Abstract
Hemophores are secreted by several gram-negative bacteria (Serratia marcescens, Pseudomonas aeruginosa, Pseudomonas fluorescens, and Yersinia pestis) and form a family of homologous proteins. Unlike the S. marcescens hemophore (HasASM), the P. fluorescens hemophore HasAPF has an additional region of 12 residues located immediately upstream from the C-terminal secretion signal. We show that HasAPF undergoes a C-terminal cleavage which removes the last 21 residues when secreted from P. fluorescens and that only the processed form is able to deliver heme to the S. marcescens outer membrane hemophore-specific receptor, HasRSM. Functional analysis of variants including those with an internal deletion of the extra C-terminal domain show that the secretion signal does not inhibit the biological activity, whereas the 12-amino-acid region located upstream does. This extra domain may inhibit the interaction of the hemophore with HasRSM. To localize the hemophore regions involved in binding to HasR, chimeric HasAPF-HasASM proteins were tested for biological activity. We show that residues 153 to 180 of HasAPF are necessary for its interaction with the receptor.
Bacteria have diverse high-affinity heme uptake systems for the various heme sources that they might encounter (3, 15). One such system is dependent on hemophores which bind heme with high affinity and fulfill a function similar to that of siderophores (2): they are secreted into the extracellular medium, where they scavenge free or protein-bound heme and then deliver it to a specific cell surface receptor. The Serratia marcescens hemophore-dependent heme acquisition system has been reconstituted in Escherichia coli (5). Exogenously added S. marcescens hemophore (HasASM) increases the efficiency of heme uptake via the specific receptor (HasRSM) and also makes available new heme sources (such as hemopexin and myoglobin) which are not recognized by HasR alone (S. Létoffé et al., unpublished results). The S. marcescens hemophore is a monomer which binds heme with a stoichiometry of 1 and an affinity lower than 10−9 M (8). The crystal structure of holoprotein has been solved and found to consist of a single module with two residues in interaction with heme (1). Both heme-free and heme-loaded hemophores bind to HasR with similar apparent affinities (10−10 M), indicating direct protein-protein interactions (10).
HasA-type hemophores are found in S. marcescens, Pseudomonas aeruginosa (11), Pseudomonas fluorescens (7), and Yersinia pestis (J. M. Ghigo, personal communication) and form a family of homologous proteins which do not share extensive similarity with any other known proteins. They are secreted by ABC transporters in a Sec-independent process (14). Like most proteins using this pathway, they do not have an N-terminal signal sequence but rather have an uncleavable C-terminal secretion sequence consisting of at least the last 15 residues. This extreme C-terminal signal contains one or two negatively charged residues followed by a hydrophobic stretch (6). It is unstructured and highly accessible to the solvent (16).
P. aeruginosa, P. fluorescens, and S. marcescens hemophores are secreted from E. coli by their reconstituted ABC transporters. However, they have apparent molecular weights higher than that of the proteins secreted by their natural hosts. Mass spectrometry has shown a single cleavage of the hemophore secreted from S. marcescens which removes the last 12 residues (9) and multiple cleavages of that from P. aeruginosa which remove 15 to 21 residues (11). C-terminal cleavage is not required for secretion and presumably occurs in the extracellular medium, a result of the activity of extracellular proteases produced by S. marcescens and P. aeruginosa but not by E. coli. Both uncleaved and cleaved HasASM and P. aeruginosa HasA (HasAPA) bind heme and can acquire heme from hemoglobin. Both forms of HasASM can deliver heme to the S. marcescens outer membrane receptor HasRSM in E. coli, allowing heme uptake. In a similar test performed with HasPA, we found that the recombinant form of HasAPA (uncleaved) cannot deliver heme to HasRSM whereas the cleaved form can. HasAPA has an additional region of 14 residues close to the C terminus not found in HasASM but which is removed in the processed form (11). We suggested that this additional domain close to the C terminus could inhibit heme delivery by preventing a direct interaction between HasRSM and HasAPA. However, the occurrence of multiple cleavages of HasAPA in P. aeruginosa and the difficulty of separating the different processed forms complicate the study of the interaction between HasAPA and HasRSM.
