Abstract
The NfrA protein, an oxidoreductase from the soil bacterium Bacillus subtilis, is synthesized during the stationary phase and in response to heat. Analysis of promoter mutants revealed that the nfrA gene belongs to the class III heat shock genes in B. subtilis. An approximate 10-fold induction at both the transcriptional and the translational levels was found after thermal upshock. This induction resulted from enhanced synthesis of mRNA. Genetic and Northern blot analyses revealed that nfrA and the gene downstream of nfrA are transcribed as a bicistronic transcriptional unit. The unstable full-length transcript is processed into two short transcripts encoding nfrA and ywcH. The nfrA-ywcH operon is not induced by salt stress or by ethanol. According to previously published data, the transcription of class III genes in general is activated in response to the addition of these stressors. However, this conclusion is based on experiments which lacked a valid control. Therefore, it seems possible that the transcription of all class III genes is specifically induced by heat shock.
The continuous demand of Bacillus subtilis to adapt to ever-changing conditions in its natural environment has forced the generation of complex regulatory mechanisms governing the transcription of stress-specific proteins. Stress-inducible genes from B. subtilis in general are subdivided into three groups (17, 18). Class I genes are specifically induced by heat stress (17). The well-known chaperonins GroEL, GroES, DnaK, DnaJ, and GrpE are encoded by genes belonging to this group (30, 41, 48, 54, 55). The transcription of the respective genes is regulated by HrcA, a transcription repressor which binds to the CIRCE element (43, 56, 57, 59). Genes transcribed in a ςB-dependent manner constitute class II stress-responsive genes (17, 18). ςB activity is triggered by different kinds of stress and by starvation (5, 7–9). Members of the last group of stress-induced genes, class III, are induced not by starvation but by several different stressful conditions. The transcription of class III genes is neither repressed by HrcA nor solely dependent on ςB. The regulator of the clpC operon, which encodes class III proteins, is known (11, 27). This operon is transcribed by the activity of RNA polymerases containing ςB and ςA (28). Nevertheless, transcription is not induced at the onset of the stationary phase (28), likely because of the activity of this regulator (11, 27).
Some of the stress-responsive proteins are regulated by two transcription factors. clpC, dps, trxA, opuE, and clpP (1, 4, 15, 29, 40, 46) are transcribed by RNA polymerase containing either ςA or ςB. csbB is under the additional control of ςX (22). The csb40 operon (50) and the yvyD gene (13) are transcribed from ςB and ςH promoters, respectively. This genetic organization enables the bacterial cell to modulate the regulation of the respective genes in response to additional challenges.
In this communication, we describe the transcriptional regulation of the nfrA-ywcH operon encoding an oxidoreductase (34, 58) and a putative monooxygenase. It has been shown that nfrA transcription is induced in a ςD-dependent manner at the onset of the stationary phase (34). nfrA transcription is also induced by heat stress from a ςA-dependent promoter overlapping the ςD promoter. The −35 region and the region upstream of the promoter are necessary for this regulation. Ethanol stress and salt stress do not induce nfrA transcription. We discuss the unusual induction pattern of this promoter.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used throughout this study are listed in Table 1. To construct pB49, the relevant part of the nfrA promoter region was amplified by PCR. Two primers that are partially identical to sequences of the intergenic region (ipa43Pup: 5′-GTGCAGAGAATTCCACTTTTGAGATCAC-3′; ipa43Pdown: 5′-CACAAAAACCTCCTGATCACTTTTTATC-3′) were used to amplify the promoter region. The PCR product was digested with EcoRI and cloned into EcoRI- and SnaBI-digested pDL (57). The sequences of the resulting fragment and all other fragments obtained by PCR were verified by DNA sequencing.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or relevant markers | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | hsdR17 endA1 recA1 gyrA96 thi relA1 supE44 φ80dlacZΔM15 Δ(lacZ-argF)U169 | Bethesda Research Laboratories |
| B. subtilis 168 | trpC2 | Bacillus Genetic Stock Center |
| B. subtilis DL | trpC2 amyE::(cat bgaB) | 34 |
| B. subtilis DH32M | trpC2 amyE::(cat spoVG::lacZ) | 34 |
| B. subtilis DIPA4 | trpC2 amyE::(cat nfrA-bgaB) | 34 |
| B. subtilis DXYL strains | trpC2 amyE::(cat PnfrA-PxylA–bgaB) | This work |
| B. subtilis B49 | trpC2 amyE::(cat nfrA-bgaB) | This work |
| B. subtilis DIPA4mut | trpC2 amyE::(cat nfrA-bgaB) | This work |
| B. subtilis IPA43 | trpC2 cat nfrA-spoVG::lacZ | This work |
| B. subtilis IPA44 | trpC2 cat ywcH-spoVG::lacZ | This work |
| B. subtilis IPA44E | trpC2 amyE::(cat nfrA-spoVG::lacZ) | This work |
| B. subtilis DIPA44 | trpC2 cat ywcH-bgaB | This work |
| Plasmids | ||
| pDL | bla amyE::(cat bgaB) | 57 |
| pIC20H | bla | 31 |
| pDH32M | bla cat spoVG::lacZ | 24 |
| pMUTIN2 | bla cat spoVG::lacZ | 49 |
| pKL4 | bla cat bgaB | 42 |
| pIPA6 | bla cat nfrA-ywcH | 34 |
| pIPA8 | bla nfrA | This work |
| pIPA11 | bla erm nfrA | 34 |
| pIPA14 | bla ywcH | This work |
| pDIPA4 | bla amyE::(cat nfrA-bgaB) | 34 |
| pB49 | bla amyE::(cat nfrA-bgaB) | This work |
| pDIPA4mut | bla amyE::(cat nfrA-bgaB) | This work |
| pDXYL plasmids | bla amyE::(cat PnfrA-PxylA–bgaB) | This work |
| pIPA43 | bla cat nfrA-spoVG::lacZ | This work |
| pIPA44 | bla cat ywcH-spoVG::lacZ | This work |
| pIPA44E | bla cat nfrA-spoVG::lacZ | This work |
| pDIPA44 | bla cat ywcH-bgaB | This work |
Mutations in the nfrA promoter region were obtained by PCR mutagenesis. To this end, the promoter region present in plasmid pDIPA4 was amplified in a reaction buffer containing manganese and a reduced amount of nucleotides (10) with the primers pDL (5′-GGGTAACTATTGCCGATGATAAGC-3′) and IPA43P (5′-GATTGTGTTATTGATCACAAAAACC-3′). The products were digested with EcoRI and SnaBI and ligated to plasmid pDL (57) hydrolyzed with the same enzymes. On average, 2 out of 100 nucleotides were mutated (data not shown). Portions of the PxylA-PnfrA hybrids were constructed by hybridizing two oligonucleotides with each other and Klenow filling in the protruding ends.
