composition. More recent developments of repeat-prime PCR with high-resolution pulse-field capillary electrophoresis analysis13 and long-range sequencing3 in the research setting allow for refinement of ATXN10 repeat composition.
It is very likely that the genomic composition of the ATXN10 repeat expansion leads to different clinical phenotypes and pathophysiological changes in patients and may influence the pathogenicity and penetrance of the expanded repeat allele. For example, epileptic seizures in SCA10 can present as complex partials with occasional secondary generalizations, greatly influencing morbidity and mortality of the disease. The ATCCT interruption of the ATTCT repeat expansion shows a higher risk (6.3-fold) of developing seizures.14
Here, we performed amplification-free targeted sequencing (Pacific Biosciences) and optical genome mapping (Bionano Genomics) in three generations of a large family from Mexico with SCA10 to assess repeat size and repeat composition in affected and unaffected relatives. We characterized the somatic variability of the repeat composition in skin fibroblasts from individuals with ATXN10 expansions. We show that the variable expansions translated into variable size and number of RNA foci detected by RNAScope in situ hybridization.
Subjects and methods
Ethics statement
This study was approved by the ethics committee of Stanford University School of Medicine (Stanford IRB-48895). All participants provided written informed consent.
Clinical assessment
We previously reported on this family and have expanded the recruitment of family members to include relatives of the third generation.15 The pedigree is depicted in Figure 1. Cases histories for family members that carry repeat expansions but have not been previously reported are described in the supplemental information.
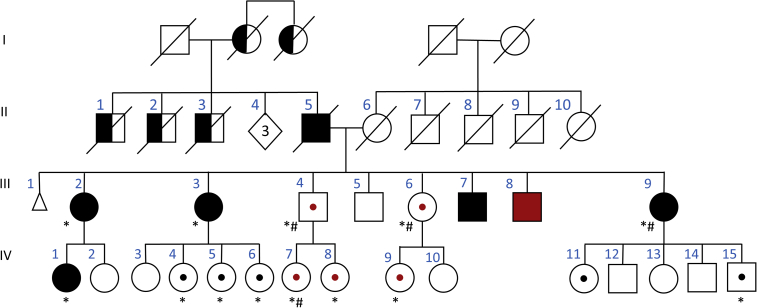
Figure 1.
ATXN10 pedigree of Mexican family
Roman numerals indicate generations to the left of each pedigree. Circles indicate female gender, squares indicate male gender, triangle indicates miscarriage, black symbols indicate affected individuals with SCA10, the red symbol indicates Parkinson's disease, half black symbol indicates SCA10 affected by medical history, and dots in the middle of the symbol mean individuals with ATXN10 repeat expansion but unaffected. Black dots indicate individual with interrupted repeat expansion, and the red dot indicates pure ATTCT repeat expansion. The age range in generation III is 57–74 years and in generation IV is 19–52 years. All individuals in the III and IV generations have been clinically evaluated by F.J.J.G. or J.F.-R. except III.1 (stillbirth) and III.5. In this study, we had DNA available for individuals labeled with an asterisk (∗) for amplification-free sequencing, and individuals labeled with a hash symbol (#) had DNA with high molecular weight quality for Saphyr optical genome mapping. For details of disease, age at disease onset as well as ATXN10 genotypes are listed in Table 1.
DNA isolation
DNA from saliva was extracted with Oragene-500 kit according to manufacturer’s instructions. For optical genome mapping, we isolated ultra-high molecular weight (UHMW) genomic DNA from blood using the Bionano Prep SP Blood & Cell Culture DNA Isolation Kit (Bionano, cat. no. 80030).
Short tandem repeat (STR) fragment analysis for identity testing
For identity testing, we used the AmpFlSTR Identifiler Plus PCR Amplification Kit (Thermo Fisher Scientific, cat. no. 4427368) according to the manufacturer’s instructions. The kit uses a five-dye fluorescent system, and the inclusion of non-nucleotide linkers allows for simultaneous amplification and efficient separation of 15 STR loci. Electropherograms of the fragments were analyzed using NCBI’s OSIRIS 2.13.1 program (Table S1).
Amplification-free targeted sequencing
DNA samples were further purified with AMPure PB beads (PacBio, cat. no. 100-265-900) before processing, and quality control (QC) on DNA samples was performed using Agilent Femto pulse analysis. Using a modified PacBio amplification-free targeted sequencing method protocol, we started with five μg input DNA for each sample and sequenced with a 20 h PacBio Sequel System/Sequencing run. More specifically, non-sequenceable single-molecule real-time bell (SMRTbell) libraries were generated from native genomic DNA digested with the restriction enzyme EcoRI-HF and a short hairpin adapter with barcode sequences. These short hairpin adapters do not contain sequencing primer complementary sequences to prevent sequencing reaction initiation. We then introduced double-strand breaks to the SMRTbell templates that contained our region of interest using Cas9 and a guide RNA (ATXN10-5′-AUACAAAGGAUCAGAAUCCC-3′) designed to be complementary to a sequence adjacent to the region of interest.16 The SMRTbell library hairpin adapter was subsequently ligated to the digested templates to form asymmetric SMRTbell libraries that allow sequencing primers to anneal with the second hairpin adapter introduced after Cas9 enzyme digestion. The library was digested with an enzyme cocktail containing exonuclease III, exonuclease VII, and four restriction enzymes (BssSαI, BlnI, SpeI, and KpnI) to remove a large portion of the non-target DNA fragments from the final library. The selected restriction enzymes are not predicted to cut inside the DNA fragments with the ATXN10 repeat expansion region. The final sequencing libraries were treated with trypsin and purified with AMPure PB beads before primer annealing, polymerase binding, and sequencing.
