Abstract
Background
Intestinal dysbiosis has been reported to be associated with celiac disease (CeD) in Western populations but little is known in other populations who have different dietary lifestyle and genetic background. The purpose of this study was to determine whether a different microbiota profile is associated with CeD in children in Saudi Arabia.
Results
Forty children with CeD, 20 healthy controls, and 19 non-CeD controls were enrolled. The median age at diagnosis was 10.3, 11.3 and 10.6 years in children with CeD, fecal, and mucosal control groups, respectively. Significant differences in microbial composition between children with CeD and controls both at fecal and mucosal level were identified. Fecal samples were more diverse and richer in bacteria as compared with mucosal samples. Proteobacteria were more abundant in duodenal mucosal samples and Firmicutes and Bacteroides were more abundant in stools. The abundance of many taxa was significantly different between children with CeD and non-CeD controls. In mucosal samples, Bifidobacterium angulatum (unadjusted p = 0.006) and Roseburia intestinalis (unadjusted p = 0.031) were examples of most significantly increased species in children with CeD and non-CeD controls, respectively. In fecal samples, there were 169 bacterial species with significantly different abundance between children with CeD and non- CeD controls.
Conclusions
To our knowledge, this is the first report on the microbial profile in a non-Western population of children with new onset CeD. The fact that mucosal and fecal samples were collected from newly diagnosed children with CeD on normal gluten-containing diet suggests strong association between the identified bacteria and CeD. The identification of many unreported bacterial species significantly associated with CeD, indicates the need for further studies from different populations to expand our understanding of the role of bacteria in the pathogenesis of CeD, hopefully leading to the discovery of new adjuvant treatment options.
Keywords: Bacteriome, Celiac disease, Children
Background
Celiac disease (CeD) is defined as an autoimmune enteropathy triggered by gluten, affecting genetically predisposed individuals (HLA DQ2 and/or DQ8) [1, 2]. Recent data show that tolerance to gluten can be lost at any time in life [3]. These findings, together with the lack of complete CeD concordance among monozygotic twins, suggest that, while genetic predisposition and gluten intake are necessary for CeD development, they are insufficient to trigger the onset of the disease [4]. Thus, other contributing factors such as changes in microbiome composition and function have been suggested to be associated with CeD.
The microbiome of a healthy individual is relatively stable by 3 years of age; however, this composition can be modulated throughout the entire lifespan by different factors, such as lifestyle, dietary choices, antibiotic treatment, stress, and other environmental components. Intestinal dysbiosis via such factors has been reported to be associated with development of CeD [5].
The Saudi population has been reported to have a high prevalence of CeD (1.5%). The high rate of CeD-predisposing HLA-DQ genotypes in the general population (52.7%) may partially account for this high prevalence, although additional external factors should also be taken into consideration [6]. The consumption of gluten-containing cereals in the diet of the Saudi population is reported to be very high as recorded by the Food and Drug Organization [7]. This high intake of cereals may directly increase the prevalence of CeD, or indirectly by altering other factors such as the microbiome composition.
Most of the literature on the microbiome in CeD were from Western populations. Cultural and dietary lifestyle in non-Western populations, mostly developing countries could affect microbiota profile and studies on microbiome in CeD from these populations may increase our understanding of the pathogenesis of CeD. Therefore, our objective was to determine whether a different microbiota profile is associated with CeD in children in Saudi Arabia.
Results
Characteristics of the study population
A total of 40 children with CeD (provided 20 tissue and 20 stool samples) and 39 controls were enrolled in this study. There were two types of controls. Twenty healthy children who provided stool samples only (fecal controls), and 19 non-CeD children who provided mucosal samples only (mucosal controls). The latter had normal endoscopy and normal duodenal mucosal histopathology. In addition, all controls had normal anti-tissue transglutaminase A values. The demographic and clinical characteristics are presented in Table 1. Briefly, males accounted for 28%, 35%, and 42% of the children with CeD, fecal, and mucosal control groups respectively. The median age at diagnosis was 10.3, 11.3 and 10.6 years in children with CeD, fecal, and mucosal control groups respectively. The number of asymptomatic children with CeD was 15/40 (38%), whereas the remainder had various combination of symptoms including anemia, growth impairment, and abdominal pain.
Table 1.
Demographic and clinical characteristics
| Variables | Celiac disease | Fecal controls | Mucosal controls |
|---|---|---|---|
| Number of children | 40 | 20 | 19 |
| Sex (% male sex) | 28% | 35% | 42% |
| Age at presentation in years: median (range) | 10.3 (7.5–15.7) | 11.3 (6.8 -15.4) | 10.6 (2–17.2) |
| Breastfeeding (%) | 85% | 68% | |
| Clinical presentation | |||
| Asymptomatic | 15 (38%) | 20 (100%) | 0 (0%) |
| Anemia | 11 (28%) | 0 (0%) | 1 (5%) |
| Diarrhea/A. distention | 7 (18%) | 0 (0%) | 0 (0%) |
| Growth impairment | 10 (25%) | 0 (0%) | 1 (5%) |
| Abdominal pain | 10 (25%) | 0 (0%) | 10 (53%) |
| Constipation | 8 (20%) | 0 (0%) | 0 (0%) |
| Vomiting | 0 (0%) | 0 (0%) | 3 (16%) |
| Dysphagia | 0 (0%) | 0 (0%) | 4 (21%) |
Alpha- and beta-diversities
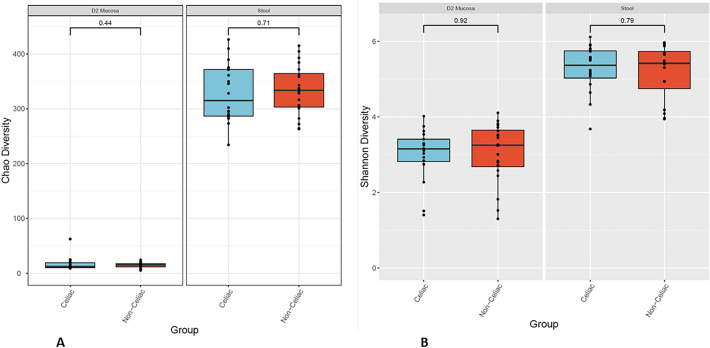
Differences in alpha diversity between the CeD and non-CeD groups were measured in both fecal and duodenal samples using the Chao and Shannon indices, an abundance-based estimators of species richness. Although not statistically significant, our analysis showed a clear difference in bacterial diversity between the mucosal and fecal samples, indicating an increased richness and variability in stools (Fig. 1A, B). Interestingly, alpha diversity did not differ between CeD and non-CeD groups, despite there being a trend toward smaller diversity in CeD stools compared with that in non-CeD stools.
