Abstract
Objective
Dicer is an enzyme that processes microRNAs (miRNAs) precursors into mature miRNAs, which have been implicated in various aspects of cancer progressions, such as clinical aggressiveness, prognosis, and survival outcomes. We previously showed that high expression of Dicer is associated with gemcitabine (GEM) resistance in pancreatic ductal adenocarcinoma (PDAC); thus, in this study, we aimed to focus on how Dicer is involved in GEM resistance in PDAC, including cancer prognosis, cell proliferation, and metabolic regulation.
Methods
We generated stable shRNA knockdown of Dicer in GEM-resistant PANC-1 (PANC-1 GR) cells and explored cell viability by MTT and clonogenicity assays. Metabolomic profiling was employed to investigate metabolic changes between parental cells, PANC-1, and PANC-1 GR cells, and further implied to compare their sensitivity to the glutaminase inhibitor, CB839, and GEM treatments. To identify putative phosphorylation site involves with Dicer and its effects on GEM resistance in PDAC cells, we further generated phosphomimetic or phosphomutant Dicer at S1016 site and examined the changes in drug sensitivity, metabolic alteration, and miRNA regulation.
Results
We observed that high Dicer levels in pancreatic ductal adenocarcinoma cells were positively correlated with advanced pancreatic cancer and acquired resistance to GEM. Metabolomic analysis indicated that PANC-1 GR cells rapidly utilised glutamine as their major fuel and increased levels of glutaminase (GLS): glutamine synthetase (GLUL) ratio which is related to high Dicer expression. In addition, we found that phosphomimetic Dicer S1016E but not phosphomutant Dicer S1016A facilitated miRNA maturation, causing an imbalance in GLS and GLUL and resulting in an increased response to GLS inhibitors.
Conclusion
Our results suggest that phosphorylation of Dicer on site S1016 affects miRNA biogenesis and glutamine metabolism in GEM-resistant pancreatic cancer.
Keywords: Dicer phosphorylation, Pancreatic ductal adenocarcinoma, Gemcitabine resistance, Glutamine metabolism, miRNA biogenesis
Highlights
-
•
Dicer expression is positively correlated with advanced pancreatic cancer.
-
•
Dicer expression is significantly correlated with high level of GLS and GLS/GLUL ratio.
-
•
Phosphomimetic Dicer S1016E enhances glutamine consumption and GLS inhibitor sensitivity.
-
•
Phosphomimetic Dicer S1016E facilitates miRNAs maturation to increase GLS/GLUL ratio.
1. Introduction
Pancreatic cancer is the fourth leading cause of cancer-related deaths in the United States [1]. The 5-year survival rate of patients with pancreatic cancer is <9%, and the median survival rate is 4–6 months [2]. The most common type of pancreatic cancer is pancreatic ductal adenocarcinoma (PDAC), which accounts for >90% of pancreatic cancers and is the most lethal major solid tumor [2]. Treatments for PDAC include surgery, chemotherapy, radiation therapy, and combination therapy. Gemcitabine (GEM), also known as 2′,2′-difluorodeoxycytidine, is a first-line standard chemotherapy drug for pancreatic cancer. GEM improved survival outcomes in 20–30% of patients with pancreatic cancer, thus presenting a substantial clinical benefit. However, because most patients with pancreatic cancer acquire resistance to GEM, mechanisms underlying GEM resistance should be investigated [3,4].
Dicer, a key cytoplasmic type III RNase, is involved in not only miRNA maturation but also miRNA biogenesis along with the proteins Drosha, Exportin-5, thyroid hormone receptor-binding protein, and Ago2. Hence, Dicer plays a critical role in the regulation of posttranscriptional gene silencing [5]. Accumulating evidence has demonstrated that the expression and activity of Dicer are involved in the dysregulation of miRNA expression in tumors that considerably alters gene expression patterns in cancer cells and ultimately contributes to tumor initiation, growth, and progression. Increased Dicer expression, for instance, has been shown to be positively associated with advanced tumor status in various cancers, such as lung, breast, and ovarian cancers, and promotes cancer stemness and invasiveness in colon cancer [[6], [7], [8], [9]]. Additionally, it was reported that silencing Dicer expression through specific RNA interference (RNAi) could increase the sensitivity of resistant human KB adenocarcinoma cells to cisplatin [10], whereas overexpression of Dicer stimulated breast cancer resistance protein, leading to cisplatin- [10] and tamoxifen-resistance MCF-7 human breast cancer cells [11]. Little is known about the mechanism underlying Dicer-induced cancer growth and drug resistance, however, the study from Burger et al. demonstrated that the afimoxifene-mediated DNA damage response increased Dicer phosphorylation at S1016 and induced nuclear Dicer accumulation in U2OS and A549 cells [12]. Dicer phosphorylation at S1016, therefore, plays a crucial role in the cell cycle and growth, which ultimately contributes to cancer progression; thus, more studies are needed to investigate how Dicer phosphorylation is regulated in cancer cells and involved in mediating drug response.
In pancreatic cancer, metabolic addiction to glucose and glutamine supports cell growth and interferes with drug responses, resulting in tumor progression and metastasis [13]. Hence, disruption of glutamine metabolism pathways might increase the efficacy of GEM treatment in GEM-resistant pancreatic cancer cells [14]. Glutaminase (GLS) is a key enzyme that catalyzes the conversion of glutamine to glutamate to begin the tricarboxylic acid (TCA) cycle, whereas glutamine synthetase (GLUL) converts glutamate and ammonia to glutamine and increases the release of glutamine into circulation [15]. Evidence has suggested that the dysregulation of metabolism in primary tumors and in circulating tumor cells is involved in the regulation of microRNA (miRNA) or miRNA-targeted metabolic proteins and molecules [[8], [9], [10]]. As a regulatory enzyme for miRNAs maturation, Dicer also has shown to play the critical role in mediating miRNAs-mediated glucose metabolism [2]. This has prompted the design of miRNA-based therapy aimed at regulating the level of miRNAs via Dicer; however, the molecular mechanisms of Dicer underlying proliferation and GEM resistance of pancreatic cancer are still remained to be further investigated.
This study explored whether Dicer phosphorylation is involved in GEM resistance and metabolic reprogramming in PDAC. We previously found that high expression of Dicer is significantly associated with GEM resistance in PANC-1 PDAC cells and its expression is positively correlated with pancreatic cancer progression [3]. In this study, we further investigated the effect of Dicer phosphorylation at S1016 and its association with glutamine metabolism and chemotherapy responses in GEM-resistant PDAC cells. The results revealed that Dicer was highly expressed in GEM-resistant PANC-1 cells and contributed to glutamine metabolic rewiring. To our knowledge, this is the first study to demonstrate that the Dicer phosphomimetic and phosphomutant S1016 selectively regulated miRNA maturation, triggering a switch in the GLS:GLUL expression ratio and thus changing the chemotherapy response of PDAC cells.
2. Material and methods
2.1. Specimen collection and patient information
Patients with pancreatic adenocarcinoma who underwent resection at National Cheng Kung University Hospital, Taipei Medical University Hospital, and Shuang Ho Hospital between June 2002 and August 2019 and patients who received GEM-based adjuvant chemotherapy in the same period were retrospectively enrolled in this study. The study protocol was approved by the institutional review board of the participating institute (No. N201807081 and N201902040). In total, 48 patients with stage I–IV pancreatic adenocarcinoma (25 men and 23 women) aged 57 to 82 (median: 65) years were recruited. Of the 48 patients, 8, 25, and 15 had grade 1 (G1), grade 2 (G2), and grade 3 (G3) disease, respectively. All the patients were followed up and underwent imaging in accordance with hospital guidelines, usually every 3 months. The cut-off date for the analysis was December 2019. Recurrence-free survival was defined as the period from surgery until tumor recurrence or death.
2.2. Immunohistochemistry
Formalin-fixed, paraffin-embedded tissue sections (5 μm) were dewaxed in xylene and hydrated in graded ethanol concentrations. Endogenous peroxidase was blocked with 3% hydrogen peroxide and then with 10% normal goat serum for 1 h. Subsequently, the sections were incubated with the primary antihuman Dicer antibody ab14601 (1:100 dilution; Abcam, Cambridge, UK) at 4 °C for 12 h. The sections were rinsed with phosphate-buffered saline and incubated with a biotinylated secondary antibody for 1 h and then with horseradish peroxidase-conjugated streptavidin for 30 min at room temperature. The sections were immunostained with 3′,3′-diaminobenzidine tetrahydrochloride for 3 min and counterstained with hematoxylin. Dicer expression was scored and stratified into three groups (Dicer−, no or weak staining in 1%–10% of cells; Dicer+, moderate staining in 10%–30% of cells, and Dicer++, strong staining in more than 30% of cells).
2.3. Cell culture
PANC-1 and GEM-resistant PANC-1 (PANC-1 GR) cells were kindly provided by Dr. Wun-Shaing Wayne Chang and Dr. Li-Tzong Chen (National Health Research Institutes, Miaoli, Taiwan). To develop PANC-1 GR cells, PANC-1 cells were treated in a stepwise manner with increasing doses of GEM for a long period. The resulting PANC-1 GR cells were maintained in a culture medium containing GEM at a final maximum concentration of 5 μM. Both the PANC-1 and PANC-1 GR cells were maintained in Dulbecco's Modified Eagle's Medium (DMEM, 23-10-013-CM, Corning®) containing high glucose (4500 mg/L), l-Glutamine (4 mM), and sodium pyruvate (1 mM) with 10% fetal bovine serum and 1% penicillin-streptomycin. The cells were incubated at 37 °C with humidified 5% CO2, and the medium was replaced every 3 days until 70% confluency was reached. These cells were free of mycoplasma contamination, and their identity was confirmed through short tandem repeat profiling at the Bioresource Collection and Research Center (Hsinchu, Taiwan) and Center for Genomic Medicine, National Cheng Kung University Hospital (Tainan, Taiwan).
2.4. Cell viability assay
The cells were seeded in 96-well plates overnight (approximately 5 × 103 cells per well) and subsequently treated with different concentrations of a GEM or GLS inhibitor (CB839). After 72 h of treatment, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) solution (1 g/mL) was added to each well, and the cells were incubated for another 4 h at 37 °C. The medium was then discarded, and 100 μL of dimethyl sulfoxide was added for 10 min at room temperature to dissolve formazan crystals. Absorbance was measured using an EPOCH2 microplate reader (BioTek, Winooski, VT, USA) at a wavelength of 570 nm. The percentage of cell viability is displayed relative to that of untreated cells.
