Abstract
Advances in pharmacomicrobiomics have shed light on the pathophysiology of drug‐induced enteropathy associated with the therapeutic use of certain non‐steroidal anti‐inflammatory drugs, anticancer chemotherapies and immunosuppressants. The toxicity pathway results from the post‐glucuronidation release and digestive accumulation of an aglycone generated in the context of intestinal dysbiosis characterized by the expansion of β‐glucuronidase‐expressing bacteria. The active aglycone could trigger direct or indirect inflammatory signaling on the gut epithelium. Therefore, taming bacterial β‐glucuronidase (GUS) activity is a druggable target for preventing drug‐induced enteropathy. In face of the limitations of antibiotic strategies that can worsen intestinal dysbiosis and impair immune functions, we hereby propose the use of a recombinant probiotic capable of mimicking repressive conditions of GUS through an inducible plasmid vector.
Keywords: bacterial β‐glucuronidase, drug‐induced enteropathy, gut microbiome, intestinal dysbiosis, pharmacomicrobiomics, recombinant probiotic
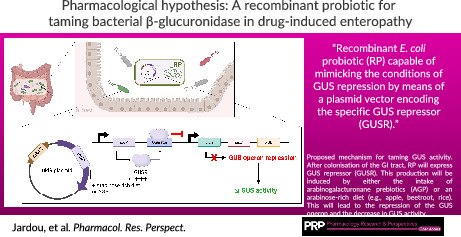
Recombinant E. coli probiotic (RP) capable of mimicking the conditions of GUS repression by means of a plasmid vector encoding the specific GUS repressor (GUSR).
Abbreviations
- AGP
arabinogalacturonanes prebiotics
- GI
gastrointestinal
- GUS
gut microbiome β‐glucuronidase
- GUSR
GUS repressor
- MPA
mycophenolic acid
- NSAID
non‐steroidal anti‐inflammatory drugs
- PBAD
arabinose operon promoter
- RP
recombinant probiotic
- UGTs
UDP‐glucuronosyltransferases
1. INTRODUCTION
Pharmacomicrobiomics is an emerging field at the frontiers of pharmacology, microbiology and genomics, which focuses on microbiome impacts on drug disposition, action and toxicity. 1 Advances in pharmacomicrobiomics have shed light on the pathophysiology of several gastrointestinal (GI) diseases including inflammatory bowel diseases and drug‐induced enteropathy. 2 A common toxic pathway involving enterohepatic recirculation and gut microbiome β‐glucuronidase (GUS) activity, has been described for GI adverse effects and histological damages associated with the use of certain non‐steroidal anti‐inflammatory drugs (e.g., diclofenac, indomethacin, ketoprofen), 3 , 4 anticancer chemotherapies (e.g., irinotecan, regorafenib) 5 , 6 and immunosuppressants (e.g., mycophenolate mofetil). 7 The therapeutic use of these drugs is associated with intestinal dysbiosis characterized by the expansion of GUS‐expressing bacteria. 8 This bacterial enzyme is involved in the back‐transformation of inactive glucuronides produced in the liver and excreted in the GI tract through bile. Once excreted, they interact with the gut microbial community where GUS hydrolyses them to their parent, or phase I metabolite, forms. 8 The interplay between the gut microbiome and GUS activity is crucial for GI damages to show up. Therefore, GUS is a druggable target and taming its activity is an option to prevent drug‐induced enteropathy. Several preventative and/or therapeutic approaches targeting GUS are under evaluation. 9 Among them, the antibiotic approach intended to decrease the populations of GUS‐expressing bacteria, unfortunately promotes dysbiosis, favors short‐ and long‐term risks of opportunistic infection, impairs the beneficial effects of the microbiota on the immune system homeostasis and increases the risk of introducing drug resistance. 7 , 10 Despite the promising results obtained in preclinical models for non‐antibiotic small inhibitory molecules on specific GUS activity, the discovery and development of these inhibitors are quite challenging. 6 , 11 This can be explained by the universal distribution of GUS in human gut microbiota, their varied substrate preference, and their distinct inhibition properties. 12 We herein conceive a novel biotechnological approach using recombinant probiotic mimicking repressive conditions of GUS synthesis with an inducible plasmid vector.