Fig. 1.
Alpha diversity. Illustration of alpha diversity measured by Chao index (A) and by Shannon index (B) for bacterial communities in duodenal and fecal samples of CeD patients and non-CeD controls
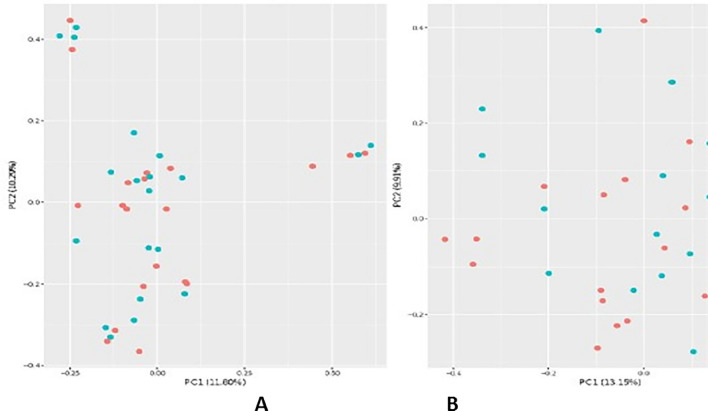
For bacterial beta diversity, Bray–Curtis PCoA analysis did not show any significant clustering patterns in samples from the duodenal mucosa or stools of the CeD and non-CeD groups (Fig. 2A, B). However, in the analysis of bacterial fecal samples, there were small clusters characteristic of either CeD or non-CeD groups.
Fig. 2.
Beta diversity. Bray–Curtis-based bacterial beta-diversity analysis of mucosal (A) and fecal samples (B) from patients with CeD (pink dots) or non-CeD controls (blue dots)
Overall bacterial composition
The overall bacterial composition of fecal and mucosal samples was analyzed in both CeD and non-CeD groups; this was represented through heatmap (Fig. 3). As expected, the bacterial richness in stools was higher than that in duodenal samples, and in both sets of samples, Firmicutes and Bacteroidetes were the most abundant phyla. In duodenal samples, an increased percentage of Proteobacteria species was detected, whereas overall, the stools were characterized by increased abundance of Verrucomicrobia species.
Fig. 3.
Heatmap: Representing bacterial microbiome composition in duodenal and fecal samples of patients with the CeD and non-CeD. The bacterial richness in stools was higher than that in duodenal samples, Firmicutes and Bacteroidetes were the most abundant phyla. Actinobacteria abundance was reduced
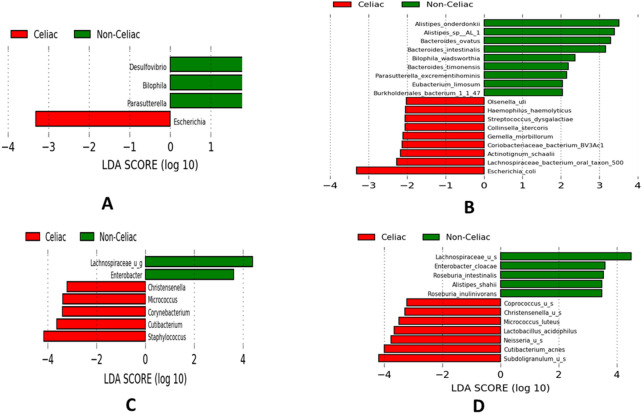
LDA effect size
The LDA effect size (LEfSe) plot revealed statistically significant different bacterial composition in fecal samples between children with CeD and non-CeD controls. For example, there was an increase of Escherichia in the CeD group and an increase of Desulfovibrio in the non-CeD group at the genus level (decreased in the CeD group) (Fig. 4A). Similarly, at the species level, there was a statistically-significant difference between the CeD and non-CeD group. For example, in the CeD group there was an increase of E. coli and Lachnospiraceae_bacterium_oral; whereas several species of Bacteroides were significantly increased in fecal samples of non-CeD controls (decreased in CeD) (Fig. 4B). In mucosal samples, although not statistically different by standard criteria, there were important differences in abundance of several taxa between CeD and non- CeD mucosal samples. For example, Lactobacillus acidophilus, Neisseria and Coprococcus species were increased in the CeD group; whereas Roseburia and Lachnospiraceae species were increased in non-CeD group (decreased in the CeD group) (Fig. 4C, D).
Fig. 4.
LEfSE LDA scores: A and B show statistically significant abundance difference in stool samples from patients with CeD with those of non-CeD controls at the genus (A) and species (B) level. C and D Illustrate the abundance difference, although not statistically different, of LDA scores in mucosal samples from patients with CeD with those of non-CeD controls at the genus (C) and species (D) level. Bars with a positive LDA score (green) are higher in non-celiac samples, and bars with a negative LDA score (red) are higher in celiac samples. The extensions u_g and _u_s mean unclassified genera and species respectively
DeSeq2 differential abundance analysis
DeSeq2 differential abundance analysis revealed statistically significant differences in log2 fold change abundance between CeD and non-CeD samples. Log2 fold change > 0 and < 0 indicate increased abundance in children with CeD and non-CeD respectively. Increased abundance in children with non-CeD suggests decreased abundance in children with CeD. Table 2 shows the log2 abundance change for the top 10 taxa (order, family, and genera) in mucosal and fecal samples, illustrating the different microbiota profile between mucosa and stool. For example, in mucosal samples, Flavobacteriales (p = 0.0005), Flavobacteriaceae (unadjusted p = 8.11–08), and Clostridium (unadjusted p = 0.011), were the most significantly decreased bacterial order, family, and genus levels respectively, whereas Micrococcales (unadjusted p = 0.018), Micrococcaceae (unadjusted p = 0.022), and Subdoligranulum (unadjusted p = 0.021) were the most significantly abundant bacterial order, family and genus levels respectively. In fecal samples, however, Cardiobacteriales (p = 0.01), Leuconostocaceae (p = 0.003), and Tannerella (p = 1.17–05) were the most significantly decreased bacterial order, family and genus levels respectively, whereas Planctomycetaceae (p = 0.013) and Kocuria (p = 0.003) were the most abundant family and genera levels. The top 10 most significant species abundance in mucosal samples is presented in Table 3. In these samples, Bifidobacterium angulatum (unadjusted p = 0.006) and Roseburia intestinalis (unadjusted p = 0.031), were examples of increased species in mucosal samples of children with CeD and non-CeD (decreased in CeD) respectively.
