Abstract
Formation of the asymmetrically located septum during sporulation of Bacillus subtilis results in enclosure of the origin-proximal 30% of the chromosome in the prespore compartment. The rest of the chromosome is then translocated into the prespore from the mother cell. Transcription of spoIIR is initiated in the prespore by RNA polymerase containing ςF soon after the septum is formed. The SpoIIR protein is required for the activation of the transcription program directed by ςE in the mother cell. The spoIIR locus is located at 324°, near the origin of replication (0/360°). We show here that movement of spoIIR to 28° had little effect on sporulation. However, movement to regions not in the origin-proximal part of the chromosome substantially reduced sporulation efficiency. At 283° sporulation was reduced to less than 20% of the level obtained when spoIIR was at its natural location, and movement to 190° reduced sporulation to about 6% of that level. These positional effects were also seen in the transcription of a spoIIR-lacZ fusion. In contrast, movement of other spo-lacZ fusions from 28° to 190° had little effect on their expression. These results suggest that spoIIR is the subject of “positional regulation,” in the sense that the chromosomal position of spoIIR is important for its expression and function.
During sporulation Bacillus subtilis undergoes an asymmetrically located cell division. This division is a modified form of the vegetative division (6, 16). However, formation of the sporulation septum results in enclosure of only about 30% of a chromosome in the smaller cell, the prespore (also called the forespore), that results from the division; the rest of the chromosome is then translocated from the larger cell, the mother cell, into the prespore by an active process requiring SpoIIIE (Fig. 1) (23, 25). A second copy of the chromosome remains in the mother cell. The prespores of SpoIIIE mutant cells contain only about 30% of a chromosome, with the other 70% remaining in the mother cell together with the whole of the mother cell chromosome (23). Formation of the asymmetrically located septum is followed by activation of two sporulation-specific transcription factors, ςF in the prespore and ςE in the mother cell, which specify different programs of gene expression in the two compartments (reviewed in reference 21). In a spoIIIE36 mutant the ςF-directed prespore genes that are located in the 70% of the chromosome distal to the origin of replication (for example, dacF and gpr) are not transcribed, whereas those located in the origin-proximal 30% are transcribed (for example, spoIIR and spoIIQ) (9, 12, 20, 22, 23, 25). Thus, it has been known for some time that chromosome position is important for expression of ςF-directed genes in a spoIIIE36 mutant (22). It seemed plausible that there could be some prespore-specific gene (or genes) that needed to be expressed as soon as the septum was formed and so needed to be located at the origin-proximal part of the chromosome in the parental, spo+ strain. If such a gene were to be relocated distal to the origin, its expression during sporulation might be impaired, resulting in a sporulation-deficient phenotype.
FIG. 1.
Model for chromosomal translocation through the sporulation septum showing the approximate location of loci used in this study (adapted from reference 24). The SpoIIIE protein is indicated as ovals at the position where the chromosome transverses the recently formed sporulation septum.
We considered that the spoIIR locus might be a possible candidate for this “positional” type of regulation of its activity. The spoIIR locus is near the origin and is transcribed only by RNA polymerase containing ςF (9, 13). It links the activation of ςE in the mother cell to the activation of ςF in the prespore, and it is the only ςF-directed gene needed for the ςE activation (9, 13). Its activation is thought to ensure that ςE is not activated until after the septum is formed (9), and rapid activation of ςE following septation may be important in preventing further septation (1). Thus, a delay in spoIIR expression may disrupt the complex network of transcription regulation that is necessary for spore formation. Below we describe experiments indicating that the spoIIR gene is the subject of such positional regulation.
MATERIALS AND METHODS
Media.
B. subtilis was grown in modified Schaeffer's sporulation medium (MSSM) and on Schaeffer's sporulation agar (17, 19). When required, 5-bromo-4-chloro-3-indolyl-β-d-galactoside at 40 μg/ml, chloramphenicol at 5 μg/ml, neomycin at 3 μg/ml, and erythromycin at 1 μg/ml were added.