2. BACTERIAL Β‐GLUCURONIDASE, AS A VALIDATED TARGET FOR DRUG‐INDUCED ENTEROPATHY
There is a vast diversity of GUS enzyme in the human gut microbiome termed “GUSome” with importance in normal physiology and disease through metabolism of endogenous (e.g., bilirubin, steroid hormones, dopamine and serotonin) and/or exogenous (e.g., irinotecan, regorafenib, diclofenac, and morphine) compounds. 12 These GUS catalyzes the production of glucuronic acid as an energy source for the GI microbiota. Among GUS enzyme, those implicated in glucuronides xenobiotic metabolization are encoded by the GUS operon 13 (Figure 1), mainly expressed by bacterial strains of the Enterobacteriaceae family, like Escherichia coli (E. coli). The expansion of these strains is often associated with drug‐induced enteropathy. In fact, studies have established the distinct substrate preference of bacterial GUS, and have shown that the deconjugation of irinotecan, diclofenac or regorafenib was processed mainly by E.coli GUS enzyme. 8 , 12 Drug‐induced enteropathy involved other GUS‐producing bacteria. These bacteria are members of the main gut microbiota phyla, namely phylum Firmicutes (Clostridium perfringens, Ruminococcus gnavus, Streptococcus agalactiae), and phylum Bacteroidetes (Bacteroides fragilis, Bacteroides uniformis, Bacteroides dorei). 12
FIGURE 1.
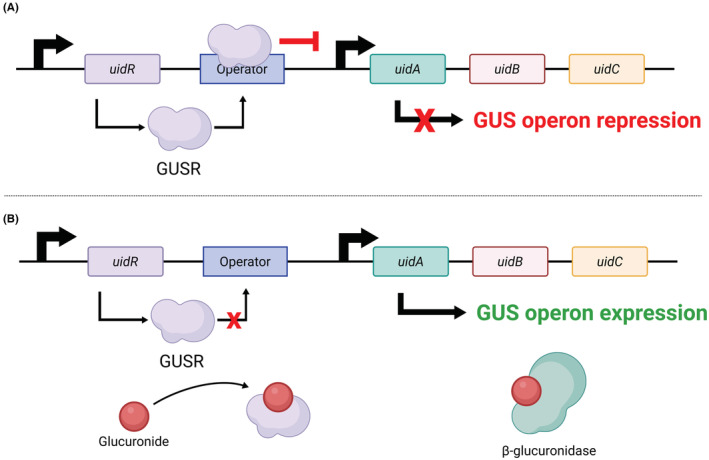
Schematic representation of the GUS operon. (A) GUS operon repression by the Glucuronide Repressor (GUSR); (B) GUS operon expression in the presence of a glucuronide ligand (e.g., non‐steroidal anti‐inflammatory drugs, anticancer chemotherapies, immunosuppressants). GUSR encoded by uidR is dissociated from the operator allowing the transcription of the proteins encoded by the GUS operon composed of the uidA, uidB, and uidC genes. uidA encodes the GUS enzyme, uidB a H+‐symporter located in the inner membrane and uidC a permease expressed in the outer membrane. uidB and uidC regulate the glucuronide‐inducible transport of glucuronides. Illustration created with BioRender.com.
GUS activity can have deleterious consequences on the pharmacokinetics and disposition of drugs. 14 It can hydrolyse inactive glucuronides generated by hepatic phase II drug‐metabolizing UDP‐glucuronosyltransferases (UGTs), releasing an active aglycone that may accumulate in the GI tract. This metabolic reversion also participates in the enterohepatic recirculation of some drugs and may activate directly or indirectly toxic signaling pathways. Many drugs with varied chemical structures and belonging to different therapeutic classes exhibit GI tract toxicity mediated by GUS (Table 1). One perfect example of drug‐induced enteropathy linked to GUS activity is irinotecan (CPT‐11), an anticancer drug whose efficacy is hampered by GI complications. 6 Another anticancer drug, regorafenib, also exhibits similar GI toxicity due to GUS activity. 5 GUS also plays a crucial role in the multi‐step pathogenesis of non‐steroidal anti‐inflammatory drugs (NSAID)‐induced enteropathy reported for diclofenac, ketoprofen and indomethacin. 16 Interestingly, these GI adverse effects may be alleviated by GUS inhibitors. 3 , 14 , 15 More recently, mycophenolic acid (MPA), a widely used immunosuppressant, frequently induces severe GI adverse effects that have been linked to intestinal dysbiosis and expansion of GUS expressing bacteria. This shift in bacterial population leads to increased intestinal GUS activity and favors the MPA‐enterohepatic cycle, thus releasing significant amounts of MPA in the intestine, responsible for the damages observed in patients. 7
TABLE 1.
