Summary
Neuronal oscillations, their inter-areal synchronization, and scale-free dynamics constitute fundamental mechanisms for cognition by regulating communication in neuronal networks. These oscillatory dynamics have large inter-individual variability that is partly heritable. We hypothesized that this variability could be partially explained by genetic polymorphisms in neuromodulatory genes. We recorded resting-state magnetoencephalography (MEG) from 82 healthy participants and investigated whether oscillation dynamics were influenced by genetic polymorphisms in catechol-O-methyltransferase (COMT) Val158Met and brain-derived neurotrophic factor (BDNF) Val66Met. Both COMT and BDNF polymorphisms influenced local oscillation amplitudes and their long-range temporal correlations (LRTCs), while only BDNF polymorphism affected the strength of large-scale synchronization. Our findings demonstrate that COMT and BDNF genetic polymorphisms contribute to inter-individual variability in neuronal oscillation dynamics. Comparison of these results to computational modeling of near-critical synchronization dynamics further suggested that COMT and BDNF polymorphisms influenced local oscillations by modulating the excitation-inhibition balance according to the brain criticality framework.
Subject areas: Biological sciences, Neuroscience, Cognitive neuroscience
Graphical abstract
Highlights
-
•
Human local oscillation dynamics is influenced by polymorphisms in COMT and BNDF
-
•
COMT and BDNF influence oscillation amplitudes and long-range temporal correlations
-
•
BDNF polymorphism affected the strength of large-scale synchronization
-
•
Framework of brain criticality links COMT and BDNF with local E/I-balance
Biological sciences; Neuroscience; Cognitive neuroscience
Introduction
Neuronal oscillations and their inter-areal synchronization play fundamental mechanistic roles in cognitive functions, regulating neuronal processing and communication across distributed brain areas (Fries, 2015; Singer, 1999; Womelsdorf et al., 2014). In humans, dynamics of neuronal oscillations and their long-range phase synchronization, observed non-invasively in magneto- (MEG) and electroencephalography (EEG) recordings, serve fundamental roles in a variety of sensory, motor, and cognitive functions (Fell and Axmacher, 2011; Jensen et al., 2014; Klimesch et al., 2007; Marzetti et al., 2019; Palva and Palva, 2012; Samaha et al., 2020; Siegel et al., 2012; Thut et al., 2012). During rest, brain activity involves frequency-specific inter-areal correlation structures throughout the 1–100 Hz range (Arnulfo et al., 2020; Betti et al., 2018; Brookes et al., 2011; Foster et al., 2016; Hipp and Siegel, 2015; Marzetti et al., 2013; Siebenhühner et al., 2020; Zhigalov et al., 2017). The intrinsic spatial organization of spontaneous resting-state functional connectivity (FC) observed with MEG (Betti et al., 2018; Oswald et al., 2017; Siebenhühner et al., 2020), intra-cranial EEG (Arnulfo et al., 2020), and functional magnetic resonance imaging (fMRI) (Hahn et al., 2020; Vidaurre et al., 2021) predicts individual variability in task performance. Resting-state FC networks reflect individual trait-like behavior (Seitzmana et al., 2019) that could arise from the underlying individual brain structural organization (Cabral et al., 2014; Mostame and Sadaghiani, 2020) and neuromodulation (McCormick et al., 2020; Pfeffer et al., 2021; van den Brink et al., 2018; van den Brink et al., 2019).
Amplitude envelopes of fast (>1 Hz) oscillations are also characterized by slow (0.1–1 Hz) and infra-slow (0.01–0.1 Hz) fluctuations (Hiltunen et al., 2014; Hipp and Siegel, 2015; Monto et al., 2008). These fluctuations are scale-free and exhibit power-law autocorrelations in their strength known as long-range temporal correlations (LRTCs) (Linkenkaer-Hansen et al., 2001; Monto et al., 2008; Palva et al., 2013; Poil et al., 2012; Thiery et al., 2018; Zhigalov et al., 2015). In addition to instantaneous oscillation dynamics, inter-individual variability of LRTCs also predicts variability in task-related behavioral LRCTs (Palva et al., 2013; Smit et al., 2013). Scale-free LRTCs are hallmarks of systems with critical or near-critical dynamics, suggesting that the brain operates near a critical phase transition between disorder and order, i.e., between inadequate and excessive synchronization, respectively (Cocchi et al., 2017), which enables high flexibility and variability.
A large fraction of the variability in fast and slow oscillation dynamics could be heritable. MEG spectral power shows similarities between siblings (Leppäaho et al., 2019; Salmela et al., 2016). Twin studies indicate that frontal midline theta activity is genetically related to symptoms and response time variability in attention-deficit/hyperactivity disorder (McLoughlin et al., 2014) and that peak frequencies (Van Pelt et al., 2012) as well as LRTCs (Linkenkaer-Hansen et al., 2007) of local cortical oscillations have a genetic basis, so that up to 60% of the variance in neuronal LRTCs (Linkenkaer-Hansen et al., 2007) and up to 85% of the variance in cognitive ability (Posthuma et al., 2001) can be attributed to genetic factors. However, what these genetic factors are and how they could contribute to the inter-individual variability of synchronization dynamics, including oscillation amplitudes, large-scale synchronization, and LRTCs, have remained central unresolved questions.
We hypothesized that polymorphism in genes regulating neuromodulation is one genetic factor underlying variability in oscillatory dynamics. Compelling evidence indicates that especially the neuromodulatory effects of the catecholamines dopamine (DA) and noradrenaline (NE) (Cools, 2019; Pfeffer et al., 2021; Robbins and Arnsten, 2009; van den Brink et al., 2018) and the indolamine serotonin (5-hydroxytryptamine; 5-HT) (Harris and Thiele, 2011; Lucki, 1988; Štrac et al., 2016) contribute to variability in neuronal dynamics (Ferguson and Cardin, 2020; Harris and Thiele, 2011; Lee and Dan, 2012; van den Brink et al., 2019).
Brain DA and NE levels are regulated by their catabolism via COMT enzyme (Männistö and Kaakkola, 1999; Parkin et al., 2018) whose activity is influenced by a common SNP in the COMT gene resulting in variation substituting valine (Val) to methionine (Met) at codon 158 Val158Met (captured as rs4680). COMT Val158 homozygotes have higher COMT activity and therefore lower brain DA and NE levels than Met158 homozygotes, with activity for heterozygotes falling in the middle (Chen et al., 2004). A considerable fraction of the variance in serotonin levels, on the other hand, is explained by Val66Met (rs6265) polymorphism, resulting in a valine (Val) to methionine (Met) substitution at codon 66, of the gene coding for brain-derived neurotrophic factor (BDNF), which influences 5-HT receptor binding (Fisher et al., 2017) and the activity-dependent secretion of BDNF (Egan et al., 2003). Convincing evidence emerging from numerous fMRI studies indicates that COMT Val158Met (rs4680) polymorphism is associated with modulation of signals especially in the prefrontal cortex (PFC) and with performance of cognitive functions dependent on PFC (Barnett et al., 2007; Meyer-Lindenberg et al., 2005; Mier et al., 2010; Papenberg et al., 2020; Robbins and Arnsten, 2009), while BDNF Val66Met polymorphism is implicated in many aspects of behavior and cognition (Beste et al., 2010; Matsuo et al., 2009; Notaras et al., 2015). In addition to modulation of 5-HT levels, activity-dependent release of BDNF also influences the maintenance and formation of neuronal networks (Park and Poo, 2013; Zagrebelsky and Korte, 2014), thereby influencing cognition and behavior over lifetime.
Although the effects of COMT Val158Met and BDNF Val66Met polymorphisms on slow neuronal dynamics have been well-studied with fMRI, their influence on fast neuronal oscillations has remained uncharted. We hypothesized that COMT Val158Met and BDNF Val66Met polymorphisms could influence inter-individual variability in oscillation amplitudes, phase synchronization, and their LRTCs. Importantly, critical dynamics in the brain are thought to be controlled primarily by the excitation-inhibition (E/I) ratio so that critical dynamics emerge at balanced E/I while excess of inhibition or excitation leads to sub- or super-critical dynamics, respectively (Plenz and Thiagarajan, 2007; Poil et al., 2012; Shew et al., 2009). We thus further hypothesized that these polymorphisms would specifically exert their influence on critical brain dynamics via changes in the E/I ratio.
Genome-wide association studies (GWAS) have demonstrated the clinical relevance of genes on functional and structural MRI data (Pereira et al., 2018) and for local oscillatory activity in the brain using sensor-level EEG and MEG (Salmela et al., 2016; Smit et al., 2018). Since our specific interest here was on the neuromodulation-related genetic influences of polymorphisms in COMT Val158Met and BDNF Val66Met candidate genes on neuronal oscillations, we did not use a GWAS approach (which would require a huge sample size N >1000) nor polygenic risk scores (which do not exist for oscillation dynamics). To test our hypotheses, we measured resting-state brain dynamics with MEG and estimated local oscillation amplitudes, large-scale phase synchronization, and LRTCs from individually source-reconstructed MEG data. We then investigated their correlation with the COMT Val158Met and BDNF Val66Met polymorphisms estimated from SNP analysis (Figure 1) and compared the results with predictions from computational modeling. In line with the predictions of the brain criticality framework and computational modeling, we found that the COMT Val158Met and BDNF Val66Met polymorphisms influenced local oscillation amplitudes and LRTCs, while BDNF Val66Met polymorphism also influenced the strength of large-scale synchronization.
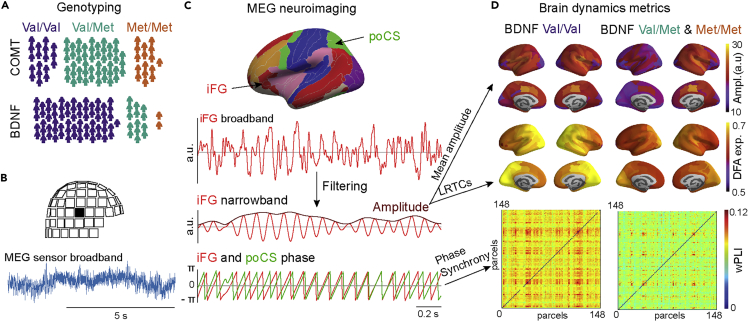
Figure 1.
A schematic of study outline and rationale
(A) Genotyping of the participants (N = 82) was used to identify the COMT Val158Met (rs4680) and BNDF Val66Met (rs6265) genetic polymorphisms.
(B) Broadband MEG data were recorded from each participant with 306-channel MEG during ∼8-min resting state.
(C) MEG data were source-reconstructed using minimum-norm estimation with individual MRI-based head and cortical surface anatomy and collapsed to cortical parcels (upper panel). Parcel-wise broadband signals (shown for inferior frontal gyrus, iFG, in second panel) were filtered to narrowband signals (signal and amplitude envelope shown here for the iFG in third panel). From the narrowband signal, instantaneous phase was extracted (shown for iFG and posterior central sulcus, poCS, in lower panel). Mean amplitudes and LRTCs were estimated for each parcel from amplitude envelope time series, and inter-parcel phase synchronization for all parcel pairs from phase time series.
(D) In group analyses, we assessed the anatomical distribution of mean amplitudes, LRTCs, and synchronization within the polymorphism groups and identified statistically significant differences between these groups (Figure 2, Figure 3, Figure 4).
Results
COMT Val158Met and BDNF Val66Met polymorphisms influence oscillation amplitudes
We first evaluated mean oscillation amplitudes between 3 and 60 Hz averaged across all cortical parcels separately for participants grouped according to the COMT Val158Met (Val/Val, Val/Met, and Met/Met) and the BDNF Val66Met (Val/Val and the combined Val/Met and Met/Met) polymorphisms. We first confirmed that age or the gender ratios did not differ significantly between the polymorphism groups (Tables 1 and 2). We then confirmed the validity of using the canonical frequency bands theta (θ, 3–7 Hz), alpha (α, 8–14 Hz), beta (β, 14–30 Hz), and gamma (γ, 30–60 Hz) by an analysis of spatial similarity across frequencies using Louvain clustering (Figure S1).