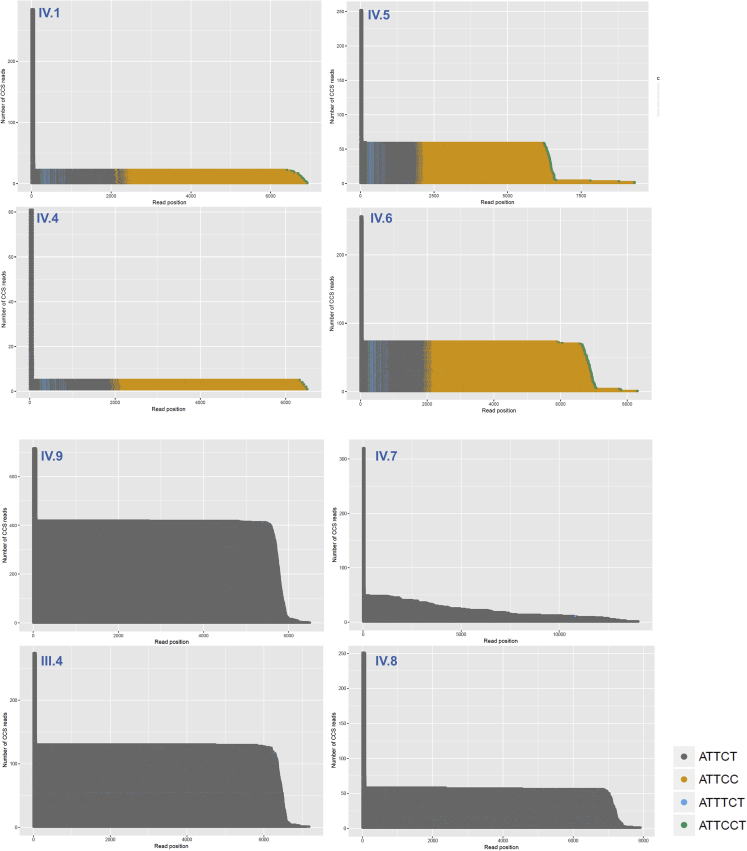
Sequencing data were first processed with PacBio SMRT analysis tool v.5.0 to identify hairpin adapters, demultiplex reads from different samples, and generate circular consensus sequencing (CCS) reads with predicted read quality above a Q20 Phred quality score. CCS reads from each sample were mapped against human reference sequence hg19, and ATXN10 repeat sequences from on-target sequencing reads were extracted from reads mapped through reference coordinates chr22:46,191,235–46,191,304. Repeat sequence length distributions and waterfall plots depicting repeat sequence variations (Figures 2 and S1) were generated using either custom software scripts or repeat analysis tools available on the GitHub website (https://github.com/PacificBiosciences/apps-scripts/tree/master/RepeatAnalysisTools) following the recommended data analysis workflow on the PacBio website (https://www.pacb.com/wp-content/uploads/Analysis-Procedure-No-Amp-Data-Preparation-and-Repeat-Analysis.pdf).
Figure 2.
Waterfall plots for amplification-free targeted sequencing of individuals with pure (ATTCT)n expansions and mixed (ATTCT)n-(ATTCC)n expansions
Examples of pure and mixed repeat expansion from different family members in generations III and IV showing repeat inter- and intra-individual variability. A row of dots represents repeat sequences in each sequencing read in this plot. Dots with different colors represent a different unit of repeat sequence context. Dark gray: ATTCT; gold: ATTCC; light blue: ATTTCT; green: ATTCCT. Circular consensus sequencing (CCS) reads are consensus sequences resulting from alignment between subreads taken from a single long-read sequence. Generating a CCS read does not include or require alignment against a reference sequence but does require at least two full-pass subreads from the insert. We detected rare hexanucleotide repeats (ATTTCT and ATTCCT) in the circular consensus reads, which were seen in several family members in a consistent pattern. For additional details, see Figure S1.
Optical genome mapping and analysis with the Saphyr system
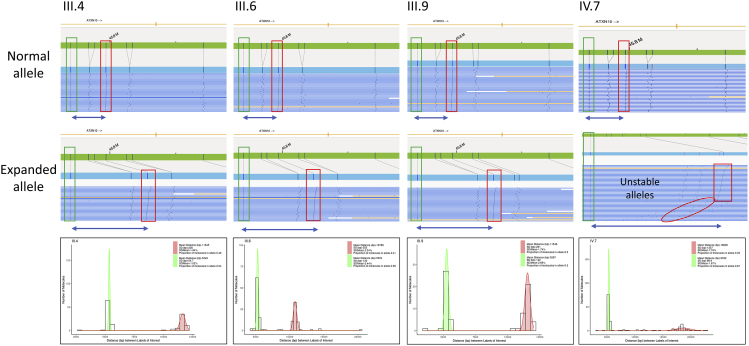
Briefly, 750 ng UHMW (>100 kb) genomic DNA was labeled with DLE-1 enzyme (Bionano Genomics) and the DNA backbone was counterstained according to Bionano Prep Direct Label and Stain protocol (document number: 30206, document revision: F) before loading on to the Bionano Chip. Loading of the chip and running of the Bionano Genomics Saphyr System were performed according to the Saphyr System User Guide (https://bionanogenomics.com/support-page/saphyr-system/). We used the Bionano de novo assembly and variant annotation pipelines to generate whole-genome assembly contigs, which span whole-chromosome arms and identify variants down to 500 bp. For single-variant (SV) detection, the consensus genome maps were aligned to reference (hg38). After running the de novo assembly pipeline using Solve v.1.7 and visualizing with Access v.3.7, we identified somatic rearrangements by selecting novel SVs to the Bionano control sample database (zero population frequency [out of 204 samples], thus potentially somatic mutations) and SVs with strong molecule support (remove false positives due to assembly errors). Somatic variability was investigated in the de novo assemblies by aligning single-molecule data to each map containing the repeat expansion. These molecules were both manually visualized and subsequently plotted by bp distance between the two labels of interest that encompass the repeat expansion to show the presence/absence of somatic mosaicism (Figure 3).