Table 2.
Log2 abundance change for bacterial order, family, and genera in children with CeD
| Level | Organism | Log2 abundance | p value | *p value | Organism | Log2 abundance | p value | *p value |
|---|---|---|---|---|---|---|---|---|
| Mucosal samples | Fecal samples | |||||||
| Order | Flavobacteriales | – 0.4366 | 1.93–05 | 0.0005 | Cardiobacteriales | – 0.4882 | 0.0001 | 0.01 |
| Order | Micrococcales | 1.0333 | 0.018 | 0.253 | Methanobacteriales | – 1.3944 | 0.0004 | 0.02 |
| Family | Flavobacteriaceae | – 0.3014 | 8.11–08 | 0.219 | Leuconostocaceae | – 2.9810 | 4.20–05 | 0.003 |
| Family | Clostridiaceae | – 1.0072 | 0.008 | 0.274 | Cardiobacteriaceae | – 0.4851 | 0.0002 | 0.010 |
| Family | Lactobacillaceae | – 0.0115 | 0.017 | 0.274 | Planctomycetaceae | 1.5023 | 0.0003 | 0.013 |
| Family | Micrococcaceae | 0.9708 | 0.022 | 0.219 | Methanobacteriaceae | – 1.2405 | 0.0004 | 0.014 |
| Genus | Clostridium | – 0.9103 | 0.011 | 0.486 | Tannerella | – 2.6849 | 4.81–08 | 1.17–05 |
| Genus | Lactobacillus | 0.0586 | 0.015 | 0.486 | Citrobacter | – 2.4701 | 7.65–05 | 0.003 |
| Genus | Subdoligranulum | 0.8853 | 0.021 | 0.486 | Methanobrevibacter | – 1.4565 | 0.0007 | 0.017 |
| Genus | Kocuria | 0.7998 | 0.036 | 0.634 | Kocuria | 1.7515 | 9.25–05 | 0.003 |
*p-value adjusted for false discovery rate
Table 3.
Log2 fold abundance change of the top 10 bacterial species in mucosal samples of children with CeD
| Organism | Log 2-fold change | p value | *p value |
|---|---|---|---|
| Bifidobacterium angulatum | 0.4177 | 0.006 | 0.460 |
| Lactobacillus acidophilus | 0.8935 | 0.011 | 0.460 |
| Kocuria rhizophila | 0.8462 | 0.021 | 0.460 |
| Roseburia intestinalis | – 0.7395 | 0.031 | 0.460 |
| Ralstonia pickettii | 0.7255 | 0.037 | 0.460 |
| Acinetobacter lwoffii | 0.7059 | 0.042 | 0.460 |
| Corynebacterium ihumii | 0.6856 | 0.047 | 0.460 |
| Corynebacterium tuberculostearicum | 0.6856 | 0.047 | 0.460 |
| Bradyrhizobium sp. DFCI-1 | 1.5177 | 0.075 | 0.601 |
| Staphylococcus aureus | 0.5959 | 0.0801 | 0.601 |
*p value adjusted for false discovery rate
The log2 fold change abundance of 169 significantly different bacterial species in fecal samples of children with CeD and non-CeD controls is depicted in Table 4. There were several species significantly decreased in children with CeD belonging to the Bifidobacterium genus, such as B. breve (p = 0.0028), B. angolatum (p = 2.24−07), B. merycicum (p = 0.012), and B. thermophilum (p = 0.027). Among Lactobacilli species, L. plantarum (p = 0.0043), was significantly less abundant in CeD samples, whereas the abundance of other lactobacilli such as L. gasseri (p = 0.033) was significantly- increased in children with CeD. Prevotella species (P. timonensis (p = 0.018); P. bergensis (p = 0.022) were significantly more abundant in stool samples of children with CeD, whereas Prevotella sp. P5-119 was significantly less abundant (p = 1.69–06). Finally, several Bacteroides species were less abundant in fecal samples from children with CeD. In contrast, different Clostridium species were increased in abundance among children with CeD.
Table 4.
Log2 fold change abundance of bacterial species in fecal samples of children with CeD
| S. no. | Organism | Log2 fold change | P value | *p value |
|---|---|---|---|---|
| 1 | Clostridium sp. L2-50 | – 2.0120 | 8.85–20 | 3.43–17 |
| 2 | Ruminococcus sp. SR1/5 | – 0.2670 | 9.89–20 | 3.43–17 |
| 3 | Streptococcus sp. SR4 | 0.0787 | 2.91–13 | 6.75–11 |
| 4 | Actinomyces sp. oral taxon 175 | 2.0207 | 1.80–12 | 3.12–10 |
| 5 | Clostridiales bacterium VE202-07 | 0.7391 | 3.68–11 | 5.11–09 |
| 6 | Actinomyces sp. ICM39 | – 1.6899 | 1.01–10 | 1.16–08 |
| 7 | Bacteroides sp. 1_1_30 | – 0.9884 | 1.39–10 | 1.37–08 |
| 8 | Alistipes inops | – 5.4881 | 4.50–10 | 3.90–08 |
| 9 | Desulfovibrio piger | – 0.7593 | 7.71–10 | 5.94–08 |
| 10 | Actinomyces sp. ICM58 | – 5.2606 | 3.41–09 | 2.24–07 |
| 11 | Bifidobacterium angulatum | – 1.3309 | 3.55–09 | 2.24–07 |
| 12 | Actinobaculum massiliense | 4.2776 | 6.03–09 | 3.49–07 |
| 13 | Bacteroides clarus | – 3.5549 | 6.67–09 | 3.56–07 |
| 14 | Oscillibacter sp. KLE 1745 | 1.8464 | 3.35–08 | 1.66–06 |
| 15 | Lactobacillus acidophilus | 0.8589 | 3.69–08 | 1.69–06 |
| 16 | Prevotella sp. P5-119 | – 3.6227 | 4.01–08 | 1.69–06 |
| 17 | Burkholderiales bacterium 1_1_47 | – 3.2591 | 4.14–08 | 1.69–06 |
| 18 | Coprobacter fastidiosus | – 4.7198 | 6.62–08 | 2.55–06 |
| 19 | Tannerella sp. 6_1_58FAA_CT1 | – 2.9135 | 7.82–08 | 2.85–06 |
| 20 | Blautia hydrogenotrophica | 4.3470 | 1.11–07 | 3.88–06 |
| 21 | Coriobacteriaceae bacterium BV3Ac1 | 2.8402 | 1.51–07 | 4.99–06 |
| 22 | Phascolarctobacterium succinatutens | 0.1041 | 2.10–07 | 6.63–06 |
| 23 | Citrobacter freundii | – 3.9256 | 3.56–07 | 1.07–05 |
| 24 | Leuconostoc pseudomesenteroides | – 4.7313 | 5.15–07 | 1.49–05 |
| 25 | Parasutterella excrementihominis | – 2.8930 | 1.38–06 | 3.84–05 |
| 26 | Coprobacillus sp. D6 | 0.7717 | 2.13–06 | 5.69–05 |
| 27 | [Ruminococcus] gnavus | 1.4824 | 2.73–06 | 7.03–05 |
| 28 | Klebsiella variicola | – 3.4290 | 3.35–06 | 8.32–05 |