Table 1.
Mean (±SD) sample characteristics by the COMT polymorphisms
| Val/Val | Val/Met | Met/Met | |
|---|---|---|---|
| N | 18 | 48 | 16 |
| Age mean, yearsa | 30.65 (9.90) | 28.53 (8.91) | 29.87 (10.69) |
| Age range | 22–55 | 19–50 | 18–53 |
| Handedness (right/left)b | 18/0 | 41/5 | 15/1 |
| Gender (Female/Male)c | 11/7 | 27/21 | 6/10 |
| Rs mean, minutesd | 7.86 (2.20) | 7.97 (3.28) | 7.49 (2.69) |
| Rs range | 5–10 | 5–24 | 5–12 |
Age information missing from three participants. The polymorphism groups did not differ in terms of age as confirmed by a one-way ANOVA.
Handedness information missing from two participants.
The percentages of female and male participants did not differ between the COMT polymorphism groups as indicated by the Chi-squared test.
Rs = resting state. The duration of the resting state recording did not differ between polymorphism groups as confirmed by a one-way ANOVA.
Table 2.
Mean (±SD) sample characteristics by the BDNF polymorphisms
| Val/Val | Val/Met + Met/Met | |
|---|---|---|
| N | 66 | 16 |
| Age mean, yearsa | 29.95 (9.26) | 26.44 (9.69) |
| Age range | 18–55 | 20–52 |
| Handedness (right/left)b | 59/6 | 15/0 |
| Gender (Female/Male)c | 35/31 | 9/7 |
| Rs mean, minutesd | 7.92 (3.07) | 7.58 (2.47) |
| Rs range | 3–24 | 5–10 |
Age information missing from three participants. The polymorphism groups did not differ in terms of age as confirmed by an independent samples t-test (2-tailed).
Handedness information missing from two participants.
The percentages of female and male participants did not differ between the BDNF polymorphism groups as indicated by the Chi-squared test.
Rs = resting state. The duration of the resting state recording did not differ between polymorphism groups as confirmed by an independent samples t-test (2-tailed).
As observed commonly in resting state, oscillation amplitudes peaked in the α band in all COMT and BDNF polymorphism groups (Figure 2A). This was similarly evident when mean oscillation amplitudes were averaged within the canonical frequency bands (Figure S2). We found no influence of COMT Val158Met and BDNF Val66Met polymorphisms on the mean oscillation amplitudes in the canonical bands using univariate ANOVAs that controlled for age effects (p > 0.0125, Bonferroni-corrected across frequency bands) (Figure S2). We repeated the same analysis at the level of individual wavelet frequencies and found no significant effects there either (Figure 2A).
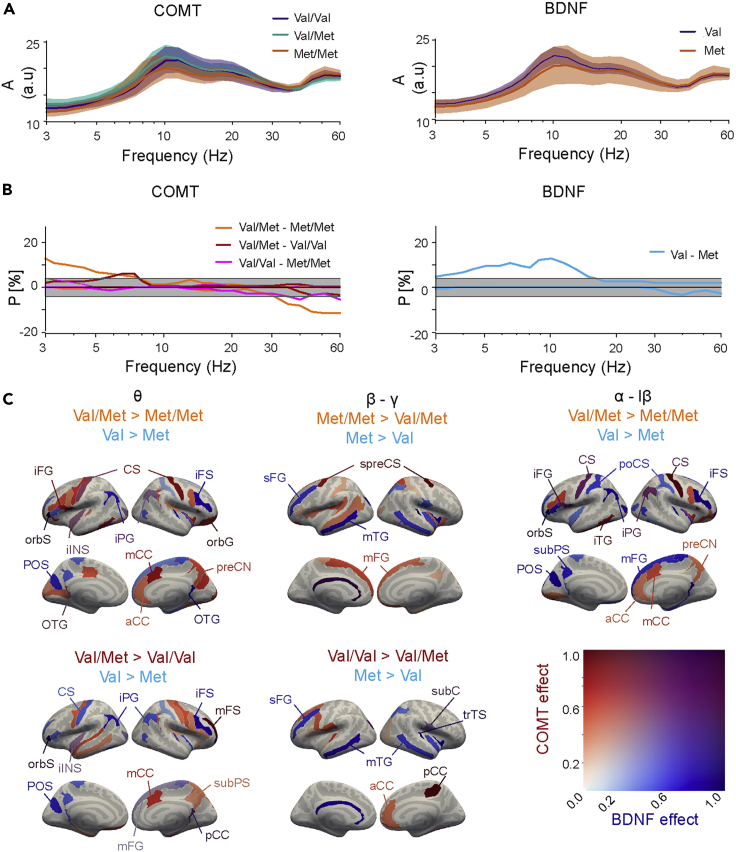
Figure 2.
COMT and BDNF polymorphisms influence oscillation amplitudes
(A) Oscillation amplitudes averaged across parcels for the COMT (Val/Val, purple, n = 18; Val/Met, turquoise; n = 48; and Met/Met, orange, n = 16) and BDNF polymorphism groups (Val/Val, purple, n = 66; Met carriers, orange, n = 16). Shaded areas represent 95% bootstrapped confidence intervals of the group means.
(B) The fraction P of parcels with a significant positive or negative pairwise difference (shown above and below zero line, respectively) between the COMT and BDNF polymorphisms groups in oscillation amplitudes (Mann–Whitney-U test, p < 0.05). The light gray area indicates the 5% alpha level for findings expected by chance under the null hypothesis. The dark gray area indicates the Q-level which defines the chance level for false positives at pq< 0.05 in any of the 26 frequencies.
(C) Cortical parcels where the amplitudes differed significantly between polymorphism groups (indicated above the brain surfaces) in the θ (3–7 Hz), β–γ (20–60 Hz), and α to low-β (7–18 Hz) frequency ranges in at least one of the individual frequencies. The color scale indicates the max-normalized amplitude differences for COMT (red) and BDNF (blue) polymorphisms independently, so that for each effect the value 1 represents the maximum observed effect, and so that parcels where the effects are co-localized have purple hues. Abbreviations are formed from a combination of i) Areas: a = anterior, ca= calcarine, d = dorsal, i = inferior, la = lateral, m = middle, mrg = marginal, opc = opercular, p = posterior, s = superior, tr = triangular and ii) Lobes: C = central/cingulate, F = frontal, P = parietal, O = occipital, T = temporal, and iii) G = Gyrus, S = Sulcus. Additional abbreviations: subC = subcentral gyrus/sulcus, preCN = precuneus.
As the polymorphism effects could also be anatomically confined, we tested significant pairwise differences between polymorphism groups for each wavelet frequency and for each individual parcel. For COMT, the amplitudes were stronger (Mann-Whitney-U test, p < 0.05, FDR and Q-level corrected) for Val/Met heterozygotes than for either Met or Val homozygotes in the θ band (3–7 Hz) in up to 13% and 6% of cortical parcels, respectively (Figure 2B), whereas in the high-β to γ (20–60 Hz) bands, amplitudes were larger for Met homozygotes in 12% of parcels compared to Val/Met, and 6% compared to Val/Val. For BDNF, oscillation amplitudes were stronger for the Val homozygotes than for Met carriers in θ and in the α to low-β bands (up to 13% of parcels). We next visualized significant differences in oscillation amplitudes between the polymorphism groups in the cortical anatomy (Figure 2C). Stronger θ and α-low-β band oscillation amplitudes for the COMT Val/Met heterozygotes were localized to the central sulcus (CS), anterior, mid-, and posterior cingulate cortex (aCC, mCC, and pCC), inferior frontal gyrus (iFG), right-hemispheric middle frontal gyrus (mFG), and inferior parietal gyrus (iPG), inferior part of the insula (iINS), and precuneus (preCN) (Figure 2C) these parcels mostly belonging to the frontoparietal network (FPN) and default mode networks (DMN) (Figure S3). Of these, iPG and left iINS and left CS showed also larger amplitudes for BDNF Val homozygotes than for Met carriers. In the β–γ bands, larger amplitudes for COMT Met homozygotes compared to Val/Met heterozygotes were observed in superior precentral sulcus (spreCS), corresponding to frontal eye fields and in temporal cortices while stronger amplitudes for the Val homozygotes were found in aCC and pCC. For BDNF Val homozygotes, oscillation amplitudes were larger than in Met carriers in iFS, parieto-occipital sulcus (POS), and postcentral sulcus (poCS) in θ to low-β bands. COMT and BDNF jointly affected θ and α to low-β band oscillation amplitudes in the CS and iPG.
COMT Val158Met and BDNF Val66Met polymorphisms influence DFA scaling exponents
We quantified long-range temporal correlations (LRTCs) in oscillation amplitude fluctuations with detrended fluctuation analysis (DFA) (Hardstone et al., 2012) and estimated the mean DFA scaling exponents in all wavelet frequencies (3–60 Hz). DFA exponents were first averaged across all parcels separately for each COMT Val158Met and BDNF Val66Met polymorphism group for each frequency (Figure 3A). Using univariate ANOVAs that controlled for age, we observed a difference of group mean whole-brain DFA exponents between COMT polymorphism in the canonical γ band [F(2, 75) = 3.80, p = 0.027, ηp2 = 0.092] and for BDNF, Val homozygotes showed significantly higher exponents than Met carriers in the canonical α band [F(1, 76) = 4.19, p = 0.044, ηp2 = 0.052] (Figure S2). However, neither of these effects were significant after Bonferroni correction over frequency bands (p > 0.0125).
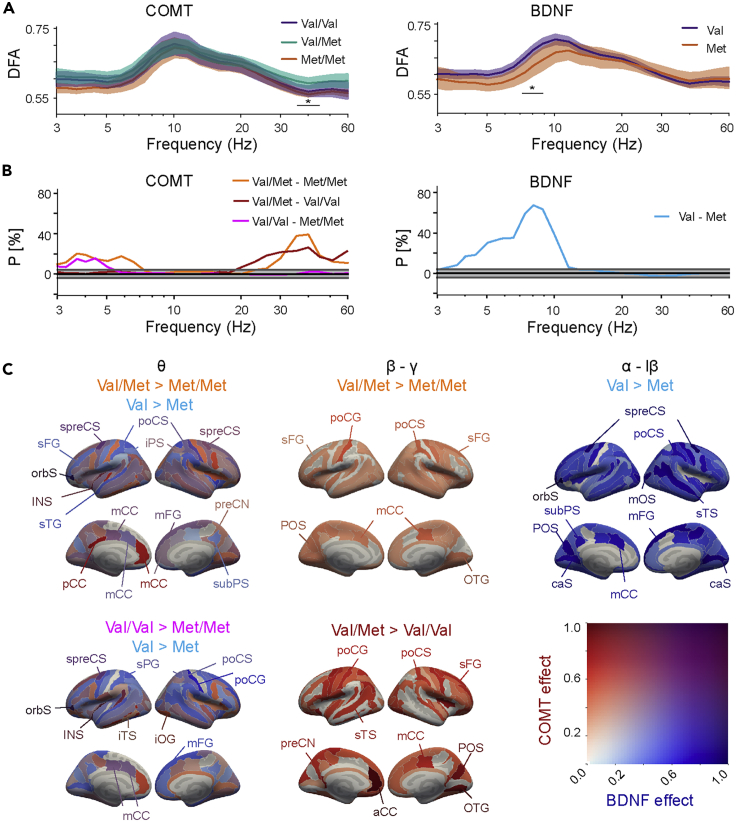
Figure 3.
Influence of COMT and BDNF polymorphisms on LRTCs
(A) Detrended fluctuation analysis (DFA) exponents averaged across parcels for the COMT and BDNF polymorphism groups. Shaded areas represent 95% bootstrapped confidence intervals of group means. Black bar with asterisk denotes the range of frequencies with significant differences between the COMT and BDNF polymorphism groups (univariate ANOVA, with age as covariate, ∗: p < 0.05, controlled for multiple comparisons with cluster-based permutation statistics).
(B) Fractions (P) of parcels with a significant positive or negative difference between COMT and BDNF polymorphism groups in the DFA exponents (Mann–Whitney-U test, p < 0.05), as in Figure 2B.