For the clonogenicity assay, the cells were seeded into six-well culture plates at a density of 200 cells/well. The medium was replaced every 3 days, and the cells were allowed to grow for 2 weeks. Subsequently, colonies were fixed with 10% formalin for 30 min and stained with 0.5% crystal violet in 2% methanol for 1 h for enumeration. Photographs were taken using a digital camera, and cell colonies were counted using ImageJ software.
2.5. RNA isolation and quantitative RT-PCR
To examine the Dicer level in pancreatic tumor tissues, total RNA was extracted from whole samples by using the NucleoSpin total RNA FFPE XS kit (REF 740969.50, Macherey-Nage, Düren, Germany) in accordance with the manufacturer's instructions. To examine the Dicer level in pancreatic cancer cells, total RNA was isolated using NucleoZOL (REF 740404.200, Macherey-Nage) and employed as a template for reverse transcription to cDNA, which was performed using the MMLV reverse transcript kit (Invitrogen) in accordance with the manufacturer's instructions. Quantitative RT-PCR (qRT-PCR) was performed using the Lightcycler 480 system (Roche). Specific primers are listed in Supplementary Table 1. The CP values were calculated using Lightcycler 480 software. The relative levels of mRNA expression were normalized to the mean levels of GAPDH and β-actin. The relative levels of gene expression are expressed as ΔCP = CP of the tested gene – CP of the reference gene.
Mature miRNA sequences were obtained from the Sanger Center miRNA Registry (http://microrna.sanger.ac.uk/sequences/), and stem-loop RT primers were designed using methods employed by Chen et al. [16], as shown in Supplementary Table 1. For mature microRNA detection, qRT-PCR was performed using forward and reverse primers at 0.5 μM concentrations, 1 μM Universal ProbeLibrary Probe #21 (Roche), 1 × LightCycler TaqMan Master mix, and 2 μL cDNA. Amplification curves were generated through initial denaturation at 95 °C for 10 min, followed by 65 cycles at 95 °C, 60 °C, and 72 °C (5, 10, and 1 s in duration, respectively). RNU6B and U47 small nuclear RNAs were used as reference genes. The relative gene expression levels are presented as ΔCP = CP of the target gene − CP of the reference gene, and the fold change in gene expression was calculated as 2−ΔΔCP.
2.6. Western blotting
After the knockdown of Dicer (shDicer) and control (shCtrl) cells, PANC-1 GR cells were lysed in radioimmunoprecipitation assay buffer containing a protease inhibitor cocktail and a phosphatase inhibitor cocktail (Roche, Boston, MA, USA). Equal amounts of total protein were resolved through sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes. The membranes were blocked with 5% skimmed milk in Tris-buffered saline containing 0.1% Tween-20 for 30 min and then incubated overnight at 4 °C with the indicated primary antibodies: β-actin (1:5000; Millipore, Billerica, MA, USA) and Dicer (1:1000; ab14601, Abcam). The membranes were washed with Tris-buffered saline containing 0.1% Tween-20 and incubated for 1 h at room temperature with appropriate secondary antibodies conjugated to horseradish peroxidase. Subsequently, the membranes were washed, and signals from immunoreactive bands were detected using an electrochemiluminescence reagent (WBLUF0500; Burlington, MA, USA). The resulting bands were generated using the UVP biochemical system and VisionWorks LS software (VisionWorks, Cedar Rapids, IA, USA).
2.7. Clinical relationship between Dicer and GLS:GLUL expression ratio in pancreatic cancer
The Oncomine (http://www.oncomine.org) database is used as a web-based tool to compare mRNA expressions of GLS and GLUL in pancreatic cancer patients based on their histological grades. Differences in mRNA expression between pancreatic cancer tissues with differential histological grades were calculated using the following threshold parameters: “cancer versus cancer,” “grade,” and “p < 0.05.” Pearson's correlation was used for multiclass ordinal analyses [17].
PROGgeneV2 database (http://genomics.jefferson.edu/proggene) is used as a web-based tool to perform survival analyses based on the expressions of the GLS and GLUL. The survival curve comparing pancreatic cancer patients with high/high (red) and low/low (green) of GLS/GLUL expression was plotted using the PROGgeneV2 to investigate the prognostic significance of the co-occurrence of two genes. Survival analysis was made by fitting cox proportional hazards model and log rank p value was retrieved from the fitted model [18].
UALCAN cancer database (ualcan.path.uab.edu) is used as a web-based tool to explore the correlation between gene expressions of Dicer vs. GLS and GLUL. Pearson's correlation was used for multiclass ordinal analyses.
2.8. Lentiviral knockdown and phosphomimetic Dicer constructs
The pLKO.1-puro-based lentiviral vectors TRCN0000290426 (shDicer#1), and TRCN0000290489 (shDicer#2), and the control plasmids TRC025.shLKO (shCtrl#1), and TRC2. Scramble (shCtrl#2), were purchased from the National RNAi Core Facility at Academia Sinica, Taipei, Taiwan. HEK293T cells were cotransfected with the lentivirus expression plasmid, packaging plasmid (pCMV-dR8.91), and envelope plasmid (VSV-G expressing plasmid, pMD2.G) with polyethyleneimine (Merck) for 48 h. We generated a recombinant lentivirus from the culture medium. The cells were infected with lentiviruses combined with 8 μg/mL polybrene, and stable cells were selected using 0.5–1 μg/mL puromycin.
To overexpress Dicer and phosphomimetic Dicer, the Dicer WT plasmid (pCAGGS-Flag-hsDicer plasmid #41584) was purchased from Addgen, and then Dicer was subcloned into the pcDNA6/myc-His vector. The pcDNA6-Dicer S1016A and S1016E mutations were achieved using the Q5 site-directed mutagenesis kit (NEB E0554S) and specific primers (Supplementary Table 1). Cells were transfected with a Dicer WT, S1016A, or S1016E plasmid or a control vector (pcDNA6) for 48 h by using the DreamFect Gold transfection reagent (DG80500, OZ Biosciences, France) in accordance with the manufacturer's instructions, and stable cell lines were selected using 5 μg/mL blasticidin (ant-bl-05; InvivoGen, USA).
2.9. Metabolomic analysis
PANC-1 and PANC-1 GR cells that achieved approximately 80% confluence were cultured in a medium containing 10% fetal bovine serum for 1 day, and the supernatant was collected after the cells were centrifuged at 3000 rpm for 10 min at 4 °C. Ten milliliters of the supernatant were mixed with 40 mL of cold methanol through gentle shock for 30 min at 4 °C. The clear supernatant was collected, lyophilized, and stored at −80 °C for further analysis. We compared the metabolite profiles of both types of cells and performed ultraperformance liquid chromatography (Acquity UPLC System, Waters Corporation, Milford, MA, USA) coupled with time-of-flight mass spectrometry (Xevo TOF MS, Waters Corporation). The quadrupole-time of flight mass spectrometry system was operated in the positive electrospray ionization mode with a mass resolution of >10,000. Mass spectrometry data were acquired and analyzed using MarkerLynx software (Waters Corporation) to convert raw data into exact mass–retention time pairs. The pairs with a P-value of <0.05 and a factor of change of >1.5 were selected for further analysis. These mass-to-charge ratios (m/z) values were searched using the Biomolecules database in MarkerLynx and The Human Metabolome Database (version 3.6), and the fold change was shown as metabolite levels of PANC-1 GR cells with respect to PANC-1 cells (PANC-1 GR/PANC-1 ratio) (Supplementary Table 2). Mass spectrometry analyses were performed at the Taipei Medical University Core Facility Center (Facilities for Proteomics and Structural Genomics) in Taipei, Taiwan.
2.10. Lactate production assay
Lactate production in PDAC cells was detected using the lactate colorimetric assay kit (K607-100, BioVision, Inc., Milpitas, CA, USA) in accordance with the manufacturer's instructions. Cells were seeded in a 96-well plate at a density of 1 × 104 cells/well 1 day prior to the assay, which was performed in triplicate. In the assay, 5–20 μL of the sample was placed at a density of 50 μL/well on a 96-well plate containing the lactate assay buffer and incubated for 30 min at room temperature and away from light. Absorbance was measured using a microplate reader at 570 nm.
2.11. Glutamine consumption and glutamate secretion assay
The glutamine consumption and glutamate secretion of transfected cells were analyzed using the glutamine/glutamate-Glo assay kit (Promega Corporation). The transfected cells were seeded into a 96-well plate at a density of 5000 cells/well in Dulbecco's modified Eagle's medium containing 5 mM glucose, 2 mM glutamine, and 10% fetal bovine serum. The 2-μL aliquots of the medium were removed and diluted in 98 μL of phosphate-buffered saline. Subsequently, 2 × 12.5-μL aliquots were transferred to a 96-well assay plate. Next, 12.5 μL of GLS buffer or GLS enzyme solution was added, and the cells were incubated for 30 min at room temperature. Thereafter, 25 μL of glutamate detection reagent was added, and after 60 min of incubation at room temperature, luminescence was measured using a luminometer (Varioskan Flash, Thermo Fisher Scientific, USA).
2.12. Animal studies
All animal experimental protocols were approved by the Institutional Animal Care and Use Committee of Taipei Medical University (No. LAC-2018-0185 and LAC-2019-0418). Four-to six-week-old NOD/CB-17 severe combined immunodeficient (NOD/SCID, NOD. CB17-Prkdcscid/NcrCrl) male mice, supplied by the National Laboratory Animal Center (Taipei, Taiwan), were used in xenograft tumor growth studies. Cells at a density of 5 × 106 cells/100 μL in phosphate-buffered saline mixed with an equal volume of Matrigel (BD Biosciences) were subcutaneously injected into the backs of each mouse. Tumor length and width were determined using calipers, and tumor volume was calculated using the following formula: 1/2 [length × width2]). When the tumor volume reached approximately 100–150 mm3, the mice were randomly allocated to four groups (n = 5/group) in which they either received 10 mg/kg CB839 or vehicle through intraperitoneal injection once a week for 4 weeks. The tumor volume was measured every 3 days.
2.13. Statistical analysis
All statistical analyses were performed using Prism 6 software (La Jolla, CA, USA). Data are presented as the means ± standard errors of the mean (SEMs), and statistical significance was examined using a two-tailed Student's t-test or two-way analysis of variance, followed by a two-sided Tukey's test. A P value of <0.05 was considered statistically significant.