Strains.
B. subtilis 168 strain BR151 trpC2 metB10 lys-3 and B. subtilis ZB307 SPβc2Δ2::Tn917::pSK10Δ6 were used as the parent strains. These and the other B. subtilis strains used are listed in Table 1. Escherichia coli strain DH5α (GIBCO/BRL) was used to maintain plasmids.
TABLE 1.
B. subtilis strains
| Strain | Relevant characteristics | Origin or reference |
|---|---|---|
| BR151 | trpC2 metB10 lys-3 | Laboratory stock |
| ZB307 | SPβc2Δ2::Tn917pSK10Δ6 | P. Youngman (26, 30) |
| SL7205 | SPβc2Δ2::Tn917pSK10Δ6 amyE::spoIIR-gusA spoIIR::spoIIR-lacZ | This study |
| SL7213 | amyE::spoIIE-gusA SPβc2Δ2::Tn917pSK10Δ6::spoIIE-lacZ | This study |
| SL7256 | SPβc2Δ2::Tn917pSK10Δ6::spoIIR spoIIR::neo | This study |
| SL7258 | SPβc2Δ2::Tn917pSK10Δ6 spoIIR::neo amyE::spoIIR | This study |
| SL7303 | amyE::spoIIR-gusA SPβc2Δ2::Tn917pSK10Δ6::spoIIR-lacZ | This study |
| SL7304 | amyE::spoIIR-gusA SPβc2Δ2::Tn917pSK10Δ6::spoIIR-lacZ spoIIAC561 | This study |
| SL7310 | trpC2 metB10 lys-3 amyE::IIR-gusA thrC::spoIIR-lacZ | This study |
| SL7313 | trpC2 metB10 lys-3 spoIIR::neo thrC::spoIIR | This study |
| SL7321 | SPβc2Δ2::Tn917pSK10Δ6 spoIID-lacZ@spoIIDa | This study |
| SL7322 | SPβc2Δ2::Tn917pSK10Δ6::spoIIR spoIIR::neo spoIID-lacZ@spoIID | This study |
| SL7340 | trpC2 metB10 lys-3 spoIIR::neo amyE::spoIIR | This study |
| SL7344 | trpC2 metB10 spoIIR::neo SPβc2Δ2::Tn917pSK10Δ6::spoIIR | This study |
| SL8344 | trpC2 metB10 lys-3 spoIIQ-gfp | This study |
| SL8345 | trpC2 metB10 lys-3 spoIIR::neo spoIIQ-gfp | This study |
| SL8378 | trpC2 metB10 lys-3 spoIIR::neo amyE::spoIIR spoIIQ-gfp | This study |
| SL8380 | trpC2 metB10 lys-3 spoIIR::neo SPβc2Δ2::Tn917pSK10Δ6::spoIIR spoIIQ-gfp | This study |
| SN178 | SPβc2Δ2::Tn917pSK10Δ6 amyE::spoIIR-gusA spoIIR-lacZ@spoIIR spoIIAC561 | This study |
@ indicates the fusion is located at the locus by a Campbell-like recombination.
The spoIIR promoter region was cloned as a NotI-HindIII fragment (9), via pBluescript to provide additional sites, into the following plasmids: pMLK83, a neo gusA fusion vector designed for the integration of constructs by a double-recombination event at the amyE locus (10); pDG793, an erm lacZ fusion vector designed for the integration of constructs by a double-recombination event at the thrC locus (a gift from P. Stragier, Institut de Biologie Physico Chimique, Paris, France); pGV34 (4, 26), a cat lacZ fusion vector designed for the integration of constructs by a double-recombination event at the SPβ locus, and also used for Campbell-like recombination at spoIIR. An intact copy of spoIIR was cloned as a 1.2-kb NotI-XhoI fragment into the same plasmids. The genetic linkage was verified for each chromosomal insertion.