| 29 | Actinomyces sp. HPA0247 | 0.3485 | 4.56–06 | 0.0001 |
| 30 | [Clostridium] spiroforme | – 0.9840 | 8.71–06 | 0.0002 |
| 31 | Bacteroides eggerthii | – 3.4963 | 1.24–05 | 0.0002 |
| 32 | Megasphaera massiliensis | 2.4114 | 2.15–05 | 0.0004 |
| 33 | Clostridiales bacterium VE202-26 | 2.0418 | 2.29–05 | 0.0004 |
| 34 | Megasphaera sp. BL7 | – 3.4443 | 2.52–05 | 0.0005 |
| 35 | Actinotignum schaalii | 2.0700 | 3.82–05 | 0.0007 |
| 36 | Enterococcus avium | 0.8707 | 7.96–05 | 0.0015 |
| 37 | Eggerthella sp. 1_3_56FAA | – 0.3357 | 8.57–05 | 0.0016 |
| 38 | Corynebacterium pyruviciproducens | 5.6575 | 8.87–05 | 0.0016 |
| 39 | Eubacterium limosum | – 0.2074 | 0.0001 | 0.0018 |
| 40 | Bifidobacterium breve | – 0.6821 | 0.0001 | 0.0028 |
| 41 | Mitsuokella jalaludinii | 0.7105 | 0.0001 | 0.0028 |
| 42 | Weissella confusa | – 2.7298 | 0.0002 | 0.0037 |
| 43 | Streptococcus pneumoniae | 1.8511 | 0.0002 | 0.0042 |
| 44 | Lactobacillus plantarum | – 1.0534 | 0.0002 | 0.0043 |
| 45 | Actinomyces cardiffensis | – 0.1851 | 0.0004 | 0.0068 |
| 46 | Bacteroides faecichinchillae | – 2.1328 | 0.0004 | 0.0068 |
| 47 | Lactobacillus mucosae | 0.6968 | 0.0005 | 0.0077 |
| 48 | Alistipes sp. HGB5 | 0.9527 | 0.0005 | 0.0081 |
| 49 | Peptoniphilus harei | 2.6088 | 0.0006 | 0.0083 |
| 50 | Kocuria rhizophila | 1.5647 | 0.0006 | 0.0083 |
| 51 | Haemophilus haemolyticus | 1.8040 | 0.0007 | 0.0098 |
| 52 | Bacteroides gallinarum | – 0.9100 | 0.0007 | 0.0098 |
| 53 | [Eubacterium] siraeum | – 1.1823 | 0.0008 | 0.0103 |
| 54 | Scardovia inopinata | 2.6347 | 0.0008 | 0.0104 |
| 55 | Dialister succinatiphilus | – 0.6384 | 0.001 | 0.011 |
| 56 | Eggerthella sp. YY7918 | – 2.9586 | 0.001 | 0.012 |
| 57 | Bifidobacterium merycicum | – 1.2398 | 0.001 | 0.012 |
| 58 | Alistipes onderdonkii | – 1.8638 | 0.001 | 0.012 |
| 59 | Lachnospiraceae bacterium oral taxon 500 | 2.4268 | 0.001 | 0.013 |
| 60 | Alistipes sp. AL-1 | – 2.0624 | 0.001 | 0.013 |
| 61 | Kandleria vitulina | – 1.1850 | 0.001 | 0.014 |
| 62 | Actinomyces turicensis | 1.7418 | 0.001 | 0.015 |
| 63 | Kallipyga massiliensis | 1.4601 | 0.001 | 0.015 |
| 64 | Prevotella timonensis | 2.3958 | 0.002 | 0.018 |
| 65 | Alistipes indistinctus | – 0.9069 | 0.002 | 0.018 |
| 66 | Streptococcus parauberis | – 1.4411 | 0.002 | 0.021 |
| 67 | Streptococcus dysgalactiae | 3.0429 | 0.002 | 0.021 |
| 68 | Acinetobacter junii | 1.3684 | 0.002 | 0.022 |
| 69 | Prevotella bergensis | 2.2901 | 0.002 | 0.022 |
| 70 | Anaerococcus prevotii | 1.3590 | 0.002 | 0.023 |
| 71 | Bifidobacterium biavatii | 0.1734 | 0.002 | 0.023 |
| 72 | Weissella cibaria | – 3.1430 | 0.003 | 0.026 |
| 73 | Peptostreptococcus anaerobius | 1.7918 | 0.003 | 0.026 |
| 74 | Gardnerella vaginalis | 1.5445 | 0.003 | 0.026 |
| 75 | Eggerthia catenaformis | – 1.3356 | 0.003 | 0.027 |
| 76 | Bifidobacterium thermophilum | – 0.8901 | 0.003 | 0.027 |
| 77 | Bacteroides finegoldii | – 2.9953 | 0.003 | 0.027 |
| 78 | Atopobium vaginae | 1.5527 | 0.003 | 0.028 |
| 79 | Mitsuokella multacida | 2.0038 | 0.003 | 0.028 |
| 80 | Anaerostipes sp. 3_2_56FAA | – 4.0899 | 0.003 | 0.028 |
| 81 | Catonella morbi | 1.5910 | 0.003 | 0.028 |
| 82 | Ruminococcus gauvreauii | 1.5330 | 0.003 | 0.029 |
| 83 | Streptococcus anginosus | 1.5551 | 0.004 | 0.031 |
| 84 | Dialister invisus | – 0.8902 | 0.004 | 0.031 |
| 85 | Dialister micraerophilus | 2.4775 | 0.004 | 0.032 |
| 86 | Sharpea azabuensis | – 1.5192 | 0.004 | 0.033 |
| 87 | Lactobacillus gasseri | 2.1423 | 0.004 | 0.033 |
| 88 | Coprobacillus sp. D7 | 0.5299 | 0.004 | 0.033 |
| 89 | Escherichia sp. 1_1_43 | 0.9507 | 0.004 | 0.033 |
| 90 | Anaerococcus obesiensis | 1.2936 | 0.004 | 0.033 |
| 91 | Clostridium celatum | 1.5093 | 0.005 | 0.037 |
| 92 | Clostridium paraputrificum | 2.9282 | 0.005 | 0.037 |
| 93 | Actinomyces dentalis | 1.3394 | 0.005 | 0.038 |
| 94 | Coprococcus sp. HPP0074 | 1.9454 | 0.005 | 0.038 |
| 95 | [Clostridium] saccharogumia | 1.2495 | 0.006 | 0.040 |
| 96 | Enterobacter cloacae complex 'Hoffmann cluster IV' | 1.3726 | 0.006 | 0.040 |
| 97 | Streptococcus sinensis | 1.2504 | 0.006 | 0.042 |
| 98 | Mageeibacillus indolicus | – 1.2756 | 0.007 | 0.047 |
| 99 | Bacteroides stercorirosoris | – 1.5318 | 0.007 | 0.048 |
| 100 | Bifidobacterium saguini | 2.2018 | 0.007 | 0.048 |
| 101 | Lactobacillus ultunensis | 2.0939 | 0.008 | 0.048 |
| 102 | Klebsiella sp. 10,982 | – 1.2026 | 0.008 | 0.048 |
| 103 | Klebsiella michiganensis | 5.7397 | 0.008 | 0.048 |
| 104 | Megasphaera micronuciformis | – 0.3100 | 0.008 | 0.050 |
| 105 | Anaeroglobus geminatus | 2.7566 | 0.009 | 0.054 |
| 106 | Corynebacterium pseudodiphtheriticum | 1.1915 | 0.009 | 0.055 |
| 107 | Clostridiales bacterium VE202-09 | – 3.922 | 0.010 | 0.059 |
| 108 | Christensenella minuta | – 0.3566 | 0.010 | 0.059 |
| 109 | Leuconostoc mesenteroides | 0.0528 | 0.010 | 0.061 |
| 110 | Bacteroides barnesiae | – 2.0367 | 0.011 | 0.062 |
| 111 | Corynebacterium argentoratense | – 0.7247 | 0.012 | 0.071 |
| 112 | Intestinimonas butyriciproducens | 0.3570 | 0.013 | 0.075 |