Here, we found that HasAPF has properties very similar to those of HasAPA: it is cleaved when secreted from P. fluorescens; both the processed and unprocessed forms bind heme, whereas only the processed form can deliver heme to HasRSM. However, unlike HasAPA, it undergoes a single cleavage in P. fluorescens. This prompted us to construct variants of HasAPF and HasAPF-HasASM chimeras and to study their biological activity and binding to HasRSM to localize hemophore domains involved in binding to HasRSM.
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. coli C600 (F− thr leu fhuA lacY thi supE) and E. coli POP 3 hemA (araD139 ΔlacU169 rpsL relA thi hemA) were from the laboratory collection. P. fluorescens 33 was a gift from Haruo Kumura. pR10K carries hasRSM on pBGS18 (5). Plasmid pSYC134 is pUC18 carrying hasASM, and pSYC150 is pACYC184 carrying hasD and hasE (11). pFXF-HasA carries hasAPF, pFXM-HasA encodes the HasAPF1-153-CterSM148-188, polypeptide, and pFBΔF-HasA encodes the HasAPFΔ181-192 polypeptide; they are derived from pUC18 and described in reference 7.
Plasmid constructions.
pUC/HasAPF1-180-CterSM148-188 was created as follows. A DNA fragment encoding amino acid residues 1 to 180 of HasAPF was produced by inserting the phosphorylated synthetic oligonucleotide linkers 5′-GATCTCTGCTGGGTTGGCCTGCA-3′ and 5′-GGCCAACCCAGCAGA-3′ between the BglII and PstI sites of pUC18 encoding HasAPF, resulting in pUC/HasAPF-PstI. The DNA fragment coding for amino acid residues 148 to 188 of HasASM was generated by PCR with the oligonucleotide 5′-GGGCTGCAGAGACCGCGCTGAACGGCATC-3′, the universal primer M4 5′-GTTTTCCCAGTCACGAC-3′, and pUC/HasASM as a template DNA. The amplified fragment was digested with PstI and BamHI and then introduced into the corresponding sites of pUC18. The PstI-BamHI fragment of ca. 0.1 kb was blunt ended and inserted between the PstI and HindIII (blunt-ended) sites of pUC/HasAPF-PstI. The resultant plasmid was digested with PstI, treated with T4 DNA polymerase, and then ligated to destroy the PstI site, generating pUC/HasAPF1-180-CterSM148-188.
The plasmid encoding a chimeric HasA protein consisting of residues 1 to 180 of HasAPF and residues 174 to 188 of HasASM was created by inserting oligonucleotides. The PCR product of ca. 0.1 kb amplified using the oligonucleotide 5′-GGGCATGCCGTGGGCGTGCAGCACGCC-3′, the universal primer M4, and a SphI-disrupted pUC/HasASM DNA was inserted between the SphI and HindIII sites of pUC/HasAPF. This plasmid was digested with BglII and SphI and blunt ended with T4 polymerase, and the oligonucleotides 5′-CGGCCAGGCCGGCCGA-3′ and 5′-GATCTCGGCCGGCCTGGCCGCATG-3′ were inserted, generating a plasmid encoding HasPF1-180-CterSM174-188. DNA sequencing of the hybrid genes showed that the genetic manipulations did not introduce any change in the hybrid protein amino acid sequences. DNA manipulations were carried out according to standard procedures (13).
Media.
All media and antibiotics were as described by Miller (12). LBD contained 0.2 mM 2,2′-dipyridyl, to reduce iron available to E. coli. LBD* contained 0.4 mM 2,2′-dipyridyl to chelate iron in P. fluorescens cultures. Hemin, hemin-agarose, and bovine hemoglobin was obtained from Sigma Chemical Co. Bovine hemoglobin agar plates were prepared as described in reference 5.
Protein analysis.
The HasAPF protein secreted from E. coli was prepared from the supernatant of an overnight culture (grown at 37°C in LBD) of C600 harboring plasmids encoding HasAPF (pFXF-HasA) and HasSM transporter (pSYC150). The HasAPF protein secreted from P. fluorescens was prepared from the supernatant of P. fluorescens grown in LBD* at 23°C for 96 h. The supernatants were concentrated either by precipitation with 10% trichloroacetic acid (TCA) (inactive supernatants) or by ammonium sulfate precipitation: 60% for HasASM and 80% for HasAPF (active supernatants).