3. Results
3.1. Clinical significance of Dicer expression in pancreatic cancer
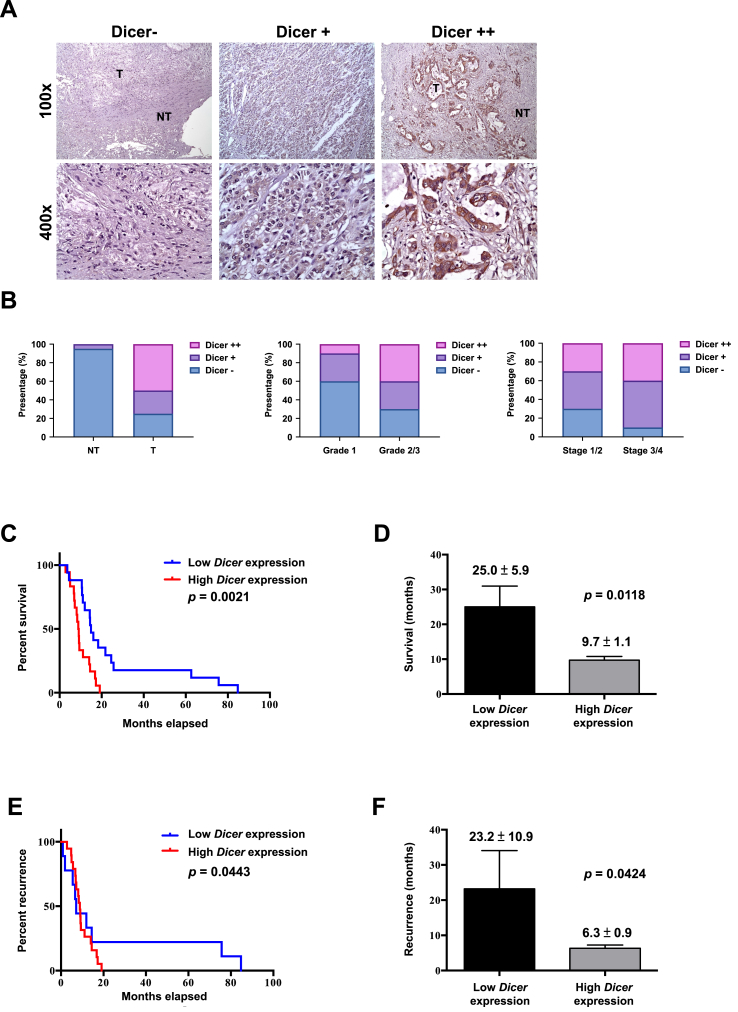
To examine the role of Dicer in pancreatic cancer progression, we detected Dicer expression in normal or tumor tissue sections through immunohistochemistry (Figure 1A). Tumor specimens were stratified on the basis of their clinically relevant Dicer expression level. Approximately 90% of normal tissue sections had lower Dicer expression levels (Dicer−), whereas approximately 50% of tumor tissue sections exhibited higher Dicer expression levels (Dicer++). Higher Dicer expression was observed in the tumor tissues of the patients with G2 or G3 and stage III or IV disease than in those with G1 and stage I or II disease (Figure 1B). Next, we analyzed the survival and recurrence rates of the 48 patients with PDAC. All the patients who received at least one chemotherapy regimen were stratified by Dicer expression (high vs. low; Figure 1C–F). The patients with higher Dicer levels had lower survival and higher recurrence rates than did those with lower Dicer levels, suggesting that Dicer expression is positively correlated with disease progression and poorer prognosis in patients with pancreatic cancer.
Figure 1.
Role of Dicer expression in pancreatic cancer progression. (A) Immunohistochemistry staining was performed to stratify Dicer expression in pancreatic cancer specimens into Dicer−, Dicer+, and Dicer++ groups. (B) Dicer expression in pancreatic cancer cells was analyzed on the basis of tumor status, with “NT” indicating nontumor and “T” indicating tumor (left panel, NT vs. T); grade status (middle panel, G1 vs. G2/3); and tumor stage (right panel, stage I/II vs. stage III/IV). (C, D) Dicer expression in pancreatic cancer cells was analyzed through qRT-PCR to determine overall survival and (E, F) recurrence-free survival outcomes in 48 patients categorized into high– and low–Dicer expression groups (log-rank test for panel C and E; student's test for panel D and F). The relative levels of gene expression are represented as ΔCP = CP of the tested gene − CP of the reference gene. Lower ΔCP values indicate higher gene expression. The median of individual ΔCP values was used as the cutoff through which high and low expression was defined. The total number of pancreatic cancer specimens was 48.
3.2. Dicer overexpression contributes to malignant behaviours in GEM-resistant PDAC cells
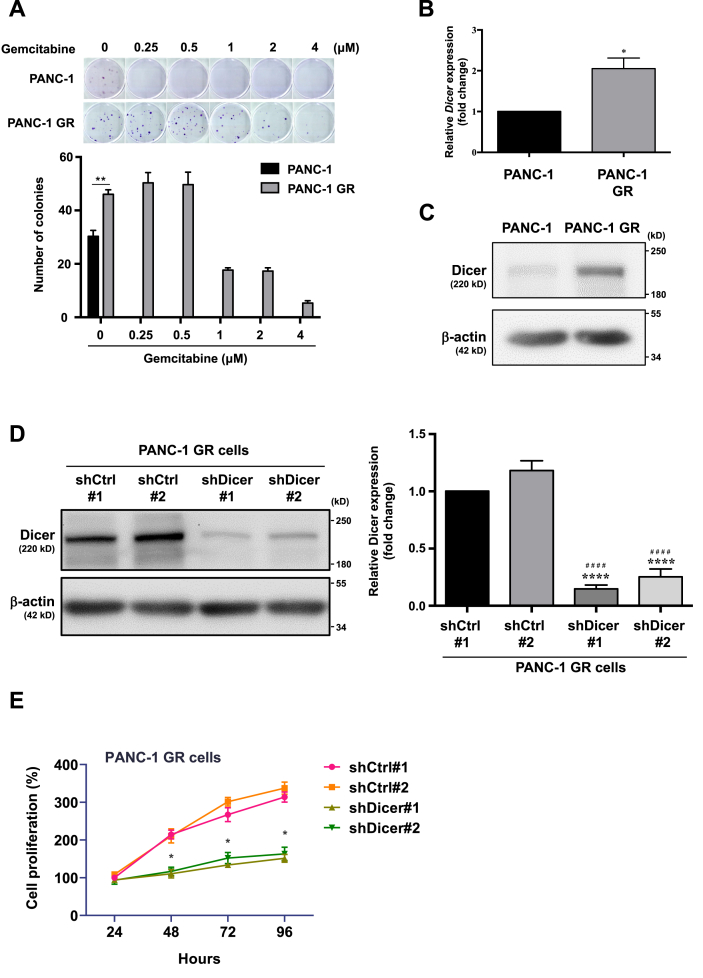
We compared clonogenicity between PANC-1 GR and PANC-1 cells after treatment with various GEM concentrations (Figure 2A). The number of colonies in PANC-1 GR cells did not change significantly while we observed that no colonies were observed in all GEM treatment groups of PANC-1 cells, suggesting that PANC-1 GR cells achieved GEM resistance. Next, we examined Dicer expression in PANC-1 and PANC-1 GR cells by performing qRT-PCR and Western blotting and observed that Dicer expression was significantly higher in PANC-1 GR cells than in PANC-1 cells at both mRNA and protein levels (Figure 2B and C). To investigate the effect of Dicer on GEM-resistant PDAC cells, we established Dicer knockdown PANC-1 GR cells (GR/shDicer; Figure 2D) and analyzed cell proliferation by performing the MTT assay (Figure 2E). GR/shDicer cell proliferation decreased to approximately 50%, 55%, and 60% of respective average shCtrl at 48, 72, and 96 h, respectively, suggesting that Dicer overexpression contributes to malignancy in GEM-resistant PDAC cells.
Figure 2.
PANC-1GR cells exhibitea high level of Dicer expression. (A) Upper panel: colony formation in PANC-1 and PANC-1 GR cells treated with different concentrations of GEM for 2 weeks. PANC-1 cells formed no colonies with GEM treatments. Bottom panel: Colonies were stained with crystal violet and counted using ImageJ software. Data are expressed as the number of colonies and as the means ± SEMs of the three experiments. ∗∗P < 0.01, one-way analysis of variance (ANOVA). (B) The mRNA expression of Dicer was analyzed through qRT-PCR. The qRT-PCR data were normalized to the β-actin level in each individual sample, and a bar plot presents fold changes in the expression of PANC-1 cells. (C) The protein expression of Dicer in PANC-1 and PANC-1 GR cells was analyzed through Western blotting. (D) Left panel: Dicer was knocked down in PANC-1 GR cells with lentivirus infection (GR/shDicer #1 and #2), and Dicer protein expression was analyzed through Western blotting. PANC-1 GR/shCtrl #1 and PANC-1 GR/shCtrl #2 cells were used for lentivirus infection control in PANC-1 GR cells. Right panel: relative quantification of Dicer expression in GR/shCtrl #1, GR/shCtrl #2, GR/shDicer #1, and GR/shDicer #2 cells was performed using ImageJ software. Data are expressed as fold changes in the expression of GR/shCtrl #1 cells. Results are presented as the means ± SEMs of the three independent experiments. ∗ indicates comparison with GR/shCtrl #1 cells; # indicates comparison with GR/shCtrl #2 cells. ∗∗∗∗P < 0.0001, ####P < 0.0001, Student's t test. (E) PANC-1 GR/shCtrl and PANC-1 GR/shDicer cells were treated with 2 μM GEM for 24, 48, 72, and 96 h, and cell viability was measured using the MTT assay. The percentage of cell proliferation is relative to that after individual 24-h treatment. Results are presented as the means ± SEMs of the three independent experiments. ∗P < 0.05, two-way ANOVA.