| 113 | Propionibacterium acidifaciens | 1.9602 | 0.014 | 0.079 |
| 114 | Klebsiella pneumoniae | – 1.3439 | 0.016 | 0.084 |
| 115 | Streptococcus constellatus | 0.6456 | 0.017 | 0.095 |
| 116 | Anaerococcus vaginalis | 1.0155 | 0.018 | 0.097 |
| 117 | Enorma massiliensis | 1.2870 | 0.018 | 0.097 |
| 118 | Lachnospiraceae bacterium 2_1_58FAA | 1.4788 | 0.019 | 0.099 |
| 119 | Ruminococcaceae bacterium D16 | – 0.8005 | 0.020 | 0.105 |
| 120 | Bacteroides salanitronis | 0.0412 | 0.020 | 0.105 |
| 121 | Akkermansia muciniphila | – 0.4313 | 0.021 | 0.111 |
| 122 | Bifidobacterium minimum | – 2.4337 | 0.022 | 0.112 |
| 123 | Candidatus Saccharibacteria oral taxon TM7x | 1.1047 | 0.022 | 0.112 |
| 124 | Shigella sp. SF-2015 | 1.7508 | 0.024 | 0.119 |
| 125 | Bifidobacterium callitrichos | – 0.0551 | 0.024 | 0.119 |
| 126 | Lachnospiraceae bacterium 1_4_56FAA | – 0.3516 | 0.025 | 0.124 |
| 127 | Bifidobacterium gallinarum | 0.2816 | 0.026 | 0.126 |
| 128 | Lachnospiraceae bacterium 9_1_43BFAA | 2.1702 | 0.027 | 0.128 |
| 129 | Turicibacter sp. HGF1 | 1.9376 | 0.027 | 0.128 |
| 130 | Leucobacter chironomi | – 0.9613 | 0.027 | 0.128 |
| 131 | Slackia exigua | 1.4279 | 0.027 | 0.128 |
| 132 | Streptococcus pyogenes | 1.9995 | 0.028 | 0.128 |
| 133 | Bulleidia extructa | 1.0910 | 0.028 | 0.129 |
| 134 | Streptococcus mutans | 1.2386 | 0.028 | 0.130 |
| 135 | bacterium LF-3 | 1.1778 | 0.030 | 0.135 |
| 136 | Morococcus cerebrosus | 0.9367 | 0.030 | 0.136 |
| 137 | Klebsiella oxytoca | 1.8890 | 0.030 | 0.136 |
| 138 | Raoultella ornithinolytica | – 4.1641 | 0.031 | 0.137 |
| 139 | Enterococcus saccharolyticus | 0.1371 | 0.031 | 0.137 |
| 140 | Mucispirillum schaedleri | – 1.0032 | 0.032 | 0.138 |
| 141 | Butyricimonas synergistica | 0.1619 | 0.032 | 0.138 |
| 142 | Eubacterium ramulus | – 0.9600 | 0.033 | 0.142 |
| 143 | Prevotella sp. BV3P1 | 4.9067 | 0.034 | 0.144 |
| 144 | Parabacteroides goldsteinii | 1.2363 | 0.035 | 0.150 |
| 145 | Haemophilus pittmaniae | 0.2372 | 0.036 | 0.152 |
| 146 | Oribacterium sinus | 0.9714 | 0.036 | 0.152 |
| 147 | Pseudoramibacter alactolyticus | 0.1447 | 0.037 | 0.153 |
| 148 | Olsenella profusa | 2.2762 | 0.038 | 0.156 |
| 149 | Bacteroides pyogenes | – 4.6019 | 0.038 | 0.158 |
| 150 | Streptococcus sp. SR1 | 0.9154 | 0.039 | 0.160 |
| 151 | candidate division TM7 single-cell isolate TM7b | 1.1954 | 0.040 | 0.161 |
| 152 | Eikenella corrodens | – 0.9100 | 0.041 | 0.165 |
| 153 | Enterobacter sp. MGH 38 | – 5.7753 | 0.042 | 0.165 |
| 154 | Streptococcus oralis | 1.0494 | 0.042 | 0.165 |
| 155 | Lactobacillus casei group | – 0.8741 | 0.043 | 0.168 |
| 156 | Desulfovibrio desulfuricans | – 1.1163 | 0.043 | 0.169 |
| 157 | Escherichia fergusonii | – 1.4098 | 0.044 | 0.169 |
| 158 | Paraclostridium bifermentans | 0.9890 | 0.044 | 0.169 |
| 159 | Citrobacter koseri | 0.8884 | 0.045 | 0.169 |
| 160 | Granulicatella elegans | – 0.3866 | 0.045 | 0.169 |
| 161 | Succinivibrio dextrinosolvens | – 2.7856 | 0.046 | 0.171 |
| 162 | Streptococcus thermophilus | – 0.8128 | 0.046 | 0.171 |
| 163 | Prevotella buccae | 0.8820 | 0.046 | 0.171 |
| 164 | Lactobacillus jensenii | – 0.4273 | 0.046 | 0.171 |
| 165 | butyrate-producing bacterium SM4/1 | – 2.8000 | 0.047 | 0.171 |
| 166 | Atopobium sp. ICM42b | – 0.9230 | 0.047 | 0.171 |
| 167 | Bifidobacterium longum | 0.9381 | 0.048 | 0.176 |
| 168 | Parabacteroides johnsonii | 1.6349 | 0.049 | 0.176 |
| 169 | Collinsella intestinalis | 1.0310 | 0.049 | 0.177 |
*p value adjusted for false discovery rate
Discussion
The association between CeD and intestinal dysbiosis has already been described in several studies [8–11]. However, the exact role of the microbiome in CeD pathogenesis has not yet been fully elucidated, and, given the fundamental functions that the intestinal microbiota plays in regulating intestinal homeostasis, it has been suggested that specific changes in microbiome composition may contribute to CeD onset [12]. The intestinal microflora is very functionally diverse, and its composition can depend on the intestinal site considered [13, 14]. CeD is a duodenum-specific enteropathy, and changes in the small intestinal microbiome are therefore thought to be associated with its development [15]. However, several studies have also shown that patients with CeD present fecal microbiota dysbiosis [16]. These data suggest that, along with the small intestine, other parts of the gastrointestinal tract, such as the colon, may be a source of information for CeD pathogenesis.
This report, the first metagenomic analysis from a population of Saudi children, highlights several important differences between mucosal and fecal microbiome. Alpha-diversity analysis, for example, confirmed previously reported findings with fecal samples having increased bacterial richness and diversity as compared with those from mucosal samples [16]. Interestingly, we did not see any differences in alpha diversity between CeD and non-CeD groups. Microbial diversity in patients with CeD has been shown to be reduced compared with that in non-CeD controls [17], although another study found this was not the case [18]. Our analysis included a relatively small number of samples, which could account for the lack of significant differences in microbial diversity.