To obtain pDXYL10, pDXYL1035, and pDXYL35, respectively, the oligonucleotides −10EcoRI (5′-CATGAGAATTCGAAAAACTAAAAAAAATATTGAAAATACTGTTTTTTTCGGATATG-3′) and −10SnaBI (5′-GTATCACTTTTTATCATATCCGAAAAAAACAG-3′), −10/35EcoRI (5′-CATGAGAATTCGAAAAACTAAAATCACTTTTGAGATCACTTTTTTTCGG-3′) and −10/35SnaBI (5′-GTATCACTTTTTATCATATCCGAAAAAAAGTGATCTCAAAAGTG-3′), and −35EcoRI (5′-CATGAGAATTCGAAAAACTAAAATCACTTTTGAGATCACTTTTGAGG-3′) and −35SnaBI (5′-GTACTTATTTTAATCTTAAATAACCTCAAAAGTGATCTCAAAAGT-3′) were used. Double-stranded DNA was digested with EcoRI and ligated to EcoRI-SnaBI-digested plasmid pDL. Plasmids pDXYL10Δ2 and pDXYL35Δ2 were constructed by amplifying the inserts of pDXYL10 and pDXYL35 with the primers pDL and DXYL10Δ2 (5′-GTATCACTTTTTATCATATCCGAAAAAAAGTATT-3′) and the primers pDL and DXYL35Δ2 (5′-GTACTTATTTTAATCTTAAATAACCAAAAGTGATCTC-3′), respectively. The products were digested with EcoRI and ligated to EcoRI-SnaBI-digested plasmid pDL. Plasmid pDXYL was constructed by amplifying the xylA promoter region with the primers xylup (5′-GAAAAACTAAAAAAAATATTGAAAATAC-3′) and xyldown (5′-GTACTTATTTTAATCTTAAATAACCTCATC-3′). The fragment was ligated to SnaBI-digested plasmid pDL.
Plasmids pIPA8 and pIPA14 were obtained by cloning the 1.5-kbp NciI fragment or the 1-kbp AflIII fragment, respectively, into the SmaI-digested pIC20H (31). The NciI fragment encodes nfrA, and the AflIII fragment encodes the intergenic region between nfrA and ywcH and the 5′ part of ywcH. pIPA43 was constructed by amplifying the 5′ end of nfrA. pIPA44 was obtained by amplifying the 3′ end of nfrA, the intergenic region, and the 5′ end of ywcH. Internal sequences of nfrA were amplified with the primers SDIPA-Eco (5′-ACGAATTCTAAGGAGGTTTTTGTGATGAAT-3′; identical to positions 3911038 to 3911017 in reference 36) and IPA62-Sac (5′-TAATCCGCGGACAGCTCACGTTTTTTC-3′; identical to positions 3910834 to 3910858 in reference 36). The fragment used to clone pIPA44 was constructed by amplifying chromosomal DNA with the primers YWCH7 (5′-GGGGATCAGGAATTCGATGAGGATGAGG-3′; identical to positions 3910135 to 3910123 in reference 36) and YWCH8 (5′-AGCACTGTACCGCGGCAGCATGACTCC-3′; identical to positions 3930848 to 3909874 in reference 36). Both products of amplification were digested with EcoRI and SacII and cloned into pMUTIN2 (49) digested with the same enzymes. Plasmid pIPA44E was constructed by amplifying chromosomal DNA with the primers YWCH5 (5′-TTCGGAATGAATTCGACAAGGTG-3′; identical to positions 3910610 to 3910588 in reference 36) and YWCH6 (5′-TCCCTACCACGGATCCTCATCGTA-3′; identical to positions 3910099 to 3910123 in reference 36). The fragment was digested with EcoRI and BamHI and ligated to plasmid pDH32M (24) hydrolyzed with the same enzymes. Plasmid pDIPA44 was constructed by hydrolyzing pIPA6 (34) with XhoI. The 1,275-bp fragment encoding the 3′ end of nfrA, the intergenic region, and the 5′ end of ywcH was cloned into XhoI-hydrolyzed pKL4 (42).
RNA analysis.
Cells were grown as described below. RNA was isolated using an RNeasy kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. To obtain a probe for the S1 nuclease assay, we hydrolyzed plasmid pIPA8 with NcoI and labeled it with [α-32P]dATP. The radioactive fragment was digested with NruI, and a 912-bp fragment was isolated. S1 nuclease analysis was performed as described elsewhere (37) using limiting amounts of nuclease. Northern blotting was done as described previously (2). Equal amounts of RNA were loaded in each lane. As a probe, we used either (i) an internal fragment of nfrA obtained by PCR amplification with plasmid pIPA11 (34) as a template and the primers “primer I” and “primer II” described previously (34) or (ii) an internal fragment of ywcH created by amplification of the plasmid pIPA14 insert with the universal and reverse primers (Pharmacia, Freiburg, Germany). To determine the sizes of the respective fragments, the signals were compared with the RNA size marker of United States Biochemicals (Bad Homburg, Germany). All experiments were reproduced at least once.
General methods.
Unless indicated otherwise, bacteria were grown at 37°C. Nutrient broth (NB) (Oxoid, Basingstoke, United Kingdom) was used as a growth medium for B. subtilis, and Luria broth (39) was used for Escherichia coli. β-Galactosidase assays were performed as described previously (42). Activity is reported in Miller units (33). Each experiment was reproduced at least three times with independent transformants. Only the results of a single experiment are presented. Standard methods were carried out as described previously (39). Western blot analysis was done as described previously (34). Equal amounts of protein were loaded in each lane.
RESULTS
nfrA transcription is induced specifically by heat.
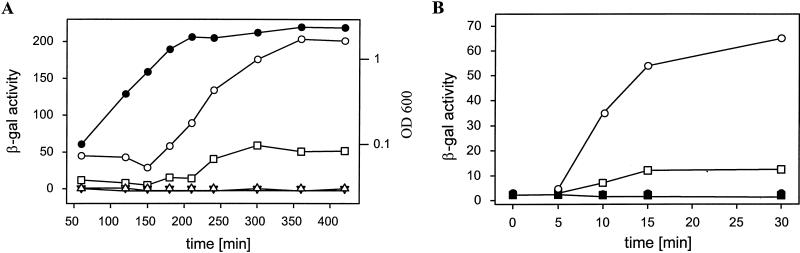
While screening for salt stress-induced genes, we isolated the nfrA gene (data not shown). We wanted to test whether NfrA is a general stress protein or is induced by specific kinds of stress. To do so, we created a fusion of PnfrA to bgaB, encoding a β-galactosidase which is not degraded in response to stress (42, 57). The resulting plasmid was integrated into the B. subtilis 168 chromosome to yield B. subtilis DIPA4 (34). This strain was grown to the early mid-log phase in NB. Stress was applied either by shifting part of the culture to 49°C or by adding ethanol or NaCl to a final concentration of 5% or 0.5 M, respectively. PnfrA-dependent BgaB activity was induced about 10-fold by heat shock, but no significant induction was obtained with ethanol or salt (Fig. 1A). Identical results were obtained using MOPSO minimal medium (26) with succinate as a C source (data not shown). Our original test for salt stress induction was based on direct quantification of mRNA as described previously (29). Obviously, the results obtained with gene fusions differed from these original data. This difference is discussed below.
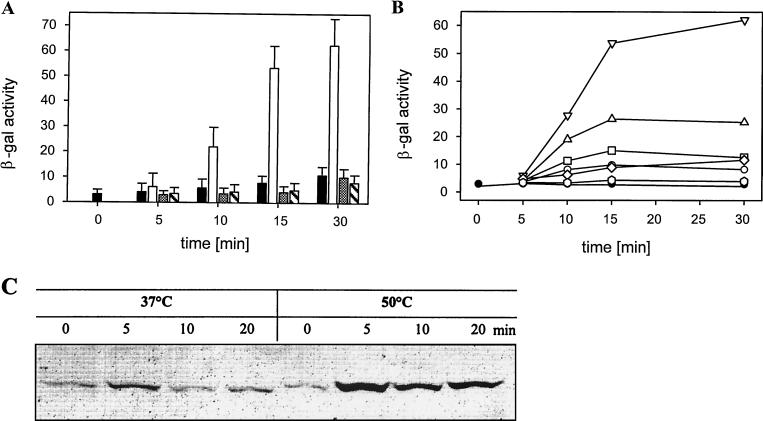
FIG. 1.