3.3. Knockdown of Dicer alters glutamine metabolism in GEM-resistant PDAC cells
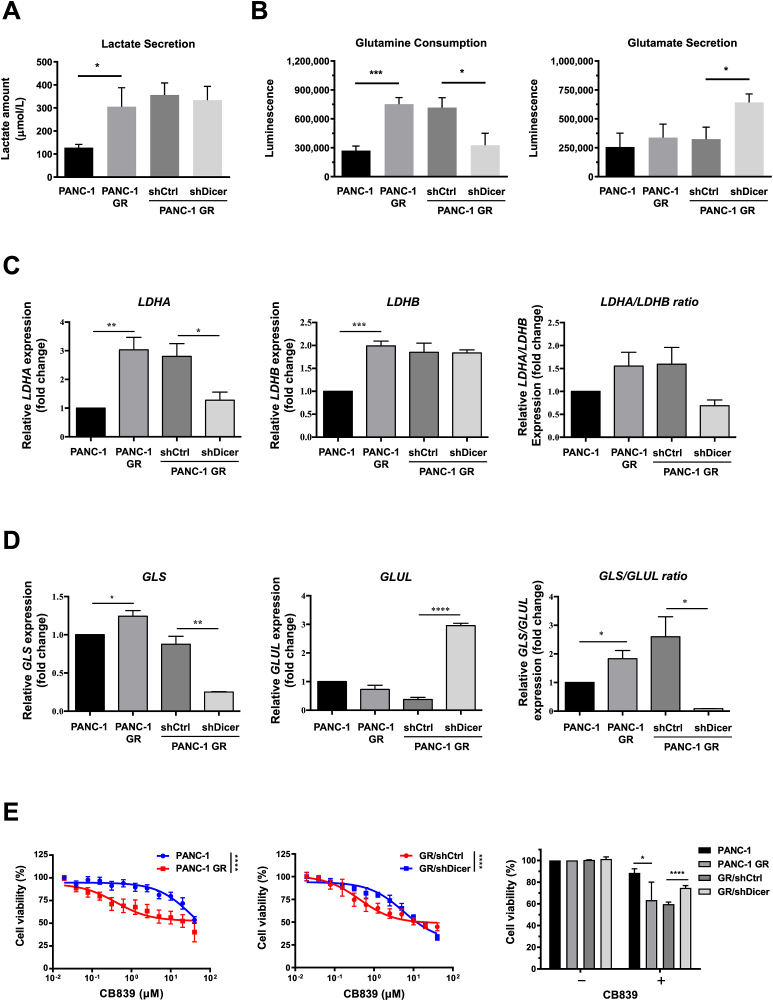
Accumulating reports indicate that cellular metabolism alterations contribute to gemcitabine resistance in pancreatic cancer [[20], [21], [22]]. The findings of liquid chromatography with tandem mass spectrometry analysis revealed that amounts of lactate, proline, ornithine, alpha-ketoglutaric acid, and glutamate metabolites significantly varied between PANC-1 GR and PANC-1 cells (Supplementary Table 2). In GR/shDicer cells, glutamate secretion increased significantly and glutamine consumption but not lactate secretion decreased significantly (Figure 3A and B). Intriguingly, we also found that PANC-1 GR cells exhibited markedly higher mRNA level of lactate dehydrogenase A (LDHA), an enzyme that catalyzes the conversion of pyruvate to lactate, than that of parental cells, whereas GR/shDicer cells reduced LDHA expression and the level was approximately similar to that of PANC-1 cells (Figure 3C). GR/shDicer cells, however, did not alter the expression of lactate dehydrogenase B (LDHB) which converts lactate back to pyruvate, which has been enhanced by PANC-1 GR cells (P < 0.001 compared to PANC-1 cells). Regarding an altered metabolism of glutamate, our results showed that PANC-1 GR increased mRNA expression of GLS:GLUL ratio compared to that of PANC-1, whereas knockdown of Dicer in PANC-1 GR cells could reduce back to the low level of mRNA GLS:GLUL ratio (Figure 3D), suggesting that Dicer mediates a metabolic switch in the expression of GLS and GLUL in GEM-resistant PDAC cells.
Figure 3.
Knockdown of Dicer in PANC-1 GR cells reduceglutamine metabolism. (A) Lactate secretion and (B) glutamine consumption (left panel) and glutamate secretion (right panel) of the indicated cells, namely PANC-1, PANC-1 GR cells, and PANC-1 GR cells with shDicer and shCtrl, were analyzed using the lactate colorimetric assay and glutamine/glutamate-Glo assay, respectively. Metabolic gene expression, including (C) the LDHA, LDHB, and LDHA:LDHB ratio and (D) GLS, GLUL, and GLS:GLUL ratio in PANC-1, PANC-1 GR, PANC-1 GR/shCtrl, and PANC-1 GR/shDicer cells were measured using qRT-PCR. The qRT-PCR data were normalized to the β-actin level in each individual sample, and a bar plot presents fold changes in the expression of PANC-1 cells. (E) The MTT assay was conducted to examine the viability of PANC-1 and PANC-1 GR cells (left panel), PANC-1 GR/shCtrl (GR/shCtrl) cells, and PANC-1 GR/shDicer (GR/shDicer) cells (middle panel) treated with various doses of the GLS inhibitor (CB839) for 72 h. Right panel, PANC-1, PANC-1 GR, GR/shCtrl, and GR/shDicer cells were incubated without (−, 0 μM) or with (+, 2.5 μM) CB839 for 72 h, and their viability was assessed using the MTT assay. The percentage of cell viability is relative to untreated controls. Results are presented as the means ± SEM of the three independent experiments. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001, two-tailed Student's t-test.
We examined the sensitivity of PANC-1 GR cells to the GLS inhibitor CB839 (Figure 3E). The results revealed that PANC-1 GR cells were more sensitive to GLS inhibitor treatment than PANC-1 cells (Figure 3E, left panel). The effects of CB839 treatment on PANC-1 GR cells, however, were reversed in PANC-1 GR/shDicer (GR/shDicer) cells, which exhibited decreased sensitivity to CB839 compared with GR/shCtrl control cells (Figure 3E, middle panel). Consistent with these observations, the viability of GR/shDicer cells treated with 2.5 μM CB839 decreased to approximately 20% that of untreated cells, whereas the viability of GR/shCtrl cells decreased to approximately 40% that of untreated cells (Figure 3E, right panel), implying that Dicer determines glutamine metabolism in PANC-1 GR cells.
3.4. Prognostic analysis of patients with pancreatic cancer on the basis of GLS:GLUL expression ratio and Dicer expression
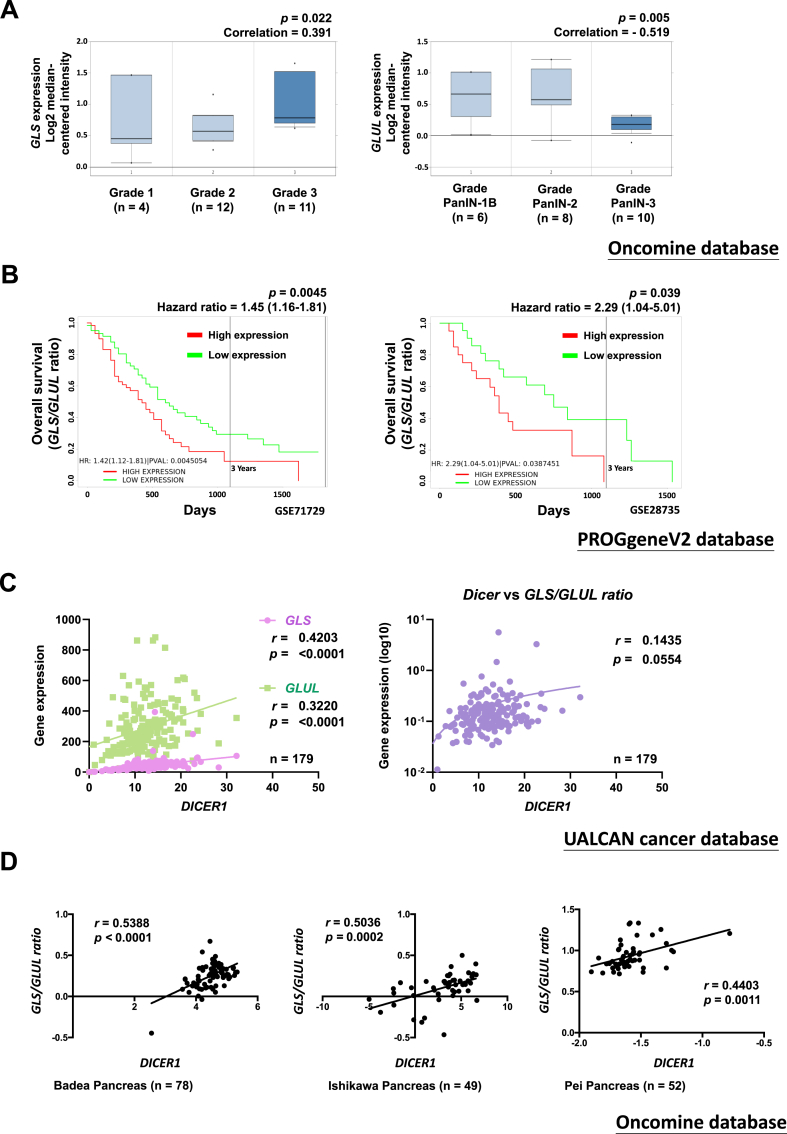
We utilised online databases to obtain insights into prognostic characteristics linked to GLS and GLUL expression. Oncomine database is used as a web-based tool to compare mRNA expressions of GLS and GLUL in pancreatic cancer. By using multiclass analyses, we found that pancreatic cancer patients with advanced histological grade (G3) exhibit augmented GLS and reduced GLUL mRNA expressions (Figure 4A). In addition, we used the PROGgeneV2 database to analyze the association of the GLS:GLUL expression ratio with the survival rate of the patients with pancreatic cancer (Figure 4B). Kaplan–Meier survival curves obtained using GSE71729 and GSE28735 datasets indicated that patients with a higher GLS:GLUL expression ratio had significantly shorter overall survival than did those with a lower GLS:GLUL expression ratio, with an increased hazard ratio of 1.45 (95% confidence interval [CI], 1.16–1.81) and 2.29 (95% CI, 1.04–5.01) in GSE71729 and GSE28735, respectively (Figure 4B). Because we noted that the knockdown of Dicer in PANC-1 GR cells induced a metabolic switch in the expression levels of GLS and GLUL (Figure 3D), we investigated the correlation between Dicer and the GLS:GLUL expression ratio by using the UALCAN cancer database (Figure 4C). We observed positive correlations between the expression of Dicer and that of either GLS (r = 0.4203; P < 0.0001) or GLUL (r = 0.3220; P < 0.0001) and between the expression of Dicer and the ratio of GLS to GLUL (r = 0.1435; P = 0.0554). The analysis of the Oncomine database revealed a significant correlation between Dicer expression and the GLS:GLUL expression ratio (Figure 4D), strengthening our hypothesis that an increase in the Dicer mRNA level may relate to an increase in the GLS level and a decrease in the GLUL level.