The spoIIE promoter region was cloned as an EcoRI-PvuII fragment in pMLK83 (10). This construct was then used to introduce the spoIIE-gusA fusion into amyE by double crossover. The spoIID-gusA fusion at amyE was derived from pMLK87 (10). The spoIID-lacZ fusion at spoIID resulted from integration of pMLK23 by a single crossover (10). P. Youngman (Millennium Pharmaceuticals, Cambridge, Mass.) kindly provided strains containing the spoIIE-lacZ and spoIID-lacZ fusions at SPβ. A strain containing a spoIIQ-gfp transcriptional fusion (12) was kindly provided by P. Stragier. The fusion was introduced by transformation into strains containing spoIIR at different chromosomal locations. Details of the construction of strains are available on request.
β-Galactosidase and β-glucuronidase assays.
Assays were performed essentially as described previously (10), using lysozyme to permeabilize the cells. Specific activity (in units) is expressed as nanomoles of o-nitrophenyl-β-d-galactoside or p-nitrophenyl-β-d-glucuronide hydrolyzed per minute per milligram of bacterial dry weight. The endogenous β-galactosidase and β-glucuronidase activities were determined in each experiment for an isogenic parental strain lacking a fusion and subtracted from the corresponding values for the fusion-containing strains.
Other methods.
B. subtilis transformation, transduction, sporulation by exhaustion in MSSM, and all genetic engineering methods were performed essentially as previously described (7, 15, 17, 28). Sporulation was assayed 18 h after the end of exponential growth by diluting cultures and determining the heat-resistant count (80°C, 20 min) and the viable count in the diluted cultures. The viable count varied somewhat from experiment to experiment: for strains with spoIIR at SPβ, SL7256, and SL7344, the range was 1.0 × 108 to 2.5 × 108 per ml; for all other strains the range was 2.5 × 108 to 6.0 × 108 per ml; there was no significant chain formation.
Cultures used for visualization of green fluorescence protein (GFP) were grown in MSSM at 33.5°C and harvested 6 h after the end of exponential growth, by which time the bulk of the population had reached the sporulation division stage. Culture samples of 10 μl of unfixed cells were transferred to 0.1% polylysine-coated slides and examined by fluorescence microscopy essentially as described previously (28).
RESULTS
Complementation of the Spo− phenotype associated with spoIIR::neo by placing the intact spoIIR gene at different chromosomal locations.
A knockout of the spoIIR gene with an insertion of a neo cassette in codon 98 of the spoIIR open reading frame (9) resulted in an asporogenous phenotype (less than 1 spore in 108 cells). To test the possibility that the chromosomal location (at 324°) of the spoIIR gene near the origin of replication (at 0/360°) might be important for its proper functioning, we chose three different chromosomal locations (11) for the integration of spoIIR: near the origin but on the other side of the origin to spoIIR (at amyE, 28°), approximately halfway from the origin to the terminus (at thrC, 283°), and near the terminus (at SPβ, 190°). The intact copy of spoIIR as a NotI-XhoI fragment was cloned in different plasmids, pDH32, pDG793, and pGV34, designed to facilitate integration of spoIIR by double crossover at amyE, thrC, and SPβ, respectively. The results of sporulation efficiency assays of strains carrying the knockout of spoIIR at its original location (324°) and integration of an intact copy of spoIIR at the different locations are summarized in Table 2. Movement of the intact copy of spoIIR to amyE had little effect on sporulation. However, movement to thrC reduced sporulation to less than 20% of the efficiency of the isogenic parent strain. Movement to SPβ reduced sporulation to about 6% of that of the parent (Table 2); the method of determining sporulation may overestimate this figure because the viable counts of strains with spoIIR at SPβ were about twofold lower than those of the other strains. The same effect of movement to SPβ was also observed with a B. subtilis strain of a different lineage, ZB307 (29). The results suggest that the efficiency of sporulation depends on the distance between the location of the spoIIR gene and the origin of replication.