(C) Cortical areas where significant differences between polymorphism groups in the DFA exponents were found in the θ, β–γ, and α– low-β bands (Mann–Whitney U test, p < 0.05, corrected). Colors and abbreviations as in Figure 2C.
At the level of individual wavelet frequencies, the age-controlled univariate ANOVAs revealed for COMT a cluster of 3 consecutive frequencies from 35 to 45 Hz that remained significant after controlling for multiple comparisons with cluster-based permutation statistics (Fcluster = 11.64 >Fperm = 6.25 at α = 0.05). For BDNF, a significant cluster was found between 7 and 9 Hz (Fcluster = 13.08 >Fperm = 8.53 at α = 0.05) (Figure 3A).
Similar to oscillation amplitudes, COMT Val158Met and BDNF Val66Met polymorphisms had a more robust effect on DFA exponents at the resolution of cortical parcels indicating large anatomical heterogeneity. DFA exponents were greater for COMT Val/Met heterozygotes than for Met homozygotes (Mann-Whitney-U test, p<0.05, FDR and Q-level corrected) in the θ band (up to 20% of parcels) and in the β–γ band (up to 40%) (Figure 3B). Furthermore, θ band DFA exponents were greater for Val/Val than for Met/Met (16%) and β–γ DFA exponents were greater in Val/Met than for Val/Val (26%) (Figure 3B). For BDNF, a very robust effect of polymorphism was found for DFA exponents (Mann-Whitney-U test, p<0.05, FDR and Q-level corrected). These were greater for Val homozygotes than Met carriers in the θ band (35%) and in α band (68%) (Figure 3B). The localization of significant differences in DFA exponents between polymorphism groups (Mann-Whitney-U test, p<0.05, FDR and Q-level corrected, Figure 3C) revealed wide-spread group differences. In the θ band, large differences between COMT Val/Met heterozygotes and Met homozygotes and between BDNF Val homozygotes and Met carriers were co-localized to preCS, mCC, and mFG (Figure 3C). DFA exponents in θ band were also greater for COMT Val/Met group in INS, pCC, and in β–γ band particularly in parcels belonging to somatomotor (SM) network and DMN (see Figure S3). BDNF Val homozygotes had greater LRTC exponents in α and low-β bands than Met carriers in nearly all parcels (Figure 3C). Generally, the effects of COMT and BDNF polymorphisms on LRCTs were widespread, in line with prior work and with the idea that LRTCs reflect global fluctuations in neuronal activity across the brain’s modular structure (Monto et al., 2008; Palva et al., 2013; Zhigalov et al., 2017).
Finally, for each polymorphism, we co-localized its effects on both oscillation amplitudes and DFA exponents. We found that joint effects on amplitudes and exponents were strongest in the parcels of PFC and PPC, and CS and anterior cingulate structures (Figure S4).
BDNF Val66Met polymorphism influences the strength of large-scale phase synchronization
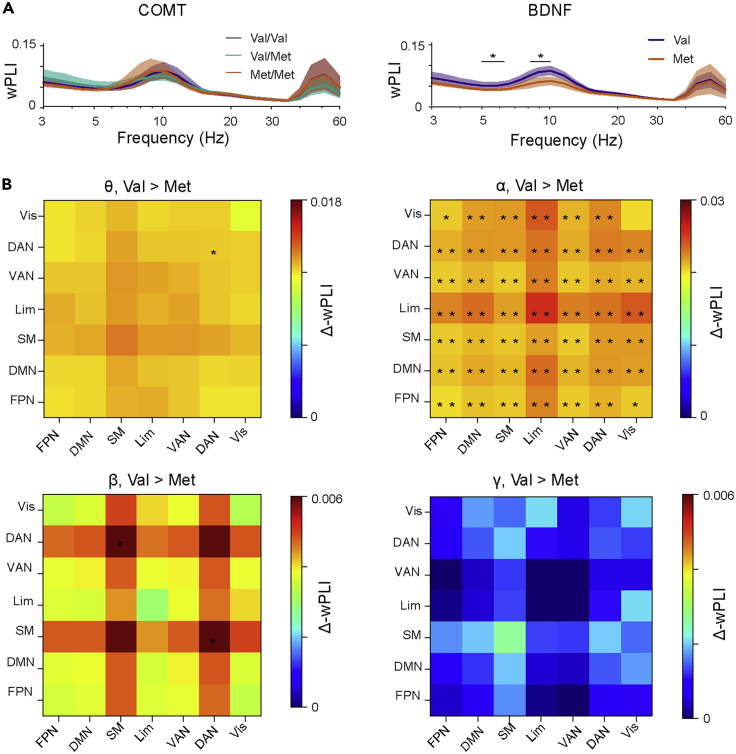
We next set out to assess the influence of COMT Val158Met and BDNF Val66Met polymorphisms on inter-areal phase synchronization of neuronal oscillations. Phase synchrony is a key characteristic of individual cortical dynamics and has fundamental influence on behavior (Brookes et al., 2011; Fell and Axmacher, 2011; Fries, 2015; Palva and Palva, 2012; Singer, 1999; Vidaurre et al., 2021). We estimated phase synchrony among all cortical parcels with the weighted phase-lag index (wPLI) that is insensitive to linear source mixing (Palva et al., 2018) and computed the mean synchronization over all parcel pairs for each of the COMT Val158Met and BDNF Val66Met polymorphism groups (Figure 4A). Both within the canonical frequency bands and in individual wavelet frequencies, we did not find any significant differences (Kruskal-Wallis test, p > 0.05) among the COMT Val158Met polymorphism groups (Figures 4A and S2). Within canonical bands, we found that mean synchronization for BDNF Val homozygotes was significantly stronger (Kruskal-Wallis test, H = 6.15, p = 0.013) than for Met carriers in the α band (Figure S2). In individual wavelet frequencies, Val homozygotes exhibited stronger synchronization (Kruskal-Wallis test, p<0.05) in two frequency clusters, from 5 to 6.6 Hz and from 8.9 to 10.1 Hz (Figure 4A), which both remained significant after cluster-based permutation correction (Hcl,1 = 14.81 and Hcl,2 = 10.57 vs Hperm = 7.14 at α = 0.05).
Figure 4.
BDNF polymorphisms influence large-scale phase synchronization
(A) The mean strength of phase synchronization averaged across the whole brain for the COMT and BDNF polymorphism groups with 95% bootstrapped confidence intervals. Black bars with asterisks denote the frequencies where a significant group difference was found (Kruskal-Wallis, ∗: p < 0.05, controlled for multiple comparisons with cluster-based permutation statistics).
(B) The differences in the mean phase synchrony (Δ-wPLI) between BDNF Val homozygotes and Met carriers within and between functional systems averaged across hemispheres and wavelet frequencies in canonical frequency bands. Red color indicates stronger synchronization for BDNF Val homozygotes and blue for Met carriers. Stars denote the brain system pairs where the group difference was significant (Kruskal-Wallis, ∗: p < 0.05, uncorrected, ∗∗: p < 0.01, Benjamini-Hochberg corrected). Abbreviations: DAN = Dorsal attention network, DMN = Default Mode network, FPN = Frontoparietal network, Lim = Limbic system, SM= Somatomotor network, VAN = Ventral attention network, Vis = Visual network.
To acquire neuroanatomical insight into the patterns of inter-areal synchronization in functional brain systems, we collapsed the parcel-wise synchronization estimates within functional systems (Yeo et al., 2011) (Figure S3) and canonical frequency bands (Figure S1). Synchronization between or within the subsystems did not differ significantly among the COMT polymorphisms (Figure S5). However, BDNF Val homozygotes exhibited stronger synchronization than Met carriers in the α band between and within nearly all subsystems (Figures 4B and S5, Kruskal-Wallis test, corrected with Benjamini-Hochberg). We also computed synchronization separately for each hemisphere and between hemispheres. Interestingly, group differences in α band synchronization were widespread and more robust for left- and inter-hemispheric connections than for the right-hemispheric connections (Figure S6). Additionally, β-band synchronization was stronger for BDNF Val homozygotes than Met carriers within and between several left-hemispheric networks, including FPN, dorsal attention network (DAN), ventral attention (VAN), and Limbic networks (Lim) and DMN (Figure S6).
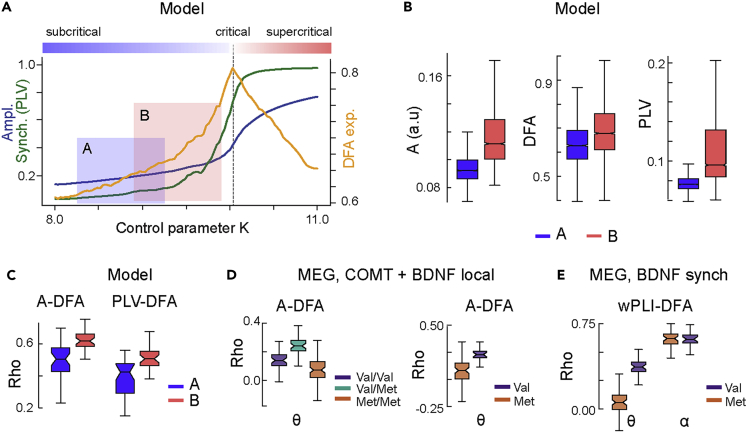
Computational model-based approach to neuromodulation effects on critical synchronization dynamics
We next set out to address the effects of COMT Val158Met and BDNF Val66Met polymorphisms in the context of brain criticality. Brain criticality is assumed to be regulated by a control parameter (K) that reflects the gross excitation/inhibition (E/I) ratio (Deco et al., 2021; Shew et al., 2009; Stephani et al., 2020). The human brain is thought to normally operate in a slightly subcritical regime (Priesemann et al., 2014), where LRTCs exhibit large inter-individual variability (Linkenkaer-Hansen et al., 2001; Palva et al., 2013; Smit et al., 2013; Zhigalov et al., 2015). We hypothesized that COMT Val158Met and BDNF Val66Met polymorphisms would bias the E/I balance and hence the “control parameter” that tunes the distance to criticality. The carriers of different variants should therefore be at different positions away from the critical point.
To assess the relationships of the observable brain dynamics measures with the control parameter, we implemented a nested Kuramoto model with separable local network (cortical parcel) and large-scale (whole-brain network) levels (Siebenhühner et al., 2020) of synchronization dynamics exhibiting a transition from sub- to super-critical dynamics (Figure 5A). We used the model to predict oscillation synchronization dynamics for two hypothetical cohorts with different values of the control parameter (K) regulating their position in the critical regime. Cohort A had lower and cohort B had higher control parameter (K) value positioning cohort B closer to the critical regime (Figure 5A) associated with stronger local amplitudes, LRTCs, and large-scale synchronization (Figure 5B). The model further predicted larger mutual correlations between DFA and oscillation amplitudes as well as between DFA and phase synchronization for cohort B with higher K (Figure 5C).
Figure 5.
Model and experimental evidence for modulation of brain criticality
(A) Computational model of local and inter-areal synchronization dynamics in the human connectome shows that both local amplitude (blue) and inter-areal synchronization (green) increase monotonically with increasing control parameter K and exhibit a phase transition between low- (subcritical) and high-synchronization (super-critical) regimes. Conversely, the DFA scaling exponent (yellow), a measure of long-range temporal correlations (LRTCs), peaks at the critical point. The model predicts that two cohorts (A and B, blue and red) with simulated subjects operating in different parts of the subcritical regime should exhibit group differences in amplitude, LRTCs, and inter-areal synchronization.
(B) Local amplitude A, DFA exponents, and inter-areal phase synchronization (PLV) in modeled data for two cohorts A and B. All metrics are greater for the cohort B that is designed to operate closer to the critical point (see panel A) than cohort A. The box ends indicate the lower quartile (Q1, 25th percentile) and upper quartile (Q3, 75th percentile) across cortical regions/parcels, notches indicate the median, and whiskers indicate the range of Q1 – 1.5 ∗ IQR and Q3 + 1.5 ∗ IQR, where IQR is the inter-quartile range (Q3-Q1).