Drug‐induced enteropathy driven by GUS activity.
| Therapeutic compounds | Therapeutic classes | GI symptoms | Key references |
|---|---|---|---|
| Irinotecan (CPT‐11) | Anticancer | Severe diarrhea, intestinal perforation, bleeding ulcers | Bhatt et al., 2020 6 |
| Regorafenib | Erwin et al., 2019 5 | ||
| Indomethacin | NSAIDs | Bleeding ulcers, gastric or intestinal perforations | Saitta et al., 2014 15 |
| Ketoprofen | |||
| Diclofenac | Saitta et al., 2014; Zhong et al.,2016 4 , 15 | ||
| Mycophenolate mofetil | Immunosuppressant | Ulceration, severe diarrhea, colonic inflammation | Taylor et al., 2019 7 |
Iatrogenic diseases associated with GUS activity in the GI tract represent a major drawback concern on the road to precision pharmacotherapy. It is worth mentioning that repression of GUS activity in c be considered due to their physiological role (e.g., dopamine, serotonin, bilirubin, steroid hormone reactivation). 12 Therefore, taming GUS activity is a promising therapeutic option for preventing or reversing drug‐induced GI adverse effects and improving patients' adherence to treatment and quality of life.
3. CURRENT THERAPEUTIC STRATEGIES
The inflammation and digestive symptoms (nausea, vomiting, diarrhea, abdominal pain) observed in drug‐induced enteropathy are combinations of multifactorial damages. In line with the pathophysiology of drug‐induced enteropathy involving GUS, different strategies could be considered (Figure 2). One of these strategies could involve the elimination of harmful bacteria by using antibiotics, while others could reduce hepatic production of glucuronides and/or block bacteria GUS activity with non‐antibiotics small inhibitory molecules.
FIGURE 2.
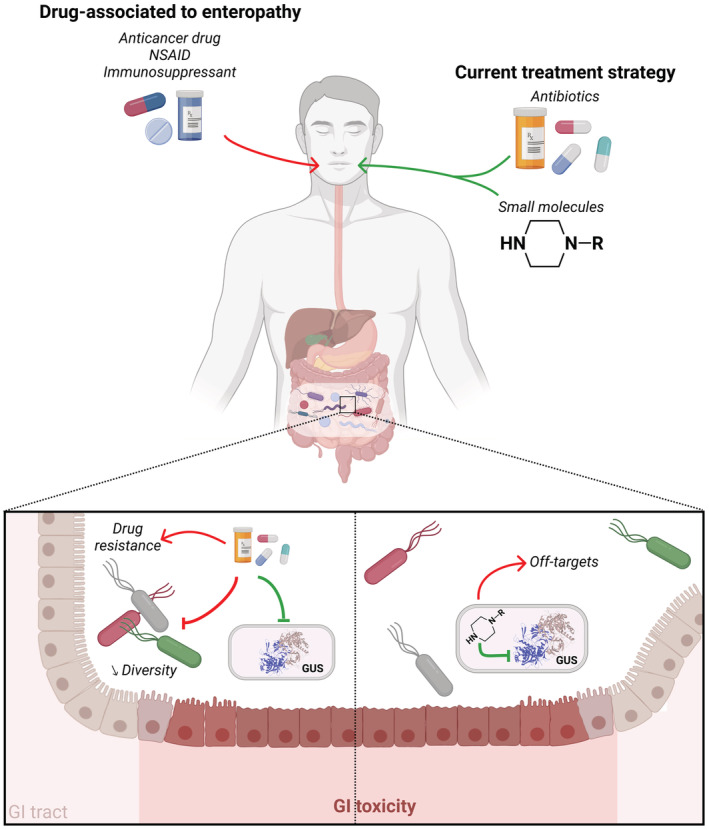
Current therapeutic strategies. Anticancer drugs, NSAID and immunosuppressants can cause GI toxicity and adverse effects through GUS activity. To counteract this toxicity, different approaches targeting this enzyme have been implemented. Antibiotics can eliminate GUS‐producing bacteria, but also commensal bacteria, favoring the selection of resistant strains and ultimately drug resistance. Some small molecules with a piperazine‐group can selectively inhibit GUS activity, but they may have off‐target effects and be involved in and drug–drug interactions. Illustration created with BioRender.com.