Proteins in P. fluorescens active supernatant were purified by hemin-agarose affinity chromatography as described previously. The bound protein (as optical density [OD] equivalents) was eluted with 200 μl of the sample buffer for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) or 200 μl of 6 M guanidine-HCl. The guanidine-eluted sample was incubated for 3 h at room temperature on a wheel and then placed on an empty disposable column with mesh to recover the eluted proteins. The elute was diluted with 200 μl of 0.2 M Tris-HCl (pH 7.5) and incubated overnight at 4°C on a wheel. After dialysis against 10 mM Tris-HCl (pH 7.5), aliquots were subjected to SDS-PAGE and stained with Coomassie blue to evaluate protein concentration and purity, and other aliquots were used for further experiments.
Amino acid sequence analysis.
N-terminal amino acid sequences were determined using the G1005A protein sequencing system (Hewlett-Packard).
Mass spectrometry.
The spectra of positive ions were recorded in linear mode with a MALDI-TOF (matrix-assisted laser desorption–ionization time-of-flight) mass spectrometer (Voyager Elite; Perceptive Biosystems) using a saturated solution of sinapinic acid (3,5-dimethoxy-4-hydroxycinnamic acid) in 30% acetonitrile as matrix. External calibration was performed with apomyoglobin and trypsinogen using the protonated ions of the monomer with average m/z ratios of 16,952.6 and 23,983, respectively.
Electrophoresis and immunological techniques.
Proteins were analyzed by SDS-PAGE followed either by Coomassie blue staining or Western blot analysis. Coimmunoprecipitations were performed as described previously (10). Anti-HasA and anti-HasR rabbit polyclonal sera, both at a 1/5,000 dilution, were used for immunodetection.
E. coli growth assays on LBD hemoglobin agar plates supplemented with purified HasA.
Growth stimulation of POP 3 hemA(pR10K) by exogenously supplied HasA was tested as follows. Cells of the HasR-producing strain were mixed with 3 ml of top agar and poured onto LBD plates supplemented with 10−6 M hemoglobin (Hb-LBD). Five-millimeter-diameter wells were cut in the agar and filled with 50 μl of serial dilutions of various HasA preparations. Growth around the wells was recorded after overnight incubation at 37°C (5).
RESULTS AND DISCUSSION
Characterization of the HasAPF proteins secreted by P. fluorescens and by E. coli expressing the HasSM transporter.
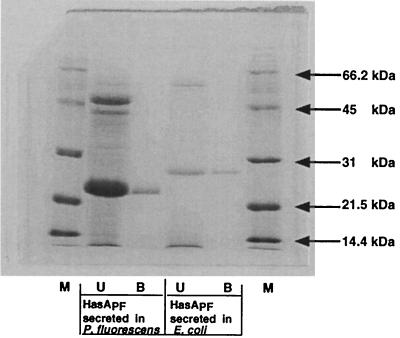
The supernatants were concentrated by TCA precipitation, resolved by SDS-PAGE, and probed with anti-HasASM antibodies. One protein band was detected in each sample. The form secreted by E. coli exhibited a molecular weight higher than that of the form secreted from P. fluorescens (data not shown). To determine the origin of the difference in molecular weight, both forms (that secreted by P. fluorescens and that secreted by E. coli expressing the HasSM transporter) were concentrated and purified by hemin-agarose chromatography. Both forms bound heme (Fig. 1). The recombinant form had an apparent molecular mass 3 kDa higher (Fig. 1). N-terminal amino acid sequencing of the proteins purified on hemin-agarose showed sequences (TISVSEAA) identical to the deduced amino acid sequence of HasAPF (amino acid residues 2 to 10). Both lacked only the first methionine at the N terminus. When analyzed by mass spectrometry, purified HasAPF from the P. fluorescens culture gave one major peak showing a molecular weight of 18,987.0. The value agreed well with the calculated molecular weight of the monoprotonated HasAPF composed of amino acid residues 2 to 185 (18,985.1). Thus, HasAPF from the P. fluorescens culture lacked also the 21 C-terminal residues (Fig. 2B).