Figure 4.
Clinical relationship between Dicer and GLS:GLUL expression ratio in pancreatic cancer. (A) Left panel: The gene expression of GLS is positively correlated with advanced PDAC (Oncomine database: Collisson et al., Nature Medicine 2011). Right panel: GLUL gene expression is negatively correlated with advanced PDAC (Oncomine database: Buchholz et al., Oncogene 2005). The Pearson correlation coefficients and P values are indicated in the box plot. (B) The GLS:GLUL expression ratio is positively correlated with the poor overall survival of patients with pancreatic cancer (PROGgeneV2 database: GSE71729 and GSE28735 gene array). The hazard ratios and P values are indicated in the box plot. (C) DICER1 is positively correlated with GLS and GLUL expression (left panel) and the GLS:GLUL ratio (right panel) in pancreatic cancer (TCGA and UALCAN databases). The Pearson correlation coefficients and P values are indicated in the box plot. (D) DICER1 expression is positively correlated with the GLS:GLUL ratio in pancreatic cancer (Oncomine database: Badea et al., Hepatogastroenterology 2008; Ishikawa et al., Cancer Science 2005; Pei et al., Cancer Cell 2009). The Pearson correlation coefficients and P values are indicated in the box plot.
3.5. Role of phosphorylation of Dicer S1016 in GEM resistance in PDAC
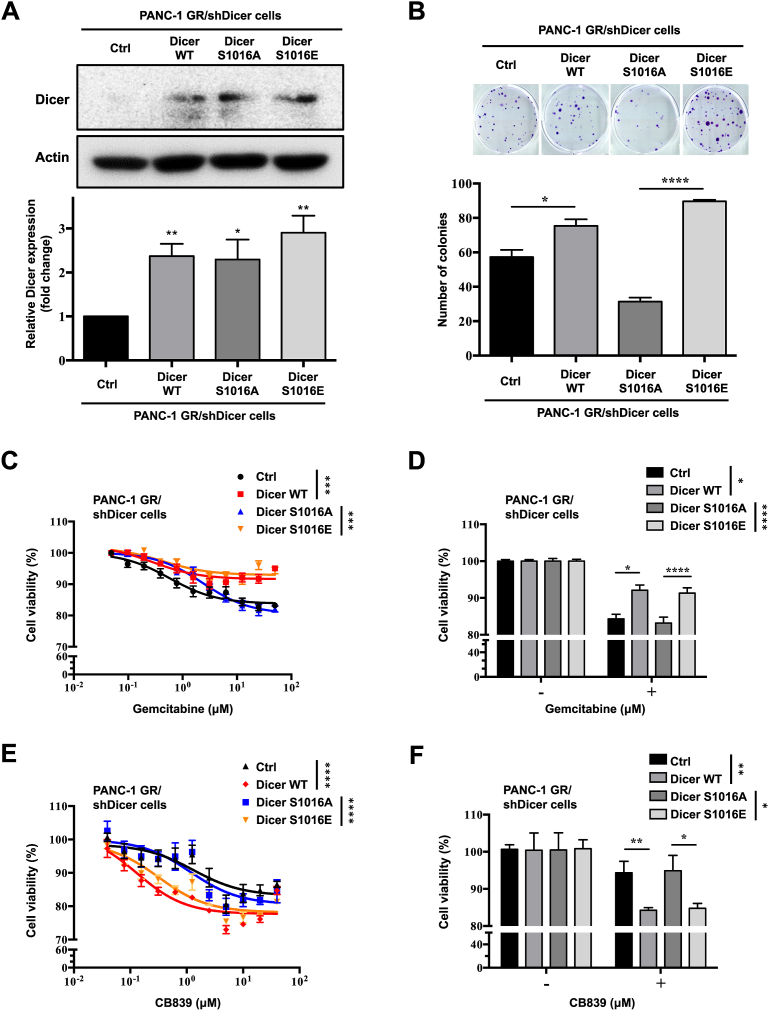
The afimoxifene-mediated DNA damage response increased Dicer phosphorylation at S1016 and triggered nuclear Dicer accumulation in U2OS and A549 cells, suggesting that Dicer phosphorylation at S1016 has crucial functions in cell cycle and growth, thus contributing to cancer progression [18]. We explored the role of Dicer phosphorylation at S1016 in GEM resistance in PDAC. We expressed phosphomimetic Dicer S1016 (S1016E) and phosphomutant Dicer S1016 (S1016A) in PANC-1 GR/shDicer cells and examined the effects of Dicer phosphorylation on colony formation, GEM resistance, and the GLS inhibitor response. The results of Western blotting revealed that the expression of Dicer S1016E and S1016A increased to the level of Dicer WT (Figure 5A). The results of the clonogenicity assay indicated that both Dicer WT and phosphomimetic Dicer S1016E rescued the clonogenic ability of GR/shDicer cells, whereas phosphomutant Dicer S1016A still exerted an inhibitory effect on the colony-forming ability of GR/shDicer cells (Figure 5B). Next, we examined the viability of GEM-treated cells and observed that both Dicer WT and Dicer S1016E in GR/shDicer cells retained resistance to GEM even after high-dose treatment (100 μM), whereas GR/shCtrl and phosphomutant Dicer S1016A increased GEM sensitivity and thus significantly reduced cell viability to a level similar to that of control cells (Figure 5C and D). We found that GR/shDicer cells exhibited lower sensitivity to the GLS inhibitor CB839 than did PANC-1 GR cells (Figure 3E). Phosphomutant Dicer S1016A cells demonstrated decreased sensitivity to CB839, whereas Dicer WT and phosphomimetic Dicer S1016E cells treated with CB839 exhibited relatively lower viability than did control and Dicer S1016A cells, respectively (Figure 5E and F).
Figure 5.
Phosphomimetic Dicer S1016E in PANC-1 GR/shDicer cells increasecell viability, GEM resistance, and GLS inhibitor response. (A) Upper panel: Dicer protein expression in phosphomimetic Dicer S1016E (Dicer S1016E), phosphomutant Dicer S1016A (Dicer S1016A), Dicer WT, and control (Ctrl) PANC-1 GR/shDicer cells was analyzed through Western blotting. β-Actin was used as a loading control. Bottom panel: The relative quantification of Dicer expression in the indicated cells was performed using ImageJ software. Western blotting data were normalized to the β-actin level in each individual sample, and a bar plot presents fold changes in control cells. The results are presented as the means ± SEMs of three independent experiments. ∗P < 0.05, ∗∗P < 0.01, two-tailed Student's t-test. (B) Upper panel: colony formation of indicated cells cultured for 2 weeks; the colonies were stained with crystal violet and counted using ImageJ software. Bottom panel: Data are expressed as the number of colonies, and results are presented as the means ± SEMs of the three independent experiments. ∗P < 0.05 and ∗∗∗∗P < 0.0001, one-way ANOVA. (C) The indicated cells were incubated with varying doses of GEM for 48 h, and cell viability was analyzed using the MTT assay. (D) Bar plot comparing results between the untreated (−, 0 μM) and treated (+, 2.5 μM) groups. (E) The indicated cells were incubated with varying doses of the GLS inhibitor CB839 for 72 h, and cell viability was analyzed using the MTT assay. (F) Bar plot comparing results between the untreated (−, 0 μM) and CB839-treated (+, 0.625 μM) groups. Results are presented as the means ± SEMs of the three independent experiments. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 and ∗∗∗∗P < 0.0001, one-way ANOVA.
3.6. Phosphorylation of Dicer at S1016 switches the GLS to GLUL ratio in GEM-resistant PDAC
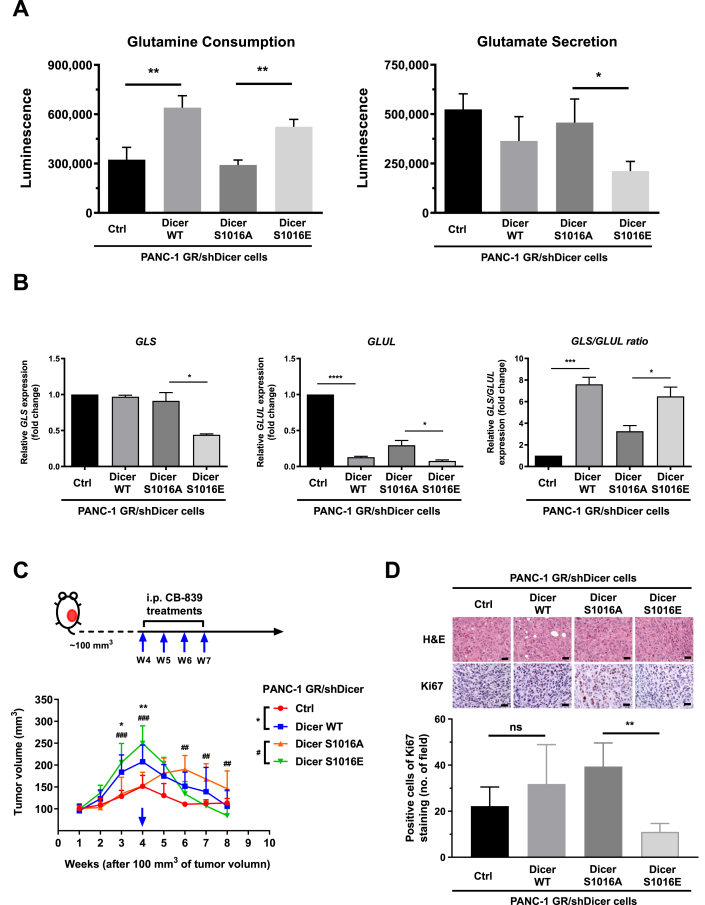
We examined whether Dicer phosphorylation at S1016 can change glutamine consumption and glutamate secretion in GR/shDicer cells (Figure 6A). We observed that phosphomimetic Dicer S1016E but not phosphomutant Dicer S1016A mutation significantly increased glutamine consumption and reduced glutamate secretion. GLS and GLUL mRNA levels were quantified through qRT-PCR, and the results revealed that both the control and phosphomutant Dicer S1016A in PANC-1 GR/shDicer cells exhibited higher GLUL expression than did Dicer WT and Dicer S1016E cells, respectively (Figure 6B). In addition, GLS expression in phosphomimetic Dicer S1016E cells was significantly lower than that in Dicer S1016A cells, leading to a high GLS:GLUL expression ratio, a pattern associated with PDAC cell growth and drug responses. By contrast, phosphomutant Dicer S1016A cells demonstrated a lower GLS:GLUL expression ratio, and the GLS:GLUL ratio significantly differed from that in Dicer S1016E cells (Figure 6B, right panel). The results of in vivo animal experiments indicated that treatment with the GLS inhibitor CB839 significantly retarded the growth of Dicer WT and phosphomimetic Dicer S1016E tumors compared with control and Dicer S1016A tumors, respectively (Figure 6C). This pattern is consistent with the results of in vitro cell experiments, in which Dicer WT and Dicer S1016E cells but not S1016A cells were more sensitive to CB839 treatment (Figure 5E and F). Next, we assessed Ki67 immunohistochemical staining in tumor sections to investigate the effect of CB839 treatment on cell proliferation (Figure 6D). Tumor sections bearing Dicer S1016A cells exhibited a significantly higher expression of Ki67 than did those bearing Dicer S1016E cells, indicating that the increased GLS:GLUL ratio by phosphomimetic Dicer S1016E is positively associated with enhancing CB839 drug sensitivity in PANC-1 GR cells.