LDA LEfSe and DeSeq2 differential abundance analyses demonstrated significant differences between CeD and non-CeD groups at both mucosal and fecal levels. Overall, samples from patients with CeD appeared to have a decreased abundance of Actinobacteria phylum that is mainly represented by bacteria belonging to the Bifidobacterium genus. Many Bifidobacteria have positive immunomodulatory functions and are therefore used as probiotics. However, the increased abundance of L. acidophilus and Coprococcus species in in children with CeD contrasts with previous reports description as “good bacteria” [19, 20]. Samples from non-CeD controls appeared to have an increased abundance of “beneficial” bacteria (decreased in CeD) such as Roseburia and Lachnospiraceae species. Roseburia species are short-chain fatty acid-producing bacteria, which modulate intestinal motility and have anti-inflammatory properties. Changes in Roseburia species abundance have been correlated to several diseases such as irritable bowel syndrome, obesity, and type 2 diabetes [21, 22]. Similarly, Lachnospiraceae are often used as probiotics because of their “beneficial” impact on overall intestinal health [23]. Finally, increased levels of Subdoligranulum species have been found in CeD samples by several groups [18, 24]. Interestingly, a recent work by Leonard et al. demonstrated an increase in this specific genus in fecal samples from infants genetically predisposed to CeD even before the onset of the disease [24]. These findings are intriguing as they suggest a causative link between dysbiosis and CeD onset. Furthermore, they also raise the possibility that fecal microbiome markers could be representative of small intestinal dysbiosis. While our findings partially confirm previously reported differences between patients with CeD and those without, the use of metagenomic technology in this study revealed many unreported species, with significantly different abundance between children with CeD and non-CeD controls. Finally, it should be noted that bacterial associations with CeD reported in this study do not imply causality, a limitation that is common to most microbiota studies.
Study limitations
The most important limitation of this study is the relatively small sample size. However, the use of shotgun metagenomic analysis and the finding of many unreported bacterial species in this population of Saudi Arab children with high prevalence CeD make the results unique. Other limitations included the unavailability of information on the diet, growth and results of laboratory investigations.
Conclusions
Although preliminary, our data from Saudi Arabia, reports new bacterial species significantly associated with CeD. The fact that mucosal and fecal samples were collected from newly diagnosed children with CeD on normal gluten-containing diet suggests strong association between the identified bacteria and CeD. In addition, the identification of many unreported taxa associated with celiac disease, indicates the need for further studies from different populations to expand our understanding of the role of bacteria in the pathogenesis of celiac disease, hopefully leading to new treatment options.
Methods
Study population
The participants were enrolled from King Khalid University Hospital, King Saud University (KSU), Al Mofarreh PolyClinic, and King Fahad Medical City Children’s Hospital, Ministry of Health. All institutions are in Riyadh, Kingdom of Saudi Arabia (KSA). Main inclusion criteria included children below 18 years of age who were on normal gluten containing diet and had no history of antibiotic intake for at least 6 months before presentation to the clinic. In addition, confirmation of CeD for cases and exclusion of CeD for controls were according to the European Society of Pediatric Gastroenterology Hepatology and Nutrition guidelines [25].
Samples collection, storage, and processing
Mucosal samples from 20 children with confirmed CeD and 19 non-CeD controls were collected from the second part of the duodenum (D2); these were then stored in cryovials without fixative or stabilizer and transported in ice to the Central Laboratory. Similarly, fecal samples were also collected in cryovials from 20 children with CeD and 20 non-CeD controls and transported in ice to the Central Laboratory at the College of Medicine, (KSU). All samples were stored at − 80 °C. At the time of analysis, all samples were retrieved and dispatched by express mail in dry ice containers with temperature control for metagenomic analysis at Cosmos ID (Rockville, MD, USA).
DNA isolation and sequencing
DNA was isolated from mucosa samples using the ZymoBiomics miniprep kit and from stool samples using QIAGEN DNeasy PowerSoil DNA kit, both according to the manufacturer’s instructions. Isolated DNA was quantified via Qubit ds DNA HS assay kit (Thermo Fisher).