(A) Induction of PnfrA by different kinds of stress. β-Galactosidase (β-gal) expression in response to different stressors was determined. β-Galactosidase activity was plotted versus time. Filled bars, control at 37°C; empty bars, heat-shocked cells; shaded bars, salt-stressed cells; hatched bars, cells grown in the presence of 5% ethanol. Samples were collected at different times after stress was applied. All experiments were done in triplicate. (B) Influence of temperature on the heat shock response. Cells were grown at 37°C before an aliquot was shifted to a new temperature. Samples were collected at different times after stress was applied. β-Galactosidase activity was plotted versus time. Filled circles, control at 37°C; empty circles, 45°C; squares, 47°C; triangles, 49°C; inverted triangles, 51°C; diamonds, 53°C; hexagons, 55°C. (C) Western blot analysis of NfrA synthesis. Equal amounts of cell extracts were subjected to Western blot analysis. Samples were taken from stressed and unstressed cells at different times after stress was applied.
To analyze the heat induction in more detail, we grew B. subtilis DIPA4 in NB and induced heat shock by incubating part of the culture at an elevated temperature. Samples were removed at short intervals, and PnfrA-dependent β-galactosidase activity was determined. The patterns of induction of promoter activity were similar at all temperatures up to 51°C, but the maximal activity was modulated in response to the severity of the heat shock. At temperatures higher than 51°C, induction was abolished (Fig. 1B).
The transcriptional start site of the nfrA transcript was mapped for unstressed cells (34). The same experiment was performed with RNA isolated from stressed cells. The start site of the transcript did not change after thermal upshock (data not shown).
To test whether the induction of transcription results in an elevated level of NfrA protein, we grew B. subtilis 168 to the early exponential phase and applied heat shock. Crude protein was isolated. Western blot analysis revealed that the amount of NfrA was increased severalfold in response to the shock (Fig. 1C).
The heat response of nfrA is mediated at the transcriptional level.
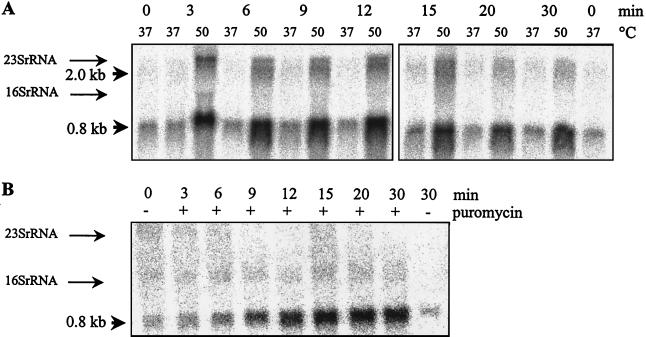
To test whether the increase in PnfrA-dependent β-galactosidase activity in response to heat shock is in fact due to an induction of PnfrA activity, we directly determined the amount of the nfrA transcript. B. subtilis DIPA4 was grown to the early log phase at 37°C and then shifted to 50°C. Cells were harvested before and at intervals after stress was applied. RNA was isolated, and equal amounts of RNA were used for Northern blot analysis. As shown in Fig. 2A two bands were obtained with a nfrA-specific probe. The first one was 0.8 kb, and the second one was 2.0 kb. Within the first 9 min after heat shock, the intensities of both bands increased about 10-fold. This result is in accordance with the results obtained with the PnfrA-bgaB fusion. The amount of specific mRNA started to decline 15 min after heat shock was applied. The addition of NaCl or ethanol had no significant effect on the nfrA mRNA amount (data not shown). In addition, we tested the effects of puromycin addition on PnfrA activity. B. subtilis DIPA4 was grown to an optical density at 600 nm (OD600) of 0.2, puromycin was added to a final concentration of 20 μg/ml, and samples were removed at intervals. RNA was isolated and used for analysis. There was a steady increase in the nfrA mRNA amount (Fig. 2B).
FIG. 2.
Northern blot analysis of nfrA transcription. Heat stress was applied. At the indicated times, cells were harvested and RNA was isolated. Equal amounts of RNA were used for the analysis. The positions of the 23S and 16S rRNAs are indicated. (A) RNA from heat-shocked cells. Time after heat shock and growth temperature are indicated. Different bands of 0.8 and 2 kb are visible. (B) RNA from puromycin-treated cells. Puromycin (20 μg/ml) (+) was added to the cell suspension; −, no puromycin. Cells were harvested at the indicated times, and RNA was isolated.
Degradation of the nfrA transcript is influenced by heat shock.
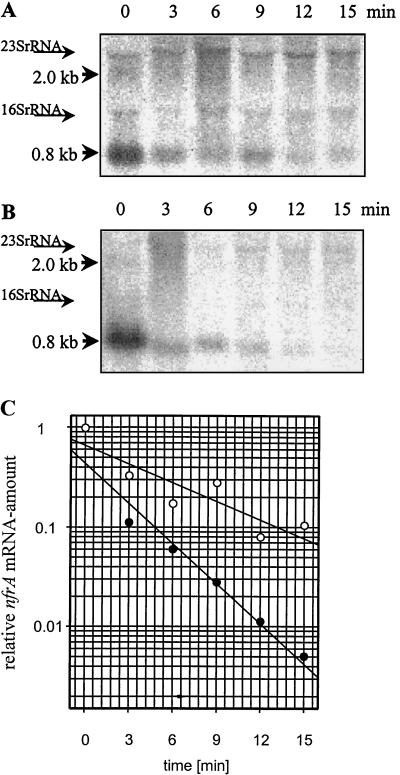
The amount of mRNA is influenced by two different parameters, the rate of RNA synthesis and the rate of RNA decay. Therefore, we wanted to test whether the stability of the nfrA mRNA is influenced by heat shock. We grew B. subtilis DIPA4 to an OD600 of 0.25 and subjected part of the culture to heat shock; the rest was allowed to grow at 37°C. Rifampin (10-μg/ml final concentration) was added to both suspensions 5 min after heat stress was applied. Samples were removed at intervals, and RNA was isolated. Northern blot analysis of both the stressed and the unstressed samples revealed that there was a modest influence of heat shock on the half-life of the nfrA mRNA. To determine this influence, the data were analyzed using the regression function of the SigmaPlot program (SPSS Inc., Chicago, Ill.). The half-life of the 0.8-kb mRNA at 37°C was about 4 min and the half-life at 50°C was about 2 min (Fig. 3). We were not able to exactly determine the half-life of the 2.0-kb mRNA because this mRNA vanished immediately after the addition of rifampin. Therefore, we conclude that the half-life of this mRNA was less than 1 min.
FIG. 3.
Decay of nfrA mRNA before and after heat shock. RNA was isolated at different times after stress was applied. (A) Decay of the nfrA mRNA at 37°C. (B) Decay of the nfrA mRNA at 50°C. (C) Decay curves for the nfrA mRNAs at 37°C (empty circles) and 50°C (filled circles). The amount of RNA at time zero was set to 1 for both graphs. Times after the addition of rifampin are given.
Mutations in the putative ςA promoter influence PnfrA activity.
Besides a putative ςA consensus sequence, the region upstream of the mRNA 5′ end includes sequences with homology to the ςB and ςD consensus sequences. Transcription of nfrA during the stationary phase is ςD dependent (34). To test which sigma factor is responsible for the heat shock induction of nfrA, we modified the sequence of the promoter region cloned into pDIPA4. First, we constructed pB49, a derivative of the promoter without the region upstream of the −35 region. The BgaB activity of the resulting B. subtilis strain, B49, was decreased; induction by heat shock was also reduced but was not abolished. These results indicate that the upstream region is necessary for full promoter activity and for some of the regulation. Therefore, we decided to modify the sequence of pDIPA4 (34) by nonspecific PCR mutagenesis (10).