(C) Correlations between amplitude and DFA exponents, and between PLV and DFA, in modeled data are also larger for cohort B than the more subcritical cohort A.
(D) Correlations of local amplitudes and DFA exponents for COMT and BDNF polymorphism groups in MEG data in θ band.
(E) Correlations between large-scale synchrony (wPLI) and DFA exponents for BDNF in MEG data differ between polymorphism groups in θ, but not in α band.
To investigate whether COMT and BDNF polymorphisms could influence synchronization dynamics via modulation of K, in line with the model predictions, we computed these dynamics separately for each polymorphism group in the canonical frequency bands (Figure S2). In line with model predictions, the elevated θ band amplitudes and LRTCs in COMT Val/Met group (Figure S2) were associated with stronger correlation of local oscillation amplitudes and LRTCs (Figure 5D, Val/Met > Met/Met in 95% and Val/Met > Val/Val in 84% of permutations). For BDNF, elevated θ band amplitudes and LRTCs in Val homozygotes (Figure S2) were associated with stronger correlation of local oscillation amplitudes and LRTCs (Figure 5D, Val > Met in 92% of permutations, for γ bands see Figure S7). In contrast, elevated α band global synchronization in BDNF Val homozygotes (Figure S2) was not associated with elevated correlation of synchronization with DFA (Figure 5E) whereas in θ band the correlation between synchronization and LRTCs was higher for BNDF Val homozygotes (Figure 5E, Val > Met in 98% of permutations).
These findings thus provide converging evidence that COMT Val158Met and BDNF Val66Met polymorphisms influence local oscillation dynamics as predicted by the framework of brain criticality under the hypothesis that these polymorphisms influence the control parameter of cortical critical dynamics. In contrast, the most robust effects of BDNF on large-scale synchronization were not in line of the model predictions, suggesting that these were mediated mainly by other factors than the control parameter of brain criticality.
Discussion
Inter-individual variability in spontaneous oscillatory dynamics influences individual behavioral performance and cognitive ability (Donhauser and Baillet, 2020; Palva and Palva, 2012, 2018; Siebenhühner et al., 2020; Vidaurre et al., 2021). While the causes of this variability are diverse, it has been shown in human MEG and EEG data that part of it is attributable to genetics (Leppäaho et al., 2019; Linkenkaer-Hansen et al., 2007; Van Pelt et al., 2012). While macroscopic oscillations can be influenced through regulation of dynamic circuit motifs on a cellular level (Leppäaho et al., 2019; Van Pelt et al., 2012; Womelsdorf et al., 2014), there has been little research on how the genetic biases of cellular mechanisms translate into alterations of macroscopic brain dynamics observable in electrophysiological neuroimaging. We tested here a hypothesis that the COMT Val158Met and BDNF Val66Met genetic polymorphisms contribute to the variability in emergent local (oscillation amplitudes and LRTCs) and global (inter-areal phase synchronization) dynamics of neuronal oscillations. We showed that both COMT and BDNF polymorphisms influenced local oscillation amplitudes and LRTCs. However, only BDNF polymorphism impacted the strength of global synchronization. Using computational modeling, we furthermore showed that the influence of COMT and BDNF on local oscillation dynamics is achieved in line with brain criticality framework. This suggests that COMT and BNDF polymorphisms influence synchronization dynamics via modulations of the brain's E/I balance in line with the established role of COMT and BDNF in the regulation of neuronal activity via DA, NE, and 5-HT.
Local oscillation dynamics are modulated by COMT and BDNF polymorphisms
Prior observations about COMT and BDNF polymorphism effects on electrophysiological activity are scarce and mainly restricted to EEG and MEG sensor level (Bodenmann et al., 2009; Bonetti et al., 2021; Garcia-Garcia et al., 2017; Rodríguez-Rojo et al., 2018; Roy et al., 2020). In these studies, COMT Val homozygotes were found to have lower α power than Met homozygotes (Bodenmann et al., 2009), and COMT polymorphism was found to influence phase-resetting of γ oscillations in attention (Garcia-Garcia et al., 2017). Furthermore, BDNF has been implicated in resting-state oscillation amplitude differences in delta (0.5–3.5 Hz), α, and β bands but not in θ band amplitudes (Roy et al., 2020) and in reduced γ band FC between anterior and posterior regions in Val/Met compared to Val/Val participants (Rodríguez-Rojo et al., 2018). We used here a data-driven approach for mapping the effects of COMT and BDNF polymorphism on both instantaneous oscillation amplitudes and their LRTCs from source-reconstructed MEG data. We found anatomically specific θ−α band oscillations amplitudes to be greater for COMT Val/Met heterozygotes than for COMT Val or Met homozygotes, and θ and α oscillation amplitudes to be greater for BDNF Val homozygotes than for Met carriers. Similar to local oscillation amplitudes, we also found that COMT and BDNF polymorphisms modulated LRTCs in oscillation amplitude dynamics and hence the variability of the slow fluctuations in oscillation strengths. Anatomically specific LRTC scaling exponents were larger for COMT Val/Met heterozygotes than for Val or Met homozygotes in θ and γ bands and larger for BDNF Val homozygotes than for Met carriers in α-low-β bands, demonstrating that near-critical dynamics of brain oscillations is influenced by COMT and BDNF polymorphisms.
Our results provide robust evidence for COMT polymorphism influencing local amplitude dynamics. As COMT Val/Met heterozygotes have intermediate COMT enzymatic activity, and therefore intermediate brain DA and NE levels compared to Val and Met homozygotes (Chen et al., 2004), and as brain dynamics operating near criticality have been proposed to be beneficial for individuals (see below), our results are consistent with previous findings showing that the impact of neuromodulation on cognition follows an “inverted-U-shape” function where both excessively low and excessively high neuromodulation impair performance (Cools and D’Esposito, 2011; Farrell et al., 2012; Harris and Thiele, 2011; Meyer-Lindenberg et al., 2005; Papenberg et al., 2020; Vijayraghavan et al., 2007).
The effects of COMT Val158Met polymorphism were localized particularly to FPN and DMN, in agreement with the role of DA in cognitive control (Cools and D’Esposito, 2011; Ott and Nieder, 2019), as well as with previous fMRI studies in which the association between the polymorphism and neuronal activity was predominantly observed in PFC (Meyer-Lindenberg et al., 2005; Mier et al., 2010; Papenberg et al., 2020; Robbins and Arnsten, 2009) which has low DA reuptake and higher COMT levels. BDNF Val homozygotes exhibited stronger α band amplitudes bilaterally in frontoparietal regions and parcels of the visual system. In line with previous observations (Palva et al., 2013), differences in LRTC exponents were widespread, suggesting that they reflect state-dependent changes affecting most of the brain. Overall, these results support the idea that COMT and BDNF influence local oscillation dynamics in a variety of cortical regions important for cognition via DA, NE, and 5-HT.
BDNF polymorphism influences whole-brain connectivity
We found that BDNF Val66Met but not COMT Val158Met polymorphism influenced large-scale global inter-areal phase synchronization. Global α band synchronization was stronger for BDNF Val homozygotes than Met carriers in all functional subsystems except within the visual system. BDNF Val homozygotes also had stronger θ synchronization within DAN and stronger left-hemispheric β synchronization between various subsystems, particularly involving DAN and DMN. This robust modulation of synchronization dynamics by BDNF polymorphism could contribute to significant differences across individuals in behavior and cognition, which have been shown to be strongly influenced by spatial and spectral characteristics of oscillatory synchronization (Betti et al., 2018; Mostame and Sadaghiani, 2020; Siebenhühner et al., 2020). However, large-scale synchronization was not influenced by COMT Val158Met polymorphism, although it has been shown that neuropharmacologically elevated levels of catecholamines are related to fMRI-based changes in functional connectivity (van den Brink et al., 2018). These differences might be explained by different effects of catecholaminergic levels on BOLD signal and electrophysiological connectivity, or by COMT polymorphism having more subtle effects on neuronal dynamics compared to neuropharmacological interventions.
COMT and BDNF influence oscillation via impact on brain critical dynamics
Near-critical synchronization dynamics have been proposed to enable transient and flexible computations that are beneficial for individuals by maximizing information storage and transmission (Cocchi et al., 2017; Daffertshofer et al., 2018; Deco and Jirsa, 2012; Kinouchi and Copelli, 2006; Palva and Palva, 2018; Priesemann et al., 2014; Shew and Plenz, 2013). This notion has been supported experimentally by behavioral data (Simola et al., 2017) and in vitro studies (Shew et al., 2009). Here, we show that genetic polymorphisms in COMT and BDNF influenced individual near-critical synchronization dynamics and proximity to the critical point. The systems-level E/I ratio (or gain) is thought to be the primary control parameter for the regulation of critical brain dynamics (Shew et al., 2009; Stephani et al., 2020). We postulated that the observed differences in neuronal synchronization dynamics are due to neuromodulation-related shifts in the system’s net E/I ratio. Comparison of experimental and computational model data showed that COMT Val158Met and BDNF Val66Met polymorphisms influenced local oscillatory dynamics and their correlations in the θ band in line with the brain criticality framework. More specifically, COMT Val158Met heterozygotes and BDNF Val66 homozygotes had larger amplitudes and LRTC exponents and stronger correlations between these than the other polymorphism groups in θ band, indicating that they are associated with a higher value of the underlying control parameter, i.e., with greater net excitatory drive or neuronal gain (Ferguson and Cardin, 2020). This is in accordance with the framework in which neuromodulatory drive influences whole-brain neural states (Braun et al., 2021; Munn et al., 2021).
In contrast, we found no evidence that the influence of BDNF Val66Met polymorphism on global α-band synchronization could be explained within the brain criticality framework. This finding implies that the strongest effects of BDNF polymorphisms on global synchronization might not be due to a shifts in global excitation, but to other mechanisms such as the influence of BDNF on maintenance, maturation, and formation of neuronal networks (Park and Poo, 2013; Zagrebelsky and Korte, 2014) putatively via receptor TrkB (neurotrophic receptor tyrosine kinase 2) signaling (Castrén and Monteggia, 2021; Winkel et al., 2021). These could impact large-scale network synchronization via changes in structural connectivity (Mostame and Sadaghiani, 2020) or via changes in brain gray matter whose density has been shown to be associated with neuroreceptor and neurotransporter availabilities (Manninen et al., 2021) and whose thickness is associated with the structure of oscillatory networks (Mahjoory et al., 2020).
Conclusions
Our data showed that COMT Val158Met and BDNF Val66Met polymorphisms contribute to the variability of oscillation dynamics, their LRTCs, and synchronization, and add on to the scarce data on the genetic basis of oscillatory neuronal dynamics. In addition to COMT and BDNF polymorphisms, many other genetic factors are likely to contribute to oscillation dynamics, as observed for the cortical structure (Grasby et al., 2020), but this remains to be charted in future studies. Because oscillation amplitudes, LRTCs, and global synchronization dynamics are predictive of behavioral variability over both long (Palva et al., 2013; Smit et al., 2013) and sub-second time scales (Palva and Palva, 2012), COMT Val158Met and BDNF Val66Met polymorphisms may underlie individual differences in trait-like behavior and cognitive performance via their effect on critical oscillatory dynamics and synchronization. Differences in these dynamics caused by genetic polymorphisms are likely to influence individuals’ cognitive and mental development and may predispose them to specific brain diseases. In line with the latter idea, alterations in the brain's E/I ratio have been suggested to predispose individuals to dementia and Alzheimer disease (Pusil et al., 2019). Our results thus point in new directions for the investigation of how genetic factors could translate to behavioral variability via modulations of the systems-level neuronal dynamics.
Limitations of the study
We did not screen participants for their polymorphisms in COMT and BDNF genes prior to the MEG recordings. Due to this, one limitation of our study is the unequal group sizes, particularly for BDNF polymorphism for which the Val/Val group was the largest while there were fewer participants for the Val/Met and only 2 participants for the Met/Met group.