Figure 6.
Phosphomimetic Dicer S1016E in PANC-1 GR/shDicer cells increaseglutamine metabolism, GLS:GLUL ratio, and GLS inhibitor response. (A) Glutamine consumption (left panel) and glutamate secretion (right panel) of the indicated cells, namely PANC-1 GR/shDicer cells with Ctrl, Dicer wild-type, Dicer S1016A, and Dicer S1016E cells, were analyzed using the glutamine/glutamate-Glo assay. Results are presented as the means ± SEMs of the three independent experiments. ∗P < 0.05 and ∗∗P < 0.01, one-way ANOVA. (B) GLS and GLUL expression and the GLS:GLUL ratios of the indicated PANC-1 GR/shDicer cells were measured through qRT-PCR. The qRT-PCR data were normalized to the β-actin level in each individual sample, and a bar plot presents fold changes in the expression of PANC-1 GR/shDicer/Ctrl cells. ∗P < 0.05, ∗∗P < 0.01 and ∗∗∗∗P < 0.0001, one-way ANOVA. (C) Phosphomimetic Dicer S1016E (Dicer S1016E), phosphomutant Dicer S1016A (Dicer S1016E), Dicer wild-type (Dicer WT), and control (Ctrl) of PANC-1 GR/shDicer cells were subcutaneously injected into the backs of each mouse. After tumor-bearing mice develop solid tumor to ∼100 mm3 tumor volume of each group, tumor-bearing mice were observed for 4 weeks and then received CB839 treatments through once-weekly intraperitoneal injection (10 mg/kg) over 4 times (blue arrow). Tumor volume in each group (n = 5) was determined by measuring the tumor length and width using calipers and calculating the volume using the formula: 1/2 (length × width2). Each point represents the mean ± standard deviation of the tumor volume of the five mice in each group. ∗, Ctrl vs. Dicer WT; #, Dicer S1016E vs. Dicer S1016A. ∗P < 0.05, ∗∗P < 0.01, ##P < 0.01 and ###P < 0.001, two-way ANOVA. (D) Upper panel: hematoxylin and eosin staining and immunohistochemical staining of mouse xenografts (after 4 weeks of CB839 treatment) were performed to analyze Ki67 expression. Bottom panel: Ki67-positive cells were quantified from five random microscopic fields. ∗∗P < 0.01, one-way ANOVA.
3.7. Phosphomimetic Dicer S1016E mediates miRNA maturation and switches glutamine metabolism in GEM-resistant PDAC cells
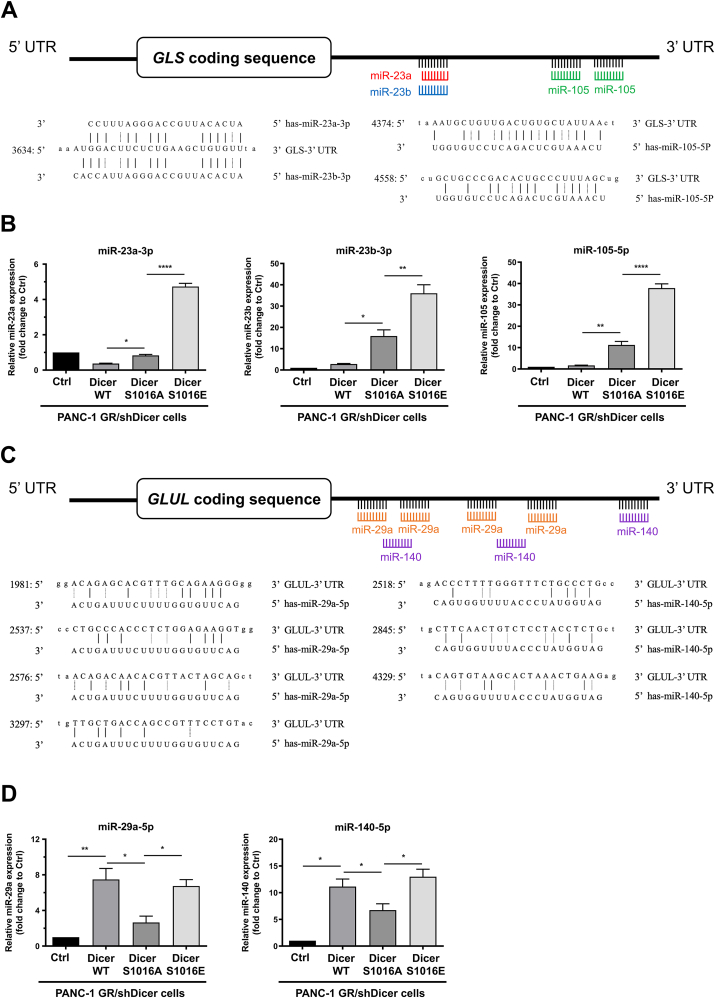
MiRNAs are small non-coding RNAs that modulate posttranscriptional regulation by binding to the 3′-UTR of target mRNAs. MiR-105 and miR-23a and b regulate GLS expression in cancer-associated fibroblasts and prostate cancer cells, respectively [23,24], whereas miR-29a and miR-140 target the 3′-UTR of GLUL to suppress the invasion and proliferation of glioma cells [25,26]. We investigated whether phosphorylated Dicer is involved in miRNA biogenesis in GEM-resistant PDAC cells and regulates glutamine metabolism in cancer cells. Using the TargetScan database, we predicted that miR-23a-3p, miR-23b-3p, and miR-105-5p targeted the 3′-UTR of GLS (Figure 7A). We found significant upregulation of miR-23a-3p, miR-23b-3p, and miR-105-5p in phosphomimetic Dicer S1016E cells compared with Dicer S1016A cells (Figure 7B). Surprisingly, the results have shown that there are statistical significant differences in GLS miRNAs expressions between Dicer WT and phosphomutant Dicer S1016A, which probably reflects that other phosphorylation sites in Dicer may simultaneously be involved in GLS-targeting miRNA regulation. Next, we identified miR-29a-5p and miR-140b-5p as potential miRNAs targeting the 3′-UTR of GLUL (Figure 7C). The results revealed that the GLUL level was suppressed in Dicer WT and phosphomimetic Dicer S1016E cells through an increase in miR-29a-5p and miR-140-5p expression in GR/shDicer cells and that the levels of these miRNAs were significantly higher than those in control and Dicer S1016A cells (Figure 7D). These results suggest that phosphomimetic Dicer S1016E affects specific miRNAs to modulate GLS:GLUL ratio and contributes to Dicer-mediated GEM resistance in PANC-1 cells.
Figure 7.
Phosphomimetic Dicer S1016E increasemiRNA maturation to regulate the 3′-UTR of GLS and GLUL. (A) Target prediction performed using TargetScan revealed that the 3′-UTR sequence of GLS contains putative binding sites for miR-23a-3p, miR-23b-3p, and miR-105-5p. (B) Expression of miR-23a-3p, miR-23b-3p, and miR-105-5p in indicated cells, namely PANC-1 GR/shDicer cells with Ctrl, Dicer wild-type (WT), Dicer S1016A, and Dicer S1016E cells, was analyzed through qRT-PCR. The qRT-PCR data were normalized to the U6B level in each individual sample, and a bar plot presents fold changes in the expression of PANC-1 GR/shDicer/Ctrl cells. Results are presented as the means ± SEMs of the three independent experiments. ∗P < 0.05, ∗∗P < 0.01 and ∗∗∗∗P < 0.0001, one-way ANOVA. (C) Target prediction performed using TargetScan revealed that the 3′-UTR sequence of GLUL contains putative binding sites for miR-29a-5p and miR-140-5p. (D) The miR-29a-5p and miR-140-5p expression in the indicated cells was analyzed through qRT-PCR. The qRT-PCR data were normalized to the U6B level in each individual sample, and a bar plot presents fold changes in the expression of PANC-1 GR/shDicer/Ctrl cells. Results are presented as the means ± SEMs of the three independent experiments. ∗P < 0.05 and ∗∗P < 0.01, one-way ANOVA.
4. Discussion
Dicer is a key processing enzyme involved in miRNA maturation. Evidence has demonstrated that a high Dicer level is associated with tumor aggressiveness and tamoxifen resistance in breast cancer [7,11], and reduced Dicer expression overcomes gefitinib tolerance in lung cancer cells [19]. Additionally, aberrance of mature miRNAs contributes to cancer stem cell features and drug resistance in hepatocellular carcinoma [[20], [21], [22], [23]] and pancreatic cancer progression [[23], [24], [25]], implying that dysregulation of miRNA maturation plays a critical role in aggressive behaviour and resistance to therapy in cancer. Our previous study indicated that Dicer expression was significantly associated with GEM resistance in PDAC cells [26] and that Dicer was highly expressed in patients with advanced PDAC and is involved in GEM sensitivity in pancreatic cancer. We thus sought to investigate further how Dicer is involved in GEM resistance in PDAC, specifically exploring its contribution to cancer prognosis, cell proliferation, and metabolic regulation. In this study, we found that phosphomimetic Dicer S1016E but not phosphomutant Dicer S1016A facilitates miRNA maturation and reprograms glutamine metabolism, leading to differential responses to chemotherapy in GEM-resistant pancreatic cancer.