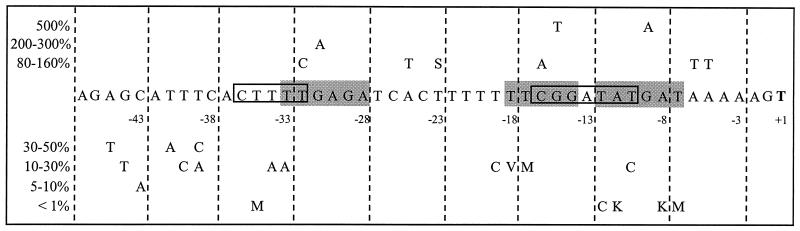
Plasmids were isolated and sequenced, and mutant plasmids were integrated into the B. subtilis 168 chromosome. The activities of different promoter variants are depicted in Fig. 4. Most mutations had only a modest influence on promoter activity. Only two positions of the proposed nfrA promoter deviated from the ςA consensus sequence. All mutations which caused a complete loss of promoter activity were either mutations within the −10 or −35 region of the putative ςA promoter or deletions in the spacer between both regions. A mutation upstream of the −35 region, C-43A, also had a severe negative effect on promoter activity. Mutations within the long poly(dT) blocks in the spacer between the −10 and −35 regions or the block overlapping the −35 region caused a 3- to 10-fold reduction in promoter activity. The same was true for two additional mutations upstream of the −35 region, T-40C and G-44T, respectively. Thirteen out of 41 mutations had only a slight effect on promoter activity. Two mutations within the putative −35 region, T-32C and G-31A, had no effect or caused even a slight increase in promoter activity. One point mutation resulted in elevated promoter activity. This mutation, G-15T, lies within the −16 region, a sequence which enhances the activity of several ςA promoters (19, 47). All promoter derivatives with significant activity during exponential growth were induced in response to heat shock.
FIG. 4.
Sequence of the PnfrA promoter region. The putative −10, −16, and −35 regions are indicated by gray backgrounds. The ςD promoter (34) is indicated by boxes. Only the effects of single mutations within this region are shown. Mutations resulting in elevated promoter activity are shown above the wild-type sequence; mutations resulting in reduced transcriptional activity are shown below. M, A or C; S, G or C; K, G or T; V, A, G, or C.
Additionally, we created B. subtilis DIPA4 derivatives harboring mutations in genes encoding putative sigma factors (36) or known regulators of stress-responsive genes (11, 27, 43, 56). We inactivated sigB, sigD, sigM, sigV, sigW, sigX, sigY, sigZ, ylaC, ykoZ, hrcA, and ctsR. None of these mutations had an effect on the heat shock induction of PnfrA (data not shown).
The −35 and upstream regions are important for heat induction.
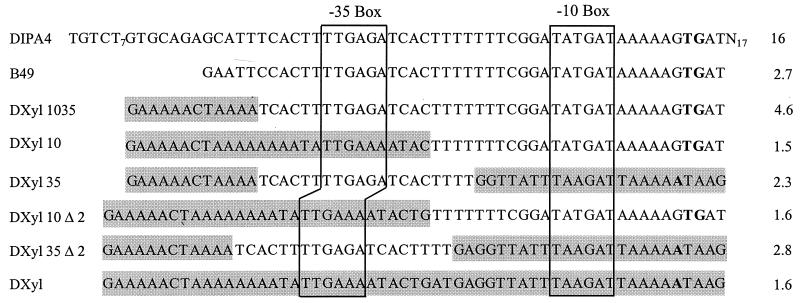
We were unable to find mutations which specifically abolished the heat shock induction of PnfrA. Our assumption was that the DNA sequences responsible for promoter activity and regulation overlap each other. Therefore, we created derivatives of the nfrA promoter by exchanging parts of this promoter with homologous parts of the B. subtilis xylA promoter (14). As described above, the upstream region influences promoter activity and heat shock induction. The sequence upstream of PxylA, which is necessary for the full activity of this promoter (25), had no positive effect on the nfrA promoter (Fig. 5). The hybrid promoter was still induced by heat shock, albeit at a reduced level. The same was true for all promoter derivatives containing the −35 region of the nfrA promoter. The −10 region of PnfrA was insufficient for heat shock induction.
FIG. 5.
Regulation of PnfrA-PxylA hybrids. The sequences of the respective promoter variants are shown. Sequences derived from PxylA are shown by a shaded background. The promoters were cloned into plasmid pDL; the resulting plasmids were integrated into the amyE gene of B. subtilis 168 to give the respective B. subtilis strains. BgaB activity was determined with cell extracts from unstressed and heat-shocked cells. The induction ratios are shown at the right. The predicted transcriptional start points of the respective RNAs are indicated by bold letters. The uninduced activities (in Miller units) of the respective promoter derivatives were as follows: DIPA4, 3 ± 0.5 U; B49, 1 ± 0.1 U; DXyl1035, 0.6 ± 0.1 U; DXyl10, 62 ± 6; DXyl35, 0.6 ± 0.2 U; DXyl10Δ2, 1 ± 0.1 U; DXyl35Δ2, 0.5 ± 0.1 U; and DXyl, 62 ± 7 U. The activity of the control, B. subtilis DL, was 0.2 ± 0.1 U (not shown).
nfrA and ywcH are transcribed as an operon.
NfrA is a flavin mononucleotide-containing oxidoreductase (34). The gene downstream of nfrA, ywcH, bears homology with a monooxygenase gene. Therefore, it seemed possible that NfrA is the electron donor for YwcH. However, the two genes are separated by an intergenic region of 177 bp containing a sequence proposed to be a transcriptional terminator (36). In order to determine whether the regulation of transcription of ywcH and nfrA is similar, we created two plasmids, pIPA44E and pIPA44, by cloning the region encoding the C-terminal part of NfrA, the intergenic region between nfrA and ywcH, and the region encoding the N-terminal part of YwcH into plasmids pDH32M (24) and pMUTIN2 (49), respectively. As a control, we created plasmid pIPA43 by cloning the region encoding the 5′ end of nfrA into plasmid pMUTIN2. pIPA44E was integrated into the amyE gene of B. subtilis 168, whereas pIPA43 and pIPA44 were integrated into the nfrA and ywcH genes of B. subtilis 168. Therefore, in B. subtilis IPA44E, β-galactosidase synthesis is controlled by promoters encoded by the fragment cloned into pDH32M, whereas in B. subtilis IPA43 and B. subtilis IPA44, β-galactosidase expression is controlled by all promoters within or upstream from the cloned fragment. The strains were grown in NB supplemented with glucose and glutamate. As described previously (34), the PnfrA-dependent β-galactosidase activity of B. subtilis IPA43 commenced at the onset of the stationary phase (Fig. 6A). The same regulatory pattern was obtained for B. subtilis IPA44, but the activity was fourfold lower. We were not able to detect β-galactosidase activity above the background in B. subtilis IPA44E.
FIG. 6.
Comparison of nfrA and ywcH induction. (A) Induction of transcriptional fusions of lacZ to the respective gene during growth in NB supplemented with glucose and glutamate. Filled circles, growth of B. subtilis IPA43, given as OD600 units; open circles, β-galactosidase (β-gal) activity of B. subtilis IPA43 (nfrA-lacZ fusion in nfrA); squares, β-galactosidase activity of B. subtilis IPA44 (ywcH-lacZ fusion in ywcH); inverted triangles, β-galactosidase activity of B. subtilis IPA44E (ywcH-lacZ fusion in amyE); triangles, β-galactosidase activity of B. subtilis DH32M (control). (B) Heat shock induction of nfrA and ywcH. Filled symbols, activity in cells grown at 37°C; empty symbols, activity of stressed cells; circles, B. subtilis DIPA4 (nfrA-bgaB); squares, B. subtilis DIPA44 (ywcH-bgaB).