Further studies are also required to verify that the influence of BDNF and COMT polymorphisms on oscillation dynamics is mediated via shifts in excitability. This could be done for example by pharmacological intervention.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| DNA extracted from split samples | This paper | N/A |
| Deposited data | ||
| MEG data to reproduce figures and analyses | This paper | https://doi.org/10.5061/dryad.3n5tb2rjp |
| Oligonucleotides | ||
| PsychArray-24 v1.1 | Illumina | https://emea.illumina.com/products/by-type/microarray-kits/infinium-psycharray.html |
| Software and algorithms | ||
| PLINK 1.9 | Chang, C.C., Chow, C.C., Tellier, L.C., Vattikuti, S., Purcell, S.M., & Lee, J.J. (2015) Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience, 4, s13742-015. | www.cog-genomics.org/plink/1.9/ |
| LabView | National Instruments | https://www.ni.com/en-us/shop/software/products/labview.html |
| Python | Python Software Foundation | https://www.python.org/ |
| MATLAB | MathWorks | https://www.mathworks.com |
| Maxfilter | Elekta Neuromag, Helsinki, Finland | https://www.elekta.com/ |
| FieldTrip | Open Source | https://www.fieldtriptoolbox.org |
| FreeSurfer | Open Source | http://surfer.nmr.mgh.harvard.edu/ |
| MNE | Open Source | https://mne.tools/0.23/index.html |
| SPSS | IBM SPSS Statistics | https://www.ibm.com/products/spss-statistics |
| Code to reproduce figures and analyses | Original code | https://github.com/palvalab/RS-Gen |
| Data underlying figures and statistics | Dryad server | https://doi.org/10.5061/dryad.3n5tb2rjp |
Resource availability
Lead contact
Requests for further information and resources should be directed to and will be fulfilled by the lead contact, Satu Palva (satu.palva@helsinki.fi).
Materials availability
Ethical restrictions apply to data and original data cannot be shared on a public server. Data underlying figures, statistics, and main conclusions will be uploaded to Dryad server upon acceptance.
Experimental model and subject details
Human subjects
Data from 82 healthy human volunteers (age 18 to 55 years old; mean age: 29 years; 6 left-handed; 44 female) were collected for this study. This sample size corresponds to a meta-analysis of neuroimaging studies for COMT effects (Mier et al., 2010) that concluded based on pooled effect size estimates that 62 is the sample size necessary to achieve 80% power to detect association with PFC activation at an α-level of 0.05 (two-tailed). The study was performed in accordance with the Declaration of Helsinki and with permission of the Ethical Committee of the Helsinki University Central Hospital. All participants gave a written informed consent prior to the recordings.
Our sample for the COMT gene consisted of 18 Val/Val, 48 Val/Met, and 16 Met/Met individuals (Table 1). A one-way ANOVA confirmed that the COMT polymorphism groups did not differ in terms of age [F(2, 46) = 0.05, p = 0.984] and the duration of the resting state recording [F(2, 46) = 0.32, p = 0.726]. A chi-square test further confirmed that the percentages of female and male participants did not differ between the COMT polymorphism groups [X2(2) = 2.11, p = 0.349]. The BDNF sample consisted of 66 Val/Val, 14 Val/Met, and 2 Met/Met individuals (Table 2). As the Met/Met polymorphism had a low frequency, we combined Met carriers (i.e., Val/Met and Met/Met polymorphisms) into one group (see also Beste et al., 2010). An independent samples t-test (2-tailed) confirmed that the BDNF polymorphisms did not differ in terms of age [t(32) = 0.20, p = 0.847] or the length of the resting state recording [t(32) = 0.76, p = 0.444]. A chi-square test showed no differences in the percentages of female and male participants between the BDNF polymorphisms [X2(1) = 0.00, p = 0.995].
Neuroimaging data
Eyes-open resting-state brain activity was recorded from all participants with 306-channel MEG (Vectorview, Elekta-Neuromag, LtD) at a sampling rate of 600 Hz for ∼8 min (duration 7.8 ± 2.9 min, mean ± SD Tables 1 and 2). T1-weighted anatomical magnetic resonance imaging (MRI) MP-RAGE scans were obtained for each participant at a resolution of 1× 1 × 1 mm using a 1.5 T MRI scanner (Siemens, Germany).
Geneticdata
Blood samples for the collection of DNA were obtained from all subjects before each MEG session and DNA was extracted from blood samples according to standard procedures at the Finnish National Institute for Health and Welfare. DNA samples were genotyped at Estonian Genome Center using Infinium PsychArray-24 v1.1 (Illumina). Quality control (QC) was performed with PLINK 1.9 (www.cog-genomics.org/plink/1.9/). Markers were removed for missingness (> 5%) and Hardy-Weinberg equilibrium (p < 1 x 10−6). Individuals were checked for missing genotypes (> 5%), relatedness (identical by descent calculation, PI_HAT > 0.2) and population stratification (multidimensional scaling). The dataset collected for this study is part of a broader project “Tunteet” that includes also other datasets and paradigms (e.g.,Bonetti et al., 2021; Kliuchko et al., 2018).
Method details
Preprocessing and source analysis of MEG data
Signal space separation method (tSSS) (Taulu et al., 2005) with the Maxfilter software (Elekta Neuromag, Helsinki, Finland) was used to suppress extracranial noise, interpolate bad channels and to co-localize the recordings in signal space individually for each participant. Next, independent component analysis (ICA, MATLAB toolbox Fieldtrip (Oostenveld et al., 2011)) was used to extract and exclude signal components that were correlated with eye movement, blink and cardiac artifacts. The preprocessed MEG data were filtered into 26 logarithmically spaced frequency bands, fmin = 3 Hz, fmax = 60 Hz, using complex Morlet Wavelets with parameter m = 5 (making a compromise between time and frequency resolution). After filtering, the time-series data were decimated to a frequency-dependent sampling rate of between 2 and 5 times the center frequency. FreeSurfer software (http://surfer.nmr.mgh.harvard.edu/) was used for volumetric segmentation of the MRI data, surface reconstruction, flattening, cortical parcellations and neuroanatomical labeling with the Destrieux atlas. We then iteratively split the largest parcel in the atlas in two, until we arrived at a resolution of 400 parcels. MNE software (https://mne.tools/stable/index.html) (Gramfort et al., 2014) was used for creating cortically constrained source models and for the preparation of the forward and inverse operators (Dale et al., 2000) using noise covariance matrices (NCMs) obtained from broad-band filtered data (125–195 Hz, with 150 Hz notch). MNE inverse operators were then computed for all wavelet frequencies and used to project the sensor-space data into source space. The source models had dipole orientations fixed to the pial surface normal, yielding 5000–8000 source vertices per hemisphere. As in previous studies (Korhonen et al., 2014; Siebenhühner et al., 2020), source-vertex time-series were then collapsed into parcel time series with individually source-reconstruction-accuracy- (fidelity-) optimized collapse operators. To enhance the possibility of detecting true connections amongst the spurious connections, we used an atlas of 400 parcels adapted from the Destrieux atlas by iteratively splitting parcels along their most elongated axis, using the same parcel splits for all subjects (Palva et al., 2010, 2011; Rouhinen et al., 2020). The 400-parcel data were collapsed to 148 parcels before statistical analyses to reduce the influence of the large inter-individual function-anatomical variability.
Estimation of oscillation dynamics
The parcel-wise narrow-band filtered time-series were used for cortex-wide mapping of induced oscillation amplitudes (A) and LRTCs in oscillation amplitude envelopes, as well as for mapping cortex-wide phase-synchronization networks. Data were averaged for each cortical parcel and frequency separately for subjects in each COMT and BDNF polymorphisms group. Oscillation amplitudes were extracted from Morlet-wavelet filtered complex-valued time-series while LRTC exponents of the amplitude envelopes were quantified with detrended fluctuation analysis (DFA) (Hardstone et al., 2012). The power-law scaling exponent β was defined as the slope of linear regression of the function F(Δt) plotted in log–log coordinates, estimated using a least-squares algorithm. To estimate cortex-wide synchronization connectomes, phase synchronization between all 400 parcels was computed using the weighted phase-lag index (wPLI) (Siebenhühner et al., 2020; Vinck et al., 2011). The wPLI is insensitive to artificial connections arising as direct effects of zero-phase lagged linear signal mixing that is a major issue in connectivity analysis using MEG/EEG data (Palva and Palva, 2012). To reduce the amount of spurious interactions, we removed from the analysis parcels with fidelity < 0.2, which led to the exclusion of 9.27% of possible parcel pairs (Korhonen et al., 2014; Siebenhühner et al., 2020). The mean connection strengths were obtained by averaging the strength of all remaining edges for each subject and then over subjects in each COMT and BDNF polymorphism group. For functional subsystem analysis, all pairwise interactions were averaged across the frequencies in one of the bands defined by clustering analyses (Figure S1) and then collapsed into the 7 functional systems of the Yeo parcellation (Yeo et al., 2011) (Figure S3).
Computational modeling
We used a nested Kuramoto model (Siebenhühner et al., 2020) to investigate the covariance and mutual correlations of local synchronization (node/parcel amplitude), inter-areal synchronization, and local LRTCs in a heuristic model of synchronization dynamics. The model was adapted from a conventional Kuramoto model so that it comprised 100 nodes (corresponding to 100 cortical parcels) and each node was modeled by a Kuramoto model of 500 oscillators. The model was defined so that for each node k, the temporal evolution of the phase θ of each oscillator i was described by:
where is the oscillation frequency of an oscillator, K is the local coupling (control) parameter, is the average phase of the node l, is the connection strength between the node k and other nodes j, and is Gaussian noise. We set all oscillators’ frequencies to a constant value (10 Hz). W was approximated from the log-scaled structural connectome of the Schaefer parcellation with 100 parcels (Schaefer et al., 2018) multiplied by a global coupling (control) parameter L at L = 0.5. Node time series mimicking the parcel time series in MEG data were obtained by averaging the complex time series of the oscillators in the node. Local synchronization was measured by the absolute value of the complex node time series (Kuramoto order) that is comparable to the amplitude of MEG parcel time series. We predicted the synchronization dynamics of neuronal oscillations for two hypothetical cohorts that differed in their control parameters regulating their position in the subcritical regime. (A, control parameter from 8.25–9.25; and B, control parameter from 8.75–9.75) We estimated pairwise synchronization here with the Phase Locking Value (PLV), since, unlike in MEG data, in modeled data there is no volume conduction. Further, in the absence of conduction delays, the coupling function used in the Kuramoto model leads exclusively to near-zero-lag phase synchrony, which is not detectable with wPLI, necessitating the use of PLV. To test the model predictions, we estimated the correlation between amplitudes and DFA exponents, and between synchronization and DFA exponents, across all nodes, with Spearman’s correlation coefficient.
Quantification and statistical analysis
Before statistical testing, time-series were collapsed into the 148 parcels of the original Destrieux atlas (Destrieux et al., 2011) to limit the number of comparisons and to decrease the influence of large function-anatomical variability. Group-level upper and lower confidence limits (2.5% and 97.5%) were computed for each polymorphism group with a bootstrapping approach, using N = 1000 resamplings with replacement of the subjects in the group.
To validate the use of canonical frequency bands, we first estimated spatial similarity across frequency-bands separately for oscillation amplitudes and DFA exponents using machine learning by Louvain community detection (Blondel et al., 2008) with gamma value of 1.09. This analysis confirmed the presence of canonical frequency bands of theta (θ, 3–7 Hz), alpha (α, 8–14 Hz), beta (β, 14–30 Hz), and gamma (γ, 30–60 Hz) based on spatial similarity (Figure S1). The effects of COMT and BDNF polymorphisms on oscillation amplitudes and DFAs over the whole brain were tested separately for each Morlet frequency (Figures 2A and 3A), as well as for the canonical frequency bands θ, α, β and γ (Figure S2), using univariate ANOVA with the polymorphisms as between-participants factors and age as a covariate. Partial eta squared (ηp2) is reported as a measure of effect size.
In order to control for multiple comparisons at the level of individual wavelet frequencies, we used cluster-based non-parametric permutation testing as described by Maris and Oostenveld (2007). Briefly, we clustered consecutive wavelet frequencies which were significant in the ANOVA, summed F-values within clusters, and computed the F-value permutation distribution (based on random assignment of participants to polymorphism groups, N = 1000) for the cluster with the largest summed F. Any cluster for which the summed F-value was larger than the 95th percentile of this permutation distribution was deemed significant.