Figure 3.
Visualization of optical genome mapping for four individuals with ATXN10 repeat expansions as molecule distance graphs and histograms
The top half of individual optical genome maps represent unexpanded molecules, and the bottom part shows expanded alleles. The green bar represents the reference allele (hg38) with marks at all locations of the sequence motif (vertical lines), which is recognized by the Bionano optical genome mapping enzyme (e.g., DLE-1, Nt.BspQI, Nb.BssSI), which occurs every 5 kbp on average. Genome maps (blue bars) are produced from images of labeled DNA molecules >150 kbp for the target DNA. We measured expanded alleles between reference label IDs 5,045 and 5,048 (ATXN10 cytogenetic location: 22q13.31, genomic coordinates [GRCh38]: 22:45,671,834–45,845,307). We measured the distance between labels 5,045 (green box) and 5,048 (red box) and calculated the expansion size. Histograms show the distribution of normal and expanded alleles for each sample. Shown are mean distance between labels, standard deviation (SD), and proportion of molecules in allele.
In situ hybridization with RNAScope
Human skin fibroblasts were derived from 4 mm skin biopsies, maintained, and banked as we previously described.17 For fibroblast-cell culture, 8-well chamber slides (Nunc Lab-Tek chamber slide, Sigma, cat. no. C7182) were coated with 0.1% gelatin for 15 min. 60,000 cells were seeded in each chamber and incubated overnight. The next day, slides were fixed with 10% neutral buffered formalin for 15 min and dehydrated by gradually increasing ethanol concentrations (50%, 70%, 100%).
In situ hybridization was performed according to the manufacturer’s instructions using the RNAscope multiplex fluorescent assay v.2 kit. In brief, slides were treated with RNAScope hydrogen peroxide (part number 322335) for 10 min at room temperature, followed by a protease III incubation (10 min, room temperature). Probe (Hs-ATXN10-ATTCT-C1, ACDBio, part ID 1001381), positive control (part number, 320861), and negative control (ACD, part number 320871) hybridizations were performed in an HybEZ oven (ACD, HybEZ II Hybridization System, part number 321710) at 40oC for 2 h. Slides were washed and subjected to three steps of amplification: AMP1 (30 min, 40oC), AMP2 (40oC), and AMP3 (15 min, 40oC). Then, slides were incubated with RNAScope HRP-C1 for 15 min at 40oC, followed by a washing cycle and incubation with TSA Plus fluorophore (1:600) for 30 min at 40oC. Lastly, slides were blocked with a horseradish peroxidase (HRP) blocker for 15 min at 40oC and counterstained with a nuclear marker (DAPI). RNAScope slides were covered with Prolong Gold Antifade reagent (Invitrogen, P36930) and a 22 × 50 mm coverslip for imaging. Imaging was performed on an ImageXpress Pico (Molecular Devices) and analyzed with CellReporterXpress Image Acquisition software (Molecular devices v.2.6.130), and RNA foci were analyzed using the “pits and vesicles” software. Statistical analysis of RNA foci counts was performed in GraphPad Prism using a one-way ANOVA (α ≤ 0.05) and a Brown-Forsythe and Welch test (95% confidence interval [CI]).
Results
We describe a four-generation Mexican family with an autosomal-dominant inheritance of an ATXN10 repeat expansion. We recently reported on this family and, interestingly, identified an individual of this family affected with typical L-dopa-responsive Parkinson's disease and a pure (ATTCT)n repeat expansion15 (Figure 1). Here, we expanded our genetic studies and included additional techniques of optical genome mapping and RNAScope in situ hybridization to understand the repeat size, genomic structure, and somatic variability in order to gain additional insight into the genetic mechanisms of the ATXN10 repeat expansion, clinical presentation, and disease trajectory.
Individuals with pure (ATTCT)n repeat expansion show reduced clinical penetrance for SCA10
We examined 22 family members from generations III (7 individuals) and IV (15 individuals) of the family (Figure 1) and collected blood, saliva, and skin biopsies from individuals who consented to the study protocol. Sixteen individuals had an ATXN10 repeat expansion (rs60726084, 7 in generation III, 9 in generation IV). We found six relatives in generation IV who had normal ATXN10 repeat expansions. Of the 16 individuals with expanded ATXN10 repeats, ten presented with the mixed (ATTCT)n-(ATTCC)n repeat expansion (Figure 1, black symbols), and six showed the pure (ATTCT)n repeat expansion (Figure 1, red symbols). Of the ten cases with a mixed repeat expansion, five were affected with typical SCA10, with an average age at onset of 41.6 years (range 35–48 years, III.2, III.3, III.7, III.9, IV.1), and three of the five had seizures (III.2, III.3, III.7). The five unaffected individuals of the mixed repeat could be pre-symptomatic at ages 20, 33, 39, 40, and 51. Further longitudinal clinical follow up will be necessary.
Of the six relatives with pure (ATTCT)n expansion, one developed Parkinson's disease at age 38, while the other five were unaffected. Two of them have passed the average age at onset in this family by over 20 years (age at last visit: 65 and 70 years), while the other three individuals with the pure (ATTCT)n repeat expansion could still be pre-symptomatic at ages 18, 38, and 44 years. None of the individuals with the pure (ATTCT)n expansion presented with medical history or symptoms compatible with the diagnosis of SCA10 (supplemental information, case history III.6).
Inheritance of pure and mixed ATXN10 repeat expansion
We observed two structurally very different repeat expansions in this family. One ATXN10 repeat presented as a pure (ATTCT)n expansion and the other as a mixed (ATTCT)n(ATTCC)n expansion. There is no clear explanation of how a repeat can mutate from one complex repeat pattern to another in one generation, e.g., from generation II to III. It is also unusual that all siblings in generation III carry an extended ATXN10 allele. Usually, in an autosomal-dominant inheritance pattern, one would expect 50% of the offspring to have an ATXN10 repeat expansion and 50% the normal allele. To exclude non-paternity, we tested 15 microsatellite markers, and all siblings in generation III tested showed matching alleles (Table S1). Furthermore, we did not detect an expanded allele in the mother II.6 with amplification-free targeted sequencing. She carried two normal alleles with 13 and 14 repeats (Table 1). DNA from the father (II.5), who died before the study began, was unavailable.