To investigate whether ywcH transcription is induced in response to heat shock, we created plasmid pDIPA44 by cloning the 1,275-bp XhoI fragment encoding the 3′ end of nfrA, the intergenic region, and the 5′ end of ywcH (36) into plasmid pKL4 (42). The plasmid was integrated into the B. subtilis 168 chromosome. Heat shock induction was determined as described above. BgaB activity was induced, but the activity was fivefold lower than the activity in B. subtilis DIPA4 (Fig. 6B).
With these results taken together, we were able to prove that the promoter driving β-galactosidase expression in B. subtilis IPA44 was not within the intergenic region between nfrA and ywcH but was upstream of the proposed transcriptional terminator. nfrA and ywcH were transcribed as a transcription unit.
The nfrA-ywcH mRNA is processed after transcription.
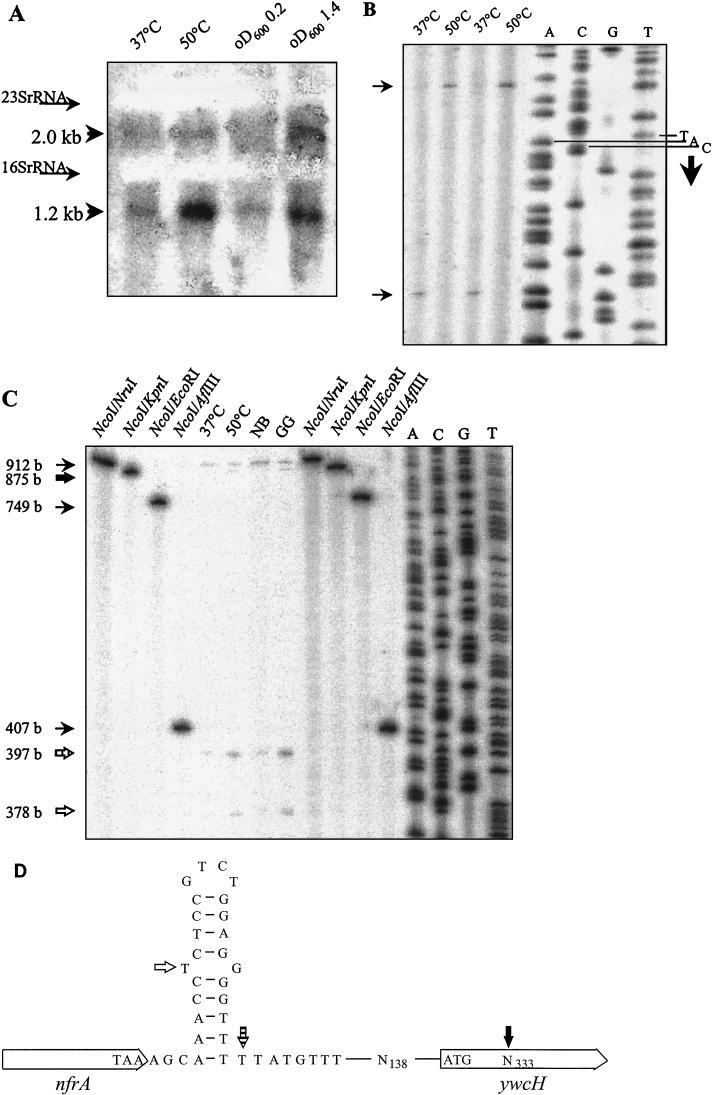
The results described above indicate that nfrA and ywcH are transcribed from a single promoter. We isolated RNA from exponentially growing B. subtilis 168, from stationary-phase cells grown in NB supplemented with glucose and glutamate (34), and from cells subjected to heat shock. Northern blot analysis was carried out using a ywcH-specific probe. The result of the experiment is shown in Fig. 7A. Two bands were obtained—a dominant band of 1.2 kb and a weaker band of 2.0 kb. Both heat shock and stationary phase resulted in an increase in the ywcH-specific RNA amount. The length of the shorter transcript corresponded to the length of the ywcH gene. Therefore, we assumed that the transcript starts in front of the ywcH gene, although we were unable to map promoter activity in that region. To test this notion, we performed a primer extension experiment. Depending on the conditions used to grow the bacteria, two different mRNA 5′ ends were mapped (Fig. 7B). mRNA isolated from heat-shocked bacteria started 11 bases in front of the ywcH gene; mRNA isolated from unstressed bacteria started 25 bases downstream of the putative translational start codon. Additionally, we mapped the 3′ end of the nfrA mRNA by S1 nuclease analysis. The 3′ part of the probe used for this experiment is complementary to the B. subtilis chromosome, whereas the 5′ end is unable to hybridize to any mRNA from B. subtilis. Therefore, we were able to detect transcripts terminating within the intergenic region and transcripts overlapping the entire intergenic region. We obtained a dominant signal just downstream of the stem-loop structure and a second, eightfold-weaker signal corresponding to the end of the homologous part of the probe (Fig. 7C).
FIG. 7.
(A) Northern blot analysis of ywcH transcription. RNA was isolated from cells before (37°C) and after (50°C) heat shock or during the exponential (OD600 = 0.2) and stationary (OD600 = 1.4) growth phases. Equal amounts of RNA were used for the analysis. The positions of the 23S and 16S rRNAs are indicated. Two bands specifically hybridizing to a ywcH probe were obtained. (B) Mapping of the ywcH 5′ end. RNA was isolated from B. subtilis 168 grown at the indicated temperatures. The sequence ladder used as a size marker was obtained by using the same primer as that used for the primer extension reaction. The putative start codon of translation is indicated (TAC). Whereas the 5′ end of the RNA is upstream from the putative translational start codon in heat-shocked cells, it is downstream from this codon in unstressed cells (arrows at left). (C) S1 nuclease mapping of the 3′ end of nfrA. RNA was isolated from cells before (37°C) and after (50°C) heat shock or from stationary-phase cells grown in NB or NB with glucose and glutamate (GG). Equal amounts of RNA were used for the analysis. Restriction fragments of known lengths and a sequence ladder of a known sequence were used as size markers. Four different signals were obtained in the S1 nuclease reaction. The 912-base fragment was derived from an unprocessed probe. The 875-base fragment (large filled arrow) was obtained by S1 nuclease processing of a probe hybridized to an unprocessed nfrA-ywcH transcript. The 397-base fragment (hatched arrow) was processed at the 5′ end of the stem-loop structure. The 378-base fragment (empty arrow) was processed at the single mismatch within the stem-loop structure. (D) Graphic illustration of the results obtained by S1 nuclease mapping and primer extension. The labeling of the arrows is like that in panel C. The 875-base fragment is not due to RNA processing but is specific for the strategy of the experiment. It indicates that readthrough occurs.
DISCUSSION
nfrA and ywcH are transcribed as an operon.
nfrA and ywcH are separated by a sequence generally used as a terminator (36). To our surprise, nfrA and ywcH nevertheless constitute an operon. The stem-loop structure may nevertheless serve as a terminator. The activity of the transcriptional fusions of ywcH to bgaB and lacZ was four- to fivefold lower than that of the respective nfrA fusions. The existence of a putative terminator within an operon was also described for the heat shock-induced dnaK operon (20, 55). We do not know whether the unusual organization of both operons serves a specific purpose. In Agrobacterium tumefaciens, the mRNA encoding the genes groES and groEL is specifically cleaved between the two genes in response to heat shock, and two monocistronic mRNAs are formed. The polycistronic mRNA is the major mRNA at a low temperature (45). This situation allows differential expression of the two proteins under stress and nonstress conditions. The processing of the full-length nfrA-ywcH transcript produces two different 5′ ends. This finding indicates the existence of additional translational control of YwcH synthesis.
Characterization of PnfrA.