Cancer is considered a metabolism-related disease [[27], [28], [29]]. Cancer cells not only increase anaerobic glycolysis to produce energy but also induce glutamine decomposition to compensate for the insufficiency of carbon sources in the TCA cycle, thus promoting cancer cell proliferation and growth [30,31]. The results of this study indicated that compared with parental PANC-1 cells, GEM-resistant PANC-1 cells exhibited marked changes in lactate and glutamate metabolites (Figure 3A and B). Although additional studies are required, our observation of a positive association between Dicer and LDHA suggests the acquisition of resistance to GEM (Figure 3C). We previously reported that LDHA-mediated lactic acid production contributes to GEM resistance in pancreatic cancer through the epigenetic regulation of FOXO3a/miR-4259 [31]. Herein, our studies emphasize the importance of future studies to further investigate whether Dicer plays the key role in the mechanism underlying LDHA and GEM resistance in pancreatic cancer.
In addition to demonstrating a preferential dependence on glycolysis, cancer cells exhibit increased consumption of glutamine [32] and reprogramming of glutaminolysis [33,34]. Glutamine provides the carbon and nitrogen required for nucleotide and nonessential amino acid biosynthesis [35] in support of the bioenergetic processing and maintenance of redox homeostasis in cancer cells [15]. Glutaminolysis driven by GLS is associated with chemoresistance in PDAC. Mukhopadhyay et al. reported that perturbing glutamine metabolism increased chemosensitivity and that the GLS inhibitor CB839 reinforced GEM efficiency in PDAC cells [34]. Consistent with this finding, we observed that PANC-1 GR cells exhibited higher glutamine consumption than did parental PANC-1 cells (Figure 3B) with an increased GLS:GLUL expression ratio (Figure 3D), and thus, were more sensitive to CB839 treatments. High expression of Dicer is associated with GEM resistance phenotype by rewiring ratio expression of GLS:GLUL and thus less sensitive to CB839 treatments (Figure 3E). Importantly, phosphorylation of Dicer at S1016E site plays a major role in mediating glutamine metabolism, which has shown to increase sensitivity to GLS inhibitors in PANC-1 GR cells and the results were correlated with high expressions in GLS:GLUL ratio. Phosphomutant Dicer S1016A, on the other hand, was shown markedly resistant to GLS inhibitors in previously Dicer knockdown cells with low expressions in GLS:GLUL ratio (Figure 6B). Our xenograft model experiments also pointed out that CB839 treatments inhibit mice bearing Dicer S1016E, not Dicer S1016A, suggesting that specific Dicer phosphorylation motifs are correlated with cancer cell sensitivity to GLS inhibitors, probably through rewiring glutamine metabolism in PANC-1 cells.
Mechanisms that modulate GLS and GLUL expression include promoter methylation [[36], [37], [38]], protein degradation [39], and miRNA dysregulation [[40], [41], [42], [43], [44]]. Bu et al. revealed that the knockdown of Dicer significantly induced G1 arrest and enhanced sensitivity to cisplatin in MCF-7 breast cancer cells through the modulation of certain miRNAs [45]. We previously reported that the ERK-mediated transcription factor Sp1 binds to the Dicer promoter region and induces Dicer expression to contribute to cell survival in GEM-resistant pancreatic cancer [26]. ERK phosphorylates Dicer at S1728 and S1852 to induce the nuclear translocation of Dicer, leading to decreased Dicer function in the female germline and small RNA repertoire [46]. Moreover, afimoxifene-mediated DNA damage response increases Dicer phosphorylation at S1016 and induced nuclear Dicer accumulation in human osteosarcoma and lung cancer cells [12], suggesting that Dicer phosphorylation plays a crucial role in cell survival and drug response of cancers.
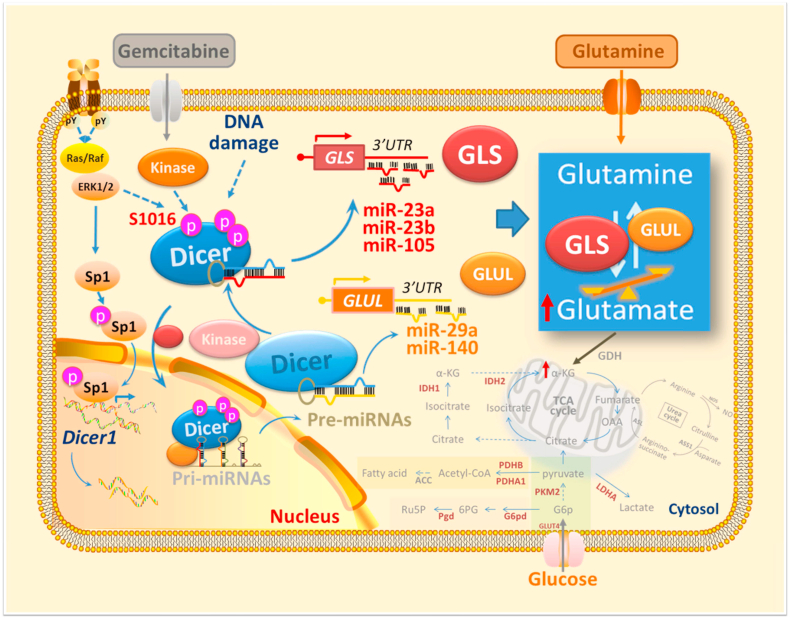
Our study further identifies that phosphomimetic Dicer S1016E exhibits high relative expressions of miR-23a-3p, miR-23b-3p, and miR-105-5p compared to that of Dicer S1016A and wild-type, which are the putative 3′-UTR sequence of GLS binding sites. Additionally, Dicer S1016E and S1016A showed opposite expression patterns of miR-29a-5p and miR-140-5p, which are the putative 3′-UTR sequence of GLUL binding sites, suggesting that Dicer phosphorylated at S1016 selectively regulates miRNA maturation to target 3′-UTR of GLS and GLUL. This is in connection with our findings that have demonstrated that phosphomimetic Dicer S1016E induces GLS:GLUL expression ratio in PANC-1 GR cells and is therefore highly sensitive to CB839 treatment (Figure 5, Figure 6C). Similar pattern was shown in phosphomutant Dicer S1016A, which exhibits low GLS:GLUL expression ratio and is therefore more sensitive to GEM treatment but not to CB839 treatments (Figure 5, Figure 6C). In Figure 7B, we also found that Dicer WT showed significantly lower miRNA expressions than those of phosphomutant Dicer S1016A, which may imply that other phosphorylation sites in Dicer may be involved in GLS-targeting miRNA regulation. Directly related, further studies are necessary to validate if predicted miRNA is genuine by performing RNA sequencing. Although more research should be completed to confirm the direct effect of miRNA modulation and GLS:GLUL ratio on PANC-1 GR cells, our study first reports that modification of phosphorylated Dicer may involve glutamine reprogramming and GEM resistance in PDAC cells, thus offering novel insight for sensitising PDAC to GEM (Figure 8).
Figure 8.
Schematic model illustrating the upregulation of glutamine metabolism by phosphomimetic Dicer S1016E in GEM-resistant pancreatic cancer cells and its effect on miRNA maturation and glutamine–glutamate balance.
5. Conclusion
This study provided the first experimental evidence that Dicer phosphorylation at S1016 affects miRNA biogenesis and glutamine metabolism in GEM-resistant pancreatic cancer. Our findings highlight the possibility of developing miRNA-based therapeutics for pancreatic cancer and provide new insights to improve the overall prognosis of patients with PDAC receiving GEM treatment. Additional studies should be conducted to understand how Dicer phosphorylation selectively regulates miRNAs in GEM-resistant PDAC.
Authors’ contributions
J.M.P.: Investigation, Formal analysis, Interpretation of data, Writing - original draft, Writing - Review & Editing; Y-S.S.: Formal analysis, Conceptualization, Interpretation of data, Funding acquisition, Writing - original draft, Performed animal experiment; C-Y.L.: Investigation, Interpretation of data, Methodology, Formal analysis; T-W.H.: Investigation, Formal analysis, Interpretation of data, Performed animal experiment; Y-H.S.: Collection of clinical samples, Data analysis; H-A.C.: Collection of clinical samples, Data analysis; Y-S.S.: Collection of clinical samples, Data analysis. J-M.P.: Methodology, Writing - original draft, Writing - Review & Editing; C.S.: Writing - Review & Editing; J-S.C.: Methodology, Writing - original draft; C-F.C.: Conceptualization, Funding acquisition, Methodology, Writing - original draft, Writing - Review & Editing, Supervision.
Funding
This research was funded by the National Science and Technology Council (Ministry of Science and Technology), Taiwan [MOST107-2320-B-038-065, MOST108-2320-B-038-015, MOST109-2314-B-866-001-MY3, MOST110-2320-B-038-071 and MOST111-2314-B-038-072]; Taipei Medical University Research Grants for Newly Hired Faculty [TMU106-AE1-B38], and the Taipei Medical University Research Center of Cancer Translational Medicine from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education in Taiwan [DP2-109-21121-03-C-08-03, DP2-110-21121-03-C-08-02 and DP2-111-21121-01-C-08-03].
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2022.101576.