Table 1.
Allele count for normal and expanded ATXN10 alleles in a Mexican family
| Pedigree ID | Disease | Sex | Age at onset | Age at blood collection | Allele 1 count | Allele 1 count range | Allele 2 count | Allele 2 count range | Repeat type |
|---|---|---|---|---|---|---|---|---|---|
| II.6 | unaffected | F | N/A | 90 | 38 | 13–14 | none | N/A | normal |
| III.2a | SCA10 | F | 35 | 71 | N/A | N/A | N/A | N/A | interrupted |
| III.3a | SCA10 | F | 48 | 68 | N/A | N/A | N/A | N/A | interrupted |
| III.4 | unaffected | M | N/A | 65 | 143 | 13–14 | 130 | 4750-7187 | pure |
| III.6a | unaffected | F | N/A | 70 | N/A | N/A | N/A | N/A | pure |
| III.7a | SCA10 | M | 48 | 65 | N/A | N/A | N/A | N/A | interrupted |
| III.8a | Parkinson’s | M | 38 | 57 | N/A | N/A | N/A | N/A | pure |
| III.9a | SCA10 | F | 37 | 53 | N/A | N/A | N/A | N/A | interrupted |
| IV.1 | SCA10 | F | 44 | 44 | 263 | 13–14 | 22 | 6422-6927 | interrupted |
| IV.4 | unaffected | F | 51 | 51 | 76 | 15–16 | 5 | 6338-6515 | interrupted |
| IV.5 | unaffected | F | 41 | 41 | 193 | 13–15 | 59 | 6244-9318 | interrupted |
| IV.6 | unaffected | F | 43 | 43 | 183 | 13–15 | 73 | 5914-8314 | interrupted |
| IV.7 | unaffected | F | N/A | 39 | 271 | 15–17 | 49 | 330-14038 | pure |
| IV.8 | unaffected | F | N/A | 19 | 193 | 16–17 | 58 | 2383-7920 | pure |
| IV.9 | unaffected | F | N/A | 44 | 297 | 13–17 | 419 | 2730-6502 | pure |
Individuals previously published in Schüle et al.15
Our interpretation from these clinical genetic findings is that the father was compound heterozygous for both the pure and the mixed extended allele, although we cannot fully exclude that he carried an unstable interrupted repeat that may have been lost the ATTCT interruption during transmission. However, the interrupted allele presents with a complex structure including dispersed repeats of ATTTCT and ATTCCT (Figure S1), which makes it unlikely that the repeat expansion lost its ATTCC repeat during transmission and expanded the ATTCT repeat in parallel and for several offspring in generation III. In a yeast model system, a proposed mechanism for repeat contractions is template skipping, whereas repeat expansions for ATTCT could be due to a template switch where the nascent leading strand might occasionally switch from its template to the nascent lagging strand.18 Clinically, the father presented with a typical SCA10 symptom complex based on his medical history, including seizures and no aggravated clinical phenotype or faster progression. He died at the age of 67.
Accurate sizing of ATXN10 pentanucleotide expansions
Previously, we used amplification-free targeted sequencing to sequence the entire ATXN10 repeat expansion and detected expansions between 1,076 and 1,363 pentanucleotide repeat expansions (5,380 to 6,815 bp) in generation III of this family.15 Long-read amplification-free targeted sequencing allows for accurate sizing in repeat composition by sequencing through the entire ATXN10 repeat without prior DNA template amplification. While we had confidence that the repeat length was correct, as the Cas9/small guide RNA recognition site is upstream of the repeat expansion, it was in stark contrast to the diagnostic sizing with PCR/southern blotting by Athena Diagnostics. The proband III.8 with Parkinson's disease presented 1,304 repeats (6,520 bp) for the expanded allele with PacBio amplification-free targeted sequencing. In contrast, diagnostic testing using Athena Diagnostics (test code 6901, PCR, southern blot) resulted in 1,968 ATTCT repeats (9,840 bp) for the extended allele, a size difference of 664 repeats (3,320 bp). Similarly, the affected brother III.7 presented 1,363 repeats (6,815 bp) with PacBio and 2,223 ATTCT repeats of the extended allele (11,115 bp) with Athena Diagnostics, a size difference of 860 repeats (4,300 bp).
It is known that Southern blotting can result in variable sizing between different diagnostic labs; hence, we wanted to address the repeat sizing with another independent technique using an amplification-free strategy. We decided to perform optical genome mapping using high-density nanochannel arrays and imaging on a Saphyr instrument (Bionano Genomics) that allows detection of structural variants greater than 500 bp. Optical genome-mapping images of long amplification-free DNA molecules fluorescently labeled at specific 6–7 bp sites preserve all genomic structural information. Optical genome mapping detects structural variants down to 1% variant allele fraction for mosaic samples.
We collected new blood samples from four family members with known repeat expansions previously detected with no-amp targeted sequencing. All four samples with an extended allele were correctly categorized as an insertion in intron 9 of the ATXN10 gene with coordinates chr22:46,191,235–46,191,304 (Hg38). Per sample, we resolved between 66 and 144 molecules (expanded alleles between 33 and 54) (Table S2). The repeat expansion sizes were comparable between the amplification-free targeted sequencing and optical genome mapping and show similar sizing and variability.