DNA libraries were prepared using the Illumina Nextera XT library preparation kit, according to the manufacturer’s protocol. Library quantity and quality were assessed using Qubit and TapeStation (Agilent Technologies, CA, USA). Libraries were then sequenced on Illumina HiSeq platform (2 × 150 bp reads). The samples were sequenced on the deeper end. They were sequenced at an average of about 20 million total reads per sample.
Bioinformatic analysis
Unassembled sequencing reads were directly analyzed using CosmosID bioinformatics platform (CosmosID Inc., Rockville, MD) for multi-kingdom microbiome analysis and quantification of organism’s relative abundance [26–29]. Briefly, the system utilizes curated genome databases and a high-performance data mining algorithm that rapidly disambiguates hundreds of millions of metagenomic sequence reads into discrete microorganisms that engender the particular sequences.
Custom analysis
Alpha-diversity boxplots
Alpha-diversity boxplots were calculated from the species-level abundance score matrices from CosmosID taxonomic analysis. Chao’s and Shannon’s alpha-diversity metrics were calculated in R using the R package Vegan. Further, t-tests were performed between each celiac and non-celiac group using the R package ggsignif. Boxplots with overlaid significance in p-value format were generated using the R package ggplot2 [30–32].
Beta-diversity principal coordinate analyses (PCoA)
Beta-diversity PCoA were calculated from the species-level relative abundance matrices from CosmosID taxonomic analysis. Bray–Curtis diversity was calculated in R using the R package Vegan with the functions vegdist; then, PCoA tables were generated using Vegan’s function PCoA. Plots were visualized using the R package ggpubr [30, 33].
Linear discriminant analysis effect size (LEfSe)
The LEfSe figures were generated using the Galaxy web application, based on relative abundance tables from CosmosID taxonomic analysis. Figures were calculated using a Kruskal–Wallis P-value of < 0.05, a Wilcoxon P-value of < 0.05, and a logarithmic linear discriminant analysis (LDA) score of ≥ 2.0 and therefore exhibited a statistically significant difference between groups. In addition, although not showing significant difference, some organisms may be functionally important. To explore this possibility, the P- values were set to < 0.2 for both Wilcoxon and Kruskal–Wallis tests, and the logarithmic LDA score of ≥ 0.05 and figures were calculated based on this threshold. In the LEfSe figures, the red bars (negative bars) indicate that the organism is more abundant in the CeD group; Whereas green bars ( positive bars) indicate greater organism abundance in the non-CeD group [34].
DeSeq2 differential abundance analysis
Differential abundance analysis used the abundance score matrices from the CosmosID taxonomic analysis. Differential abundance for organisms was calculated using DeSEQ2 from the R Phyloseq package (R Foundation for Statistical Computing, Vienna, Austria). For the mucosal and stool samples separately, the log2 fold change and associated P-values for celiac vs. non-celiac samples are displayed [35, 36]. A log2 > 0 indicates that the organism is more abundant in the CeD group; whereas a value < 0 indicates more abundance in the non-CeD group. P-values were calculated using the t-test function in R and adjusted for false discover rate. However, we also reported unadjusted p values to detect taxa not reaching the adjusted significance level but with possible biologic importance. The difference in abundance was considered significant when the adjusted P- value was < 0.05. In addition, unadjusted P- value was reported to reveal taxa that might have functional properties.
Acknowledgements
The authors extend their appreciations to the Deanship of Scientific Research at King Saud University for funding this work through Research Group No (RGP-1441-007).
Author contributions
ME designed the study and finalized the manuscript, GS drafted the manuscript and approved the final version. AA, AH, AS, MA, MM participated in enrollment of participants, reviewed and approved the manuscript. AF participated in the design, critically reviewed the manuscript. All authors read and approved the final manuscript.
Funding
This study was funded by the Deanship for Scientific Research, King Saud University, Kingdom of Saudi Arabia. Grant No. RGP- 1441–007. However, the Deanship had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the NCBI SRA. Access link: http://www.ncbi.nlm.nih.gov/bioproject/757365
Declarations
Ethics approval and consent to participate
This study was approved by the Institutional Review Board (IRB) of the College of Medicine, King Saud University in Riyadh, Kingdom of Saudi Arabia (no. 14/4464/IRB). The informed consent form approved by the IRB was presented and explained to the parents and children. Since all participants were children below 18 years of age, the informed consent was signed by one of the parents.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lebwohl B, Sanders DS, Green PHR. Coeliac disease. Lancet. 2018;391:70–81. doi: 10.1016/S0140-6736(17)31796-8. [DOI] [PubMed] [Google Scholar]
- 2.Gudeta AN, Ramelius A, Balcha TT, Girma A, Ilonen J, Agardh D. Distribution of hla-dq risk genotypes for celiac disease in ethiopian children. HLA. 2020;96:681–687. doi: 10.1111/tan.14119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Serena G, Lima R, Fasano A. Genetic and environmental contributors for celiac disease. Curr Allergy Asthma Rep. 2019;19:40. doi: 10.1007/s11882-019-0871-5. [DOI] [PubMed] [Google Scholar]
- 4.Lionetti E, Castellaneta S, Francavilla R, Pulvirenti A, Tonutti E, Amarri S, et al. Introduction of gluten, hla status, and the risk of celiac disease in children. N Engl J Med. 2014;371:1295–1303. doi: 10.1056/NEJMoa1400697. [DOI] [PubMed] [Google Scholar]