The antibiotic strategy which aims to eliminate GUS‐expressing bacteria has shown beneficial effects in various cases of drug‐induced enteropathy. 17 For instance, the gut microbiome is crucial for MPA‐induced enteropathy to show up in a preclinical model 18 and a cocktail of broad‐spectrum antibiotics or vancomycin alleviates GI damages and significantly reverses MPA‐induced weight loss and colonic inflammation. 7 In another preclinical model, irinotecan‐treated rats given antibiotics (e.g., penicillin and streptomycin) showed significant less diarrhea symptoms and caecal histological damage. 19 Although eliminating GUS‐expressing bacterial strains with antibiotics is effective and fairly easy to implement, this is an arguable strategy since the benefit can only be short‐term due to the lack of specificity. Actually, chronic use of antibiotics cannot be considered due to the major risk of devastation and functional impairement (e.g., alteration of short‐chain fatty acids production) of the GI tract microbiome, the risk of subsequent impairment of the immune system homeostasis, 10 not to mention the risk of introducing drug resistance. 20
A non‐antibiotic strategy is possible through the use of selective GUS inhibitors, such as some piperazine (e.g., amoxapine, palbociclib, vortioxetine) and piperidine (e.g., crizotinib) containing compounds. 21 In pre‐clinical models of irinotecan‐induced enteropathy, these selective GUS inhibitors successfully prevented GI toxicity without affecting the drug pharmacokinetics. However, the identification and development of these inhibitors is still challenging due to some limitations (e.g., a universal distribution of GUS in human gut microbiota, varied substrate preference, and distinct inhibition properties). 6 , 11 There is not any pan‐GUS small inhibitor identified yet. Therefore, for precision medicine purposes, we propose a strategy to tame specific bacterial GUS involved in drug‐induced enteropathy. Our strategy relies on the fact that, there are different bacterial GUS implicated into exogenous and endogenous compounds metabolism. 12 Furthermore, it is worth mentioning that monitoring of GUS activity in currently treated patients could represent a valuable diagnostic tool for identifying early‐risk patients, as recently suggested for irinotecan therapy. 22
4. CONCEPTION OF A RECOMBINANT PROBIOTIC TO TAME GUS ACTIVITY
We propose a novel biotechnological strategy specifically targeting the GUS with operon regulation mechanism. It requires developing a recombinant E. coli probiotic (RP) capable of mimicking the conditions of GUS repression by means of a plasmid vector encoding the specific GUS repressor (GUSR). More precisely, the vector contained in the RP would be composed of the uidR gene, under the dependence of the inducible arabinose operon promoter (PBAD). The plasmid vector expression would be modulated by supplying arabinogalacturonanes prebiotics (AGP). As a result, the transcription of uidA, the gene encoding GUS, would be downregulated by promoting the binding of GUSR to GUS operon (Figure 3). In a similar context, an E. coli Nissle 1917 probiotic‐associated therapeutic has been developed as a gastric pad to prevent chronic inflammatory bowel disease. 23 Our aim is to take advantage of this strategy to maintain a non‐iatrogenic microbiota in patients. The main challenge is to downregulate GUS without favoring other, more pathogenic or antibiotic‐resistant, bacterial strains.
FIGURE 3.
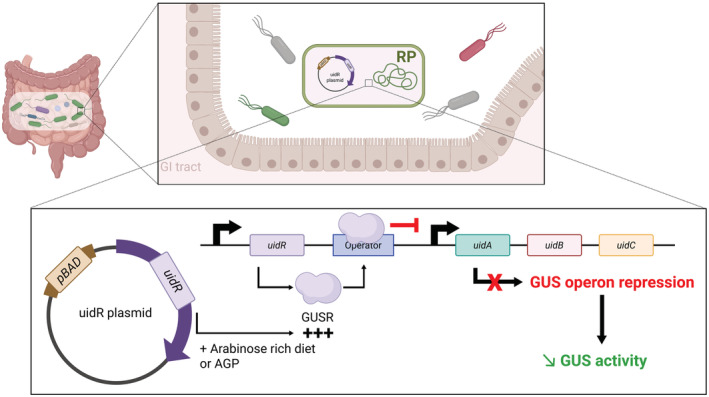
Proposed mechanism for taming GUS activity. After colonization of the GI tract, RP will express GUS repressor (GUSR). This production will be induced by either the intake of arabinogalacturonane prebiotics (AGP) or an arabinose‐rich diet (e.g., apple, beetroot, rice). This will lead to the repression of the GUS operon and the decrease in GUS activity. Illustration created with BioRender.com.
Therefore, these specifications drove us to design our RP strategy through three main approaches. A first, proof‐of‐concept, in vitro experiment would determine the most effective and viable system for reducing GUS expression. A second, ex vivo study would test the most relevant RP in a holistic system, such as an artificial microbiota. Finally, its protective effects against drug‐induced enteropathy would be evaluated in vivo, in a murine model grafted with a human‐associated microbiome.