Somatic mosaicism detected with both no-amp targeted sequencing and optical genome mapping
Next, we compared the size range of the expanded allele for PacBio amplification-free targeted sequencing and optical genome mapping. For amplification-free targeted sequencing, we sequenced, on average, 101 CCS reads for the expanded allele (range: 5–419 CCS reads). Interestingly, we found that samples with a mixed repeat show less variability in the size of the expanded allele (average of 1,539 bp; range: 177–3,074 bp) compared with the pure repeat expansion (average of 6,363 bp; range: 2,437–13,708 bp). These data indicate that the pure expansions might be less stable and exhibit more somatic variability than the mixed ATXN10 repeat expansions.
For Bionano optical genome mapping, we had two samples with a pure repeat expansion for direct comparison. While optical genome mapping cannot give precise estimates down to individual bp resolution, we estimated the consensus map size and subtracted the size (5,355 bp) between the consensus map labels to report the repeat expansion size. Both samples III.4 and III.6 showed similar repeat expansion lengths and levels of somatic variability (Figure 4N, violin plots), whereas IV.7 showed a high degree of somatic mosaicism with unstable expanded repeats (Figure 2, Figure 3 and 3), which we further evaluated in skin fibroblasts from these individuals (Figure 4).
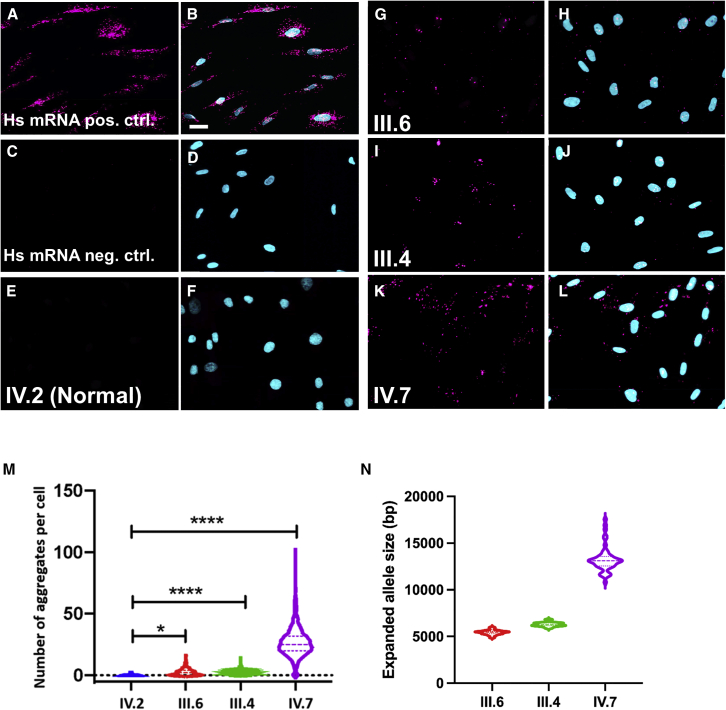
Figure 4.
RNAScope in situ hybridization of ATXN10 repeat expansion
(A and B) RNAscope human mRNA positive control.
(C and D) RNAscope human negative control.
(E and F) Healthy control.
(G and H) Individual with ATXN10 repeat expansion III.6.
(I and J) Individual with ATXN10 repeat expansion III.4.
(K and L) Individual with ATXN10 repeat expansion IV.7.
(E, G, I, and K) Hs-ATXN10-ATTCT-C1 RNAScope probe. (B, D, F, H, J, and L) Merge with DAPI. Scale bar in (B) represents 20 um and applies to all images.
(M) Violin plots of counts of RNA foci per cell using ImageXpress. Mean number of RNA signals per cells for control = 0.29, standard deviation (SD) 0.6 (1,870 nuclei); III.6 = 3.51, SD 2.33 (2,034 nuclei); III.4 = 3.89, SD 2.70 (759 nuclei); IV.7 = 26.65, SD 11.71 (2,034 nuclei).
(N) Violin plots for Bionano Saphyr optical genome mapping structural variant calls of the ATXN10 repeat expansion region for the same individual with ATXN10 repeat expansion (bp, base pairs). Normal allele size for control = 5,355 bp (normal allele); median expanded allele size III.6 = 5,459 bp ( 1,091 ATTCT repeats); III.4 = 6,260 bp ( 1,252 ATTCT repeats); IV.7 = 13,102 bp ( 2620 ATTCT repeats). p values are represented as ∗p = 001 and ∗∗∗∗p = 0.0001.
Variable ATXN10 repeat sizes correlate with varying counts of ATXN10 RNA foci in patient-derived skin fibroblasts
To investigate to what extent the somatic variability of the ATXN10 repeat size relates to (ATTCT)n repeat RNA foci within cells, we custom-designed an in situ hybridization RNAScope probe against the ATTCT repeat. It is known that spliced intronic repeat expansions can accumulate as intracellular RNA foci and contribute to cellular stress and neurodegeneration.19
We derived skin fibroblasts from three individuals with ATXN10 repeat expansions (III.4, III.6, and IV.7) and one healthy relative (IV.2) who had two normal ATXN10 alleles. We detected RNA foci within the nuclei and cytoplasm in all individuals with the expanded ATXN10 allele and none in the control (Figures 4E–4L). Positive and negative RNAScope controls were included (Figures 4A–4D). When we quantified the number of RNA aggregates in the three individuals with the expanded ATXN10 allele, we counted an average of 3.51 and 2.33 foci in cases III.4 and III.6 and 26.65 in case IV.7, with a wide range of counts that correlated to the amplification-free targeted sequencing data and the ATXN10 repeat sizing of the optical genome mapping (Table 1; Figure 4M).
Discussion
Three factors of the repeat expansion might influence clinical presentation and outcome: repeat length, repeat interruption, and somatic mosaicism.20,21
While it is known that the pathogenic repeat length for the ATXN10 repeat expansion is >850 repeats, there are significant gaps in our current knowledge about the nature and clinical relevance of repeat composition and repeat interruptions. Advanced long-read sequencing technologies and optical genome mapping allow us now to fully resolve very large repeat expansions.