We then used the Mann-Whitney-U test (p < 0.05) to test pairwise differences between the polymorphism groups separately for each wavelet frequency (Figures 2B and 3B) and for each of the 148 parcels (Figures 2C and 3C). Multiple comparisons were accounted for in two steps. First, we used false discovery rate reduction (FDR) by discarding as many least-significant observations as were predicted to be false by the alpha-level (5% at α = 0.05) for each time-frequency bin. Second, we estimated a threshold Q to define the joint probability, pq< 0.05, for residual fraction of significants that could arise by chance in any of the frequencies (see dark gray area in Figures 2 and 3B). This threshold gives an additional estimate of expected false positives.
For inter-areal synchronization, confidence intervals were computed with a bootstrapping approach (N = 1000). As the synchronization data violated the normality assumption, we tested, for the canonical frequency bands and each Morlet wavelet frequency, whether there was a significant difference between polymorphism groups with a non-parametric Kruskal-Wallis test (p < 0.05) (Figures S2 and 4A). We used the same cluster-based non-parametric permutation testing approach to control for multiple comparisons at the wavelet frequency level as we had used for amplitude and DFA. Connection density (K) of the mean connection strengths was used to index the proportion of significant edges from all possible interactions. Differences between the polymorphism groups in connectivity within and between functional subsystems (Yeo et al., 2011) (Figure S3) were then tested for statistical significance with the Kruskal-Wallis test (p < 0.05) and corrected for multiple comparisons with Benjamini-Hochberg (Figure 4B).
Finally, we estimated the correlation coefficient between amplitudes and DFA exponents and between synchronization and DFA exponents, for each parcel and wavelet frequency, across subjects within the polymorphism groups, using Spearman’s rank correlation test. Values were then averaged across all parcels and within frequency bands. Confidence limits from 2.5% to 97.5% were obtained from bootstrapping (N = 1000) as above. Significance testing for difference between polymorphism groups was done with permutation statistics (N = 1000) where the subjects were randomly assigned, without replacement, to original-sized polymorphism groups.
Acknowledgments
This work was supported by the Jane and Aatos Erkko Foundation, by the Academy of Finland (SA 1266402, 1267030 to S.P., SA 1266745, 1296304 to J.M.P., and SA 1294761 to J.S.).
Author contributions
Conceptualization, S.P. and J.M.P.; Methodology, F.S., S.P., J.M.P., and V.M.; Software, F.S., J.M.P., and V.M.; Formal Analysis (MEG), J.S., F.S., and V.M.; Formal Analysis (Genotyping), K.K. and T.P.; Investigation, E.B.; Writing – Original Draft, J.S., S.P., F.S., J.M.P., and E.B.; Writing – Review & Editing, J.S., F.S., S.P., J.M.P., and V.M.; Visualization, J.S., F.S., and V.M.; Supervision, S.P., J.M.P., and E.B.; Funding Acquisition, E.B., J.M.P., S.P., T.P., and J.S.
Declaration of interests
The authors declare no competing interests.
Published: September 16, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104985.
Contributor Information
Jaana Simola, Email: jaana.simola@helsinki.fi.
Satu Palva, Email: satu.palva@helsinki.fi.
Supplemental information
Data and code availability
-
•
The neuroimaging and genetic data reported in this study cannot be deposited in a public repository because ethical restrictions apply to original data. To request access, contact Satu Palva (satu.palva@helsinki.fi). In addition, data underlying figures, statistics and main conclusions derived from the raw data have been deposited at Dryad server and are publicly available as of the date of publication. DOIs are listed in the Key resources table.
-
•
All original code has been deposited at https://github.com/palvalab/RS-Gen and is publicly available as of the date of publication.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Arnulfo G., Wang S.H., Myrov V., Toselli B., Hirvonen J., Fato M.M., Nobili L., Cardinale F., Rubino A., Zhigalov A., et al. Long-range phase synchronization of high-frequency oscillations in human cortex. Nat. Commun. 2020;11:5363. doi: 10.1038/s41467-020-18975-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett J.H., Jones P.B., Robbins T.W., Müller U. Effects of the catechol-O-methyltransferase Val158Met polymorphism on executive function: a meta-analysis of the Wisconsin Card Sort Test in schizophrenia and healthy controls. Mol. Psychiatry. 2007;12:502–509. doi: 10.1038/sj.mp.4001973. [DOI] [PubMed] [Google Scholar]
- Beste C., Kolev V., Yordanova J., Domschke K., Falkenstein M., Baune B.T., Konrad C. The role of the BDNF Val66Met polymorphism for the synchronization of error-specific neural networks. J. Neurosci. 2010;30:10727–10733. doi: 10.1523/JNEUROSCI.2493-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betti V., Corbetta M., de Pasquale F., Wens V., Della Penna S. Topology of functional connectivity and hub dynamics in the beta band as temporal prior for natural vision in the human brain. J. Neurosci. 2018;38:3858–3871. doi: 10.1523/JNEUROSCI.1089-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel V.D., Guillaume J.-L., Lambiotte R., Lefebvre E. Fast unfolding of communities in large networks. J. Stat. Mech. 2008;2008 doi: 10.1088/1742-5468/2008/10/P10008. [DOI] [Google Scholar]
- Bodenmann S., Rusterholz T., Dürr R., Stoll C., Bachmann V., Geissler E., Jaggi-Schwarz K., Landolt H.-P. The functional Val158Met polymorphism of COMT predicts interindividual differences in brain oscillations in young men. J. Neurosci. 2009;29:10855–10862. doi: 10.1523/JNEUROSCI.1427-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonetti L., Bruzzone S.E.P., Sedghi N.A., Haumann N.T., Paunio T., Kantojärvi K., Kliuchko M., Vuust P., Brattico E. Brain predictive coding processes are associated to COMT gene Val158Met polymorphism. Neuroimage. 2021;233 doi: 10.1016/j.neuroimage.2021.117954. [DOI] [PubMed] [Google Scholar]
- Braun U., Harneit A., Pergola G., Menara T., Schäfer A., Betzel R.F., Zang Z., Schweiger J.I., Zhang X., Schwarz K., et al. Brain network dynamics during working memory are modulated by dopamine and diminished in schizophrenia. Nat. Commun. 2021;12:3478. doi: 10.1038/s41467-021-23694-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes M.J., Woolrich M., Luckhoo H., Price D., Hale J.R., Stephenson M.C., Barnes G.R., Smith S.M., Morris P.G. Investigating the electrophysiological basis of resting state networks using magnetoencephalography. Proc. Natl. Acad. Sci. USA. 2011;108:16783–16788. doi: 10.1073/pnas.1112685108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabral J., Kringelbach M.L., Deco G. Exploring the network dynamics underlying brain activity during rest. Prog. Neurobiol. 2014;114:102–131. doi: 10.1016/j.pneurobio.2013.12.005. [DOI] [PubMed] [Google Scholar]
- Castrén E., Monteggia L.M. Brain-derived neurotrophic factor signaling in depression and antidepressant action. Biol. Psychiatry. 2021;90:128–136. doi: 10.1016/j.biopsych.2021.05.008. [DOI] [PubMed] [Google Scholar]
- Chen J., Lipska B.K., Halim N., Ma Q.D., Matsumoto M., Melhem S., Kolachana B.S., Hyde T.M., Herman M.M., Apud J., et al. Functional analysis of genetic variation in catechol-O-methyltransferase (COMT): effects on mRNA, protein, and enzyme activity in postmortem human brain. Am. J. Hum. Genet. 2004;75:807–821. doi: 10.1086/425589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocchi L., Gollo L.L., Zalesky A., Breakspear M. Criticality in the brain: a synthesis of neurobiology, models and cognition. Prog. Neurobiol. 2017;158:132–152. doi: 10.1016/j.pneurobio.2017.07.002. [DOI] [PubMed] [Google Scholar]
- Cools R. Chemistry of the adaptive mind: lessons from dopamine. Neuron. 2019;104:113–131. doi: 10.1016/j.neuron.2019.09.035. [DOI] [PubMed] [Google Scholar]
- Cools R., D’Esposito M. Inverted-U–shaped dopamine actions on human working memory and cognitive control. Biol. Psychiatry. 2011;69:e113–e125. doi: 10.1016/j.biopsych.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daffertshofer A., Ton R., Kringelbach M.L., Woolrich M., Deco G. Distinct criticality of phase and amplitude dynamics in the resting brain. Neuroimage. 2018;180:442–447. doi: 10.1016/j.neuroimage.2018.03.002. [DOI] [PubMed] [Google Scholar]
- Dale A.M., Liu A.K., Fischl B.R., Buckner R.L., Belliveau J.W., Lewine J.D., Halgren E. Dynamic statistical parametric mapping: combining fMRI and MEG for high-resolution imaging of cortical activity. Neuron. 2000;26:55–67. doi: 10.1016/s0896-6273(00)81138-1. [DOI] [PubMed] [Google Scholar]
- Deco G., Jirsa V.K. Ongoing cortical activity at rest: criticality, multistability, and ghost attractors. J. Neurosci. 2012;32:3366–3375. doi: 10.1523/JNEUROSCI.2523-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deco G., Kringelbach M.L., Arnatkeviciute A., Oldham S., Sabaroedin K., Rogasch N.C., Aquino K.M., Fornito A. Dynamical consequences of regional heterogeneity in the brain’s transcriptional landscape. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abf4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Destrieux C., Fischl B., Dale A., Halgren E. Automatic parcellation of human cortical gyri and sulci using standard anatomical nomenclature. Neuroimage. 2011;53:1–15. doi: 10.1016/j.neuroimage.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donhauser P.W., Baillet S. Two distinct neural timescales for predictive speech processing. Neuron. 2020;105:385–393.e9. doi: 10.1016/j.neuron.2019.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan M.F., Kojima M., Callicott J.H., Goldberg T.E., Kolachana B.S., Bertolino A., Zaitsev E., Gold B., Goldman D., Dean M., et al. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112:257–269. doi: 10.1016/s0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- Farrell S.M., Tunbridge E.M., Braeutigam S., Harrison P.J. COMT Val158Met genotype determines the direction of cognitive effects produced by catechol-O-methyltransferase inhibition. Biol. Psychiatry. 2012;71:538–544. doi: 10.1016/j.biopsych.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fell J., Axmacher N. The role of phase synchronization in memory processes. Nat. Rev. Neurosci. 2011;12:105–118. doi: 10.1038/nrn2979. [DOI] [PubMed] [Google Scholar]
- Ferguson K.A., Cardin J.A. Mechanisms underlying gain modulation in the cortex. Nat. Rev. Neurosci. 2020;21:80–92. doi: 10.1038/s41583-019-0253-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher P.M., Ozenne B., Svarer C., Adamsen D., Lehel S., Baaré W.F.C., Jensen P.S., Knudsen G.M. BDNF val66met association with serotonin transporter binding in healthy humans. Transl. Psychiatry. 2017;7 doi: 10.1038/tp.2016.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster B.L., He B.J., Honey C.J., Jerbi K., Maier A., Saalmann Y.B. Spontaneous neural dynamics and multi-scale network organization. Front. Syst. Neurosci. 2016;10:7. doi: 10.3389/fnsys.2016.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fries P. Rhythms for cognition: communication through coherence. Neuron. 2015;88:220–235. doi: 10.1016/j.neuron.2015.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Garcia M., Via M., Zarnowiec K., SanMiguel I., Escera C., Clemente I.C. COMT and DRD2/ANKK-1 gene-gene interaction account for resetting of gamma neural oscillations to auditory stimulus-driven attention. PLoS One. 2017;12 doi: 10.1371/journal.pone.0172362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramfort A., Luessi M., Larson E., Engemann D.A., Strohmeier D., Brodbeck C., Parkkonen L., Hämäläinen M.S. MNE software for processing MEG and EEG data. Neuroimage. 2014;86:446–460. doi: 10.1016/j.neuroimage.2013.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasby K.L., Jahanshad N., Painter J.N., Colodro-Conde L., Bralten J., Hibar D.P., Lind P.A., Pizzagalli F., Ching C.R.K., McMahon M.A.B., et al. The genetic architecture of the human cerebral cortex. Science. 2020;367 doi: 10.1126/science.aay6690. [DOI] [Google Scholar]