FIG. 1.
Hemin-agarose chromatography of HasAPF secreted from P. fluorescens and E. coli. Lane 1 (M), molecular weight markers. Lane 2 (U [unbound]) was loaded with 5 OD equivalents of concentrated P. fluorescens PF33 supernatant not bound to hemin-agarose. Lane 3 (B [bound]) was loaded with hemin-bound material eluted by boiling in SDS-sample buffer corresponding to 3 OD equivalents of ammonium sulfate-concentrated supernatant. Lane 4 was loaded with 3 OD equivalents of concentrated supernatant of E. coli strain C600(pSYC150, pFXF-HasA) not bound to hemin-agarose. Lane 5 was loaded with hemin-bound material eluted by boiling in SDS-sample buffer corresponding to 1.5 OD equivalents of ammonium sulfate-concentrated supernatant. Samples were subjected to SDS-PAGE (15% gel) followed by Coomassie brilliant blue G-250 staining.
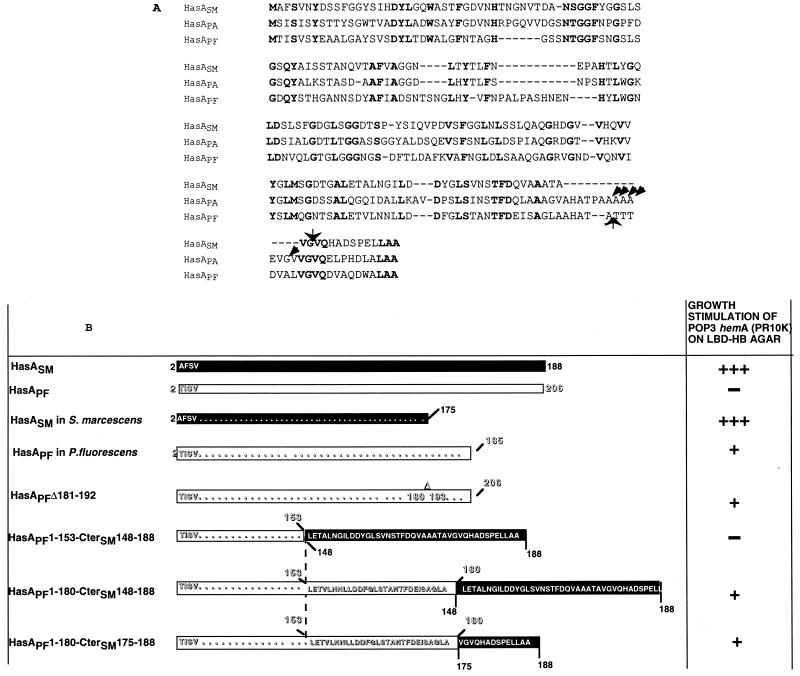
FIG. 2.
Chimeric HasA polypeptides. (A) Alignment of the amino acid sequences of S. marcescens, P. aeruginosa, and P. fluorescens HasA proteins. The residues are numbered starting from the N-terminal methionine. However, mature HasASM, HasAPA, and HasAPF polypeptides lack the N-terminal methionine. The C-terminal proteolytic cleavage sites in S. marcescens, P. aeruginosa, and P. fluorescens are indicated by arrows. Bold characters indicate identical residues in the three polypeptides. (B) Schematic representation of HasAPF variants and HasAPF-HasASM hybrid proteins. The solid and open boxes represent amino acid sequences of HasASM and HasAPF, respectively. The deletion of the region between residues 180 and 193 of HasAPF (AHATATTTDVAL) is indicated by a Δ above the corresponding box. The C-terminal amino acid sequences of the chimera are shown. The ability of each polypeptide to stimulate growth of the HasRSM-producing strain is shown on the right. The wells contained 2 to 10 μg of the HasAPF wild-type and variant proteins. Similar growth stimulation was obtained with 2 to 10 ng of HasASM. Growth around the wells was recorded after overnight incubation at 37°C. +, bacterial growth ring of 2 mm; +++, bacterial growth ring of 5 mm; —, no growth.