TABLE 2.
Efficiency of sporulation of strains carrying a single intact copy of spoIIR at different chromosomal locations
| Location of intact copy of spoIIR | % Sporulationa
|
|
|---|---|---|
| A | B | |
| spoIIR | 47, 55 (BR151) | 55, 71 (ZB307) |
| amyE | 42, 46 (SL7340) | 59, 81 (SL7258) |
| thrC | 7, 10 (SL7313) | ND, ND |
| SPβ | 3.3, 2.7 (SL7344) | 0.9, 2.9 (SL7256) |
Results are for two different experiments with BR151 (A) and ZB307 (B) as parent strains. ND, not determined.
Expression of a spoIIR-lacZ fusion in different chromosomal locations.
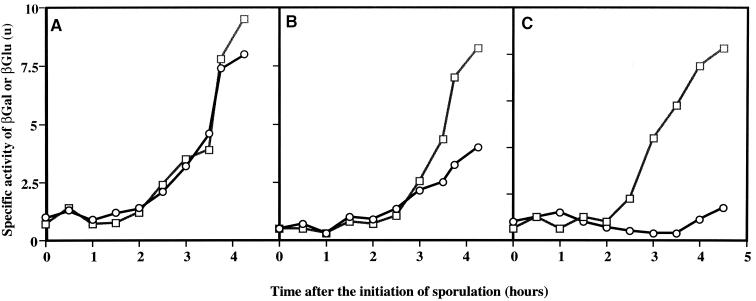
To test spoIIR transcription at the different locations, we employed a spoIIR-lacZ transcriptional fusion in strains that also contained a spoIIR-gusA fusion inserted at amyE as an internal control. No significant differences were observed in the timing or the level of expression of spoIIR-lacZ integrated at spoIIR compared to the spoIIR-gusA fusion at amyE (Fig. 2A). Expression of spoIIR-lacZ was reduced at thrC (Fig. 2B) and was barely detectable at SPβ (Fig. 2C). Introduction of an inducible copy of the gene for ςF, spoIIAC, under the control of the Pspac promoter (5) into the latter strain established that the spoIIR-lacZ at SPβ fusion was still functional (data not shown).
FIG. 2.
Expression of spoIIR at various chromosomal locations in a spoIIA+ background. □, expression of amyE::spoIIR-gusA. ○, expression of spoIIR-lacZ at spoIIR (SL7205) (A), thrC (SL7310) (B), or SPβ (SL7303) (C). SL7310 is a derivative of BR151; SL7205 and SL7303 are derivatives of ZB307. βGal, β-galactosidase; βGlu, β-glucuronidase.
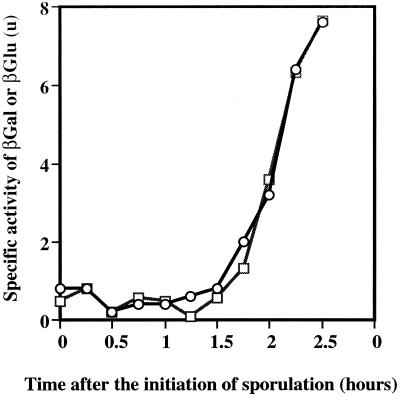
We considered it unlikely that the local context of SPβ would explain the reduced expression of spoIIR-lacZ. The SPβ system (29) has been used extensively, and we had noted no reduction in expression of the strong ςH promoter, ftsAp2, when it was located at SPβ (3). However, we considered it necessary to retest the possibility that expression of lacZ fusions at SPβ might somehow be inhibited by the local gene context. We compared the expression at amyE and SPβ of the ςA-dependent sporulation-specific gene spoIIE, which is also very weakly expressed. There was no difference in the level and pattern of expression, regardless of chromosomal position (Fig. 3). Fusions to gusA were used at amyE, and lacZ fusions were used at SPβ; previous studies had shown that the gusA and lacZ fusions gave activities similar to each other (10).
FIG. 3.