The genes induced in response to different kinds of stress have been subdivided into three groups (17, 18). Until now, no detailed promoter analysis has been undertaken for class III genes. Based on sequence similarities of the regions upstream of the mapped 5′ ends of the respective mRNAs to the consensus sequence, it was proposed that these genes are transcribed by ςA-containing RNA polymerase. Recently, a new group of genes important for survival at high temperatures was described (21, 22). These genes are preceded by promoters recognized by the alternative sigma factor ςX. nfrA transcription is specifically induced by heat. Inactivation of genes encoding putative sigma factors had no effect on the heat induction of nfrA transcription. Additionally, we mutagenized the promoter region. All mutations causing a complete loss of promoter activity resided within the important parts of the putative ςA promoter. Two mutations with strongly increased activity were obtained. Both exchanges resulted in promoter derivatives fitting the ςA consensus sequence better than the wild type does. Two mutations, T-32C and G-31A, do not fit into the emerging picture. Both are mutations in the putative −35 region but have no negative effect on promoter activity. Therefore, we think that it was not possible to prove unambiguously that PnfrA is used in a ςA-dependent manner.
PnfrA regulation is not dependent on known regulators (11, 27, 43, 56) of stress-responsive genes. It is induced by the addition of puromycin. According to the definition of class III genes (17, 18), nfrA and ywcH belong to this group of stress-responsive genes. Only recently was the creation of a new class of stress-responsive genes proposed (11). Genes whose transcription is regulated by a novel regulator of the stress and heat shock response, CtsR, constitute class III genes according to this classification. According to this new definition, nfrA and ywcH are class IV stress-responsive genes.
nfrA-ywcH transcription is specifically induced by heat.
Originally, the nfrA gene was isolated when we screened for salt stress-induced genes (O. Krispin, E. Gaul, and R. Allmansberger, unpublished data). The screen was based on direct quantification of mRNA amounts using the xynA mRNA (12, 29, 52) as a control. The unambiguous result of this experiment was that the amount of the nfrA transcript is increased in response to salt stress. To our surprise, we were unable to reproduce the results obtained by RNA dot blotting with a PnfrA-bgaB fusion. It turned out that, in contrast to a statement made previously (29), the amount of the xynA transcript is severely influenced by different kinds of stress (2). The alleged increase in nfrA mRNA was in fact a reduction of the amount of the control.
Because it is not possible to use the LacZ protein to determine heat shock induction in B. subtilis (5), a considerable amount of data concerning the stress response in B. subtilis has been collected using RNA dot blots as a method to quantify the induction of the respective genes. The control used for the first set of these experiments (12, 29, 51) is obviously invalid (2; O. Krispin and R. Allmansberger, unpublished data). Therefore, it is very likely that the interpretation of the data presented in the mentioned publications was incorrect (see below). It is indubitable that there was a relative increase in the synthesis of the respective proteins, because most of the genes were identified by sequencing of proteins whose synthesis increased in response to different kinds of stress (6, 51). However, an increase in the relative synthesis of a protein does not prove that transcription of the encoding gene is induced. For example, the same result is obtained when stress conditions reduce the translation of most other genes. The few proteins with unchanged translational efficiency would cause stronger signals on two-dimensional gels. This general effect seems possible because stress obviously reduces the mRNA stability of some genes whose stability is not influenced by specific structures (2, 57). In contrast, the half-lives of mRNAs encoding stress-induced genes are almost unaltered in response to stress (23, 57).
In response to the finding that the amount of xynA mRNA is severely influenced by stress, this control was not used any longer. Instead, mRNA quantifications were performed without an internal control (3, 5, 12, 32, 38, 40). RNA dot blotting is a multistep experiment. For example, it is a difficult task to obtain RNA preparations of identical quality. Therefore, we consider a valid internal control to be a prerequisite for accurate quantifications of RNA amounts, an opinion which is supported by the fact that the same authors who indicated that clpC transcription is induced by salt stress (29) reported in a recent publication that the regulator of clpC transcription is not induced by salt stress (27). The induction of all class III general stress proteins activated by several stressors was determined by RNA dot blotting. Two class III genes, htpG (44) and nfrA, are induced by heat shock only. Induction of these genes was determined by measuring the induction of bgaB fusions to the respective promoters. Therefore, it is our suspicion that the method used to determine stress induction in B. subtilis influences the results and the interpretation of the experiments considerably. In addition, salt stress and ethanol induction of most class III genes is rather low (3, 12, 15, 16, 29, 38, 40). It seems necessary to reproduce experiments using a different method or a valid control to determine stress induction.
Transcription of class I and several class III genes is induced by the addition of the antibiotic puromycin (29, 35, 38, 40). Class II genes are not induced by the addition of this antibiotic. It was proposed that this difference allows the conclusion that the signals for the induction of class I and class II genes are different (35). While the amount of the nfrA transcript is elevated in response to puromycin treatment, the mechanism responsible for this increase is unclear. We believe that the fact that the amounts of transcripts from class II genes are not elevated in response to puromycin treatment is insufficient to conclude the existence of different induction mechanisms for class II genes and the rest of the stress-responsive genes. The expression of class II genes depends on the activity of the gene product encoded by sigB. Transcription of the sigB gene itself is induced in response to stress (5, 7, 53). As a consequence, the amount of ςB increases and the response is amplified. It is likely that this induction of ςB synthesis is necessary to obtain strong induction of the whole regulon. Even if puromycin addition induces ςB activity, the induction of the transcription of ςB-dependent genes will be diminished, because the majority of this induction is brought about by de novo synthesis of ςB itself, which is blocked by puromycin.
The regulatory region and the promoter overlap.
Our results indicate that heat shock-induced nfrA transcription is dependent on RNA polymerase containing ςA. The underlying regulatory mechanism is unknown. Nonspecific mutagenesis did not result in mutants with altered regulation. The original −10 and −35 regions of the promoter are necessary for full heat shock induction. We tried to identify a trans-acting regulatory protein using saturating transposon mutagenesis. We obtained about 105 independent mutants, but we were unable to identify a mutant which changed the regulation of nfrA transcription (data not shown). The nfrA gene promoter for B. subtilis is unusual because the distance between the −10 and −35 regions is rather short. Therefore, it seems possible that the DNA structure itself is important for heat shock induction. For example, heat shock influences DNA supercoiling in B. subtilis (26). However, our results are insufficient to allow a definite conclusion. Additional work is necessary to elucidate the mechanism of nfrA heat shock induction.
ACKNOWLEDGMENTS
We thank W. Hillen for financial support. C.M. was supported by a personal grant from the Evangelischen Studienwerk Villigst e.V., Germany, and O.S. was supported by the DFG.
We also thank K. Oliva for editing the manuscript. We also thank M. Hecker, Greifswald, Germany, for the ctsR-negative B. subtilis strain; J. Helman for the sigX and sigW mutants; and W. Schumann, Bayreuth, Germany, for the hrcA-negative strain.