- Hahn A., Breakspear M., Rischka L., Wadsak W., Godbersen G.M., Pichler V., Michenthaler P., Vanicek T., Hacker M., Kasper S., et al. Reconfiguration of functional brain networks and metabolic cost converge during task performance. Elife. 2020;9 doi: 10.7554/eLife.52443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardstone R., Poil S.-S., Schiavone G., Jansen R., Nikulin V.V., Mansvelder H.D., Linkenkaer-Hansen K. Detrended fluctuation analysis: a scale-free view on neuronal oscillations. Front. Physiol. 2012;3:450–513. doi: 10.3389/fphys.2012.00450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris K.D., Thiele A. Cortical state and attention. Nat. Rev. Neurosci. 2011;12:509–523. doi: 10.1038/nrn3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiltunen T., Kantola J., Abou Elseoud A., Lepola P., Suominen K., Starck T., Nikkinen J., Remes J., Tervonen O., Palva S., et al. Infra-slow EEG fluctuations are correlated with resting-state network dynamics in fMRI. J. Neurosci. 2014;34:356–362. doi: 10.1523/JNEUROSCI.0276-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipp J.F., Siegel M. BOLD fMRI correlation reflects frequency-specific neuronal correlation. Curr. Biol. 2015;25:1368–1374. doi: 10.1016/j.cub.2015.03.049. [DOI] [PubMed] [Google Scholar]
- Jensen O., Gips B., Bergmann T.O., Bonnefond M. Temporal coding organized by coupled alpha and gamma oscillations prioritize visual processing. Trends Neurosci. 2014;37:357–369. doi: 10.1016/j.tins.2014.04.001. [DOI] [PubMed] [Google Scholar]
- Kinouchi O., Copelli M. Optimal dynamical range of exitable networks at criticality. Nat. Phys. 2006;2:348–351. doi: 10.1038/nphys289. [DOI] [Google Scholar]
- Klimesch W., Sauseng P., Hanslmayr S. EEG alpha oscillations: the inhibition–timing hypothesis. Brain Res. Rev. 2007;53:63–88. doi: 10.1016/j.brainresrev.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Kliuchko M., Puoliväli T., Heinonen-Guzejev M., Tervaniemi M., Toiviainen P., Sams M., Brattico E. Neuroanatomical substrate of noise sensitivity. Neuroimage. 2018;167:309–315. doi: 10.1016/j.neuroimage.2017.11.041. [DOI] [PubMed] [Google Scholar]
- Korhonen O., Palva S., Palva J.M. Sparse weightings for collapsing inverse solutions to cortical parcellations optimize M/EEG source reconstruction accuracy. J. Neurosci. Methods. 2014;226:147–160. doi: 10.1016/j.jneumeth.2014.01.031. [DOI] [PubMed] [Google Scholar]
- Lee S.-H., Dan Y. Neuromodulation of brain states. Neuron. 2012;76:209–222. doi: 10.1016/j.neuron.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppäaho E., Renvall H., Salmela E., Kere J., Salmelin R., Kaski S. Discovering heritable modes of MEG spectral power. Hum. Brain Mapp. 2019;40:1391–1402. doi: 10.1002/hbm.24454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkenkaer-Hansen K., Nikouline V.V., Palva J.M., Ilmoniemi R.J. Long-range temporal correlations and scaling behavior in human brain oscillations. J. Neurosci. 2001;21:1370–1377. doi: 10.1523/JNEUROSCI.21-04-01370.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkenkaer-Hansen K., Smit D.J.A., Barkil A., van Beijsterveldt T.E.M., Brussaard A.B., Boomsma D.I., van Ooyen A., de Geus E.J.C. Genetic contributions to long-range temporal correlations in ongoing oscillations. J. Neurosci. 2007;27:13882–13889. doi: 10.1523/JNEUROSCI.3083-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucki I. The spectrum of behaviors influenced by serotonin. Biol. Psychiatr. 1998;44:151–162. doi: 10.1016/s0006-3223(98)00139-5. [DOI] [PubMed] [Google Scholar]
- Mahjoory K., Schoffelen J.-M., Keitel A., Gross J. The frequency gradient of human resting- state brain oscillations follows cortical hierarchies. Elife. 2020;9 doi: 10.7554/eLife.53715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manninen S., Karjalainen T., Tuominen L.J., Hietala J., Kaasinen V., Joutsa J., Rinne J., Nummenmaa L. Cerebral grey matter density is associated with neuroreceptor and neurotransporter availability: a combined PET and MRI study. Neuroimage. 2021;235 doi: 10.1016/j.neuroimage.2021.117968. [DOI] [PubMed] [Google Scholar]
- Männistö P.T., Kaakkola S. Catechol-O-methyltransferase (COMT): biochemistry, molecular biology, pharmacology, and clinical efficacy of the new selective COMT inhibitors. Pharmacol. Rev. 1999;51:593–628. [PubMed] [Google Scholar]
- Maris E., Oostenveld R. Nonparametric statistical testing of EEG- and MEG-data. J. Neurosci. Methods. 2007;164:177–190. doi: 10.1016/j.jneumeth.2007.03.024. [DOI] [PubMed] [Google Scholar]
- Marzetti L., Basti A., Chella F., D’Andrea A., Syrjälä J., Pizzella V. Brain functional connectivity through phase coupling of neuronal oscillations: a perspective from magnetoencephalography. Front. Neurosci. 2019;13:964. doi: 10.3389/fnins.2019.00964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzetti L., Della Penna S., Snyder A.Z., Pizzella V., Nolte G., de Pasquale F., Romani G.L., Corbetta M. Frequency specific interactions of MEG resting state activity within and across brain networks as revealed by the multivariate interaction measure. Neuroimage. 2013;79:172–183. doi: 10.1016/j.neuroimage.2013.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo K., Walss-Bass C., Nery F.G., Nicoletti M.A., Hatch J.P., Frey B.N., Monkul E.S., Zunta-Soares G.B., Bowden C.L., Escamilla M.A., Soares J.C. Neuronal correlates of brain-derived neurotrophic factor Val66Met polymorphism and morphometric abnormalities in bipolar disorder. Neuropsychopharmacology. 2009;34:1904–1913. doi: 10.1038/npp.2009.23. [DOI] [PubMed] [Google Scholar]
- McCormick D.A., Nestvogel D.B., He B.J. Neuromodulation of brain state and behavior. Annu. Rev. Neurosci. 2020;43:391–415. doi: 10.1146/annurev-neuro-100219-105424. [DOI] [PubMed] [Google Scholar]
- McLoughlin G., Palmer J.A., Rijsdijk F., Makeig S. Genetic overlap between evoked frontocentral theta-band phase variability, reaction time variability, and Attention-Deficit/Hyperactivity Disorder symptoms in a twin study. Biol. Psychiatry. 2014;75:238–247. doi: 10.1016/j.biopsych.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Lindenberg A., Kohn P.D., Kolachana B., Kippenhan S., McInerney-Leo A., Nussbaum R., Weinberger D.R., Berman K.F. Midbrain dopamine and prefrontal function in humans: interaction and modulation by COMT genotype. Nat. Neurosci. 2005;8:594–596. doi: 10.1038/nn1438. [DOI] [PubMed] [Google Scholar]
- Mier D., Kirsch P., Meyer-Lindenberg A. Neural substrates of pleiotropic action of genetic variation in COMT: a meta-analysis. Mol. Psychiatry. 2010;15:918–927. doi: 10.1038/mp.2009.36. [DOI] [PubMed] [Google Scholar]
- Monto S., Palva S., Voipio J., Palva J.M. Very slow EEG fluctuations predict the dynamics of stimulus detection and oscillation amplitudes in humans. J. Neurosci. 2008;28:8268–8272. doi: 10.1523/JNEUROSCI.1910-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostame P., Sadaghiani S. Oscillation-based connectivity architecture is dominated by an intrinsic spatial organization, not cognitive state or frequency. J. Neurosci. 2020;41:179–192. doi: 10.1523/JNEUROSCI.2155-20.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn B.R., Müller E.J., Wainstein G., Shine J.M. The ascending arousal system shapes neural dynamics to mediate awareness of cognitive states. Nat. Commun. 2021;12 doi: 10.1038/s41467-021-26268-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notaras M., Hill R., van den Buuse M. The BDNF gene Val66Met polymorphism as a modifier of psychiatric disorder susceptibility: progress and controversy. Mol. Psychiatry. 2015;20:916–930. doi: 10.1038/mp.2015.27. [DOI] [PubMed] [Google Scholar]
- Oostenveld R., Fries P., Maris E., Schoffelen J.-M. FieldTrip: open source software for advanced analysis of MEG, EEG, and invasive electrophysiological data. Comput. Intell. Neurosci. 2011;2011 doi: 10.1155/2011/156869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald V., Zerouali Y., Boulet-Craig A., Krajinovic M., Laverdière C., Sinnett D., Jolicoeur P., Lippé S., Jerbi K., Robaey P. Spontaneous brain oscillations as neural fingerprints of working memory capacities: a resting-state MEG study. Cortex. 2017;97:109–124. doi: 10.1016/j.cortex.2017.09.021. [DOI] [PubMed] [Google Scholar]
- Ott T., Nieder A. Dopamine and cognitive control in prefrontal cortex. Trends Cogn. Sci. 2019;23:213–234. doi: 10.1016/j.tics.2018.12.006. [DOI] [PubMed] [Google Scholar]
- Palva J.M., Monto S., Kulashekhar S., Palva S. Neuronal synchrony reveals working memory networks and predicts individual memory capacity. Proc. Natl. Acad. Sci. USA. 2010;107:7580–7585. doi: 10.1073/pnas.0913113107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palva J.M., Wang S.H., Palva S., Zhigalov A., Monto S., Brookes M.J., Schoffelen J.-M., Jerbi K. Ghost interactions in MEG/EEG source space: a note of caution on inter-areal coupling measures. Neuroimage. 2018;173:632–643. doi: 10.1016/j.neuroimage.2018.02.032. [DOI] [PubMed] [Google Scholar]
- Palva J.M., Zhigalov A., Hirvonen J., Korhonen O., Linkenkaer-Hansen K., Palva S. Neuronal long-range temporal correlations and avalanche dynamics are correlated with behavioral scaling laws. Proc. Natl. Acad. Sci. USA. 2013;110:3585–3590. doi: 10.1073/pnas.1216855110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palva S., Kulashekhar S., Hämäläinen M., Palva J.M. Localization of cortical phase and amplitude dynamics during visual working memory encoding and retention. J. Neurosci. 2011;31:5013–5025. doi: 10.1523/JNEUROSCI.5592-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palva S., Palva J.M. Discovering oscillatory interaction networks with M/EEG: challenges and breakthroughs. Trends Cogn. Sci. 2012;16:219–230. doi: 10.1016/j.tics.2012.02.004. [DOI] [PubMed] [Google Scholar]
- Palva S., Palva J.M. Roles of brain criticality and multiscale oscillations in temporal predictions for sensorimotor processing. Trends Neurosci. 2018;41:729–743. doi: 10.1016/j.tins.2018.08.008. [DOI] [PubMed] [Google Scholar]
- Papenberg G., Karalija N., Salami A., Rieckmann A., Andersson M., Axelsson J., Riklund K., Lindenberger U., Lövdén M., Nyberg L., Bäckman L. Balance between transmitter availability and dopamine D2 receptors in prefrontal cortex influences memory functioning. Cereb. Cortex. 2020;30:989–1000. doi: 10.1093/cercor/bhz142. [DOI] [PubMed] [Google Scholar]
- Park H., Poo M.m. Neurotrophin regulation of neural circuit development and function. Nat. Rev. Neurosci. 2013;14:7–23. doi: 10.1038/nrn3379. [DOI] [PubMed] [Google Scholar]
- Parkin G.M., Udawela M., Gibbons A., Scarr E., Dean B. Catechol-O-methyltransferase (COMT) genotypes are associated with varying soluble, but not membrane-bound COMT protein in the human prefrontal cortex. J. Hum. Genet. 2018;63:1251–1258. doi: 10.1038/s10038-018-0511-2. [DOI] [PubMed] [Google Scholar]