4.1. Proof‐of‐concept experiments
During this step, the most relevant inducer/probiotic combination with significant inhibition of GUS activity will be selected and oriented by pharmacological perspectives (e.g., influence on drug metabolism and disposition). This repression of GUS is possible owing to the non‐essential role of this enzyme, since uidA mutations would not affect bacterial survival. 24 When implementing this approach, huge attention will be paid to the safety parameters regarding virulence, pathogenicity, and antibiotic resistance.
For this purpose, the bacterial strain must be carefully chosen as some commensal E. coli strains used for probiotic applications are subject to virulence. Eight main phylogenetic commensal groups in E. coli (A, B1, B2, C, D, E, F, I) have been identified. Among them, the B2 group is the most diverse but its members present huge virulence properties. It has been shown that E. coli Nissle 1917, a member of this B2 group with well‐documented probiotic effects, produces the colibactin toxin responsible for double‐stranded breaks in the DNA associated with chromosomal abnormalities in intestinal epithelial cells. 25 , 26 In contrast, E. coli K‐12 MG1655 (from the phylogroup A) and E. coli CEC15 (from the phylogroup C) have shown beneficial effects on the intestinal microenvironment homeostasis. The K‐12 strain has been associated with improved intestinal epithelium integrity (evaluated through occludin expression). 27 The CEC15 strain has been involved in colonic epithelium remodeling by promoting mucus production and ionic movements. 28 As a matter of fact, these properties could be synergistic with our RP beneficial effect in patients prone to GI toxicity.
An auxotrophic cassette (e.g., glycine A, lysine) could be another option for the RP selection, so as to limit dissemination of antibiotic‐resistance genes through conjugative transfers. 29 This implies that the candidate strain should be mono‐auxotrophic for the selected amino acid. The selection is performed through the functional complementation provided by the plasmid vector. Besides, the plasmid should be non‐episomal to avoid modifications in the bacterial genome, which could generate off‐target effects. A reporter gene such as luciferase could be implemented into the vector to monitor the inserted uidR expression. GUS activity could also be assessed using a chromogenic (e.g., 4‐nitrophenyl β‐D‐glucuronide) or fluorescent (e.g., fluorescein‐di‐β‐D‐glucuronide) glucuronide substrate 17 to get a first insight into the efficiency of this system in isolated conditions or in the presence of a relatively complex bacterial community. Co‐cultures in oxygen‐deprived conditions to mimic anaerobic conditions of the human intestine with either Freiter's chambers (using gas mixtures), jars or plastic pouch (using chemical catalysts) or an intestine‐on‐chip system 30 could be used to investigate the interplay between the RP and commensal gut bacteria.
4.2. Validation in artificial microbiota
The effectiveness of the selected inducer/RP combination could be assessed ex vivo, in a more complex environment close to the actual gut microbiota. An artificial microbiota could be used to investigate the evolution of this system and its adaptation to the GI tract environment. 31 Indeed, it is important to consider the interaction of this recombinant system with endogenous bacteria able to coordinate their collective behavior within their transcriptional niche, by means of quorum sensing. 32 The quorum sensing system allows cell maintenance, biofilm formation and horizontal gene transfer through the expression of various proteins mediated by high bacterial density. 33 For example, there are some protease (alkaline protease), lipase (LipA) and adhesin (LapA) proteins that are expressed in the context of quorum sensing. 34 The most common quorum sensing signals are N‐acyl homoserine lactones in Gram‐negative bacteria. 35 In this sense, knowledge associated with quorum sensing signals termed microbial languages for intraspecies (auto‐inducing peptides, cholera inducer, photopyrones) and interspecies (autoinducer, indole) could be exploited to engineer the artificial microbiota around the RP. 36 In this context, meta‐analyses based on RNA‐seq coupled with functional enrichment analyses could provide information about the impact of the RP on differential expression of microbial genes and on gut homeostasis.
4.3. Evaluation in a preclinical model
The safety of the system will be evaluated and pharmacokinetic interactions explored in a drug‐induced enteropathy murine model with human‐associated microbiome. In this sense, it would be of great interest to screen the causal drug metabolites and their involvement in the enterohepatic recirculation. Based on our expertise in MPA metabolism, 37 we could perform untargeted determination of MPA metabolites in fecal and blood samples from treated mice. In addition, oral administration of a fluorescent glucuronide would help to evaluate the efficiency of the system after induction of uidR expression in mice. 17 All these preliminary data would help us to determine the optimal RP dosing regimen and its effectiveness in preventing drug toxicity.