Somatic mosaicism of (ATTCT)n and (ATTCT)n(ATTCC)n repeat expansions
The advantage of amplification-free targeted sequencing and optical genome mapping is that these techniques directly use genomic DNA without introducing amplification errors, recombination effects, or cell culture artifacts through prior amplification.
On the one hand, we found that repeat interruptions, as in the mixed (ATTCT)n(ATTCC)n repeat expansions, can stabilize the repeat as been previously reported (Figure 2).22 On the other hand, the individuals with the pure (ATTCT)n expansion show more variability in the size of the expanded alleles (average of 6,363 bp; range: 2,437–13,708 bp) compared with the mixed repeat expansion alleles (average of 1,539 bp; range: 177–3,074 bp). In particular, we identified one individual (IV.7) with a very unstable (ATTCT)n allele ranging from 66 to 2,782 (ATTCT)n repeats. This variability is present with PacBio amplification-free targetd sequencing and Bionano optical genome mapping (Figure 2, Figure 3 and 3).
This unstable repeat expansion also translates into variable counts of intracellular inclusions or RNA foci in primary skin fibroblasts from these cases. However, we do not know if and how these RNA foci translate to cellular stress, degeneration, and the development of clinical symptoms. Still, it has been shown that specific RNA-binding proteins get sequestered by the repeat expansion with particular sequence motifs, e.g., heterogeneous nuclear ribonucleoprotein K binds to the ATTCT repeat.23
Reduced disease penetrance in individuals with pure ATTCT expansions
In this Mexican kindred, we identified two distinct ATXN10 repeat expansions. One expansion was a pure ATTCT(n) expansion with no interruptions and a repeat length between 1,278 and 2,782 repeats (Figure S1). The second repeat is a mixed repeat with (ATTCT)n(ATTCC)n clinically associated with SCA10, whereas the pure repeat expansion shows reduced penetrance for SCA10.
Of the six individuals with pure ATXN10 repeat expansions, none showed clinical signs that would allow for the diagnosis of SCA10. However, as we reported earlier, one relative presented with Parkinson's disease.15 While three individuals with expansions might be pre-symptomatic (at ages 18, 38, and 44 years), two relatives from the third generation were 65 and 70 years at the time of assessment. They did not show clinical signs of SCA10 (assessed by F.J.J.G. and A.C.-P.; clinical examination III.6 in the supplemental information).
Notably, the seven siblings tested in generation III carry either a pure or a mixed repeat expansion, and the mother II.6 did not carry any ATXN10 expansion, suggesting that their father II.5 was compound heterozygous for the pure and mixed ATXN10 repeat expansion. By history, he had the typical presentation of SCA10 and died at age 67.
Both results suggest that the pure originally described (ATTCT)n expansion is not pathogenic, whereas repeat insertions or interruptions cause SCA10 symptoms. The ability to sequence through the entire repeat expansion from genomic DNA with long-read sequencing techniques makes it possible to resolve the complete sequence of the ATXN10 repeat expansion—a feat not possible using Southern blot analysis, PCR electrophoresis, or repeat-prime PCR.13
Characterizations of ATXN10 expansions in additional SCA10 families, using the methodology presented in this paper, are needed to better understand if (1) all SCA10 individuals have ATTCC interruptions, (2) large pure ATTCT repeat expansions are present in the unaffected population, or (3) other genetic/epigenetic modifiers may prevent SCA10 clinical presentation.
Our results indicate that ATXN10 repeat expansions fall into a category of repeat expansions that only become clinically symptomatic when repeat interruptions are present. Similar genetic loci have recently been reported for spinocerebellar ataxia 31 (SCA31 [MIM:117210 ]),24 spinocerebellar ataxia 37 (SCA37 [MIM:615945]),25,26 and three loci for benign adult familial myoclonic epilepsy 1 (BAFME1 [MIM:601068]),27 where repeat interruptions are inserted in non-pathogenic repetitive motifs. In SCA31, which is caused by a complex intronic pentanucleotide repeat thatcontains (TGGAA)n, (TAGAA)n, (TAAAA)n, and (TAAAATAGAA)n repeats in the gene called brain expressed, associated with Nedd4 (BEAN1 [MIM:612051]), only the (TGGAA)n repeat drives pathogenicity.24 An (ATTTC)n insertion causes SCA37 within a polymorphic ATTTT repeat in the non-coding 5′ untranslated region of the DAB adapter protein 1 gene (DAB1 [MIM:603448]).25,26 Lastly, BAFME1 is caused by intronic (TTTCA)n and (TTTTA) n repeats in the sterile α-motif domain-containing protein 12 gene (SAMD12 [MIM:618073]), the trinucleotide repeat-containing gene 6A (TNRC6A [MIM:610739]), and the Rap guanine nucleotide-exchange factor 2 gene (RAPGEF2 [MIM:609530]), where the same repeat (TTTCA)n and (TTTTA) n is driving pathogenicity regardless of the locus or gene in which the repeats are located.27
We propose that the pure (ATTCT)n is non-pathogenic or has only low penetrance with a mild disease course. In contrast, repeat interruptions mainly drive the clinical symptoms and presumably neuropathology. Longitudinal follow-up and in vitro functional studies will be essential to understand the ATXN10 repeat expansion pathophysiology further. Mechanistically, the insertion of repeats into the (ATTCT)n might be a driving factor for disease causation in SCA10 by sequestering certain RNA-binding proteins that do not bind to the (ATTCT)n repeat.
In summary, amplification-free targeted sequencing technology and optical genome mapping allow for accurate sizing of the repeat expansion and composition and can reveal somatic mosaicism, which is impossible with current diagnostic methods. While repeat-prime PCR is available at a lower cost and can delineate some of the genomic structure, it cannot resolve the complete length of the repeat expansion and can only detect known repeat motifs with pre-designed primers.13 We developed a 15-gene multiplex panel to detect repeat expansion disorders using no-amp targeted sequencing,28 which could serve as a platform technology for clinical diagnostics.