REFERENCES
- 1.Akbar S, Price C W. Isolation and characterization of csbB, a gene controlled by Bacillus subtilis general stress transcription factor sigma B. Gene. 1996;177:123–128. doi: 10.1016/0378-1119(96)00287-9. [DOI] [PubMed] [Google Scholar]
- 2.Allmansberger R. Degradation of the Bacillus subtilis xynA transcript is accelerated in response to stress. Mol Gen Genet. 1996;251:108–112. doi: 10.1007/BF02174351. [DOI] [PubMed] [Google Scholar]
- 3.Antelmann H, Engelmann S, Schmid R, Hecker M. General and oxidative stress responses in Bacillus subtilis: cloning, expression, and mutation of the alkyl hydroperoxide reductase operon. J Bacteriol. 1996;178:6571–6578. doi: 10.1128/jb.178.22.6571-6578.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antelmann H, Engelmann S, Schmid R, Sorokin A, Lapidus A, Hecker M. Expression of a stress- and starvation-induced dps/pexB-homologous gene is controlled by the alternative sigma factor sigma B in Bacillus subtilis. J Bacteriol. 1997;179:7251–7256. doi: 10.1128/jb.179.23.7251-7256.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benson A K, Haldenwang W G. The sigma B-dependent promoter of the Bacillus subtilis sigB operon is induced by heat shock. J Bacteriol. 1993;175:1929–1935. doi: 10.1128/jb.175.7.1929-1935.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernhardt J, Völker U, Völker A, Antelmann H, Schmid R, Mach H, Hecker M. Specific and general stress proteins in Bacillus subtilis—a two-dimensional protein electrophoresis study. Microbiology. 1997;143:999–1017. doi: 10.1099/00221287-143-3-999. [DOI] [PubMed] [Google Scholar]
- 7.Boylan S A, Redfield A R, Brody M S, Price C W. Stress-induced activation of the sigma B transcription factor of Bacillus subtilis. J Bacteriol. 1993;175:7931–7937. doi: 10.1128/jb.175.24.7931-7937.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boylan S A, Redfield A R, Price C W. Transcription factor sigma B of Bacillus subtilis controls a large stationary-phase regulon. J Bacteriol. 1993;175:3957–3963. doi: 10.1128/jb.175.13.3957-3963.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boylan S A, Rutherford A, Thomas S M, Price C W. Activation of Bacillus subtilis transcription factor sigma B by a regulatory pathway responsive to stationary-phase signals. J Bacteriol. 1992;174:3695–3706. doi: 10.1128/jb.174.11.3695-3706.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cadwell R C, Joyce G F. Mutagenic PCR. In: Dieffenbach C W, Dveksler G D, editors. PCR primer: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1995. pp. 583–589. [Google Scholar]
- 11.Derré I, Rapoport G, Msadek T. CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in Gram-positive bacteria. Mol Microbiol. 1999;31:117–131. doi: 10.1046/j.1365-2958.1999.01152.x. [DOI] [PubMed] [Google Scholar]
- 12.Deuerling E, Päslack B, Schumann W. The ftsH gene of Bacillus subtilis is transiently induced after osmotic and temperature upshift. J Bacteriol. 1995;177:4105–4112. doi: 10.1128/jb.177.14.4105-4112.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drzewiecki K, Eymann C, Mittenhuber G, Hecker M. The yvyD gene of Bacillus subtilis is under dual control of ςB and ςH. J Bacteriol. 1998;180:6674–6680. doi: 10.1128/jb.180.24.6674-6680.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gärtner D, Geissendorfer M, Hillen W. Expression of the Bacillus subtilis xyl operon is repressed at the level of transcription and is induced by xylose. J Bacteriol. 1988;170:3102–3109. doi: 10.1128/jb.170.7.3102-3109.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerth U, Krüger E, Derré I, Msadek T, Hecker M. Stress induction of the Bacillus subtilis clpP gene encoding a homologue of the proteolytic component of the Clp protease and the involvement of ClpP and ClpX in stress tolerance. Mol Microbiol. 1998;28:787–802. doi: 10.1046/j.1365-2958.1998.00840.x. [DOI] [PubMed] [Google Scholar]
- 16.Gerth U, Wipat A, Harwood C R, Carter N, Emmerson P T, Hecker M. Sequence and transcriptional analysis of clpX, a class-III heat-shock gene of Bacillus subtilis. Gene. 1996;181:77–83. doi: 10.1016/s0378-1119(96)00467-2. [DOI] [PubMed] [Google Scholar]
- 17.Hecker M, Schumann W, Völker U. Heat-shock and general stress response in Bacillus subtilis. Mol Microbiol. 1996;19:417–428. doi: 10.1046/j.1365-2958.1996.396932.x. [DOI] [PubMed] [Google Scholar]
- 18.Hecker M, Völker U. Non-specific, general and multiple stress resistance of growth-restricted Bacillus subtilis cells by the expression of the ςB regulon. Mol Microbiol. 1998;29:1129–1136. doi: 10.1046/j.1365-2958.1998.00977.x. [DOI] [PubMed] [Google Scholar]
- 19.Helmann J D. Compilation and analysis of Bacillus subtilis sigma A-dependent promoter sequences: evidence for extended contact between RNA polymerase and upstream promoter DNA. Nucleic Acids Res. 1995;23:2351–2360. doi: 10.1093/nar/23.13.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Homuth G, Masuda S, Mogk A, Kobayashi Y, Schumann W. The dnaK operon of Bacillus subtilis is heptacistronic. J Bacteriol. 1997;179:1153–1164. doi: 10.1128/jb.179.4.1153-1164.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang X, Decatur A, Sorokin A, Helmann J D. The Bacillus subtilis sigma(X) protein is an extracytoplasmic function sigma factor contributing to survival at high temperature. J Bacteriol. 1997;179:2915–2921. doi: 10.1128/jb.179.9.2915-2921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang X, Helmann J D. Identification of target promoters for the Bacillus subtilis sigma X factor using a consensus-directed search. J Mol Biol. 1998;279:165–173. doi: 10.1006/jmbi.1998.1765. [DOI] [PubMed] [Google Scholar]
- 23.Jürgen B, Schweder T, Hecker M. The stability of mRNA from the gsiB gene of Bacillus subtilis is dependent on the presence of a strong ribosome binding site. Mol Gen Genet. 1998;258:538–545. doi: 10.1007/s004380050765. [DOI] [PubMed] [Google Scholar]
- 24.Kraus A, Hueck C, Gärtner D, Hillen W. Catabolite repression of the Bacillus subtilis xyl operon involves a cis element functional in the context of an unrelated sequence, and glucose exerts additional XylR-dependent repression. J Bacteriol. 1994;176:1738–1745. doi: 10.1128/jb.176.6.1738-1745.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kreuzer P, Gärtner D, Allmansberger R, Hillen W. Identification and sequence analysis of the Bacillus subtilis W23 xylR gene and xyl operator. J Bacteriol. 1989;171:3840–3845. doi: 10.1128/jb.171.7.3840-3845.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krispin O, Allmansberger R. Changes in DNA supertwist as a response of Bacillus subtilis towards different kinds of stress. FEMS Microbiol Lett. 1995;134:129–135. doi: 10.1111/j.1574-6968.1995.tb07926.x. [DOI] [PubMed] [Google Scholar]
- 27.Krüger E, Hecker M. The first gene of the Bacillus subtilis clpC operon, ctsR, encodes a negative regulator of its own operon and other class III heat shock genes. J Bacteriol. 1998;180:6681–6688. doi: 10.1128/jb.180.24.6681-6688.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krüger E, Msadek T, Hecker M. Alternate promoters direct stress-induced transcription of the Bacillus subtilis clpC operon. Mol Microbiol. 1996;20:713–723. doi: 10.1111/j.1365-2958.1996.tb02511.x. [DOI] [PubMed] [Google Scholar]