- 5.Olshan KL, Leonard MM, Serena G, Zomorrodi AR, Fasano A. Gut microbiota in celiac disease: Microbes, metabolites, pathways and therapeutics. Expert Rev Clin Immunol. 2020;16:1075–1092. doi: 10.1080/1744666X.2021.1840354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Al-Hussaini A, Alharthi H, Osman A, Eltayeb-Elsheikh N, Chentoufi A. Genetic susceptibility for celiac disease is highly prevalent in the saudi population. Saudi J Gastroenterol. 2018;24:268–273. doi: 10.4103/sjg.SJG_551_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rostami K, Malekzadeh R, Shahbazkhani B, Akbari MR, Catassi C. Coeliac disease in middle eastern countries: A challenge for the evolutionary history of this complex disorder? Dig Liver Dis. 2004;36:694–697. doi: 10.1016/j.dld.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 8.Chibbar R, Dieleman LA. The gut microbiota in celiac disease and probiotics. Nutrients. 2019;11:10. doi: 10.3390/nu11102375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krishnareddy S. The microbiome in celiac disease. Gastroenterol Clin North Am. 2019;48:115–126. doi: 10.1016/j.gtc.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 10.Cenit MC, Olivares M, Codoner-Franch P, Sanz Y. Intestinal microbiota and celiac disease: Cause, consequence or co-evolution? Nutrients. 2015;7:6900–6923. doi: 10.3390/nu7085314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kho ZY, Lal SK. The human gut microbiome - a potential controller of wellness and disease. Front Microbiol. 2018;9:1835. doi: 10.3389/fmicb.2018.01835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dominguez-Bello MG, Godoy-Vitorino F, Knight R, Blaser MJ. Role of the microbiome in human development. Gut. 2019;68:1108–1114. doi: 10.1136/gutjnl-2018-317503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez-Guryn K, Leone V, Chang EB. Regional diversity of the gastrointestinal microbiome. Cell Host Microbe. 2019;26:314–324. doi: 10.1016/j.chom.2019.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valitutti F, Cucchiara S, Fasano A. Celiac disease and the microbiome. Nutrients. 2019;11:10. doi: 10.3390/nu11102403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Golfeyz S. Celiac disease and fecal microbiota transplantation: A new beginning? Am J Gastroenterol. 2018;113:1256. doi: 10.1038/s41395-018-0094-8. [DOI] [PubMed] [Google Scholar]
- 16.Li G, Yang M, Zhou K, Zhang L, Tian L, Lv S, et al. Diversity of duodenal and rectal microbiota in biopsy tissues and luminal contents in healthy volunteers. J Microbiol Biotechnol. 2015;25:1136–1145. doi: 10.4014/jmb.1412.12047. [DOI] [PubMed] [Google Scholar]
- 17.Panelli S. Capelli E, Lupo GFD, Schiepatti A, Betti E, Sauta E, et al. Comparative study of salivary, duodenal, and fecal microbiota composition across adult celiac disease. J Clin Med. 2020;9:1109. doi: 10.3390/jcm9041109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zafeiropoulou K, Nichols B, Mackinder M, Biskou O, Rizou R, Karanikolou A, et al. Alterations in intestinal microbiota of children with celiac disease at the time of diagnosis and on a gluten-free diet. Gastroenterology. 2020;159(2039–51):e2020. doi: 10.1053/j.gastro.2020.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duncan SH, Holtrop G, Lobley GE, Calder AG, Stewart CS, Flint HJ. Contribution of acetate to butyrate formation by human faecal bacteria. Br J Nutr. 2004;91:915–923. doi: 10.1079/BJN20041150. [DOI] [PubMed] [Google Scholar]
- 20.Esmaeili SA, Mahmoudi M, Rezaieyazdi Z, Sahebari M, Tabasi N, Sahebkar A, et al. Generation of tolerogenic dendritic cells using lactobacillus rhamnosus and lactobacillus delbrueckii as tolerogenic probiotics. J Cell Biochem. 2018;119:7865–7872. doi: 10.1002/jcb.27203. [DOI] [PubMed] [Google Scholar]
- 21.Debedat J, Clement K, Aron-Wisnewsky J. Gut microbiota dysbiosis in human obesity: Impact of bariatric surgery. Curr Obes Rep. 2019;8:229–242. doi: 10.1007/s13679-019-00351-3. [DOI] [PubMed] [Google Scholar]
- 22.Liu T, Gu X, Li LX, Li M, Li B, Cui X, et al. Microbial and metabolomic profiles in correlation with depression and anxiety co-morbidities in diarrhoea-predominant ibs patients. BMC Microbiol. 2020;20:168. doi: 10.1186/s12866-020-01841-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J, Song L, Wang Y, Liu C, Zhang L, Zhu S, et al. Beneficial effect of butyrate-producing lachnospiraceae on stress-induced visceral hypersensitivity in rats. J Gastroenterol Hepatol. 2019;34:1368–1376. doi: 10.1111/jgh.14536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leonard MM, Karathia H, Pujolassos M, Troisi J, Valitutti F, Subramanian P, et al. Multi-omics analysis reveals the influence of genetic and environmental risk factors on developing gut microbiota in infants at risk of celiac disease. Microbiome. 2020;8:130. doi: 10.1186/s40168-020-00906-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Husby S, Koletzko S, Korponay-Szabó IR, Mearin ML, Phillips A, Shamir R, et al. European Society for Pediatric Gastroenterology, Hepatology, and Nutrition Guidelines for the Diagnosis of Coeliac Disease. J Pediatr Gastroenterol Nutr. 2012;54:136–160. doi: 10.1097/MPG.0b013e31821a23d0. [DOI] [PubMed] [Google Scholar]
- 26.Ottesen A, Ramachandran P, Reed E, White JR, Hasan N, Subramanian P, et al. Enrichment dynamics of Listeria monocytogenes and the associated microbiome from naturally contaminated ice cream linked to a listeriosis outbreak. BMC Microbiol. 2016;16:275. doi: 10.1186/s12866-016-0894-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hasan NA, Young BA, Minard-Smith AT, Saeed K, Li H, Heizer EM, et al. Microbial community profiling of human saliva using shotgun metagenomic sequencing. PLoS ONE. 2014;9:e97699. doi: 10.1371/journal.pone.0097699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lax S, Smith DP, Hampton-Marcell J, Owens SM, Handley KM, Scott NM, et al. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science. 2014;345:1048–1052. doi: 10.1126/science.1254529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ponnusamy D, Kozlova EV, Sha J, Erova TE, Azar SR, Fitts EC, et al. Crosstalk among flesh-eating Aeromonas hydrophila strains in mixed infection leading to necrotizing fasciitis. PNAS. 2016;113:722–727. doi: 10.1073/pnas.1523817113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, et al. vegan: Community Ecology Package. R package version 2.5–6; 2019. https://cran.r-project.org, https://github.com/vegandevs/vegan.
- 31.Ahlmann-Eltze C. ggsignif: significance Brackets for ’ggplot2’. R package version 0.6.0; 2019. https://CRAN.R-project.org/package=ggsignif.
- 32.Wickham H. ggplot2: elegant graphics for data analysis. New York: Springer; 2016. [Google Scholar]
- 33.Kassambara A. ggpubr: ‘ggplot2’ Based Publication Ready Plots. R package version 0.2.5; 2020. https://CRAN.R-project.org/package=ggpubr.
- 34.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett Ws, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. [DOI] [PMC free article] [PubMed]
- 35.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the NCBI SRA. Access link: http://www.ncbi.nlm.nih.gov/bioproject/757365