Comparison of the biological activity of the recombinant and processed HasAPF.
Growth stimulation of POP3 hemA(pR10K) by addition of serial dilutions of concentrated HasAPF preparations was tested on Hb-LBD. The unprocessed form did not show any growth stimulation, whereas the processed form had significant activity (Fig. 2B). This suggests that the 21 C-terminal residues interfere with the heterologous complementation. The 14 C-terminal residues of this region can promote (alone or fused to passenger proteins) efficient secretion via the HasSM transporter and therefore constitute the secretion signal (7). This 14-amino-acid secretion signal is well conserved in the three hemophores HasAPF, HasASM, and HasAPA (Fig. 2A). In contrast, the 12 amino acids in HasAPF (from residues 181 to 192) located just upstream from this secretion signal are absent from HasASM (Fig. 2A). To determine whether the presence of this nonhomologous region was inhibiting the heterologous complementation, we tested a hasAPF variant (encoded by pFBΔF-HasA) consisting of an internally deleted protein in which the N-terminal 180 amino acids are fused directly to the C-terminal 14 amino acids (HasAPFΔ181-192 [Fig. 2B]) (7). This construct lacks the extra 12-residue domain and is therefore similar to the active HasASM produced by E. coli.
Biological activity of HasAPFΔ181-192.
The HasAPFΔ181-192 variant was secreted by the HasSM transporter (Fig. 3). Added exogenously, this protein showed the same growth stimulation of POP3 hemA(pR10K) as did the naturally processed protein on Hb-LBD plates (Fig. 2B). Hence, the deletion of the 12 amino acids restored biological activity in the heterologous complementation test, indicating that it is not the secretion signal per se which blocks the hemophore function.
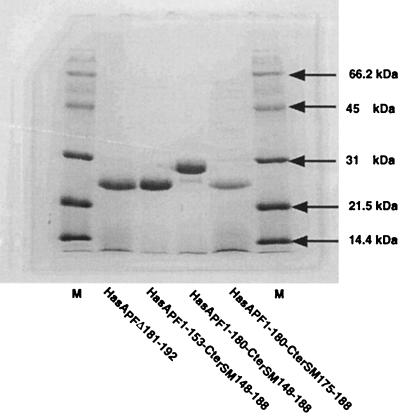
FIG. 3.
Secretion of HasAPF chimeras and variants. C600 carrying pSYC150 and the various pUC derivatives encoding the HasAPF polypeptides were grown overnight. The proteins in the culture supernatants were concentrated by TCA precipitation and subjected to SDS-PAGE analysis. The gels were stained with Coomassie brilliant blue G-250. Each lane was loaded with 3 OD equivalents of concentrated supernatant.
As both the naturally processed form (at residue 185) and the internally deleted variant lacking residues 181 to 192 are active, the first 180 amino acids of HasAPF bear all of the determinants required for heme binding and delivery to the receptor HasRSM. To map more precisely the domain required for heme delivery to HasRSM, we constructed chimeric HasAPF having either the 153 or 180 N-terminal amino acids of HasAPF fused to the 41-amino-acid C-terminal secretion signal of HasASM.
Biological activity of chimeric HasAPF-HasASM proteins.