Deciphering the composition of repeat expansion will allow us to distinguish between pathogenic and non-pathogenic repeats, unravel disease mechanisms, and develop targeted therapies. These advanced genetic technologies are critical not only for implementing better diagnostic testing to diagnose disease more precisely but also for facilitating accurate genetic counseling as variation in the repeat can affect disease penetrance, symptoms, and disease trajectory.
Acknowledgments
We are indebted to the family members for their participation in this study and their commitment to helping the understanding of underlying genetic causes of neurodegenerative disorders. We thank Jose Miranda for supporting study recruitment. Start-up funding and Chan Zuckerberg Initiative Supplement funds to B.S. supported the research. We received in-kind donations from PacBio and Bionano in the form of sequencing and optical genome mapping.
Declaration of interests
I.M., Y.-C.T., and J.Z. are employees and shareholders of Pacific Biosciences. K.H., J. Lai, J. Lee, and M.D.G. are employees and shareholders of Bionano Genomics. B.S., F.J.J.G., J.P.V., F.Z., C.A.M.T., F.T., T.A., J.F.-R., A.C.-P., and A.O.R.-M. declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2022.100137.
Web resources
Online Mendelian Inheritance in Man, http://www.omim.org.
Supplemental information
Data and code availability
The Bionano data from five samples (III.4, III.6, III.9, IV.7, and IV.10) generated during this study are available at the NCBI BioProject database under BioProject: PRJNA838989 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA838989).
References
- 1.Rodríguez-Labrada R., Martins A.C., Magaña J.J., Vazquez-Mojena Y., Medrano-Montero J., Fernandez-Ruíz J., Cisneros B., Teive H., McFarland K.N., Saraiva-Pereira M.L., et al. Founder effects of spinocerebellar ataxias in the American continents and the caribbean. Cerebellum. 2020;19:446–458. doi: 10.1007/s12311-020-01109-7. [DOI] [PubMed] [Google Scholar]
- 2.Matsuura T., Yamagata T., Burgess D.L., Rasmussen A., Grewal R.P., Watase K., Khajavi M., McCall A.E., Davis C.F., Zu L., et al. Large expansion of the ATTCT pentanucleotide repeat in spinocerebellar ataxia type 10. Nat. Genet. 2000;26:191–194. doi: 10.1038/79911. [DOI] [PubMed] [Google Scholar]
- 3.McFarland K.N., Liu J., Landrian I., Godiska R., Shanker S., Yu F., Farmerie W.G., Ashizawa T. SMRT sequencing of long tandem nucleotide repeats in SCA10 reveals unique insight of repeat expansion structure. PLoS One. 2015;10:e0135906. doi: 10.1371/journal.pone.0135906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moro A., Munhoz R.P., Moscovich M., Arruda W.O., Raskin S., Silveira-Moriyama L., Ashizawa T., Teive H.A.G. Nonmotor symptoms in patients with spinocerebellar ataxia type 10. Cerebellum. 2017;16:938–944. doi: 10.1007/s12311-017-0869-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santos L.R., Teive H.A.G., Lopes Neto F.D.N., Macedo A.C.B.d., Mello N.M.d., Zonta M.B. Quality of life in individuals with spinocerebellar ataxia type 10: a preliminary study. Arq. Neuropsiquiatr. 2018;76:527–533. doi: 10.1590/0004-282X20180077. [DOI] [PubMed] [Google Scholar]
- 6.Moro A., Munhoz R.P., Camargo C.H., Moscovich M., Farah M., Teive H.A.G. Is fatigue an important finding in patients with spinocerebellar ataxia type 10 (SCA10)? J. Clin. Neurosci. 2020;71:150–152. doi: 10.1016/j.jocn.2019.08.097. [DOI] [PubMed] [Google Scholar]
- 7.London E., Camargo C.H.F., Zanatta A., Crippa A.C., Raskin S., Munhoz R.P., Ashizawa T., Teive H.A.G. Sleep disorders in SCA10. J. Sleep Res. 2018;27:e12688. doi: 10.1111/jsr.12688. [DOI] [PubMed] [Google Scholar]
- 8.Moscovich M., Munhoz R.P., Moro A., Raskin S., McFarland K., Ashizawa T., Teive H.A.G., Silveira-Moriyama L. Olfactory function in SCA10. Cerebellum. 2019;18:85–90. doi: 10.1007/s12311-018-0954-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schultz D.B., Nascimento F.A., Camargo C.H.F., Ashizawa T., Teive H.A.G. Cancer frequency in patients with SCA10. Parkinsonism Relat. Disord. 2020;76:1–2. doi: 10.1016/j.parkreldis.2020.05.032. [DOI] [PubMed] [Google Scholar]
- 10.Hernandez-Castillo C.R., Diaz R., Vaca-Palomares I., Torres D.L., Chirino A., Campos-Romo A., Ochoa A., Rasmussen A., Fernandez-Ruiz J. Extensive cerebellar and thalamic degeneration in spinocerebellar ataxia type 10. Parkinsonism Relat. Disord. 2019;66:182–188. doi: 10.1016/j.parkreldis.2019.08.011. [DOI] [PubMed] [Google Scholar]
- 11.Bastepe M., Xin W. Huntington disease: molecular diagnostics approach. Curr. Protoc. Hum. Genet. 2015;87:9.26.1–9.26.23. doi: 10.1002/0471142905.hg0926s87. [DOI] [PubMed] [Google Scholar]
- 12.Viguera E., Canceill D., Ehrlich S.D. In vitro replication slippage by DNA polymerases from thermophilic organisms. J. Mol. Biol. 2001;312:323–333. doi: 10.1006/jmbi.2001.4943. [DOI] [PubMed] [Google Scholar]