5. CONCLUSION
Recent advances in pharmacomicrobiomics have clearly established the gut microbiome as a “metabolic organ” involved in drug biotransformation and have shed light on the pathophysiology of drug‐induced enteropathy driven by GUS activity. Therefore, targeting GUS could prevent or suppress severe GI toxicity and improve patient adherence to treatment. Due to the limitations and/or adverse effects of the chronic use of antibiotics, new, innovative and effective therapy is needed. Moreover, regarding the universal distribution of GUS in human gut microbiota, the varied substrate preference, and the distinct inhibition properties, the development of small inhibitory molecules capable of targeting GUS is quite challenging. Recombinant probiotics with GUS repressor activity owing to the insertion of a plasmid vector would offer great perspectives. The expected benefits could probably be observed only in early symptomatic patients with intestinal dysbiosis, as there is no reason to conceptually believe that AGP would lead to such successful competition away from microbes that are responding to dozens of glucuronidated endogenous and xenobiotics in intestinal normobiosis. However, our RP could efficiently alleviate damages associated with intestinal dysbiosis. Furthermore, GUS activity could serve as a predictive biomarker to early identify patients at risk of developing drug‐induced enteropathy. In addition, it will be of interest to identify for each glucuronidated xenobiotics the predominant GUS involved in its deconjugation in order to adapt the design of the RP.
6. NOMENCLATURE OF TARGETS AND LIGANDS
Key protein targets and ligands in this article are hyperlinked to corresponding entries in http://www.guidetopharmacology.org, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY, 38 and are permanently archived in the Concise Guide to PHARMACOLOGY 2019/20. 39
AUTHORS' CONTRIBUTIONS
All authors contributed in designing the study, drafting the manuscript and approved the final manuscript.
FUNDING INFORMATION
No funding was received for the publication of this manuscript.
CONFLICT OF INTEREST
The authors have no conflicts of interest to declare.
ETHICS STATEMENT
Not required.
CONSENT STATEMENT
Not required.
ACKNOWLEDGMENTS
We would like to acknowledge Ms Karen Poole for manuscript editing and Dr Thomas Jové (Univ. Limoges, Inserm U1092, RESINFIT, F‐87000, Limoges, France) and Prof. Pierre Marquet (Univ. Limoges, Inserm U1248, Pharmacology & Transplantation, F‐87000, Limoges, France) for their valuable inputs and discussions during the preparation of this manuscript.
Jardou M, Brossier C, Guiyedi K, Faucher Q, Lawson R. Pharmacological hypothesis: A recombinant probiotic for taming bacterial β‐glucuronidase in drug‐induced enteropathy. Pharmacol Res Perspect. 2022;10:e00998. doi: 10.1002/prp2.998
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- 1. Aziz RK, Rizkallah MR, Saad R, ElRakaiby MT. Translating pharmacomicrobiomics: three actionable challenges/prospects in 2020. OMICS. 2020;24(2):60‐61. doi: 10.1089/omi.2019.0205 [DOI] [PubMed] [Google Scholar]
- 2. Ananthakrishnan AN, Luo C, Yajnik V, et al. Gut microbiome function predicts response to anti‐integrin biologic therapy in inflammatory bowel diseases. Cell Host Microbe. 2017;21(5):603‐610.e3. doi: 10.1016/j.chom.2017.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. LoGuidice A, Wallace BD, Bendel L, Redinbo MR, Boelsterli UA. Pharmacologic targeting of bacterial β‐glucuronidase alleviates nonsteroidal anti‐inflammatory drug‐induced enteropathy in mice. J Pharmacol Exp Ther. 2012;341(2):447‐454. doi: 10.1124/jpet.111.191122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhong ZY, Sun BB, Shu N, et al. Ciprofloxacin blocked enterohepatic circulation of diclofenac and alleviated NSAID‐induced enteropathy in rats partly by inhibiting intestinal β‐glucuronidase activity. Acta Pharmacol Sin. 2016;37(7):1002‐1012. doi: 10.1038/aps.2016.