Expression of spoIIE at different chromosomal locations. □, amyE::spoIIE-gusA; ○, SPβ::spoIIE-lacZ (SL7213). βGal, β-galactosidase; βGlu, β-glucuronidase.
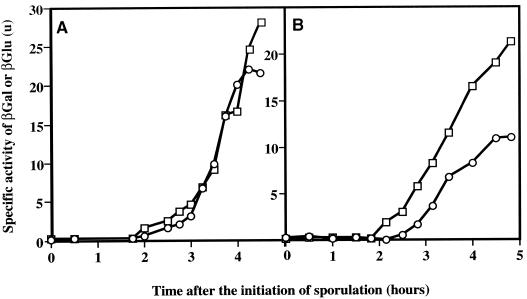
The transcription of spoIIR is normally weak and is substantially higher in spoIIAC(P) mutants (9). Consequently we checked spoIIR-lacZ expression at the different locations in a spoIIAC561 background in order to visualize more clearly possible differences in expression patterns. The spoIIAC561 mutation is a V233M change in the 4.2 promoter recognition region of ςF and does not affect regulation of ςF activity (14, 27). It curtails transcription of some ςF-directed genes (14, 27) but enhances transcription of spoIIR (9). Again, no significant differences were observed in the timing or level of expression of spoIIR-lacZ at spoIIR compared to spoIIR-gusA at amyE (Fig. 4). When the fusion was integrated at SPβ, there was a delay in expression of spoIIR-lacZ, and expression was also reduced compared to that of spoIIR-gusA at amyE. The delay was difficult to measure accurately; in different experiments it was about 15 to 20 min. These data indicate that chromosome position is important for the expression of spoIIR; they suggest that the SpoIIIE-mediated active transport of the distal 70% of the chromosome through the sporulation septum into the prespore (25) may take 15 to 20 min.
FIG. 4.
Expression of spoIIR at different chromosomal locations in a spoIIA561 background. □, expression of amyE::spoIIR-gusA; ○, expression of spoIIR-lacZ at spoIIR (SN178) (A) or SPβ (SL7304) (B). βGal, β-galactosidase; βGlu, β-glucuronidase.
Activity of ςE is significantly decreased when the spoIIR chromosomal location is altered.
Having the level of spoIIR expression at SPβ significantly decreased and delayed, it was reasonable to expect that expression of ςE-dependent genes would be impaired. To test this expectation, we introduced a spoIID-lacZ fusion into strain ZB307 and into a derivative of ZB307 carrying a single functional copy of the spoIIR gene at SPβ. The pattern of spoIID-lacZ expression in cultures of these two strains is shown in Fig. 5. Relocation of the spoIIR gene to the terminus region resulted in reduction in expression by about 80%, and this was accompanied by a delay compared to spoIID-lacZ expression in the parent strain. It seems plausible that the sporulation-deficient phenotype of strains containing spoIIR located only at SPβ is the result of a decrease in ςE-dependent gene expression.
FIG. 5.
Effect of spoIIR location on expression of spoIID-lacZ. □, spoIIR at spoIIR (SL7321); ○, spoIIR at SPβ (SL7322). βGal, β-galactosidase.
The major role of spoIIR is thought to be to ensure that activation of ςE requires prior activation of ςF, and so ςE activation follows formation of the sporulation septum (9). Mutations in the structural gene for ςE, spoIIGB, result in the abortively disporic phenotype in which a sporulation division has occurred near both cell poles, and it is inferred that a role of ςE during sporulation is to prevent the formation of the second, asymmetrically located division septum (1). Mutation of spoIIR also results in this abortively disporic phenotype (in which each of the prespores contains a nucleoid, whereas the mother cell is nucleoid free), although at a slightly reduced frequency (9) (our unpublished results). Appearance (or lack thereof) of the abortively disporic phenotype is used here as a separate test of the effect on ςE activation of moving spoIIR. The prespore-specific expression of a spoIIQ-gfp transcriptional fusion (12) was used as an indicator of prespore formation. In this system, a spoIIGB mutant gave 45% disporic and 55% monosporic organisms displaying GFP fluorescence 6 h after the start of sporulation at 33.5°C. When an intact copy of spoIIR was located at SPβ, the strain exhibited an abortively disporic phenotype, similar to that of a spoIIR null mutant, with about 30% of fluorescing organisms displaying the disporic pattern (Table 3); a similar pattern was obtained for strains constructed in the ZB307 background (data not shown). Disporic forms were very rare when spoIIR was located at spoIIR or at amyE (Table 3).