- Pereira L.P., Köhler C.A., Stubbs B., Miskowiak K.W., Morris G., de Freitas B.P., Thompson T., Fernandes B.S., Brunoni A.R., Maes M., et al. Imaging genetics paradigms in depression research: systematic review and T meta-analysis. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 2018;86:102–113. doi: 10.1016/j.pnpbp.2018.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer T., Ponce-Alvarez A., Tsetsos K., Meindertsma T., Gahnström C.J., van den Brink R.L., Nolte G., Engel A.K., Deco G., Donner T.H. Circuit mechanisms for the chemical modulation of cortex-wide network interactions and behavioral variability. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abf5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plenz D., Thiagarajan T.C. The organizing principles of neuronal avalanches: cell assemblies in the cortex? Trends Neurosci. 2007;30:101–110. doi: 10.1016/j.tins.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Poil S.-S., Hardstone R., Mansvelder H.D., Linkenkaer-Hansen K. Critical-state dynamics of avalanches and oscillations jointly emerge from balanced excitation/Inhibition in neuronal networks. J. Neurosci. 2012;32:9817–9823. doi: 10.1523/JNEUROSCI.5990-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posthuma D., de Geus E.J., Boomsma D.I. Perceptual speed and IQ are associated through common genetic factors. Behav. Genet. 2001;31:593–602. doi: 10.1023/a:1013349512683. [DOI] [PubMed] [Google Scholar]
- Priesemann V., Wibral M., Valderrama M., Pröpper R., Le Van Quyen M., Geisel T., Triesch J., Nikolić D., Munk M.H.J. Spike avalanches in vivo suggest a driven, slightly subcritical brain state. Front. Syst. Neurosci. 2014;8:108. doi: 10.3389/fnsys.2014.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusil S., López M.E., Cuesta P., Bruña R., Pereda E., Maestú F. Hypersynchronization in mild cognitive impairment: the ‘X’ model. Brain. 2019;142:3936–3950. doi: 10.1093/brain/awz320. [DOI] [PubMed] [Google Scholar]
- Robbins T.W., Arnsten A.F.T. The neuropsychopharmacology of fronto-executive function: monoaminergic modulation. Annu. Rev. Neurosci. 2009;32:267–287. doi: 10.1146/annurev.neuro.051508.135535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Rojo I.C., Cuesta P., López M.E., de Frutos-Lucas J., Bruña R., Pereda E., Barabash A., Montejo P., Montenegro-Peña M., Marcos A., et al. BDNF Val66Met polymorphism and gamma band disruption in resting state brain functional connectivity: a magnetoencephalography study in cognitively intact older females. Front. Neurosci. 2018;12:684. doi: 10.3389/fnins.2018.00684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhinen S., Siebenhühner F., Palva J.M., Palva S. Spectral and anatomical patterns of large-scale synchronization predict human attentional capacity. Cereb. Cortex. 2020;30:5293–5308. doi: 10.1093/cercor/bhaa110. [DOI] [PubMed] [Google Scholar]
- Roy N., Barry R.J., Fernandez F.E., Lim C.K., Al-Dabbas M.A., Karamacoska D., Broyd S.J., Solowij N., Chiu C.L., Steiner G.Z. Electrophysiological correlates of the brain-derived neurotrophic factor (BDNF) Val66Met polymorphism. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-74780-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmela E., Renvall H., Kujala J., Hakosalo O., Illman M., Vihla M., Leinonen E., Salmelin R., Kere J. Evidence for genetic regulation of the human parieto-occipital 10-Hz rhythmic activity. Eur. J. Neurosci. 2016;44:1963–1971. doi: 10.1111/ejn.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaha J., Iemi L., Haegens S., Busch N.A. Spontaneous brain oscillations and perceptual decision-making. Trends Cogn. Sci. 2020;24:639–653. doi: 10.1016/j.tics.2020.05.004. [DOI] [PubMed] [Google Scholar]
- Schaefer A., Kong R., Gordon E.M., Laumann T.O., Zuo X.-N., Holmes A.J., Eickhoff S.B., Yeo B.T.T. Local-global parcellation of the human cerebral cortex from intrinsic functional connectivity MRI. Cereb. Cortex. 2018;28:3095–3114. doi: 10.1093/cercor/bhx179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitzman B.A., Gratton C., Laumann T.O., Gordon E.M., Adeyemo B., Dworetsky A., Kraus B.T., Gilmore A.W., Berg J.J., Ortega M., et al. Trait-like variants in human functional brain networks. Proc. Natl. Acad. Sci. USA. 2019;116:22851–22861. doi: 10.1073/pnas.1902932116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shew W.L., Plenz D. The functional benefits of criticality in the cortex. Neuroscientist. 2013;19:88–100. doi: 10.1177/1073858412445487. [DOI] [PubMed] [Google Scholar]
- Shew W.L., Yang H., Petermann T., Roy R., Plenz D. Neuronal avalanches imply maximum dynamic range in cortical networks at criticality. J. Neurosci. 2009;29:15595–15600. doi: 10.1523/JNEUROSCI.3864-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenhühner F., Wang S.H., Arnulfo G., Lampinen A., Nobili L., Palva J.M., Palva S. Genuine cross-frequency coupling networks in human resting-state electrophysiological recordings. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel M., Donner T.H., Engel A.K. Spectral fingerprints of large-scale neuronal interactions. Nat. Rev. Neurosci. 2012;13:121–134. doi: 10.1038/nrn3137. [DOI] [PubMed] [Google Scholar]
- Simola J., Zhigalov A., Morales-Muñoz I., Palva J.M., Palva S. Critical dynamics of endogenous fluctuations predict cognitve flexibility in the Go/NoGo task. Sci. Rep. 2017;7:2909. doi: 10.1038/s41598-017-02750-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer W. Neuronal synchrony: a versatile code for the definition of relations? Neuron. 1999;24:49–65. doi: 10.1016/s0896-6273(00)80821-1. 111-125. [DOI] [PubMed] [Google Scholar]
- Smit D.J.A., Linkenkaer-Hansen K., de Geus E.J.C. Long-range temporal correlations in resting-state alpha oscillations predict human timing-error dynamics. J. Neurosci. 2013;33:11212–11220. doi: 10.1523/JNEUROSCI.2816-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit D.J.A., Wright M.J., Meyers J.L., Martin N.G., Ho Y.Y.W., Malone S.M., Zhang J., Burwell S.J., Chorlian D.B., de Geus E.J.C., et al. Genome-wide association analysis links multiple psychiatric liability genes to oscillatory brain activity. Hum. Brain Mapp. 2018;39:4183–4195. doi: 10.1002/hbm.24238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephani T., Waterstraat G., Haufe S., Curio G., Villringer A., Nikulin V.V. Temporal signatures of criticality in human cortical excitability as probed by early somatosensory responses. J. Neurosci. 2020;40:6572–6583. doi: 10.1523/JNEUROSCI.0241-20.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Štrac Š.D., Pivac N., Mück-Šeler D. The serotonergic system and cognitive function. Transl. Neurosci. 2016;7:35–49. doi: 10.1515/tnsci-2016-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taulu S., Simola J., Kajola M. Applications of the signal space separation method. IEEE Trans. Signal Process. 2005;53:3359–3372. doi: 10.1109/TSP.2005.853302. [DOI] [Google Scholar]
- Thiery T., Lajnef T., Combrisson E., Dehgan A., Rainville P., Mashour G.A., Blain-Moraes S., Jerbi K. Long-range temporal correlations in the brain distinguish conscious wakefulness from induced unconsciousness. Neuroimage. 2018;179:30–39. doi: 10.1016/j.neuroimage.2018.05.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thut G., Miniussi C., Gross J. The functional importance of rhythmic activity in the brain. Curr. Biol. 2012;22:R658–R663. doi: 10.1016/j.cub.2012.06.061. [DOI] [PubMed] [Google Scholar]
- van den Brink R.L., Nieuwenhuis S., Donner T.H. Amplification and suppression of distinct brainwide activity patterns by catecholamines. J. Neurosci. 2018;38:7476–7491. doi: 10.1523/JNEUROSCI.0514-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Brink R.L., Pfeffer T., Donner T.H. Brainstem modulation of large-scale intrinsic cortical activity correlations. Front. Hum. Neurosci. 2019;13:340. doi: 10.3389/fnhum.2019.00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Pelt S., Boomsma D.I., Fries P. Magnetoencephalography in twins reveals a strong genetic determination of the peak frequency of visually induced gamma-band synchronization. J. Neurosci. 2012;32:3388–3392. doi: 10.1523/JNEUROSCI.5592-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidaurre D., Llera A., Smith S.M., Woolrich M.W. Behavioural relevance of spontaneous, transient brain network interactions in fMRI. Neuroimage. 2021;229 doi: 10.1016/j.neuroimage.2020.117713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayraghavan S., Wang M., Birnbaum S.G., Williams G.V., Arnsten A.F.T. Inverted-U dopamine D1 receptor actions on prefrontal neurons engaged in working memory. Nat. Neurosci. 2007;10:376–384. doi: 10.1038/nn1846. [DOI] [PubMed] [Google Scholar]
- Vinck M., Oostenveld R., van Wingerden M., Battaglia F., Pennartz C.M.A. An improved index of phase-synchronization for electrophysiological data in the presence of volume-conduction, noise and sample-size bias. Neuroimage. 2011;55:1548–1565. doi: 10.1016/j.neuroimage.2011.01.055. [DOI] [PubMed] [Google Scholar]
- Winkel F., Ryazantseva M., Voigt M.B., Didio G., Lilja A., Llach Pou M., Steinzeig A., Harkki J., Englund J., Khirug S., et al. Pharmacological and optical activation of TrkB in Parvalbumin interneurons regulate intrinsic states to orchestrate cortical plasticity. Mol. Psychiatry. 2021;26:7247–7256. doi: 10.1038/s41380-021-01211-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Womelsdorf T., Valiante T.A., Sahin N.T., Miller K.J., Tiesinga P. Dynamic circuit motifs underlying rhythmic gain control, gating and integration. Nat. Neurosci. 2014;17:1031–1039. doi: 10.1038/nn.3764. [DOI] [PubMed] [Google Scholar]
- Yeo B.T.T., Krienen F.M., Sepulcre J., Sabuncu M.R., Lashkari D., Hollinshead M., Roffman J.L., Smoller J.W., Zöllei L., Polimeni J.R., et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J. Neurophysiol. 2011;106:1125–1165. doi: 10.1152/jn.00338.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagrebelsky M., Korte M. Form follows function: BDNF and its involvement in sculpting the function and structure of synapses. Neuropharmacology. 2014;76:628–638. doi: 10.1016/j.neuropharm.2013.05.029. [DOI] [PubMed] [Google Scholar]
- Zhigalov A., Arnulfo G., Nobili L., Palva S., Palva J.M. Relationship of fast-and slow-timescale neuronal dynamics in human MEG and SEEG. J. Neurosci. 2015;35:5385–5396. doi: 10.1523/JNEUROSCI.4880-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhigalov A., Arnulfo G., Nobili L., Palva S., Palva J.M. Modular co-organization of functional connectivity and scale-free dynamics in the human brain. Netw. Neurosci. 2017;1:143–165. doi: 10.1162/netn_a_00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The neuroimaging and genetic data reported in this study cannot be deposited in a public repository because ethical restrictions apply to original data. To request access, contact Satu Palva (satu.palva@helsinki.fi). In addition, data underlying figures, statistics and main conclusions derived from the raw data have been deposited at Dryad server and are publicly available as of the date of publication. DOIs are listed in the Key resources table.
-
•
All original code has been deposited at https://github.com/palvalab/RS-Gen and is publicly available as of the date of publication.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.