- 29.Krüger E, Völker U, Hecker M. Stress induction of clpC in Bacillus subtilis and its involvement in stress tolerance. J Bacteriol. 1994;176:3360–3367. doi: 10.1128/jb.176.11.3360-3367.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li M, Wong S L. Cloning and characterization of the groESL operon from Bacillus subtilis. J Bacteriol. 1992;174:3981–3992. doi: 10.1128/jb.174.12.3981-3992.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marsh J L, Erfle M, Wykes E J. The pIC plasmid and phage vectors with versatile cloning sites for recombinant selection by insertional inactivation. Gene. 1984;32:481–485. doi: 10.1016/0378-1119(84)90022-2. [DOI] [PubMed] [Google Scholar]
- 32.Maul B, Völker U, Riethdorf S, Engelmann S, Hecker M. Sigma B-dependent regulation of gsiB in response to multiple stimuli in Bacillus subtilis. Mol Gen Genet. 1995;248:114–120. doi: 10.1007/BF02456620. [DOI] [PubMed] [Google Scholar]
- 33.Miller J. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 34.Moch C, Schrögel O, Allmansberger R. The ςD-dependent transcription of the ywcG gene from Bacillus subtilis is dependent on an excess of glucose and glutamate. Mol Microbiol. 1998;27:889–898. doi: 10.1046/j.1365-2958.1998.00734.x. [DOI] [PubMed] [Google Scholar]
- 35.Mogk A, Völker A, Engelmann S, Hecker M, Schumann W, Völker U. Nonnative proteins induce expression of the Bacillus subtilis CIRCE regulon. J Bacteriol. 1998;180:2895–2900. doi: 10.1128/jb.180.11.2895-2900.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moszer I, Glaser P, Danchin A. SubtiList: a relational database for the Bacillus subtilis genome. Microbiology. 1995;141:261–268. doi: 10.1099/13500872-141-2-261. [DOI] [PubMed] [Google Scholar]
- 37.Müller B, Allmansberger R, Klein A. Termination of a transcription unit comprising highly expressed genes in the archaebacterium Methanococcus voltae. Nucleic Acids Res. 1985;13:6439–6445. doi: 10.1093/nar/13.18.6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riethdorf S, Völker U, Gerth U, Winkler A, Engelmann S, Hecker M. Cloning, nucleotide sequence, and expression of the Bacillus subtilis lon gene. J Bacteriol. 1994;176:6518–6527. doi: 10.1128/jb.176.21.6518-6527.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Scharf C, Riethdorf S, Ernst H, Engelmann S, Völker U, Hecker M. Thioredoxin is an essential protein induced by multiple stresses in Bacillus subtilis. J Bacteriol. 1998;180:1869–1877. doi: 10.1128/jb.180.7.1869-1877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmidt A, Schiesswohl M, Völker U, Hecker M, Schumann W. Cloning, sequencing, mapping, and transcriptional analysis of the groESL operon from Bacillus subtilis. J Bacteriol. 1992;174:3993–3999. doi: 10.1128/jb.174.12.3993-3999.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schrögel O, Allmansberger R. Optimisation of the BgaB reporter system: determination of transcriptional regulation of stress responsive genes in Bacillus subtilis. FEMS Microbiol Lett. 1997;153:237–243. doi: 10.1111/j.1574-6968.1997.tb10488.x. [DOI] [PubMed] [Google Scholar]
- 43.Schulz A, Schumann W. hrcA, the first gene of the Bacillus subtilis dnaK operon, encodes a negative regulator of class I heat shock genes. J Bacteriol. 1996;178:1088–1093. doi: 10.1128/jb.178.4.1088-1093.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schulz A, Schwab S, Homuth G, Versteeg S, Schumann W. The htpG gene of Bacillus subtilis belongs to class III heat shock genes and is under negative control. J Bacteriol. 1998;180:3103–3109. doi: 10.1128/jb.179.10.3103-3109.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Segal G, Ron E Z. The groESL operon of Agrobacterium tumefaciens: evidence for heat shock-dependent mRNA cleavage. J Bacteriol. 1995;177:750–757. doi: 10.1128/jb.177.3.750-757.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spiegelhalter F, Bremer E. Osmoregulation of the opuE proline transport gene from Bacillus subtilis: contributions of the sigma A- and sigma B-dependent stress-responsive promoters. Mol Microbiol. 1998;29:285–296. doi: 10.1046/j.1365-2958.1998.00929.x. [DOI] [PubMed] [Google Scholar]
- 47.Tatti K M, Moran C P., Jr Promoter recognition by sigma-37 RNA polymerase from Bacillus subtilis. J Mol Biol. 1984;175:285–297. doi: 10.1016/0022-2836(84)90349-8. [DOI] [PubMed] [Google Scholar]
- 48.Tozawa Y, Yoshikawa H, Kawamura F, Itaya M, Takahashi H. Isolation and characterization of the groES and groEL genes of Bacillus subtilis Marburg. Biosci Biotechnol Biochem. 1992;56:1995–2002. doi: 10.1271/bbb.56.1995. [DOI] [PubMed] [Google Scholar]
- 49.Vagner V, Dervyn E, Ehrlich S D. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology. 1998;144:3097–3104. doi: 10.1099/00221287-144-11-3097. [DOI] [PubMed] [Google Scholar]
- 50.Varon D, Brody M S, Price C W. Bacillus subtilis operon under the dual control of the general stress transcription factor sigma B and the sporulation transcription factor sigma H. Mol Microbiol. 1996;20:339–350. doi: 10.1111/j.1365-2958.1996.tb02621.x. [DOI] [PubMed] [Google Scholar]
- 51.Völker U, Engelmann S, Maul B, Riethdorf S, Völker A, Schmid R, Mach H, Hecker M. Analysis of the induction of general stress proteins of Bacillus subtilis. Microbiology. 1994;140:741–752. doi: 10.1099/00221287-140-4-741. [DOI] [PubMed] [Google Scholar]
- 52.Völker U, Riethdorf S, Winkler A, Weigend B, Fortnagel P, Hecker M. Cloning and characterization of heat-inducible promoters of Bacillus subtilis. FEMS Microbiol Lett. 1993;106:287–293. doi: 10.1111/j.1574-6968.1993.tb05978.x. [DOI] [PubMed] [Google Scholar]
- 53.Völker U, Völker A, Maul B, Hecker M, Dufour A, Haldenwang W G. Separate mechanisms activate sigma B of Bacillus subtilis in response to environmental and metabolic stresses. J Bacteriol. 1995;177:3771–3780. doi: 10.1128/jb.177.13.3771-3780.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wetzstein M, Schumann W. Nucleotide sequence of a Bacillus subtilis gene homologous to the grpE gene of E. coli located immediately upstream of the dnaK gene. Nucleic Acids Res. 1990;18:1289. doi: 10.1093/nar/18.5.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wetzstein M, Völker U, Dedio J, Lobau S, Zuber U, Schiesswohl M, Herget C, Hecker M, Schumann W. Cloning, sequencing, and molecular analysis of the dnaK locus from Bacillus subtilis. J Bacteriol. 1992;174:3300–3310. doi: 10.1128/jb.174.10.3300-3310.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yuan G, Wong S L. Isolation and characterization of Bacillus subtilis groE regulatory mutants: evidence for orf39 in the dnaK operon as a repressor gene in regulating the expression of both groE and dnaK. J Bacteriol. 1995;177:6462–6468. doi: 10.1128/jb.177.22.6462-6468.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yuan G, Wong S L. Regulation of groE expression in Bacillus subtilis: the involvement of the sigma A-like promoter and the roles of the inverted repeat sequence (CIRCE) J Bacteriol. 1995;177:5427–5433. doi: 10.1128/jb.177.19.5427-5433.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zenno S, Kobori T, Tanokura M, Saigo K. Purification and characterization of NfrA1, a Bacillus subtilis nitro/flavin reductase capable of interacting with the bacterial luciferase. Biosci Biotechnol Biochem. 1998;62:1978–1987. doi: 10.1271/bbb.62.1978. [DOI] [PubMed] [Google Scholar]
- 59.Zuber U, Schumann W. CIRCE, a novel heat shock element involved in regulation of heat shock operon dnaK of Bacillus subtilis. J Bacteriol. 1994;176:1359–1363. doi: 10.1128/jb.176.5.1359-1363.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]