TABLE 3.
Frequency of monosporic and disporic phenotypes in strains having spoIIR at different chromosomal locationsa
| Strain | Location of intact copy of spoIIR | No. of organisms with expression pattern of spoIIQ-gfp
|
|
|---|---|---|---|
| Disporic | Monosporic | ||
| SL8344 | spoIIR | 0 | 200 |
| SL8378 | amyE | 1 | 199 |
| SL8380 | SPβ | 54 | 146 |
| SL8345 | None | 61 | 139 |
The phenotypes were assayed for each strain by scoring 200 organisms that were displaying fluorescence from GFP resulting from prespore-specific expression of a spoIIQ-gfp transcriptional fusion (12). Organisms were sampled 6 h after the end of exponential growth at 33.5°C in MSSM; similar results were obtained with samples taken at 5 and 8 h (data not shown). The strains are in the BR151 genetic background.
DISCUSSION
An early stage in sporulation of B. subtilis is an asymmetric cell division that forms the prespore and the mother cell (1, 16, 21). Shortly after the division, ςF becomes active and governs gene expression in the prespore (21). However, the asymmetrically located division septum, when first formed, traps the origin-distal 70% of the prespore-destined chromosome in the mother cell (25) (Fig. 1); movement of the rest of the chromosome into the prespore is an active process requiring the membrane-associated DNA translocase SpoIIIE (23). Thus, a ςF-dependent gene whose activity is required early in the prespore may need to be located near the chromosome origin. Our results demonstrate impaired sporulation when the ςF-dependent spoIIR locus is moved away from the origin to either the thrC locus (283°; less than 20% of the sporulation of the isogenic parent with spoIIR at its natural position) or SPβ (190°; about 6%) (Table 2). This phenotype can be explained by the observed impairment in spoIIR expression at the origin-distal locations. With a spoIIAC561 background, which enhances spoIIR transcription (9), we were able to observe that spoIIR-lacZ expression was delayed when the fusion was located at SPβ (Fig. 4). This delay is thought to represent the time required for SpoIIIE-dependent chromosome translocation through the sporulation septum. The 15- to 20-min estimate agrees with that of Pogliano et al. (18) obtained from microscopy studies. These observations are in agreement with the hypothesis that chromosome partitioning during sporulation is an active, unidirectional, and time-requiring process (23, 25). Frandsen et al. (2) have utilized the transient gene asymmetry resulting from this slow chromosome partitioning to engineer activation of ςF independent of its normal regulators, SpoIIAA, SpoIIAB, and SpoIIE.
Why may a delay in the prespore localization of the spoIIR gene result in significantly lower expression? The native spoIIR promoter is weak (9, 13). Thus, when spoIIR is located near the terminus, one possibility is that the observed decrease in its expression results from competition with other ςF-directed genes that now precede spoIIR into the prespore. In this regard, Ju et al. (8) have shown that expression of another ςF-directed gene is higher when the gene is located nearer to the origin. The role of SpoIIR in spore formation is to activate ςE in the mother cell (9, 13), and a consequence of ςE activation is to block further septation (1). It is thought that spoIIR needs to be expressed very soon after the septum is formed (9). Reducing and/or delaying spoIIR expression is presumably sufficient to disrupt the delicate balance of controls that coordinate transcription between mother cell and prespore (21). The positional type of transcription regulation for spoIIR may thus be critical to the complex sporulation process.