Conflict of interest
The authors declare no conflicts of interest.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2020. CA: A Cancer Journal for Clinicians. 2020;70(1):7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Garrido-Laguna I., Hidalgo M. Pancreatic cancer: from state-of-the-art treatments to promising novel therapies. Nature Reviews Clinical Oncology. 2015;12(6):319–334. doi: 10.1038/nrclinonc.2015.53. [DOI] [PubMed] [Google Scholar]
- 3.Oettle H., Post S., Neuhaus P., Gellert K., Langrehr J., Ridwelski K., et al. Adjuvant chemotherapy with gemcitabine vs observation in patients undergoing curative-intent resection of pancreatic cancer: a randomized controlled trial. JAMA. 2007;297(3):267–277. doi: 10.1001/jama.297.3.267. [DOI] [PubMed] [Google Scholar]
- 4.Yoneyama H., Takizawa-Hashimoto A., Takeuchi O., Watanabe Y., Atsuda K., Asanuma F., et al. Acquired resistance to gemcitabine and cross-resistance in human pancreatic cancer clones. Anti-Cancer Drugs. 2015;26(1):90–100. doi: 10.1097/CAD.0000000000000165. [DOI] [PubMed] [Google Scholar]
- 5.Lund E., Dahlberg J.E. Substrate selectivity of exportin 5 and Dicer in the biogenesis of microRNAs. Cold Spring Harbor Symposia on Quantitative Biology. 2006;71:59–66. doi: 10.1101/sqb.2006.71.050. [DOI] [PubMed] [Google Scholar]
- 6.Merritt W.M., Lin Y.G., Han L.Y., Kamat A.A., Spannuth W.A., Schmandt R., et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. New England Journal of Medicine. 2008;359(25):2641–2650. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grelier G., Voirin N., Ay A.S., Cox D.G., Chabaud S., Treilleux I., et al. Prognostic value of Dicer expression in human breast cancers and association with the mesenchymal phenotype. British Journal of Cancer. 2009;101(4):673–683. doi: 10.1038/sj.bjc.6605193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karube Y., Tanaka H., Osada H., Tomida S., Tatematsu Y., Yanagisawa K., et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Science. 2005;96(2):111–115. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iliou M.S., da Silva-Diz V., Carmona F.J., Ramalho-Carvalho J., Heyn H., Villanueva A., et al. Impaired DICER1 function promotes stemness and metastasis in colon cancer. Oncogene. 2014;33(30):4003–4015. doi: 10.1038/onc.2013.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pouliot L.M., Shen D.W., Suzuki T., Hall M.D., Gottesman M.M. Contributions of microRNA dysregulation to cisplatin resistance in adenocarcinoma cells. Experimental Cell Research. 2013;319(4):566–574. doi: 10.1016/j.yexcr.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Selever J., Gu G., Lewis M.T., Beyer A., Herynk M.H., Covington K.R., et al. Dicer-mediated upregulation of BCRP confers tamoxifen resistance in human breast cancer cells. Clinical Cancer Research. 2011;17(20):6510–6521. doi: 10.1158/1078-0432.CCR-11-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burger K., Schlackow M., Potts M., Hester S., Mohammed S., Gullerova M. Nuclear phosphorylated Dicer processes double-stranded RNA in response to DNA damage. The Journal of Cell Biology. 2017;216(8):2373–2389. doi: 10.1083/jcb.201612131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qin C., Yang G., Yang J., Ren B., Wang H., Chen G., et al. Metabolism of pancreatic cancer: paving the way to better anticancer strategies. Molecular Cancer. 2020;19(1):50. doi: 10.1186/s12943-020-01169-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen R., Lai L.A., Sullivan Y., Wong M., Wang L., Riddell J., et al. Disrupting glutamine metabolic pathways to sensitize gemcitabine-resistant pancreatic cancer. Scientific Reports. 2017;7(1):7950. doi: 10.1038/s41598-017-08436-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hensley C.T., Wasti A.T., DeBerardinis R.J. Glutamine and cancer: cell biology, physiology, and clinical opportunities. Journal of Clinical Investigation. 2013;123(9):3678–3684. doi: 10.1172/JCI69600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rahman M., Hasan M.R. Cancer metabolism and drug resistance. Metabolites. 2015;5(4):571–600. doi: 10.3390/metabo5040571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mahavisno R.D.K.-S.S., Briggs V.V.R.Y.J., Kincead B.B.T.A.M., Oncomine B.C.K.P. 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goswami C.P., Nakshatri H. PROGgene: gene expression based survival analysis web application for multiple cancers. Journal of Clinical Bioinformatics. 2013;3(1):1–9. doi: 10.1186/2043-9113-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen J.C., Su Y.H., Chiu C.F., Chang Y.W., Yu Y.H., Tseng C.F., et al. Suppression of Dicer increases sensitivity to gefitinib in human lung cancer cells. Annals of Surgical Oncology. 2014;21(Suppl 4):S555–S563. doi: 10.1245/s10434-014-3673-y. [DOI] [PubMed] [Google Scholar]
- 20.Liu K., Liu S., Zhang W., Ji B., Wang Y., Liu Y. miR222 regulates sorafenib resistance and enhance tumorigenicity in hepatocellular carcinoma. International Journal of Oncology. 2014;45(4):1537–1546. doi: 10.3892/ijo.2014.2577. [DOI] [PubMed] [Google Scholar]
- 21.Xia H., Ooi L.L., Hui K.M. MicroRNA-216a/217-induced epithelial-mesenchymal transition targets PTEN and SMAD7 to promote drug resistance and recurrence of liver cancer. Hepatology. 2013;58(2):629–641. doi: 10.1002/hep.26369. [DOI] [PubMed] [Google Scholar]
- 22.Meng F., Glaser S.S., Francis H., DeMorrow S., Han Y., Passarini J.D., et al. Functional analysis of microRNAs in human hepatocellular cancer stem cells. Journal of Cellular and Molecular Medicine. 2012;16(1):160–173. doi: 10.1111/j.1582-4934.2011.01282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yonemori K., Kurahara H., Maemura K., Natsugoe S. MicroRNA in pancreatic cancer. Journal of Human Genetics. 2017;62(1):33–40. doi: 10.1038/jhg.2016.59. [DOI] [PubMed] [Google Scholar]
- 24.Morris J.P.t., Greer R., Russ H.A., von Figura G., Kim G.E., Busch A., et al. Dicer regulates differentiation and viability during mouse pancreatic cancer initiation. PLoS One. 2014;9(5):e95486. doi: 10.1371/journal.pone.0095486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu Y.F., Hannafon B.N., Ding W.Q. microRNA regulation of human pancreatic cancer stem cells. Stem Cell Investigation. 2017;4:5. doi: 10.21037/sci.2017.01.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su Y.H., Hsu T.W., Chen H.A., Su C.M., Huang M.T., Chuang T.H., et al. ERK-mediated transcriptional activation of Dicer is involved in gemcitabine resistance of pancreatic cancer. Journal of Cellular Physiology. 2020;236(6):4420–4434. doi: 10.1002/jcp.30159. [DOI] [PubMed] [Google Scholar]
- 27.Seyfried T.N., Flores R.E., Poff A.M., D'Agostino D.P. Cancer as a metabolic disease: implications for novel therapeutics. Carcinogenesis. 2014;35(3):515–527. doi: 10.1093/carcin/bgt480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hainaut P., Plymoth A. Cancer as a metabolic disease. Current Opinion in Oncology. 2012;24(1):56–57. doi: 10.1097/CCO.0b013e32834e388a. [DOI] [PubMed] [Google Scholar]
- 29.Dang C.V. Links between metabolism and cancer. Genes & Development. 2012;26(9):877–890. doi: 10.1101/gad.189365.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsu P.P., Sabatini D.M. Cancer cell metabolism: warburg and beyond. Cell. 2008;134(5):703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 31.Choi Y.K., Park K.G. Targeting glutamine metabolism for cancer treatment. Biomolecules & Therapeutics (Seoul) 2018;26(1):19–28. doi: 10.4062/biomolther.2017.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liberti M.V., Locasale J.W. The warburg effect: how does it benefit cancer cells? (vol 41, pg 211, 2016) Trends in Biochemical Sciences. 2016;41(3) doi: 10.1016/j.tibs.2015.12.001. 287–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rong Y., Wu W., Ni X., Kuang T., Jin D., Wang D., et al. Lactate dehydrogenase A is overexpressed in pancreatic cancer and promotes the growth of pancreatic cancer cells. Tumor Biology. 2013;34(3):1523–1530. doi: 10.1007/s13277-013-0679-1. [DOI] [PubMed] [Google Scholar]
- 34.Zhao H., Duan Q., Zhang Z., Li H., Wu H., Shen Q., et al. Up-regulation of glycolysis promotes the stemness and EMT phenotypes in gemcitabine-resistant pancreatic cancer cells. Journal of Cellular and Molecular Medicine. 2017;21(9):2055–2067. doi: 10.1111/jcmm.13126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dai S., Peng Y., Zhu Y., Xu D., Zhu F., Xu W., et al. Glycolysis promotes the progression of pancreatic cancer and reduces cancer cell sensitivity to gemcitabine. Biomedicine & Pharmacotherapy. 2020;121:109521. doi: 10.1016/j.biopha.2019.109521. [DOI] [PubMed] [Google Scholar]
- 36.Chiu C.-F. Abstracts of the 76th annual meeting of the Japanese cancer association, Yokohama, Japan. 2018. Epigenetic regulation of LDHA dictates the gemcitabine resistance in pancreatic cancer; p. 1146. [Google Scholar]
- 37.Son J., Lyssiotis C.A., Ying H., Wang X., Hua S., Ligorio M., et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496(7443):101–105. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sousa C.M., Kimmelman A.C. The complex landscape of pancreatic cancer metabolism. Carcinogenesis. 2014;35(7):1441–1450. doi: 10.1093/carcin/bgu097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mukhopadhyay S., Goswami D., Adiseshaiah P.P., Burgan W., Yi M., Guerin T.M., et al. Undermining glutaminolysis bolsters chemotherapy while NRF2 promotes chemoresistance in KRAS-driven pancreatic cancers. Cancer Research. 2020;80(8):1630–1643. doi: 10.1158/0008-5472.CAN-19-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y., Bai C., Ruan Y., Liu M., Chu Q., Qiu L., et al. Coordinative metabolism of glutamine carbon and nitrogen in proliferating cancer cells under hypoxia. Nature Communications. 2019;10(1):201. doi: 10.1038/s41467-018-08033-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao P., Tchernyshyov I., Chang T.C., Lee Y.S., Kita K., Ochi T., et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458(7239):762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan W., Wu X., Zhou W., Fong M.Y., Cao M., Liu J., et al. Cancer-cell-secreted exosomal miR-105 promotes tumour growth through the MYC-dependent metabolic reprogramming of stromal cells. Nature Cell Biology. 2018;20(5):597–609. doi: 10.1038/s41556-018-0083-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Q., Souba W.W., Croce C.M., Verne G.N. MicroRNA-29a regulates intestinal membrane permeability in patients with irritable bowel syndrome. Gut. 2010;59(6):775–784. doi: 10.1136/gut.2009.181834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang R., Zhu J.C., Hu H., Lin Q.Y., Shao W., Ji T.H. MicroRNA-140-5p suppresses invasion and proliferation of glioma cells by targeting glutamate-ammonia ligase (GLUL) Neoplasma. 2020;67(2):371–378. doi: 10.4149/neo_2020_190514N432. [DOI] [PubMed] [Google Scholar]
- 45.Bu Y., Lu C., Bian C., Wang J., Li J., Zhang B., et al. Knockdown of Dicer in MCF-7 human breast carcinoma cells results in G1 arrest and increased sensitivity to cisplatin. Oncology Reports. 2009;21(1):13–17. [PubMed] [Google Scholar]
- 46.Drake M., Furuta T., Suen K.M., Gonzalez G., Liu B., Kalia A., et al. A requirement for ERK-dependent Dicer phosphorylation in coordinating oocyte-to-embryo transition in C. elegans. Developmental Cell. 2014;31(5):614–628. doi: 10.1016/j.devcel.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.