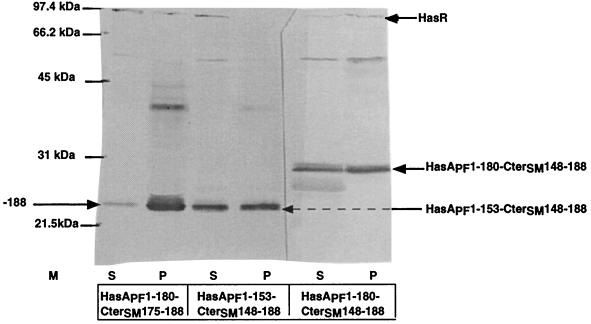
HasAPF1-180-CterSM148-188 and HasAPF1-153-CterSM148-188 were both efficiently secreted by E. coli expressing the HasSM transporter (Fig. 3). Both bound heme (data not shown). Only HasAPF1-180-CterSM148-188 had activity in the heterologous complementation test (Fig. 2B). The absence of biological activity of the shorter HasAPF hybrid could be due to its failure to interact with the receptor. As direct interactions between HasRSM and HasASm have been previously demonstrated by coimmunoprecipitation (10), we used this technique to determine the binding capacity of the two chimeras. Both chimeras were immunoprecipitated with a monoclonal antibody directed against the C-terminal 50 amino acids of HasASM. HasR was detected only in the immunoprecipitate of the longer fusion (Fig. 4). This suggests that the HasAPF residues 153 to 180 are required for the interaction between HasAPF and HasRSM. The CterSM148-188 region shares 50% identity with the corresponding region of HasAPF (Fig. 2A). However, the inactivity of HasAPF1-153-CterSM148-188 indicated that the fusion did not lead to the reconstitution of a functional binding domain. To confirm that the CterSM148-188 domain does not contain the elements necessary for binding, we tested the activity of a chimera with a shorter CterSM of 15 amino acids.
FIG. 4.
Coimmunoprecipitation of HasA and HasR with a mouse monoclonal anti-Cter-HasA antibody. Western blots were probed with rabbit anti-HasA and anti-HasR antisera, both at a 1/5,000 dilution. Approximately 40 OD culture equivalents of solubilized whole membrane proteins were incubated with 10 μg of various HasAPF polypeptides as indicated at the bottom. S, supernatant of the immunoprecipitate (4 OD culture equivalents); P, immunoprecipitated proteins (20 OD culture equivalents).
HasAPF1-180-CterSM174-188 was efficiently secreted from E. coli by the HasSM transporter (Fig. 3) and was active for heterologous complementation (Fig. 2B). HasRSM was coimmunoprecipitated with this last chimera (Fig. 4).
As HasASM residues 174 to 188 can be deleted without changing the properties of HasA, the activity of HasAPF1-180-CterSM174-188 is clearly not dependent on the added HasASM domain. Thus, residues 153 to 180 of HasAPF are necessary for its biological activity and in particular for its binding to the receptor. HasAPF shares 41% identity with HasASM. This degree of similarity all along the protein sequence suggests that the two proteins have similar secondary and tertiary structures (4). The last 14 residues of the HasASM secretion signal are not seen in the three-dimensional structure, and nuclear magnetic resonance spectroscopy indicates that they are poorly structured (9). The region located just upstream from the secretion signal forms two α helices (residues 144 to 155 and 167 to 174) on the side of the globular HasA molecule opposite the heme pocket (1). Similarly, the corresponding region of HasAPF, between residues 144 and 180, is also likely to be on the opposite face of the protein relative to the heme pocket. Possibly, recognition of the heterologous receptor by HasAPF does not involve the heme binding site but rather uses a C-terminal domain of the molecule located upstream from the secretion signal.
We have identified a 12-amino-acid sequence (181 to 192) in HasAPF located immediately upstream from the 14 C-terminal secretion signal which is absent from HasASM and which interferes with heterologous complementation (heme delivery via HasAPF to HasRSM). The small size of the inhibitory region suggests that it inhibits by masking the functionally important neighboring domain (amino acids 150 to 180).
The physiological significance of the C-terminal cleavage that all three studied hemophores undergo is unclear. The unprocessed and in vitro processed forms of HasASM appear to have the same biological activity when exogenously added to S. marcescens hemA or hasA mutants or to E. coli hemA mutants expressing HasRSM (data not shown). Thus, the inhibitory effect is revealed only in the heterologous systems. Cleavage of the HasA C termini when secreted from their natural hosts might simply be a consequence of their high accessibility to the extracellular proteases also produced by these species.
ACKNOWLEDGMENTS
We are grateful to Philippe Delepelaire, Jean-Marc Ghigo, Laurent Debarbieux, Pascal Arnoux, and Mirjam Czjzek for helpful discussions.