ACKNOWLEDGMENTS
We thank W. Harling for help in an early part of the study. We thank M. L. Karow, P. Stragier, and P. Youngman for plasmids and strains used in this study. We are especially grateful to A. Wolfson for many helpful discussions.
This work was supported by Public Health Service grant GM43577 from the National Institutes of Health.
REFERENCES
- 1.Errington J. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol Rev. 1993;57:1–33. doi: 10.1128/mr.57.1.1-33.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frandsen N, Barák I, Karmazyn-Campelli C, Stragier P. Transient gene asymmetry during sporulation and establishment of cell specificity in Bacillus subtilis. Genes Dev. 1999;13:394–399. doi: 10.1101/gad.13.4.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gholamhoseinian A, Shen Z, Wu J-J, Piggot P. Regulation of transcription of the cell division gene ftsA during sporulation of Bacillus subtilis. J Bacteriol. 1992;174:4647–4656. doi: 10.1128/jb.174.14.4647-4656.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guzman P, Westpheling J, Youngman P. Characterization of the promoter region of the Bacillus subtilis spoIIE operon. J Bacteriol. 1988;170:1598–1609. doi: 10.1128/jb.170.4.1598-1609.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henner D. Inducible expression of regulatory genes in Bacillus subtilis. Methods Enzymol. 1990;185:223–228. doi: 10.1016/0076-6879(90)85022-g. [DOI] [PubMed] [Google Scholar]
- 6.Hitchins A D, Slepecky R A. Bacterial sporulation as a modified procaryotic cell division. Nature. 1969;223:804–807. doi: 10.1038/223804a0. [DOI] [PubMed] [Google Scholar]
- 7.Hoch J A. Genetic analysis in Bacillus subtilis. Methods Enzymol. 1991;204:305–320. doi: 10.1016/0076-6879(91)04015-g. [DOI] [PubMed] [Google Scholar]
- 8.Ju J, Luo T, Haldenwang W G. Forespore expression and processing of the SigE transcription factor in wild-type and mutant Bacillus subtilis. J Bacteriol. 1998;180:1673–1681. doi: 10.1128/jb.180.7.1673-1681.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karow M L, Glaser P, Piggot P J. Identification of a gene, spoIIR, that links the activation of ςE to the transcriptional activity of ςF during sporulation in Bacillus subtilis. Proc Natl Acad Sci USA. 1995;92:2012–2016. doi: 10.1073/pnas.92.6.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karow M L, Piggot P J. Construction of gusA transcriptional fusion vectors for Bacillus subtilis and their utilization for studies of spore formation. Gene. 1995;163:69–74. doi: 10.1016/0378-1119(95)00402-r. [DOI] [PubMed] [Google Scholar]
- 11.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 12.Londoño-Vallejo J-A, Fréhel C, Stragier P. spoIIQ, a forespore-expressed gene required for engulfment in Bacillus subtilis. Mol Microbiol. 1997;24:29–39. doi: 10.1046/j.1365-2958.1997.3181680.x. [DOI] [PubMed] [Google Scholar]
- 13.Londoño-Vallejo J-A, Stragier P. Cell-cell signaling pathway activating a developmental transcription factor in Bacillus subtilis. Genes Dev. 1995;9:503–508. doi: 10.1101/gad.9.4.503. [DOI] [PubMed] [Google Scholar]
- 14.Margolis P, Driks A, Losick R. Establishment of cell type by compartmentalized activation of a transcription factor. Science. 1991;254:562–565. doi: 10.1126/science.1948031. [DOI] [PubMed] [Google Scholar]
- 15.Piggot P J. Mapping of asporogenous mutations of Bacillus subtilis: a minimum estimate of the number of sporulation operons. J Bacteriol. 1973;114:1241–1253. doi: 10.1128/jb.114.3.1241-1253.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piggot P J, Bylund J E, Higgins M L. Morphogenesis and gene expression during sporulation. In: Piggot P J, Moran C P Jr, Youngman P, editors. Regulation of bacterial differentiation. Washington, D.C.: American Society for Microbiology; 1994. pp. 113–137. [Google Scholar]