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ervin SM, Hanley RP, Lim L, et al. Targeting regorafenib‐induced toxicity through inhibition of gut microbial β‐glucuronidases. ACS Chem Biol. 2019;14(12):2737‐2744. doi: 10.1021/acschembio.9b00663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bhatt AP, Pellock SJ, Biernat KA, et al. Targeted inhibition of gut bacterial β‐glucuronidase activity enhances anticancer drug efficacy. Proc Natl Acad Sci U S A. 2020;117(13):7374‐7381. doi: 10.1073/pnas.1918095117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Taylor MR, Flannigan KL, Rahim H, et al. Vancomycin relieves mycophenolate mofetil–induced gastrointestinal toxicity by eliminating gut bacterial β‐glucuronidase activity. Sci Adv. 2019;5(8):eaax2358. doi: 10.1126/sciadv.aax2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dashnyam P, Mudududdla R, Hsieh TJ, et al. β‐Glucuronidases of opportunistic bacteria are the major contributors to xenobiotic‐induced toxicity in the gut. Sci Rep. 2018;8(1):16372. doi: 10.1038/s41598-018-34678-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cully M. Microbiome therapeutics go small molecule. Nat Rev Drug Discov. 2019;18(8):569‐572. doi: 10.1038/d41573-019-00122-8 [DOI] [PubMed] [Google Scholar]
- 10. Peterson CT, Sharma V, Elmén L, Peterson SN. Immune homeostasis, dysbiosis and therapeutic modulation of the gut microbiota. Clin Exp Immunol. 2015;179(3):363‐377. doi: 10.1111/cei.12474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Roberts AB, Wallace BD, Venkatesh MK, Mani S, Redinbo MR. Molecular insights into microbial β‐glucuronidase inhibition to abrogate CPT‐11 toxicity. Mol Pharmacol. 2013;84(2):208‐217. doi: 10.1124/mol.113.085852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang P, Jia Y, Wu R, Chen Z, Yan R. Human gut bacterial β‐glucuronidase inhibition: an emerging approach to manage medication therapy. Biochem Pharmacol. 2021;190:114566. doi: 10.1016/j.bcp.2021.114566 [DOI] [PubMed] [Google Scholar]
- 13. Little MS, Pellock SJ, Walton WG, Tripathy A, Redinbo MR. Structural basis for the regulation of β‐glucuronidase expression by human gut Enterobacteriaceae. Proc Natl Acad Sci U S A. 2018;115(2):E152‐E161. doi: 10.1073/pnas.1716241115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Awolade P, Cele N, Kerru N, Gummidi L, Oluwakemi E, Singh P. Therapeutic significance of β‐glucuronidase activity and its inhibitors: a review. Eur J Med Chem. 2020;187:111921. doi: 10.1016/j.ejmech.2019.111921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Maseda D, Ricciotti E. NSAID–gut microbiota interactions. Front Pharmacol. 2020;11:1153. doi: 10.3389/fphar.2020.01153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Saitta KS, Zhang C, Lee KK, Fujimoto K, Redinbo MR, Boelsterli UA. Bacterial β‐glucuronidase inhibition protects mice against enteropathy induced by indomethacin, ketoprofen or diclofenac: mode of action and pharmacokinetics. Xenobiotica. 2014;44(1):28‐35. doi: 10.3109/00498254.2013.811314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen M, Cheng KW, Chen YJ, et al. Real‐time imaging of intestinal bacterial β‐glucuronidase activity by hydrolysis of a fluorescent probe. Sci Rep. 2017;7:3142. doi: 10.1038/s41598-017-03252-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Flannigan KL, Taylor MR, Pereira SK, et al. An intact microbiota is required for the gastrointestinal toxicity of the immunosuppressant mycophenolate mofetil. J Heart Lung Transplant. 2018;37(9):1047‐1059. doi: 10.1016/j.healun.2018.05.002 [DOI] [PubMed] [Google Scholar]
- 19. Takasuna K, Hagiwara T, Hirohashi M, et al. Involvement of β‐glucuronidase in intestinal microflora in the intestinal toxicity of the antitumor Camptothecin derivative irinotecan hydrochloride (CPT‐11) in rats. Cancer Res. 1996;56(16):3752‐3757. [PubMed] [Google Scholar]
- 20. Kim S, Covington A, Pamer EG. The intestinal microbiota: antibiotics, colonization resistance, and enteric pathogens. Immunol Rev. 2017;279(1):90‐105. doi: 10.1111/imr.12563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pellock SJ, Creekmore BC, Walton WG, et al. Gut microbial β‐glucuronidase inhibition via catalytic cycle interception. ACS Cent Sci. 2018;4(7):868‐879. doi: 10.1021/acscentsci.8b00239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chamseddine AN, Ducreux M, Armand JP, et al. Intestinal bacterial β‐glucuronidase as a possible predictive biomarker of irinotecan‐induced diarrhea severity. Pharmacol Ther. 2019;199:1‐15. doi: 10.1016/j.pharmthera.2019.03.002 [DOI] [PubMed] [Google Scholar]