REFERENCES
- 1.Arnoux P, Haser R, Izadi N, Lecroisey A D, Wandersman C, Czjzek M. The crystal structure of HasA, a hemophore secreted by Serratia marcescens. Nature Struct Biol. 1999;6:516–520. doi: 10.1038/9281. [DOI] [PubMed] [Google Scholar]
- 2.Braun V, Killmann H. Bacterial solutions to the iron-supply problem. Trends Biochem Sci. 1999;24:104–109. doi: 10.1016/s0968-0004(99)01359-6. [DOI] [PubMed] [Google Scholar]
- 3.Cope L, Thomas S, Hrkal Z, Hansen E. Binding of heme-hemopexin complexes by soluble HxuA protein allows utilization of this complexed heme by Haemophilus influenzae. Infect Immun. 1998;66:4511–4516. doi: 10.1128/iai.66.9.4511-4516.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Creighton T. The folded conformation of globular proteins. W. H. New York, N.Y: Freeman and Company; 1993. [Google Scholar]
- 5.Ghigo J M, Létoffé S, Wandersman C. A new type of hemophore-dependent heme acquisition system of Serratia marcescens reconstituted in Escherichia coli. J Bacteriol. 1997;179:3572–3579. doi: 10.1128/jb.179.11.3572-3579.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghigo J M, Wandersman C. A carboxyl-terminal four-amino acid motif is required for secretion of the metalloprotease PrtG through the Erwinia chrysanthemi protease secretion pathway. J Biol Chem. 1994;269:8979–8985. [PubMed] [Google Scholar]
- 7.Idei A, Kawai E, Akatsuka H, Omori K. Cloning and characterization of the Pseudomonas fluorescens ATP binding cassette exporter, HasDEF, for the heme acquisition protein HasA. J Bacteriol. 1999;181:7545–7551. doi: 10.1128/jb.181.24.7545-7551.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Izadi N, Henri Y, Haladjan J, Goldberg M E, Wandersman C, Delepierre M, Lecroisey A. Purification and characterization of an extra cellular heme binding protein, HasA, involved in heme iron acquisition. Biochemistry. 1997;36:7050–7057. doi: 10.1021/bi962577s. [DOI] [PubMed] [Google Scholar]
- 9.Izadi-Pruneyre N, Wolff N, Redeker V, Wandersman C, Delepierre M, Lecroisey A. NMR studies of the C-terminal secretion signal of the heme binding protein HasA. J Biochem. 1999;261:562–568. doi: 10.1046/j.1432-1327.1999.00305.x. [DOI] [PubMed] [Google Scholar]
- 10.Létoffé S, Nato F, Goldberg M E, Wandersman C. Interactions of HasA, a bacterial haemophore, with haemoglobin and with its outer membrane receptor HasR. Mol Microbiol. 1999;33:546–555. doi: 10.1046/j.1365-2958.1999.01499.x. [DOI] [PubMed] [Google Scholar]
- 11.Létoffé S, Redeker V, Wandersman C. Isolation and characterisation of an extracellular heme binding protein from Pseudomonas aeruginosa that shares function and sequence similarities with the Serratia marcescens HasA hemophore. Mol Microbiol. 1998;28:1223–1234. doi: 10.1046/j.1365-2958.1998.00885.x. [DOI] [PubMed] [Google Scholar]
- 12.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. [Google Scholar]
- 13.Sambrook J, Fritsch F, Maniatis T E. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 14.Wandersman C. Protein and peptide secretion by ABC exporters. Res Microbiol. 1998;149:163–170. doi: 10.1016/s0923-2508(98)80076-4. [DOI] [PubMed] [Google Scholar]
- 15.Wandersman C, Stojiljkovic I. Bacterial heme sources: role of hemophores and receptors. Curr Opin Microbiol. 2000;3:215–220. doi: 10.1016/s1369-5274(00)00078-3. [DOI] [PubMed] [Google Scholar]
- 16.Wolff N, Delepierre P, Ghigo J, Delepierre M. Spectroscopic studies of the C-terminal secretion signal of the Serratia marcescens haem acquisition protein (HasA) in various membrane-mimetic environments. Eur J Biochem. 1997;15:400–407. doi: 10.1111/j.1432-1033.1997.0400a.x. [DOI] [PubMed] [Google Scholar]