- 17.Piggot P J, Curtis C A M. Analysis of the regulation of gene expression during Bacillus subtilis sporulation by manipulation of the copy number of spo-lacZ fusions. J Bacteriol. 1987;169:1260–1266. doi: 10.1128/jb.169.3.1260-1266.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pogliano J, Osborne N, Sharp M D, Abanes-De Hello A, Parez A, Sun Y-L, Pogliano K. A vital stain for studying membrane dynamics in bacteria: a novel mechanism controlling septation during Bacillus subtilis sporulation. Mol Microbiol. 1999;31:1149–1159. doi: 10.1046/j.1365-2958.1999.01255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schaeffer P, Millet J, Aubert J-P. Catabolic repression of bacterial sporulation. Proc Natl Acad Sci USA. 1965;54:704–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schuch R, Piggot P J. The dacF-spoIIA operon of Bacillus subtilis, encoding ςF, is autoregulated. J Bacteriol. 1994;176:4104–4110. doi: 10.1128/jb.176.13.4104-4110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet. 1996;30:297–341. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- 22.Sun D, Fajardo-Cavazos P, Sussman M D, Tovar-Rojo F, Cabrera-Martinez R-M, Setlow P. Effect of chromosome location of Bacillus subtilis forespore genes on their spo gene dependence and transcription by EςF: identification of features of good EςF-dependent promoters. J Bacteriol. 1991;173:7867–7874. doi: 10.1128/jb.173.24.7867-7874.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu L J, Errington J. Bacillus subtilis SpoIIIE protein required for segregation during asymmetric cell division. Science. 1994;264:572–575. doi: 10.1126/science.8160014. [DOI] [PubMed] [Google Scholar]
- 24.Wu L J, Errington J. Septal localization of the SpoIIIE chromosome partitioning protein in Bacillus subtilis. EMBO J. 1997;16:2161–2169. doi: 10.1093/emboj/16.8.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu L J, Lewis P J, Allmansberger R, Hauser P M, Errington J. A conjugation-like mechanism for prespore chromosome partitioning during sporulation in Bacillus subtilis. Genes Dev. 1995;9:1316–1326. doi: 10.1101/gad.9.11.1316. [DOI] [PubMed] [Google Scholar]
- 26.Youngman P. Use of transposons and integrational vectors for mutagenesis and construction of gene fusions in Bacillus species. In: Harwood C R, Cutting S M, editors. Molecular biological methods for Bacillus. Chichester, England: John Wiley & Sons; 1990. pp. 221–256. [Google Scholar]
- 27.Yudkin M D. Structure and function in a Bacillus subtilis sporulation-specific sigma factor: molecular nature of mutations in spoIIAC. J Gen Microbiol. 1987;133:475–481. doi: 10.1099/00221287-133-3-475. [DOI] [PubMed] [Google Scholar]
- 28.Zhang L, Higgins M L, Piggot P J, Karow M L. Analysis of the role of prespore gene expression in the compartmentalization of mother cell-specific gene expression during sporulation of Bacillus subtilis. J Bacteriol. 1996;178:2813–2817. doi: 10.1128/jb.178.10.2813-2817.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zuber P, Losick R. Use of a lacZ fusion to study the role of the spoO genes of Bacillus subtilis in developmental regulation. Cell. 1983;35:275–283. doi: 10.1016/0092-8674(83)90230-1. [DOI] [PubMed] [Google Scholar]
- 30.Zuber P, Losick R. Role of AbrB in Spo0A- and Spo0B-dependent utilization of a sporulation promoter in Bacillus subtilis. J Bacteriol. 1987;169:2223–2230. doi: 10.1128/jb.169.5.2223-2230.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]