- 23. Praveschotinunt P, Duraj‐Thatte AM, Gelfat I, et al. Coli Nissle 1917 for the delivery of matrix‐tethered therapeutic domains to the gut. Nat Commun. 2019;10(1):5580. doi: 10.1038/s41467-019-13336-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Novel M, Novel G. Regulation of beta‐glucuronidase synthesis in Escherichia coli K‐12: constitutive mutants specifically derepressed for uidA expression. J Bacteriol. 1976;127(1):406‐417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Massip C, Branchu P, Bossuet‐Greif N, et al. Deciphering the interplay between the genotoxic and probiotic activities of Escherichia coli Nissle 1917. PLoS Pathog. 2019;15(9):e1008029. doi: 10.1371/journal.ppat.1008029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pradhan S, Weiss AA. Probiotic properties of Escherichia coli Nissle in human intestinal organoids. mBio. 2020;11(4):e01470‐20. doi: 10.1128/mBio.01470-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Becker HM, Apladas A, Scharl M, Fried M, Rogler G. Probiotic Escherichia coli Nissle 1917 and commensal E. coli K12 differentially affect the inflammasome in intestinal epithelial cells. Digestion. 2014;89(2):110‐118. doi: 10.1159/000357521 [DOI] [PubMed] [Google Scholar]
- 28. Escribano‐Vazquez U, Verstraeten S, Martin R, et al. The commensal Escherichia coli CEC15 reinforces intestinal defences in gnotobiotic mice and is protective in a chronic colitis mouse model. Sci Rep. 2019;9(1):11431. doi: 10.1038/s41598-019-47611-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vandermeulen G, Marie C, Scherman D, Préat V. New generation of plasmid backbones devoid of antibiotic resistance marker for gene therapy trials. Mol Ther. 2011;19(11):1942‐1949. doi: 10.1038/mt.2011.182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jalili‐Firoozinezhad S, Gazzaniga FS, Calamari EL, et al. A complex human gut microbiome cultured in an anaerobic intestine‐on‐a‐chip. Nat Biomed Eng. 2019;3(7):520‐531. doi: 10.1038/s41551-019-0397-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cordonnier C, Thévenot J, Etienne‐Mesmin L, et al. Dynamic in vitro models of the human gastrointestinal tract as relevant tools to assess the survival of probiotic strains and their interactions with gut microbiota. Microorganisms. 2015;3(4):725‐745. doi: 10.3390/microorganisms3040725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Metagenomics of the Human Intestinal Tract (MetaHIT) Consortium , Plichta DR, Juncker AS, Bertalan M, et al. Transcriptional interactions suggest niche segregation among microorganisms in the human gut. Nat Microbiol. 2016;1(11):16152. doi: 10.1038/nmicrobiol.2016.152 [DOI] [PubMed] [Google Scholar]
- 33. Pena RT, Blasco L, Ambroa A, et al. Relationship between quorum sensing and secretion systems. Front Microbiol. 2019;10:1100. doi: 10.3389/fmicb.2019.01100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kanonenberg K, Schwarz CKW, Schmitt L. Type I secretion systems ‐ a story of appendices. Res Microbiol. 2013;164(6):596‐604. doi: 10.1016/j.resmic.2013.03.011 [DOI] [PubMed] [Google Scholar]
- 35. Li Z, Nair SK. Quorum sensing: how bacteria can coordinate activity and synchronize their response to external signals? Protein Sci. 2012;21(10):1403‐1417. doi: 10.1002/pro.2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu S, Feng J, Liu C, et al. Machine learning aided construction of the quorum sensing communication network for human gut microbiota. Nat Commun. 2022;13(1):3079. doi: 10.1038/s41467-022-30741-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Picard N, Ratanasavanh D, Prémaud A, Le Meur Y, Marquet P. Identification of the UDP‐glucuronosyltransferase isoforms involved in mycophenolic acid phase II metabolism. Drug Metab Dispos. 2005;33(1):139‐146. doi: 10.1124/dmd.104.001651 [DOI] [PubMed] [Google Scholar]
- 38. Harding SD, Sharman JL, Faccenda E, et al. The IUPHAR/BPS guide to PHARMACOLOGY in 2018: updates and expansion to encompass the new guide to IMMUNOPHARMACOLOGY. Nucleic Acids Res. 2018;46(D1):D1091‐D1106. doi: 10.1093/nar/gkx1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Alexander SPH, Fabbro D, Kelly E, et al. THE CONCISE GUIDE TO PHARMACOLOGY 2021/22: enzymes. Br J Pharmacol. 2021;178(S1):S313‐S411. doi: 10.1111/bph.15542 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