- 13.Hashem V., Tiwari A., Bewick B., Teive H.A.G., Moscovich M., Schüele B., Bushara K., Bower M., Rasmussen A., Tsai Y.C., et al. Pulse-Field capillary electrophoresis of repeat-primed PCR amplicons for analysis of large repeats in Spinocerebellar Ataxia Type 10. PLoS One. 2020;15:e0228789. doi: 10.1371/journal.pone.0228789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McFarland K.N., Liu J., Landrian I., Zeng D., Raskin S., Moscovich M., Gatto E.M., Ochoa A., Teive H.A.G., Rasmussen A., Ashizawa T. Repeat interruptions in spinocerebellar ataxia type 10 expansions are strongly associated with epileptic seizures. Neurogenetics. 2014;15:59–64. doi: 10.1007/s10048-013-0385-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schüle B., McFarland K.N., Lee K., Tsai Y.C., Nguyen K.D., Sun C., Liu M., Byrne C., Gopi R., Huang N., et al. Parkinson's disease associated with pure ATXN10 repeat expansion. NPJ Parkinsons Dis. 2017;3:27. doi: 10.1038/s41531-017-0029-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsai Y.-C., Greenberg D., Powell J., Höijer I., Ameur A., Strahl M., Ellis E., Jonasson I., Mouro Pinto R., Wheeler V.C., et al. Amplification-free, CRISPR-Cas9 targeted enrichment and SMRT sequencing of repeat-expansion disease causative genomic regions. bioRxiv. 2017 Preprint at. [Google Scholar]
- 17.Vangipuram M., Ting D., Kim S., Diaz R., Schüle B. Skin punch biopsy explant culture for derivation of primary human fibroblasts. J. Vis. Exp. 2013:e3779. doi: 10.3791/3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cherng N., Shishkin A.A., Schlager L.I., Tuck R.H., Sloan L., Matera R., Sarkar P.S., Ashizawa T., Freudenreich C.H., Mirkin S.M. Expansions, contractions, and fragility of the spinocerebellar ataxia type 10 pentanucleotide repeat in yeast. Proc. Natl. Acad. Sci. USA. 2011;108:2843–2848. doi: 10.1073/pnas.1009409108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang N., Ashizawa T. RNA toxicity and foci formation in microsatellite expansion diseases. Curr. Opin. Genet. Dev. 2017;44:17–29. doi: 10.1016/j.gde.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mangin A., de Pontual L., Tsai Y.C., Monteil L., Nizon M., Boisseau P., Mercier S., Ziegle J., Harting J., Heiner C., et al. Robust detection of somatic mosaicism and repeat interruptions by long-read targeted sequencing in myotonic dystrophy type 1. Int. J. Mol. Sci. 2021;22:2616. doi: 10.3390/ijms22052616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Depienne C., Mandel J.-L. 30 years of repeat expansion disorders: what have we learned and what are the remaining challenges? Am. J. Hum. Genet. 2021;108:764–785. doi: 10.1016/j.ajhg.2021.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Villate O., Ibarluzea N., Maortua H., de la Hoz A.B., Rodriguez-Revenga L., Izquierdo-Álvarez S., Tejada M.I. Effect of AGG interruptions on FMR1 maternal transmissions. Front. Mol. Biosci. 2020;7:135. doi: 10.3389/fmolb.2020.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.White M.C., Gao R., Xu W., Mandal S.M., Lim J.G., Hazra T.K., Wakamiya M., Edwards S.F., Raskin S., Teive H.A.G., et al. Inactivation of hnRNP K by expanded intronic AUUCU repeat induces apoptosis via translocation of PKCdelta to mitochondria in spinocerebellar ataxia 10. PLoS Genet. 2010;6:e1000984. doi: 10.1371/journal.pgen.1000984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishikawa K., Nagai Y. Molecular mechanisms and future Therapeutics for spinocerebellar ataxia type 31 (SCA31) Neurotherapeutics. 2019;16:1106–1114. doi: 10.1007/s13311-019-00804-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seixas A.I., Loureiro J.R., Costa C., Ordóñez-Ugalde A., Marcelino H., Oliveira C.L., Loureiro J.L., Dhingra A., Brandão E., Cruz V.T., et al. A pentanucleotide ATTTC repeat insertion in the non-coding region of DAB1, mapping to SCA37, causes spinocerebellar ataxia. Am. J. Hum. Genet. 2017;101:87–103. doi: 10.1016/j.ajhg.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loureiro J.R., Oliveira C.L., Mota C., Castro A.F., Costa C., Loureiro J.L., Coutinho P., Martins S., Sequeiros J., Silveira I. Mutational mechanism for DAB1 (ATTTC)n insertion in SCA37: ATTTT repeat lengthening and nucleotide substitution. Hum. Mutat. 2019;40:404–412. doi: 10.1002/humu.23704. [DOI] [PubMed] [Google Scholar]
- 27.Ishiura H., Doi K., Mitsui J., Yoshimura J., Matsukawa M.K., Fujiyama A., Toyoshima Y., Kakita A., Takahashi H., Suzuki Y., et al. Expansions of intronic TTTCA and TTTTA repeats in benign adult familial myoclonic epilepsy. Nat. Genet. 2018;50:581–590. doi: 10.1038/s41588-018-0067-2. [DOI] [PubMed] [Google Scholar]
- 28.Tsai Y.-C., Zafar F., McEachin Z.T., McLaughlin I.J., Van Blitterswijk M., Ziegle J., Schüle B. In: Genomic Structural Variants in Nervous System Disorders. Proukakis C., editor. Springer US; 2022. Multiplex CRISPR/Cas9-Guided No-Amp targeted sequencing panel for spinocerebellar ataxia repeat expansions; pp. 95–120. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Bionano data from five samples (III.4, III.6, III.9, IV.7, and IV.10) generated during this study are available at the NCBI BioProject database under BioProject: PRJNA838989 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA838989).